Abstract
In our previous phase 1/2 study aimed at controlling graft-versus-host disease, 12 patients received Herpes simplex virus thymidine kinase (HSV-tk+)/neomycin phosphotransferase (NeoR+)–expressing donor gene-modified T cells (GMCs) and underwent an HLA-identical sibling T-cell–depleted bone marrow transplantation (BMT). This study's objective was to follow up, to quantify, and to characterize persistently circulating GMCs more than 10 years after BMT. Circulating GMCs remain detectable in all 4 evaluable patients. However, NeoR- and HSV-tk–polymerase chain reaction (PCR) differently quantified in vivo counts, suggesting deletions within the HSV-tk gene. Further experiments, including a novel “transgene walking” PCR method, confirmed the presence of deletions. The deletions were unique, patient-specific, present in most circulating GMCs expressing NeoR, and shown to occur at time of GMC production. Unique patient-specific retroviral insertion sites (ISs) were found in all GMCs capable of in vitro expansion/cloning as well. These findings suggest a rare initial gene deletion event and an in vivo survival advantage of rare GMC clones resulting from an anti–HSV-tk immune response and/or ganciclovir treatment. In conclusion, we show that donor mature T cells infused with a T-cell–depleted graft persist in vivo for more than a decade. These cells, containing transgene deletions and subjected to significant in vivo selection, represent a small fraction of T cells infused at transplantation.
Introduction
Donor T cells can recognize a host as dangerous, resulting in graft-versus-host disease (GvHD).1 However, T cells also play a major role in tumor eradication2 and, when infused with a hematopoietic graft, in engraftment and immune reconstitution. Adequate regulation of alloreactivity remains elusive, and innovation in this area is needed. Use of donor T cells expressing a suicide gene such as herpes simplex thymidine kinase (HSV-tk) could allow for selective in vivo depletion in GvHD. Before hand, and perhaps permanently in the absence of GvHD, the recipient could enjoy the full benefit of donor T cells. In addition, compared with the broad immunosuppressive agents presently used for GvHD prophylaxis and treatment, conditional selective depletion of alloreactive donor T cells could be less toxic
Based on in vitro3-6 and in vivo7,8 preclinical studies, we9 and others10 previously demonstrated that this approach is feasible and has no apparent major side effects. Between 1996 and 1998, we conducted a phase 1/2 clinical trial involving ex vivo retrovirally modified donor primary T cells expressing HSV-tk. Twelve high-risk GvHD patients with a human leukocyte antigen (HLA)–identical sibling graft received a small number (2 × 105-2 × 106 cells/kg body weight) of gene-modified T cells (GMCs) with a T-cell–depleted bone marrow graft.9 This clinical study confirmed that in instances of GvHD, cells expressing HSV-tk can be controlled only by ganciclovir (GCV) treatment. Using a molecular quantification method,11 the presence of GMCs was confirmed in all patients after their administration at the time of transplantation. Moreover, after an early increase in the counts of circulating GMCs, counts progressively decreased over the first year. GCV treatment resulted in a rapid 80% to 98% decrease in the number of GMCs and efficient treatment of acute and chronic GvHD.9
Despite these encouraging results, we showed the impairment of GMCs' anti–Epstein-Barr virus (EBV) competence and demonstrated that it is linked to activation-induced cell death and toxicity of the NeoR-based selection process.12,13 Furthermore, a fraction of GMCs containing a truncated nonfunctional HSV-tk gene (ΔHSV-tk),14,15 resulting from an alternative splicing,14 underwent an increase relative to the whole GMC compartment after GCV treatment.
In the field of ex vivo gene therapy, recent experimental studies using murine animal models16-18 and clinical trials19,20 have reported significant complications regarding retroviral-mediated gene-modified hematopoietic stem cells, namely, the induction of target cell transformations by gene transfer.21-23 Moreover, mounting of immune responses against cells expressing HSV-tk has been reported.24-27 These phenomena should be considered in the design of future gene therapy trials. In this regard, molecular characterization of persistently circulating GMCs would help elucidate these phenomena28,29 and inform the pursuit of promising gene therapy approaches.
Taking advantage of our long-term follow-up (more than 10 years) of HSV-tk cells in our clinical trial, we have better characterized circulating GMCs in transplant recipients. Here, we report for the first time the results of long-term quantitative follow-up of gene-modified donor T cells more than 10 years after their infusion with a T-cell–depleted bone marrow graft. In addition, we report the results of molecular investigations revealing transgene alterations within long-term circulating GMCs. We precisely identify these molecular deletions, their time of occurrence, and the possible underlying mechanism.
Patients, materials, and methods
Approval was obtained from the Besançon ethics committee for these studies. Informed consent was obtained in accordance with the Declaration of Helsinki.
Clinical protocol and patient characteristics
Details of the protocol and clinical results were reported previously.9,30 GMCs were prepared as previously described6 with the G1Tk1SvNa vector (GTI, Gaithersburg, MD) carrying the HSV-tk gene and the neomycin phosphotransferase II (NeoR) gene under the 5′LTR and internal SV40 promoter, respectively.
There were 4 patients (3 chronic myelogenous leukemia and 1 acute lymphocytic leukemia Philadelphia positive) who received 6 × 105 donor CD3+ GMCs/kg (2 of whom developed GvHD that was successfully treated with GCV) and were still alive more than 10 years after bone marrow transplantation (BMT). At the time of this study, these patients were in complete remission, off immunosuppressants, and free of GvHD (Table 1).
Characteristics of patients, engraftment, GvHD occurrence, and GCV treatment, outcome, and immune response
| Patient . | Age, y . | Diagnosis . | Clinical status at time of BMT . | CD3+/tk+, ×105/kg . | GvHD . | Grade . | Response . | Follow-up, y (mo) after graft; remission . | Donor chimerism, % of mononuclear cells . | GMC-specific immune response . |
|---|---|---|---|---|---|---|---|---|---|---|
| 6 | 51 | CML | CP | 6 | aGvHD (d 20) | II | CR (GCV + steroids) | 10.1 (121); CR | 80 | Yes |
| 7 | 61 | ALL-Ph+ | First CR1 | 6 | — | — | — | 10.1 (121); CR* | 80 | Not detectable |
| 8 | 53 | CML | CP | 6 | — | — | — | 9.8 y (119); CR | 100 | Yes |
| 9 | 52 | CML | CP | 6 | cGvHD (d 136) | — | CR (GCV) | 9.5 (115); CR | 100 | Yes |
| Patient . | Age, y . | Diagnosis . | Clinical status at time of BMT . | CD3+/tk+, ×105/kg . | GvHD . | Grade . | Response . | Follow-up, y (mo) after graft; remission . | Donor chimerism, % of mononuclear cells . | GMC-specific immune response . |
|---|---|---|---|---|---|---|---|---|---|---|
| 6 | 51 | CML | CP | 6 | aGvHD (d 20) | II | CR (GCV + steroids) | 10.1 (121); CR | 80 | Yes |
| 7 | 61 | ALL-Ph+ | First CR1 | 6 | — | — | — | 10.1 (121); CR* | 80 | Not detectable |
| 8 | 53 | CML | CP | 6 | — | — | — | 9.8 y (119); CR | 100 | Yes |
| 9 | 52 | CML | CP | 6 | cGvHD (d 136) | — | CR (GCV) | 9.5 (115); CR | 100 | Yes |
CML indicates chronic myelogenous leukemia; CP, chronic phase; aGvHD, acute GvHD; CR, complete remission; ALL, acute lymphoblastic leukemia; cGvHD, chronic GvHD; —, not available.
Reappearance of a low level of Bcr/Abl transcripts 92 months after BMT.
Posttransplantation molecular monitoring of patients
Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article) reports the primer-pair combinations, reaction conditions, and polymerase chain reaction (PCR) product lengths for each PCR and fluorescent probe (for quantitative real-time PCR [QPCR]).
Analysis of Bcr/Abl fusion gene transcripts was routinely performed using qualitative reverse-transcription (RT)–PCR31 or quantitative QRTPCR32 detection.
Routine chimerism analyses were performed on marrow, peripheral blood, and CD3+ T cells using short tandem repeat (STR) quantitative analysis (since 2000).33
Investigation of T-cell clonality was performed using genomic DNA (gDNA) from patients' peripheral blood mononuclear cells (PBMCs) and from G418-reselected GMCs using the multiplexed fluorescent standardized European Biomed-2 protocol.34
Briefly, PCR assays were used to detect complete or partial DNA rearrangements of the T-cell receptor TCRγ chain (Vγ/Jγ, 2 multiplex PCR mixes A&B), with a sensitivity of 10−2 to 10−3
Peripheral blood samples were routinely cell-numbered and CD3, CD4, CD8, CD56 phenotyped.
Ex vivo reselection and cloning of circulating GMCs
Starting with 50 mL blood, 20 × 106 PBMCs were Ficoll harvested and expanded 7 days in culture in the presence of magnetic beads coated with CD3 and CD28 monoclonal antibodies (Xcyte Therapies, Seattle, WA) at a 1:1 ratio of beads-cells and 500 IU/mL recombinant IL-2 (rIL-2). This procedure allowed a 10- to 15-fold increase in the number of cells in culture. On day 7, GMCs were selected for 5 days in presence of 800 μg/mL G418 (Sigma, St Quentin Fallavier, France). At day 12, dead cells were removed by Ficoll centrifugation.
Reselected GMCs were lysed for DNA extraction or cloned by limiting dilution in the presence of 75 Gy–irradiated allogeneic EBV-transformed B-cell lines, 40 Gy–irradiated allogeneic PBMCs, 1 μg/mL phytohemagglutinin (PHA), and 500 UI/mL rIL-2, as previously described.35
Qualitative and quantitative PCR of NeoR and HSV-tk genes
gDNA was extracted from PBMCs using a standard salting-out procedure. Simple or nested PCR for retroviral-carried transgenes NeoR and HSV-tk was performed on posttransplantation PBMCs, G418-selected GMCs, and cloned T cells, as previously described.14,36 This enabled screening of resistant G418 T-cell clones obtained after G418-mediated reselection of GMCs, and identification of both wild-type (functional) and truncated (nonfunctional) forms of the HSV-tk gene.
QPCR (targeting NeoR, HSV-tk, and reference GAPDH) was performed in duplicate, on a lightcycler (Roche Diagnostics, Meylan, France), from 500 ng gDNA.37 The ΔΔ cycle threshold method38 was used to determine the amount of target relative to the amount of reference or calibrator (DNA extract with a known percentage [range, 100-0.01%] of NeoR+/HSV-tk+ GMCs). The NeoR and HSV-tk QPCR assays have comparable sensitivity (Figure S2).
Throughout the paper, the percentage of GMCs is calculated using transgene copy number obtain by QPCR, assuming that there is one transgene copy number per GMC. The absolute number of GMCs was estimated using white blood cell counts and assuming that circulating lymphocytes represent 2% of the total lymphocyte pool.39
Macromolecular localization of G1Tk1SvNa retroviral sequence deletions
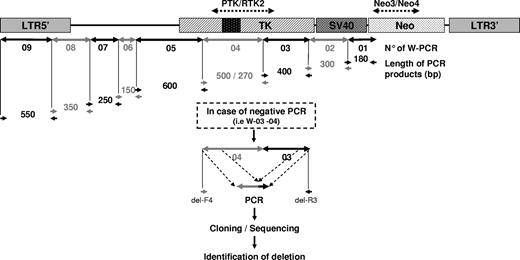
Starting from the region of the NeoR gene targeted by the NeoR-PCR assay and known to be expressed within the G418 reselected cloned GMCs, 9 different consecutive PCR assays in cascade (called “transgene walking” PCR) were designed, generating 9 PCR products (W-01 to W-09) differing in length. The PCR product W-04 enabled determination of wild-type HSV-tk/spliced-tk (ΔHSV-tk). The absence of PCR product in one or many successive PCRs indicated the presence of deletions (Figure 1). The PCR reactions and amplification conditions for the 9 assays were identical: 35 cycles of denaturation for 1 minute at 95°C, annealing for 1 minute, and extension for 1 minute at 72°C.
Transgene walking PCR assay. Starting from the NeoR gene, known to be present and expressed, 9 successive PCRs in sequence (W-01 to W-09) were designed to cover all transgene sequences to the LTR. PCR W-04 enabled detection of the spliced truncated HSV-tk gene. Different lengths enabled rapid determination of the 9 PCR products. In the case of negative PCRs (as illustrated for W-03 to W-04), secondary PCR products, obtained using transgene walking primers flanking the detected deletion area (del-F4 and del-R3 in this example), were cloned and sequenced.
Transgene walking PCR assay. Starting from the NeoR gene, known to be present and expressed, 9 successive PCRs in sequence (W-01 to W-09) were designed to cover all transgene sequences to the LTR. PCR W-04 enabled detection of the spliced truncated HSV-tk gene. Different lengths enabled rapid determination of the 9 PCR products. In the case of negative PCRs (as illustrated for W-03 to W-04), secondary PCR products, obtained using transgene walking primers flanking the detected deletion area (del-F4 and del-R3 in this example), were cloned and sequenced.
Identification of deletions and sensitive deletion-specific PCR design
After screening the DNA integrity of transgenes using transgene walking PCR, which enabled the detection of the absence of a PCR product in one or more successive PCRs, we amplified gDNA using primers of the neighbor's flanking PCR reactions that yielded a positive PCR signal (Figure 1).
After purification, generated PCR products were cloned in pCR-TOPO 2.1 TA cloning vector (InVitrogen, Cergy Pontoise, France). Plasmid DNA extracted from 4 recombinant transfected bacteria colonies was sequenced, on a 3130 automated sequencer in both orientations using Big Dye terminator chemistry (Applied Biosystems, Courtaboeuf, France).
Based on the sequencing, we designed a sensitive nested PCR: the first run (generic) used 2 outer primers (del-F6 or del-F7 and del-R1, NeoR-R4, or del-R2) flanking the deleted area. The second run used an inner reverse primer spanning the junction area (del-F5a or del-F6 associated with del-TK1-R, del-TK2-R, del-R1, or del-TK4-R) (Table S1).
Analysis of 5′-LTR integration sites
The locations of integration sites were determined using the highly sensitive linear amplification–mediated–PCR (LAM-PCR), as previously described.40,41 Briefly, gDNA was amplified using linear amplification with a G1TK1SvNa 5′LTR reverse-biotinylated primer LTR-Biot. PCR products were purified using magnetic selection. After enzymatic restriction (Tsp509I; BioLabs, Ipswich, United Kingdom) a 5′ fragment ligation (LigaFast Rapid DNA Ligation System; Promega, Charbonnières-les-bains, France) of a double-stranded asymmetric linker cassette was performed. The nested PCR was performed with GSP-1/CAS-1 for the first run and GSP-5/CAS-2 for the second run, using linker-cassette primers and consensus LTR newly designed primers specific to our retroviral vector, G1TK1SvNa.
Purified PCR products were cloned in a pCR-TOPO 2.1 T/A cloning vector (InVitrogen) for subsequent sequencing. Unknown genomic parts of the sequence, identified between the known 5′LTR and the ligated known prefabricated double-DNA strand cassette sequence, were aligned on a genome database using Blat online software (http://genome.ucsc.edu/cgi-bin/hgBlat, University of California Santa Cruz).
Based on sequencing data, we designed 4 site integration-specific PCR assays using 4 forward primers (Si-1-F, Si-3-F, Si-9-F, and Si-13-F) that were complementary to previously identified genomic integrated DNA and a reverse primer complementary to the 5′ LTR (Si-LTR-R).
Detection of deletions and time course analysis
After generic PCR using primers flanking the junction area, deletion-specific nested PCR was performed on gDNA extracted from the packaging cell line that produced the retroviral G1TK1SvNa supernatant used for the clinical study. Primers overlapping the junction sequence were used. Genomic DNA from the infused donor GMCs of the 4 patients was also screened with each PCR deletion assay for the presence of the previously identified deletions. Deletions in patients' PBMCs were examined at 1, 3, 6, and 9 years after transplantation.
Results
Gene-modified T cells circulate more than 10 years after infusion
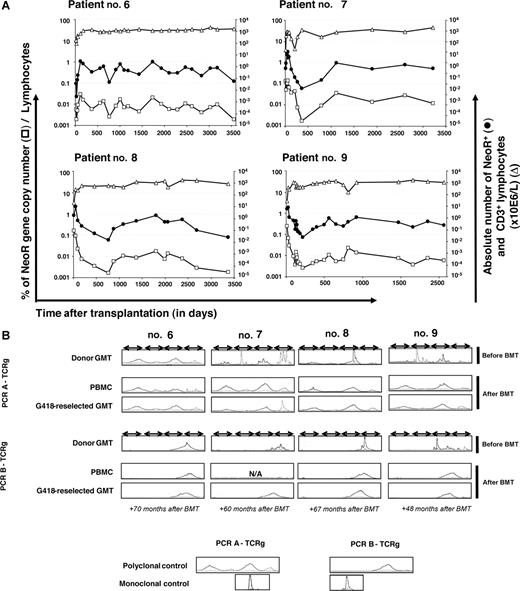
The presence of persistently circulating GMCs was quantified by QPCR of the NeoR gene (assuming that there is one transgene copy of per cell) in posttransplantation PBMC samples from the 4 patients. Gene-modified cells could be detected at a low frequency up to 10.1 years after transplantation. For the last available quantification of NeoR after transplantation (median, 9.9 years; range, 9.5-10.1 years), the mean number of NeoR+ GMCs was 0.150 × 106 cells/L (range, 0.03-0.283 × 106; Figure 2A). All 4 patients received the same dose of GMCs (6 × 105 GMCs/kg body weight). The patients' mean weight was 76.1 kg (range, 44.5-93 kg), and the mean estimated absolute number of GMCs was 37.5 × 106 (range, 7.5-70.1 × 106). Thus, comparing to the mean absolute number of GMCs initially infused, 45.7 × 106 (range, 26.7-55.8 × 106), it is clear that nearly the same quantity of GMCs was present in the patients 10 years after transplantation.
Monitoring of NeoR+ persistently circulating GMCs. (A) Kinetics of persistently circulating NeoR+ GMCs in 4 patients with a follow-up of more than 10 years. △ and ● indicate the absolute number of lymphocytes and GMCs, respectively (right y-axis); □ indicates the percentage of GMCs among circulating lymphocytes (assuming that there is one transgene copy of per cell) (left y-axis). The last analysis date was day 3557 (118.5 months), 3228 (118.5 months), 3379 (112.6 months), and 2063 (68.76 months) for patients 6, 7, 8, and 9, respectively. (B) Absence of T-cell clonality. Clonality was determined by PCR analysis of TCRγ gene rearrangements within donor GMCs before infusion, within PBMCs, and within GMCs G418-reselected at different time points after BMT, using 2 Vγ-Jγ PCR mixes (PCR A-TCRg: V γ 1f + V γ 10 − J γ 1.1/2.1 + J γ 1.3/2.3; PCR B-TCRg: V γ 9 + V γ 11 − J γ 1.1/2.1 + J γ 1.3/2.3). Both PCRs used consensus-specific primers from Vγ families, allowing coverage of most of the TCRγ rearrangements (sensitivity, 10−2 to 10−3). The Gaussian profile confirms the absence of detectable clonal expansion. Multiple peaks without Gaussian distribution were in favor of an oligoclonal T-cell population. Polyclonal and monoclonal controls were DNA extracted from tonsil and Jurkat cell lines, respectively. N/A indicates not analyzable.
Monitoring of NeoR+ persistently circulating GMCs. (A) Kinetics of persistently circulating NeoR+ GMCs in 4 patients with a follow-up of more than 10 years. △ and ● indicate the absolute number of lymphocytes and GMCs, respectively (right y-axis); □ indicates the percentage of GMCs among circulating lymphocytes (assuming that there is one transgene copy of per cell) (left y-axis). The last analysis date was day 3557 (118.5 months), 3228 (118.5 months), 3379 (112.6 months), and 2063 (68.76 months) for patients 6, 7, 8, and 9, respectively. (B) Absence of T-cell clonality. Clonality was determined by PCR analysis of TCRγ gene rearrangements within donor GMCs before infusion, within PBMCs, and within GMCs G418-reselected at different time points after BMT, using 2 Vγ-Jγ PCR mixes (PCR A-TCRg: V γ 1f + V γ 10 − J γ 1.1/2.1 + J γ 1.3/2.3; PCR B-TCRg: V γ 9 + V γ 11 − J γ 1.1/2.1 + J γ 1.3/2.3). Both PCRs used consensus-specific primers from Vγ families, allowing coverage of most of the TCRγ rearrangements (sensitivity, 10−2 to 10−3). The Gaussian profile confirms the absence of detectable clonal expansion. Multiple peaks without Gaussian distribution were in favor of an oligoclonal T-cell population. Polyclonal and monoclonal controls were DNA extracted from tonsil and Jurkat cell lines, respectively. N/A indicates not analyzable.
The NeoR transgene is expressed late after transplantation
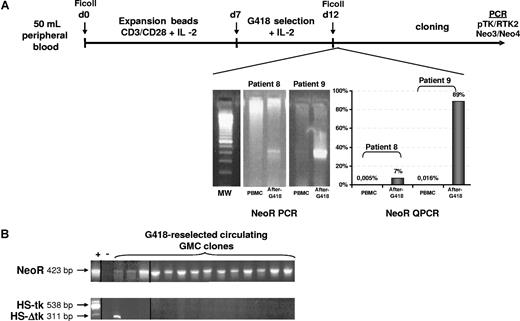
Posttransplantation PBMCs harvested from patients at different time points after BMT were activated and expanded (mean level of expansion, 19.5 ± 13.84-fold) before G418 selection (Figure 3A). When possible, G418 reselection performance was quantified using NeoR QPCR assay before cloning. Starting with 0.005% and 0.016% NeoR+ GMCs for patients 8 and 9, respectively, we were able to enrich the levels by 7% and 89%, respectively. However because of the low number of G418-resistant cells recovered after reselection for patients 6 and 7, NeoR QPCR was not performed in order to allow T-cell cloning of the cell suspension.
The NeoR transgene is expressed late after transplantation. (A) Synopsis of ex vivo GMC G418 reselection and cloning. The process consists of a 7-day polyclonal (CD3-CD28 beads + IL-2) expansion followed by a 5-day selection in the presence of G418 before cloning. A representative agarose gel electrophoresis of qualitative NeoR PCR is shown. When possible, QPCR for the NeoR gene was applied after the selection step to quantify the selection efficiency. After cloning, screening was performed with both qualitative PCR assays to confirm the presence of the NeoR and HSV-tk genes. (B) Representative 2% agarose gel electrophoresis of screening. Except for 1 clone positive for the truncated form of the HSV-tk gene (311 bp of PCR product), all clones were HSV-tk−/NeoR+. Vertical lines have been inserted to indicate a repositioned gel lane.
The NeoR transgene is expressed late after transplantation. (A) Synopsis of ex vivo GMC G418 reselection and cloning. The process consists of a 7-day polyclonal (CD3-CD28 beads + IL-2) expansion followed by a 5-day selection in the presence of G418 before cloning. A representative agarose gel electrophoresis of qualitative NeoR PCR is shown. When possible, QPCR for the NeoR gene was applied after the selection step to quantify the selection efficiency. After cloning, screening was performed with both qualitative PCR assays to confirm the presence of the NeoR and HSV-tk genes. (B) Representative 2% agarose gel electrophoresis of screening. Except for 1 clone positive for the truncated form of the HSV-tk gene (311 bp of PCR product), all clones were HSV-tk−/NeoR+. Vertical lines have been inserted to indicate a repositioned gel lane.
The absolute number of GMCs recovered after selection was insufficient to evaluate the functionality of the HSV-tk transgene through a GCV sensitivity assay based on GCV-mediated inhibition of TH3 incorporation.
To assess the clonality of circulating T cells and detect potentially emerging T-cell clones, we used a TCRγ gene rearrangement DNA PCR. Based on our previous study of T-cell repertoire,37,42 we found, as expected, oligoclonal profiles for 3 of 4 donors' preinfusion GMCs (GMCs for patient 6 presented a polyclonal profile). All patients' posttransplantation PBMCs were polyclonal (Figure 2B). To further assess for clonality within GMCs circulating long term after infusion, we analyzed for TCR rearrangement in PBMCs after G418 treatment. The profile of such cells was polyclonal as well in all 4 patients.
Partial transgene deletions occur in posttransplantation circulating G418-reselected GMCs
During long-term GMC monitoring as part of our clinical trial, we noted that a nested PCR was required to detect the HSV-tk gene in long-term posttransplantation PBMC samples, while the NeoR gene was detected after a single PCR run. This was why monitoring of GMCs was preferentially performed using NeoR QPCR. Thesimilar sensitivity of NeoR-specific and HSV-tk–specific qualitative PCR assays and NeoR-specific and HSV-tk–specific quantitative PCR assays (Figure S2) suggested that transgene alterations may occur in GMCs.
To further confirm this result, G418-reselected GMCs obtained from patients 6, 7, 8, and 9 at 70, 106, 67, and 76 months after transplantation, respectively, were cloned and analyzed by qualitative PCR for the presence of the NeoR and HSV-tk transgenes. A total of 876 clones (136, 612, 83, and 45 for patients 6, 7, 8, and 9, respectively, Table 2) was harvested at a seeding density of 10 cells/well and screened by PCR. The cloning efficiency at this seeding density was 12.3% (± 13.12%; range, 7.5%-31.9%), demonstrating clonality according to Poisson law. No clones were obtained at a lower seeding density. Among these 876 clones, 3.7% (n = 32) (n = 12, patient 6; n = 3, patient 7; n = 3, patient 8; n = 14, patient 9) were positive for NeoR. However, only 1 of these 32 NeoR+ clones was positive for the HSV-tk gene, a clone from patient 8 that carried the truncated spliced form of the HSV-tk gene14 (Table 2; Figure 3B). Qualitative PCR (W-09) targeting the 5′LTR part of the vector was performed on NeoR-negative/HSV-tk–negative clones of patients 8 (n = 79) and 9 (n = 31). All of the tested clones (n = 110) were found to be negative for this part of the vector sequence. Thus, partial transgene alterations may occur within the transgene, at least in the hybridization area of the HSV-tk PCR primers. Moreover, for patient 8, we harvested a clone that was positive for HSV-tk PCR (full-length HSV-tk) but negative for the presence of NeoR gene. This NeoR−/HSV-tk+ clone was not included in our deletion study but is discussed in “Discussion.”
PCR detection of NeoR, wild-type HSV-tk, and ΔHSV-tk (truncated) transgenes in GMC clones
| Patient; no. of clones . | 6; 136 . | 7; 612 . | 8; 83 . | 9; 45 . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mo after BMT and GMC infusion . | 70 . | 106 . | 67 . | 76 . | ||||||
| * | * | * | * | * | * | |||||
| NeoR+ | − | + | − | + | − | + | + | − | − | + |
| HSV-tk+ | − | − | − | − | − | − | − | + | − | − |
| ΔHSV-tk+ | − | − | − | − | − | − | + | − | − | − |
| 5′ LTR PCR (W-09) | ND | + | ND | + | − | + | + | + | − | + |
| Total, no. | 124 | 12 | 609 | 3 | 79 | 2 | 1 | 1 | 31 | 14 |
| Patient; no. of clones . | 6; 136 . | 7; 612 . | 8; 83 . | 9; 45 . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mo after BMT and GMC infusion . | 70 . | 106 . | 67 . | 76 . | ||||||
| * | * | * | * | * | * | |||||
| NeoR+ | − | + | − | + | − | + | + | − | − | + |
| HSV-tk+ | − | − | − | − | − | − | − | + | − | − |
| ΔHSV-tk+ | − | − | − | − | − | − | + | − | − | − |
| 5′ LTR PCR (W-09) | ND | + | ND | + | − | + | + | + | − | + |
| Total, no. | 124 | 12 | 609 | 3 | 79 | 2 | 1 | 1 | 31 | 14 |
ND indicates not done; +, present; and −, absent.
Data in these columns represent gene-modified clones, positive for LTR PCR.
Transgene walking PCR as a tool for transgene deletions identification
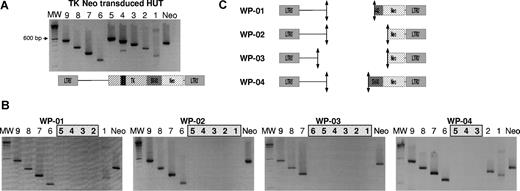
We validated a molecular tool on a HUT cell line transduced with the G1TK1SvNa vector and selected by G418. Nine PCRs, all performed with the same conditions, enabled generation of 9 PCR products of different lengths, ranging from 150 bp (PCR W-06) to 600 bp (PCR W-05). Remarkably, PCR W-04 generated 2 PCR products of 500 bp and 270 bp, corresponding to the full-length and truncated forms of the HSV-tk gene. Thus, the electrophoresis gel analysis shown in Figure 4A is the walking PCR signature of the entire undeleted transgene.
Deletions occur within the HSV-tk gene. (A) Agarose gel electrophoresis of PCR performed on DNA of an integral transgene. The 9 PCRs were performed on DNA extracted from 100% G1TKSvNa-transduced HUT cells. As expected, PCR W-04 produced 2 PCR products corresponding to the full-length and truncated HSV-tk gene. (B) Different deletion profiles (WP-01 to WP-04) obtained from clones of different patients. The absence of PCR products in successive PCRs delimited the deletion area. MW indicates molecular weight marker 100-bp DNA ladder. (C) Schematic representation of deletion areas on the G1TKSvNa vector linear map.
Deletions occur within the HSV-tk gene. (A) Agarose gel electrophoresis of PCR performed on DNA of an integral transgene. The 9 PCRs were performed on DNA extracted from 100% G1TKSvNa-transduced HUT cells. As expected, PCR W-04 produced 2 PCR products corresponding to the full-length and truncated HSV-tk gene. (B) Different deletion profiles (WP-01 to WP-04) obtained from clones of different patients. The absence of PCR products in successive PCRs delimited the deletion area. MW indicates molecular weight marker 100-bp DNA ladder. (C) Schematic representation of deletion areas on the G1TKSvNa vector linear map.
Performing this PCR assay on the 31 NeoR+/HSV-tk− clones enabled identification of 4 different PCR profiles based on the absence of PCR products within different successive PCRs, such as W-02 to W-05 (walking profile WP-01), W-01 to W-05 (WP-02), W-01 to W-06 (WP-03), and W-03 to W-05 (WP-04) (Figure 4B). All the clones from the same patient carried the same deletion walking profile. Based on these PCR profiles, we demonstrated that in all clones the entire HSV-tk gene is deleted together with flanking sequences, as schematically represented in Figure 4C.
Recombination motifs within the transgene sequence are involved in the deletion mechanism
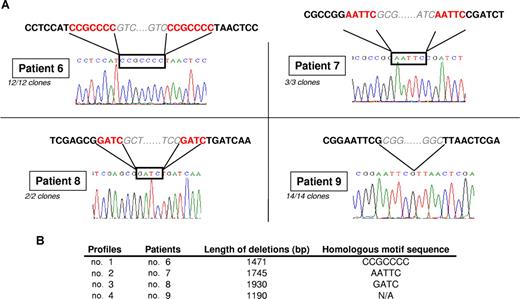
To precisely define the exact junction of the deletion, we sequenced PCR products obtained using flanking primers of the deletion area. As shown in Figure 5A, all NeoR+/HSV-tk− clones from the same patient carried exactly the same deletion. This enabled determination of deletions of 1471 bp, 1745 bp, 1930 bp, and 1190 bp for clones of patients 6, 7, 8, and 9, respectively, confirming the deletion of the entire HSV-tk gene associated with deletion of the SV40 internal promoter or packaging signal ψ sequence. Thus, deletions within the HSV-tk gene are different from the spliced HSV-tk form previously reported.14
Recombination motif sequences may explain the deletion mechanism. (A) Sequencing showed that the junction area corresponded to the homologous motif sequence for 3 of 4 patients and that every clone from the same patient carried the same deletion. (B) Size of deletion and precise homologous motif sequence for each patient.
Recombination motif sequences may explain the deletion mechanism. (A) Sequencing showed that the junction area corresponded to the homologous motif sequence for 3 of 4 patients and that every clone from the same patient carried the same deletion. (B) Size of deletion and precise homologous motif sequence for each patient.
For 3 of 4 deletions, examination of the junction sequence established the involvement of specific patterns of sequence homology in this deletion's mechanism: CCGCCC, AATTC, and GATC for patients 6, 7, and 8, respectively (Figure 5B). No remarkable sequence motifs were identified for patient 9. The identification of such homologous sequence patterns in the junction areas strongly suggests their involvement in a recombination event resulting from reverse-transcriptase errors.43
Clones derive from the same circulating GMC
Since we determined that all the clones from the same patient carried the same deletion, we sought to determine whether the clones derived from different persistent GMCs carrying the same deletion in vivo or from 1 circulating GMC amplified during the ex vivo CD3/CD28 expansion and G418 reselection phases before cloning (Figure S3). We identified, using LAM-PCR, the retroviral insertion sites (ISs) for all 4 patients and determined that all clones from the same patient harbor the same retroviral IS, which was not detected in any other patient. Thus, all clones harvested from a patient were derived from the same persistent circulating GMC that was amplified during the ex vivo process of amplification and G418 reselection.
Blat alignments revealed that for patient 6 retroviral sequences were inserted within the noncoding sequence, located in chromosome (ch) no. 12. The closest gene (located at 64 802 bp) encodes the TMEM5 protein. For patients 7 and 9, transgenes were inserted within intron 1 of the TNFRSF8 (CD30) gene and exon 2 of the RBM8A (RNA-binding motif protein 8A) gene, respectively, both located on ch no. 1. For patient 8, the IS was located within a hypothetical gene on ch no. 7. The Ephrin gene is the closest identified gene (at 566299 bp).
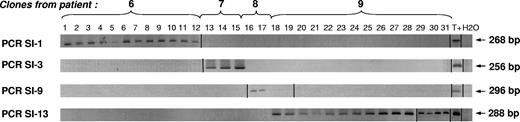
To confirm these results and to search for a patient-specific IS within the clones of the other patients, we designed site integration–specific PCR assays with a forward primer–specific genome sequence (Figure 6).
All the NeoR+/HSV-tk− clones from the same patient descend from the same circulating GMC. All 31 NeoR+/HSV-tk− clones were screened with the 4 retroviral insertion-specific PCRs. PCRs were performed with a LTR-specific reverse primer and a forward primer complementary to the genomic sequence where the retroviral sequence was inserted, previously identified after cloning/sequencing of the LAM PCR product. A PCR product of the expected length was detected in every clone of 1 patient and was absent in the 3 others. Vertical lines have been inserted to indicate a repositioned gel lane.
All the NeoR+/HSV-tk− clones from the same patient descend from the same circulating GMC. All 31 NeoR+/HSV-tk− clones were screened with the 4 retroviral insertion-specific PCRs. PCRs were performed with a LTR-specific reverse primer and a forward primer complementary to the genomic sequence where the retroviral sequence was inserted, previously identified after cloning/sequencing of the LAM PCR product. A PCR product of the expected length was detected in every clone of 1 patient and was absent in the 3 others. Vertical lines have been inserted to indicate a repositioned gel lane.
Deletions occur during the ex vivo production of retroviral GMCs
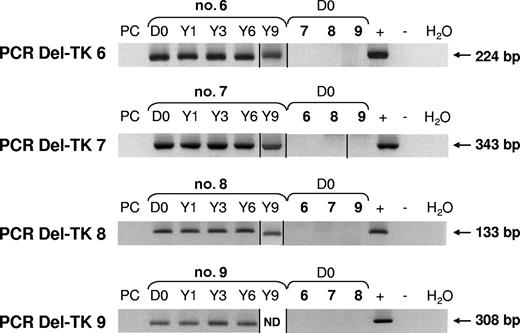
Using a newly designed nested PCR assay, with a second run using a primer spanning the junction area of the deletion, we searched for deletions within the DNA of the packaging cell line that produced the retroviral supernatant used in our clinical trial. For all of the identified deletions, no positive PCR signal was detected within the packaging cell line, but positive signals were detected within GMCs before infusion and until 10 years after grafting. This suggests that the deletions occurred during the ex vivo production of GMCs during the period between retroviral transduction and end of selection. Once again, we confirmed that a deletion found in 1 patient was never detected in any other patient (Figure 7). This is in agreement with the hypothesis that a slippage of reverse transcriptase is involved in the deletion mechanism.
Deletions occurred during the ex vivo production of GMCs. Using nested PCR assays with a second run using a specific primer spanning the junction sequence, no deletions within packaging cell line DNA (PC) were detected. PCR products were detected within GMC products before infusion (D0), as well as 1, 3, 6, and 9 years (Y) after infusion. Specific PCR was applied to GMC samples of the 4 patients, enabling demonstration that deletions were patient specific. ND indicates not done. Vertical lines have been inserted to indicate a repositioned gel lane.
Deletions occurred during the ex vivo production of GMCs. Using nested PCR assays with a second run using a specific primer spanning the junction sequence, no deletions within packaging cell line DNA (PC) were detected. PCR products were detected within GMC products before infusion (D0), as well as 1, 3, 6, and 9 years (Y) after infusion. Specific PCR was applied to GMC samples of the 4 patients, enabling demonstration that deletions were patient specific. ND indicates not done. Vertical lines have been inserted to indicate a repositioned gel lane.
Discussion
To date, the suicide gene therapy approach to control post-BMT alloreactivity represents one of the widest clinical applications in the field of T-cell–based gene transfer. Indeed, more than 100 patients have received HSV-tk GMCs worldwide,44 demonstrating the feasibility and safety9,10,45,46 as well as the efficiency47 of this approach. However, to increase the efficacy of suicide gene therapy, several studies have resulted in technologic improvements, involving the T-cell activation,48,49 transduction,50 and selection steps51,52 of GMC production, as well as in alternatives to the HSV-tk/GCV suicide gene system.52-54
In this context, our study establishes that mature allogeneic donor T cells having undergone gene modification and infused at the time of BMT survive in vivo for more than 10 years. After an initial rise following transplantation, circulating GMC blood counts progressively decline9 to a level constituting 0.001% to 0.01% of all lymphocytes. However, assuming that 2% of the total pool of lymphocytes circulates in the blood,39 the absolute number of persisting GMCs is approximately equal to the number of GMCs initially infused.
Long-term follow-up of genetically engineered stem cells or primary T cells was reported for the first gene therapy (adenosine deaminase) clinical trial55,56 and for large-animal models.57 Muul et al reported that autologous adenosine deaminase gene-corrected T cells were detectable in 1 patient at a frequency of 20% of total lymphocytes more than 10 years after infusion55 with efficient expression of the adenosine deaminase transgene. These cells have a survival advantage over nontransduced cells, due to increased clearance efficiency of soluble deleterious metabolites. In our study, HSV-tk–transduced cells were not expected to have a survival advantage over nontransduced cells. In phase 1 trials of donor lymphocyte administration of T cells transduced with the HSV-tk gene after allogeneic HLA genoidentical and haploidentical human stem cell transplantation, it was reported that GMCs survived more than 10 years47,58,59 and still expressed the ΔLNGFR selection gene. However, the frequency of circulating GMCs was not indicated. In both studies, immune responses against GMCs were reported26 and hypothesized to impair the long-term survival of GMCs.
In our study, it was possible to reselect GMCs from blood samples harvested 67 to 106 months after transplantation based on ex vivo resistance to G418. This demonstrates that some GMCs continued to express a functional NeoR transgene late after transplantation. Evaluation by NeoR QPCR revealed that efficiency of G418 reselection differed between both patients (7% in patient 8 and 89% in patient 9), with nevertheless a significant 3- and 4-log GMC enrichment in both patients, respectively. Several factors may have contributed to this difference between both patients, including a different frequency of GMCs in the starting amount of circulating GMCs (0.005% vs 0.016% for patients 8 and 9, respectively). Furthermore, despite an 89% selection efficiency in patient 9, most of the clones obtained subsequently were non–gene modified, thus suggesting that even in patient 9, clonable non–gene-modified cells survived G418 selection in significant numbers. Interestingly in all 4 patients, most clones were derived from non–gene-modified cells having survived ex vivo G418 selection. This could be at least partly expected in patient 8 in whom selection efficiency was low but less so in patient 9 in whom the postselection precloning material comprised 89% gene-modified cells. This finding suggests that the NeoR expression donor GMC having circulated more than 10 years after BMT had a lower cloning efficiency than the non–gene-modified T cells having survived the G418 treatment.
Analysis of TCRγ rearrangement determined that posttransplantation PBMCs from the 4 patients were polyclonal. Considering the method's limit of sensitivity (10−2), this suggests that there was no clonal proliferation within circulating PBMCs. The profile of G418-reselected cells was also polyclonal. This polyclonal profile may reflect contamination by the polyclonal repertoire of residual non-GMCs that escaped G418 reselection because of the low G418 reselection efficiency—not polyclonality of the GMC repertoire itself. Indeed, oligoclonal profiles were found within GMCs before infusion (for 3 of 4 donors), which is in agreement with our previous studies demonstrating that the gene transfer process can skew the TCRβV repertoire of infused GMCs.37,42 However, the T-cell repertoire profile of G418-reselected GMCs of patient 9 remained polyclonal, with 89% NeoR+ cell enrichment, suggesting again that there was no monoclonal expansion in these GMCs.
Due to an insufficient number of cells at the end of ex vivo G418 reselection, it was not possible to perform a GCV sensitivity assay based on GCV-induced inhibition of proliferation. Therefore, cells retrieved after G418 reselection were cloned. Thirty-three GMCs were obtained from 4 patients, of which 31 were NeoR+/HSV-tk−, 1 was NeoR+/ΔHSV-tk+, and 1 was NeoR−/HSV-tk+. The latter clone was obtained due to partial reselection efficiency. Indeed, only 3.7% of the total number (n = 876) of harvested clones were NeoR+. Surprisingly, for each patient, all clones presented the same patient-specific deletion—except for 2 clones of patient 8—suggesting that they derived from 1 originating cell. Sequencing of the IS after LAM-PCR confirmed that all clones within each patient were identical. A deletion found in the clones of a given patient was also retrieved in his circulating GMCs and the infused donor's GMCs at the time of BMT, but was not found in other patients' cells, or in the packaging cell line used to produce the retroviral vector. This demonstrates that each deletion was patient specific and occurred during GMC production. The mechanisms involved in such HSV-tk gene deletions have not been elucidated but could possibly result from reverse-transcriptase slippage, as previously reported during minus-strand DNA synthesis.43,60,61 Indeed, the presence of direct repeats, and thus potential stem-loop structures, in the retroviral genome was reported to increase the mutation rate 50-fold, leading to deletions in retroviral sequences.61 Of interest, we found direct repeat sequences in flanking deletion areas in 3 of 4 patients. To our knowledge, our study is the first in vivo description of such deletions.
As analysis of TCRγ rearrangement did not demonstrate monoclonal proliferation in reselected GMCs before cloning, our results raise questions regarding the representativeness of GMC clones. It is possible that multiple GMC clones circulate long-term in vivo and may be able to expand during the 2-week ex vivo reselection process (leading to an oligoclonal profile of TCRγ rearrangements), but not during the cloning step that requires 1 to 3 weeks of additional culture. This is consistent with the observation that HSV-tk–specific QPCR gives a positive signal, demonstrating that some HSV-tk+ GMC clones circulate in vivo. These HSV-tk+ GMCs were not reselected, possibly because of deletions in the NeoR gene. Of interest, in the case of patient 8, 1 NeoR−/HSV-tk+ clone escaped G418 reselection, demonstrating that deletions in the NeoR gene may occur but cannot be analyzed, as GMCs can be purified only based on the presence and functional expression of the NeoR gene. However, the fact that the NeoR QPCR performed on circulating PBMCs yielded higher signals than HSV-tk QPCR strongly argues in favor of a higher frequency of circulating NeoR+/HSV-tk+ cells than NeoR−/HSV-tk+ cells.
In light of the low cloning efficiency obtained after reselection, few GMC clones may be able to persist through the whole process of reselection and cloning. Most circulating GMCs may be exhausted by the expansion induced during the 12 days of ex vivo culture required for gene transfer at the time of GMC production and/or by the peripheral expansion observed early after BMT.9 After ex vivo and in vivo expansions, GMCs may have shorter telomeres than do bone marrow–derived T cells recovered from lymphopoiesis.
The preferential deletion of the HSV-tk gene may provide a survival advantage to GMCs, compared with HSV-tk+ GMCs, especially in the setting of GCV administration upon GvHD treatment. We previously showed that GCV administration led to elimination of HSV-tk+ GMCs, but that a fraction of GMCs expressing a truncated nonfunctional ΔHSV-tk gene survived GCV administration. This ΔHSV-tk gene resulted from an alternative splicing that occurred during the production of the retroviral vector.14 ΔHSV-tk gene–transduced GMCs represented approximately 10% of total GMCs at the time of infusion and became the major form of GMCs after GCV administration.14 Surprisingly, only 1 NeoR+/ΔHSV-tk+ clone was obtained among 33 GMC clones.
Complete deletion of the HSV-tk gene may provide additional advantages such as an escape advantage in the setting of immune GMC-specific response. Indeed, occurrence of an immune response against retrovirally transduced GMCs has been reported after single infusion (enhanced after multiple infusion) of anti–human immunodeficiency virus (HIV) GMC clones expressing a hygromycin resistance (Hy)/HSV-tk fusion gene in immunocompetent HIV patients.24 The same was reported after delayed infusion of polyclonal GMCs expressing Hy/HSV-tk25 or HSV-tk/ΔLNGFR10 in postrelapse immunoreconstituted allogeneic hematopoietic stem cell (HSC) patients. GMC-specific immune responses were mediated by CD8+ cells, HLA class I restricted,24,25 and directed toward both HSV-tk and Hy.25
In the present clinical trial, using the interferon-γ enzyme-linked immunosorbent spot (Elispot) assay,27 we also detected immune responses against GMCs. Despite these immune responses, circulating GMCs were detected long term after transplantation. The finding that most long-term–derived GMC clones were NeoR+ HSV-tk− suggests that complete deletion of HSV-tk may provide a beneficial survival advantage over GMCs expressing the HSV-tk or ΔHSV-tk gene. Therefore, immune responses could be preferentially directed against HSV-tk, rather than the NeoR gene, in accordance with our Elispot assays.27 It should be noted that one patient7 also had GMC clones with deletion in HSV-tk, in the absence of a detectable immune response and GCV administration, suggesting that these selection pressures may not be necessary to allow emergence of NeoR+/HSV-tk− GMCs, which were present at a low frequency (< 10−3) in the infused GMC product. The preferential cloning of such rare NeoR+/HSV-tk− GMCs raises questions regarding the possible long-term toxicity of HSV-tk gene expression in human T cells. We can hypothesize that long-term HSV-tk gene expression within a T cell affects the stasis levels of purine nucleotide pools and normal cellular metabolism, imparting a negative growth advantage leading, over time, to the elimination of these cells.
Occurrence of immune responses was associated with complete elimination of GMCs in studies by Riddell et al24 and Berger et al25 In the Riddell et al study, GMCs were quantified using a Hy-specific PCR assay, but multiple GMC infusions were performed, accounting for GMC elimination. In the Berger et al study, GMCs were quantified using a Hy/HSV-tk–specific QPCR encompassing the junction of the Hy and HSV-tk genes and were undetectable 1 month after a single GMC infusion. Therefore, this assay may underestimate the true amount of GMCs in the case of HSV-tk deletions.
Long-term persistence of a very low frequency of GMCs carrying HSV-tk deletions could raise questions regarding the control of posttransplantation alloreactivity with the suicide gene approach. However, despite a possible survival advantage allowing these cells to escape immune responses or GCV administration, the presence of such deletions was not deleterious, as acute and chronic GvHD occurring up to 135 days after transplantation9 was efficiently controlled by GCV administration.
In conclusion, this work suggests the occurrence of an extremely rare initial gene deletion event and a possible in vivo growth/survival advantage of rare GMC clones carrying transgene deletions. Overall, circulating gene-modified T cells persist in vivo for more than a decade after infusion in allogeneic settings. Such cells have undergone significant in vivo selection and represent only a very small percentage of the infused T cells at the time of transplantation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Program Hospitalier de Recherche Clinique (PHRC nos. 950898 and RC39), the Ministère de l'Enseignement Supérieur et de la Recherche (Center Réseau de Développement des Thérapies Géniques [CRTG]), Genetic Therapy, Novartis, and the European Community (Biomed contract no. CT97-2074). M.D. has benefited from fellowships of the Ligue Contre le Cancer, Comité du Jura.
We thank the technical staff of the molecular biology laboratory of Etablissement Français du Sang Bourgogne/Franche-Comté (EFSB/F-C) for the clonality study, as well as Anne Duperrier for her work in GMC quantification.
Authorship
Contribution: M.D. designed and executed the molecular study and wrote the original draft of the paper; P.M.-L., C.P., and C.H. performed the in vitro and cellular cloning; J.-M.C. collected samples and performed monitoring; B.L., J.-Y.C., and E.D. provided clinical data and patient samples; P.T. and J.Y.C. initiated the clinical study and contributed to its execution and writing of the paper; E.R. and C.F. directed the study and contributed to writing the paper.
M.D. and P.M.-L. contributed equally to this work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Christophe Ferrand, Laboratoire de Thérapeutique Immuno-Moléculaire, UMR INSERM-university U645/IFR133, Etablissement Français du Sang–Bourgogne/Franche-Comté, 1, Bd Alexandre Fleming, 25020 Besançon cedex, France; e-mail: christophe.ferrand@efs.sante.fr.