The T-cell receptor β (TCRβ)/pre-TCRα (pTα) pre-TCR complex (pre-TCR) signals the expansion and differentiation of de-veloping thymocytes. Functional pro-perties of the pre-TCR rely on its unique pTα chain, which suggests the participation of specific intracellular adaptors. However, pTα-interacting molecules remain unknown. Here, we identified a polyproline-arginine sequence in the human pTα cytoplasmic tail that interacted in vitro with SH3 domains of the CIN85/CMS family of adaptors, and mediated the recruitment of multiprotein complexes involving all (CMS, CIN85, and CD2BP3) members. Supporting the physiologic relevance of this interaction, we found that 1 such adaptor, CMS, interacted in vivo with human pTα, and its expression was selectively up-regulated during human thymopoiesis in pre-TCR–activated thymocytes. Upon activation, pre-TCR clustering was induced, and CMS and polymerized actin were simultaneously recruited to the pre-TCR activation site. CMS also associated via its C-terminal region to the actin cytoskeleton in the endocytic compartment, where it colocalized with internalized pTα in traffic to lysosomal degradation. Notably, deletion of the pTα CIN85/CMS-binding motif impaired pre-TCR–mediated Ca2+ mobilization and NFAT transcriptional activity, and precluded activation induced by overexpression of a CMS-SH3 N-terminal mutant. These results provide the first molecular evidence for a pTα intracellular adaptor involved in pre-TCR function.
Introduction
Intrathymic differentiation of αβ T lymphocytes is a complex process regulated at 2 consecutive checkpoints through the pre–T-cell receptor (TCR) and the αβ TCR.1,2 Surface expression of the TCR β–pre-TCRα (pTα) pre-TCR heterodimer (pre-TCR) signals the expansion and further differentiation of developing pre-T cells, a process called β-selection.3,–5 Thereafter, replacement of the pre-TCR by the mature TCRαβ allows for positive and negative selection of developing thymocytes.1,2
The similar biochemical composition of the pre-TCR and the TCRαβ initially supported the view that pTα is simply a “surrogate” TCRα chain. However, pTα and TCRα are not interchangeable partners of TCRβ during T-cell development.6,7 Rather, the pTα molecule endows the pre-TCR with unique functional properties, such as constitutive clustering and ligand-independent activation,5,8,,–11 which likely occur through oligomerization mediated by the pTα extracellular (EC) domain.12 Other pTα structural features that could account for the unique functional properties of the pre-TCR rely on the presence of a long cytoplasmic tail that could mediate pre-TCR signaling. However, despite being highly conserved at the EC and transmembrane (TM) domains, mouse and human pTα differ significantly at the cytoplasmic (CT) domain (30 and 114 amino acids, respectively). This difference, together with experimental data in mice, initially diminished the functional relevance of the pTα CT domain.13,14 In contrast, the human pTα CT domain was shown to serve an endoplasmic reticulum retention function that regulated pre-TCR assembly and expression.15 Moreover, constitutive pre-TCR internalization and degradation16,17 was also dependent on the pTα CT domain in humans.16 Despite these initial discrepancies, more recent data have pointed to an essential and previously unappreciated functional role for the CT tail of pTα also in mice, since the C-terminal (C-term) portion of murine pTα was proved to be crucial for intrathymic development.18 What exactly the pTα CT domain does to promote pre-TCR signaling remains an open question, but proper pre-TCR function seems to require the contribution of 2 proline-rich sequences present in its C-term portion.18 It is thus likely that polyproline sequences within the pTα tail directly interact with molecules that could propagate signals emanating from the pre-TCR. Therefore, the characterization of those intracellular adaptors would be essential to understand the basis of the unique signaling properties of the pre-TCR. However, proteins that associate with the pTα cytoplasmic domain have not been identified as yet.
The proline-rich tandem repeat (PPTHR and PPSRK) present within the murine pTα CT domain is conserved as a single sequence (PPGRK) in humans.4 Although different from the consensus PXXP or PPLP SH3-binding motifs, this sequence displays some homology with 2 tandem PPGHR sequences within the CT tail of the costimulatory molecule CD2.4,18 In mature T cells, this region is responsible for the direct interaction of CD2 with the intracellular adaptor CD2BP2, which is involved in signal transduction.19 Additional molecules interact with the CD2 CT tail through distinct proline-rich domains, including the CD2BP1 adaptor, implicated in actin polymerization and T-cell adhesion, motility, and activation20,–22 ; and CMS and CIN85, 2 members of the CMS/CIN85 adaptor family shown to regulate cytoskeletal rearrangements and T-cell polarization.23,24
We show in this study that, besides the conserved CD2-like proline-rich sequence, the human pTα CT domain carries a polyproline-arginine sequence that fits the atypical recognition consensus recently reported for SH3 domains of the CIN85/CMS family of adaptors.25,26 This sequence mediates binding of CIN85/CMS adaptors to multiple signaling molecules, including the CD2 coreceptor in T cells,23 and regulates multiple functions in different cell types,27 suggesting a role in pre-TCR function as well. We provide evidence that the polyproline motif identified here in the pTα CT tail is in fact an indispensable binding site for CIN85/CMS adaptors involved in pre-TCR signaling. Therefore, our data identify CIN85/CMS proteins as the first known intracellular adaptors of human pTα required for pre-TCR function.
Materials and methods
Cell lines and transfections
cDNA constructs
cDNAs encoding human CIN85 or CMS were amplified by polymerase chain reaction (PCR) from thymocyte cDNA. PCR products were cloned into either the pcDNA3-Flag or the pcDNA3-HA vector. Flag-tagged CMS and CIN85/CD2BP3 truncated forms were generated by PCR using CMS-Flag and CIN85-Flag, respectively. GST (glutathione S-transferase) fusion proteins for the full-length pTα CT domain (GST-pTα), or for the truncated proline-rich ΔPro1 and ΔPro2 pTα mutants (GST-pTαΔPro1, GST-pTαΔPro2) were PCR-amplified using a pcDNA3-pTα vector15 and cloned into pGEX-4T1 (Amersham Biosciences, Arlington Heights, IL). The GST-pTαΔPro3 construct generated from the GST-pTα construct was subcloned into pGEX-4T3 (Amersham Biosciences). Green fluorescent protein (GFP)–tagged CMS constructs (CMS-GFP, SH3ABC-GFP, and C-term–GFP) generated by PCR using CMS-Flag were subcloned into pEGFP-N1 (BD Biosciences, Palo Alto, CA). Cloned constructs were DNA sequenced. Specific primer combinations used are shown in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
GST fusion proteins and GST binding assays
GST fusion proteins were affinity-purified using glutathione-Sepharose beads (Amersham Biosciences). COS7 cells were transiently transfected with either Flag- or HA-tagged CIN85 or CMS constructs, lysed in phosphate-buffered saline (PBS) containing 1% Triton X-100 (Sigma-Aldrich, St Louis, MO), protease inhibition cocktail (Roche, Indianapolis, IN), 1 mM PMSF, and 50 mM NaF, and incubated with GST fusion proteins coupled to gluthatione-Sepharose beads. Bound proteins were resolved by SDS-PAGE under reducing conditions. Immunoblotting was performed using anti-Flag (M2; Sigma-Aldrich) or anti-HA (12CA5; Roche) monoclonal antibodies (mAbs) plus horseradish peroxidase–anti-mouse IgG (Jackson Immunoresearch, West Grove, PA).
Immunoprecipitations and immunoblotting
SupT1 cells transfected with CMS-Flag were lysed in 0.3% Brij96V (Sigma-Aldrich) buffer. Immunoprecipitation was performed as described16 with a polyclonal rabbit antibody against the human pTα cytoplasmic domain,15 and immunoblotting was performed with anti-Flag. JRpTαWT- and JRpTαΔPro1-transduced cells were subjected to immunoprecipitation and immunoblotting using anti-Flag. Western immunoblotting of endogenous CMS and transfected Flag-tagged CMS and CMS-SH3ABC forms was performed with anti-CMS/CD2AP (H-290; Santa Cruz Biotechnology, Santa Cruz, CA) and anti-Flag, respectively.
Retroviral infections and flow cytometry
JR3.11 cells were transduced as described30 using the MigR1.1-GFP retroviral bicistronic vector31 encoding Flag-tagged full-length pTαWT or pTαΔPro1. Primer combinations used for subcloning are described in Table S1. Sorted GFP+-transduced cells were stained with an anti-CD3ϵ (UCHT1) mAb plus APC–anti-mouse Igs (BD Biosciences) and analyzed in a FACSCalibur (BD Biosciences). Background staining was determined with an irrelevant isotype-matched mAb plus APC–anti-mouse Igs.
Thymocyte subset isolation and Northern blotting
Postnatal thymocytes were isolated from thymus samples of patients aged 1 month to 3 years after informed consent and approval by the Consejo Superior de Investigaciones Científicas (CSIC) Review Board was provided. The following cell subsets were isolated from the very same thymus sample by Percoll (Amersham Pharmacia Biotech, Piscataway, NJ) density fractionation and AutoMacs (Miltenyi Biotec, Bergisch Gladbach, Germany) sorting:30,32 pre-β–selected (CD4+ CD8− CD3−), pre-TCR+ (CD4+ CD8+ CD3+ TCRαβ−), post-β–selected (CD4+ CD8+ CD3−), double-positive (DP) TCRαβ+ (CD4+ CD8+ CD3+), and single-positive (SP) TCRαβ+ (CD3+ CD4+ or CD8+). Total RNA (10 μg) was resolved by formaldehyde-agarose gel electrophoresis and blotted onto nitrocellulose membranes (Zeta-Probe B; Bio-Rad). Hybridization was performed as described.33
Confocal microscopy
SupT1 cells transiently transfected with CMS-GFP, SH3ABC-GFP, or C-term–GFP were seeded onto poly-L-lysine (Sigma-Aldrich)–precoated coverslips, and fixed with 4% paraformaldehyde for 30 minutes at room temperature (RT). Labeling with phalloidin-TRITC (Sigma-Aldrich), phalloidin-Alexa647 (Invitrogen, Carlsbad, CA), anti-CD63 (Developmental Studies Hybridoma Bank, Iowa City, IA), anti-EEA1, and anti-Lamp1 (BD Biosciences) was performed in the presence of 0.05% saponin (Sigma-Aldrich). Secondary antibodies included FITC–anti-IgG2a (Southern Biotechnology, Birmingham, AL), Alexa555–anti-mouse IgGs, Alexa647–anti-IgG1, Alexa555-IgG1, and Alexa488–anti-FITC (Invitrogen).
For pre-TCR activation, cells were incubated with anti-CD3ϵ plus Alexa555–anti-mouse IgGs on ice, washed, and incubated for 2 minutes at 37°C prior to fixation. Pre-TCR stimulation with anti-CD3ϵ–coated magnetic beads (Dynabeads; Dynal, Oslo, Norway) was performed for 5 or 15 minutes at 37°C (1:1 ratio) prior to fixation. To analyze pTα internalization, cells were incubated with an anti-pTα mAb10 for 15 minutes at 37°C, washed, and fixed or incubated for additional 15 minutes at 37°C prior to fixation. Confocal microscopy was performed in a Radiance 2000 confocal microscope (Bio-Rad) coupled to an Axiovert S100TV inverted microscope (Zeiss, Oberkochen, Germany) using Methamorph 6.1R6 software (Universal Imaging, Downingtown, PA). Images are single XY sections extracted from a Z-series of optical sections recorded at 0.5-μm intervals (63×/1.4 oil Plan-Apochromat objective).
Ca2+ mobilization assay
JR3.11-transduced cells were loaded with 2 μg/mL Indo-1 AM (Molecular Probes, Eugene, OR) in cell loading medium (Hanks balanced salt solution [HBSS] containing 1 mM each CaCl2 and MgCl2 and 1% FCS) for 30 minutes at 30°C in the dark. Washed cells were incubated in cell loading medium for 5 minutes at 37°C, and 510-nm (FL4 channel) and 400-nm (FL5 channel) emissions were analyzed in a FACSCVantage SE Cell Sorter (BD Biosciences) at different time points both before and after stimulation with anti-CD3ϵ. Ionophore (Sigma-Aldrich) treatment was used to control for cell viability and intact calcium stores. The data were processed for calculating the FL4/FL5 emission ratio using the Three Star FlowJo software (BD Biosciences).
NFAT transcriptional activation assay
JR3.11-transduced cells were transfected with a luciferase reporter plasmid containing 3 tandem copies of the composite NFAT/AP1-response element from the human IL-2 gene promoter, together with a pRL-CMV Renilla reporter construct (Promega, Madison, WI), as described.34 After 24 hours, cells were cultured for 6 hours with or without plate-bound anti-CD3ϵ or anti-pTα mAbs and lysed using the Dual luciferase assay Kit (Promega). Firefly luciferase and Renilla luciferase activities were determined in duplicates. The luciferase activity was normalized by the Renilla luciferase activity and expressed as fold-induction relative to the basal activity seen in nonstimulated cells.
Results
CIN85 and CMS interact with the cytoplasmic domain of human pTα through different SH3 domains
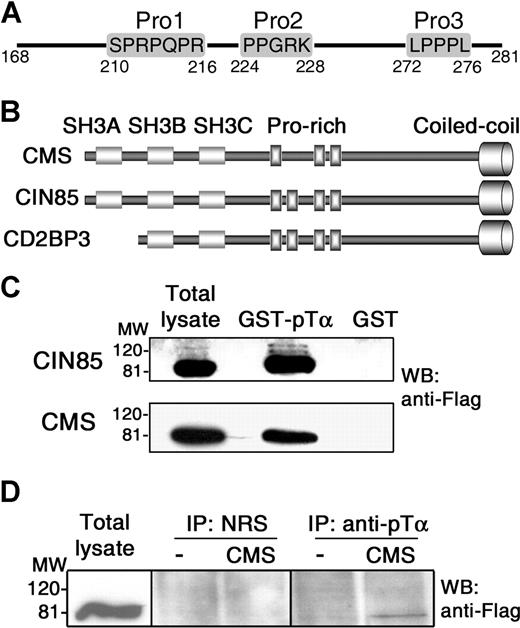
Besides the reported CD2-like proline-rich sequence (Figure 1A; Pro2) that is reiterated in mouse, the human pTα CT domain was found to carry 2 additional proline-based sequences: a C-term noncanonical proline-rich sequence (LPPPL; Pro3), and a polyproline-arginine sequence (SPRPQPR; Pro1) placed directly upstream of Pro2 (Figure 1A), which fits the PX(P/A)XXR or PXXXPR recognition consensus of CIN85/CMS SH3 domains identified in multiple signaling molecules.25,26 CIN85 and CMS adaptors share a similar structure with 3 SH3 domains in the amino end, a central proline-rich region, and a C-term coiled-coil domain27 (Figure 1B). A third member of the family, CD2BP3, is a CIN85 isoform which lacks the first SH3 domain due to an alternative splicing.24,27
In vitro and in vivo interactions of CIN85/CMS adaptors with the human pTα cytoplasmic tail. (A) Proline-rich motifs within the cytoplasmic domain of human pTα. (B) Domain organization of CMS, CIN85, and CD2BP3 members of the CIN85/CMS adaptor family. (C) CMS and CIN85 interact in vitro with the human pTα cytoplasmic domain. Cell lysates from COS7 cells transfected with Flag-tagged CMS or CIN85 were pulled-down with a GST fusion protein of the pTα tail (GST-pTα) or with GST alone. Immunoblotting was carried out with an anti-Flag mAb. (D) CMS interacts in vivo with the human pTα cytoplasmic domain. Total-cell lysates of SupT1 cells transfected with Flag-tagged CMS or with an empty vector were immunoprecipitated with a rabbit antibody against the human pTα cytoplasmic tail or with preimmune rabbit serum (NRS), and analyzed by Western blotting with anti-Flag. Data are representative of at least 3 independent experiments. Vertical lines have been inserted to indicate a repositioned gel lane.
In vitro and in vivo interactions of CIN85/CMS adaptors with the human pTα cytoplasmic tail. (A) Proline-rich motifs within the cytoplasmic domain of human pTα. (B) Domain organization of CMS, CIN85, and CD2BP3 members of the CIN85/CMS adaptor family. (C) CMS and CIN85 interact in vitro with the human pTα cytoplasmic domain. Cell lysates from COS7 cells transfected with Flag-tagged CMS or CIN85 were pulled-down with a GST fusion protein of the pTα tail (GST-pTα) or with GST alone. Immunoblotting was carried out with an anti-Flag mAb. (D) CMS interacts in vivo with the human pTα cytoplasmic domain. Total-cell lysates of SupT1 cells transfected with Flag-tagged CMS or with an empty vector were immunoprecipitated with a rabbit antibody against the human pTα cytoplasmic tail or with preimmune rabbit serum (NRS), and analyzed by Western blotting with anti-Flag. Data are representative of at least 3 independent experiments. Vertical lines have been inserted to indicate a repositioned gel lane.
To investigate whether CIN85/CMS adaptors physically interact with human pTα, a GST-pTα fusion protein containing the pTα CT domain (GST-pTα) was used in pull-down assays of COS7 cells transfected with Flag-tagged full-length CIN85, CMS, or CD2BP3 proteins. Immunoblotting with anti-Flag revealed that both CIN85 and CMS specifically associate with the CT tail of pTα (Figure 1C), while interaction of CD2BP3 was hardly detected (see “Recruitment of multiprotein complexes involving CMS, CIN85, and CD2BP3 adaptors to the cytoplasmic domain of the human pTα chain”). Next, CMS and CIN85 binding to pTα was assessed in vivo in SupT1 pre-T cells, which naturally express physiologic levels of surface pre-TCR.15 Immunoprecipitations carried out with an anti-pTα tail antibody15 after transfection with CMS-Flag confirmed that CMS was specifically coimmunoprecipitated with pTα from nonstimulated SupT1 cells at low amounts, as expected from the low surface pre-TCR expression levels (Figure 1D), and similar CMS levels were observed upon anti-CD3ϵ activation (not shown). Therefore, CMS specifically associates both in vitro and in vivo with the CT tail of human pTα. Equivalent experiments failed to reveal a CIN85-pTα association in vivo, even under activating conditions (data not shown), suggesting that CIN85 has a weaker affinity for pTα than CMS in SupT1 pre-T cells.
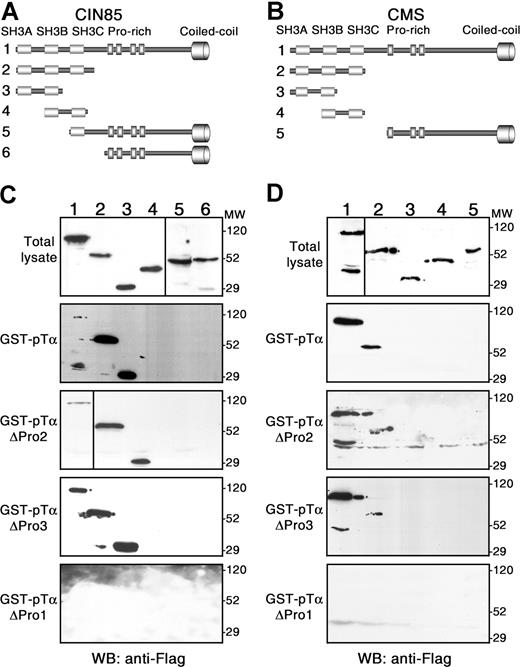
To identify the specific CIN85/CMS domains involved in pTα binding, GST-pTα pull-downs were performed using Flag-tagged truncated forms of either CIN85 or CMS (Figure 2A,B, respectively). CIN85-SH3ABC and CIN85-SH3AB truncated proteins were precipitated with GST-pTα at higher amounts than full-length CIN85 (Figure 2C; lanes 1-3). However, no binding of CIN85-SH3BC and neither of the CIN85 forms lacking the SH3AB N-terminal (N-term) region was detected (Figure 2C; lanes 4-6), suggesting that the SH3A domain is essential for specific interaction of CIN85 with the pTα tail. Accordingly, CD2BP3 bound very inefficiently to pTα (see “Recruitment of multiprotein complexes involving CMS, CIN85, and CD2BP3 adaptors to the cytoplasmic domain of the human pTα chain”; Figure 3A). In contrast to CIN85, full-length CMS was coprecipitated with pTα more efficiently than the CMS-SH3ABC truncated form (Figure 2D; lanes 1-2), and at higher amounts than full-length CIN85 (compare Figure 2D, lane 1 with Figure 2C, lane 1). No binding of the C-term CMS fragment lacking the SH3 domains was observed (Figure 2D; lane 5), and deletion of either the SH3C or SH3A domains also abolished the interaction of the remaining SH3 domains (Figure 2D; lanes 3-4), suggesting that both SH3C and SH3A are simultaneously required for pTα binding. Although we cannot formally exclude that the folding of the truncated proteins could account for the observed results, as a whole, our data suggest that CIN85 and CMS bind to pTα through different SH3 domains, as previously reported for binding to other proteins such as Cbl.24
CMS and CIN85 interact with the polyproline-arginine sequence within the human pTα tail through different SH3 domains. Schematic representation of Flag-tagged CIN85 (A) and CMS (B) forms used in pull-down assays shown in panels C and D, respectively. Pull-down assays were performed with GST fusion proteins of either the wild-type pTα cytoplasmic tail (GST-pTα) or the indicated proline-rich deletion mutants (GST-pTαΔPro1, GST-pTαΔPro2, and GST-pTαΔPro3). Precipitated proteins were analyzed by Western blotting with an anti-Flag mAb. Data are representative of at least 3 independent experiments. Vertical lines have been inserted to indicate a repositioned gel lane.
CMS and CIN85 interact with the polyproline-arginine sequence within the human pTα tail through different SH3 domains. Schematic representation of Flag-tagged CIN85 (A) and CMS (B) forms used in pull-down assays shown in panels C and D, respectively. Pull-down assays were performed with GST fusion proteins of either the wild-type pTα cytoplasmic tail (GST-pTα) or the indicated proline-rich deletion mutants (GST-pTαΔPro1, GST-pTαΔPro2, and GST-pTαΔPro3). Precipitated proteins were analyzed by Western blotting with an anti-Flag mAb. Data are representative of at least 3 independent experiments. Vertical lines have been inserted to indicate a repositioned gel lane.
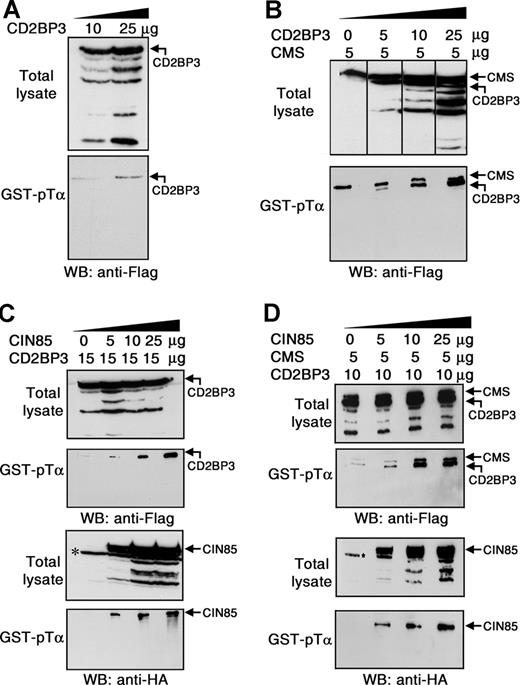
Multiprotein complexes involving CMS, CIN85, and CD2BP3 can be recruited to the cytoplasmic domain of human pTα. Total lysates from COS7 cells transfected with the indicated amounts of CD2BP3-Flag (A) or CMS-Flag and CD2BP3-Flag (B) were subjected to pull-down with GST-pTα. Precipitated proteins were analyzed by Western blotting with an anti-Flag mAb. COS7 cells were transiently cotransfected with the indicated amounts of CD2BP3-Flag and CIN85-HA (C) or CD2BP3-Flag, CMS-Flag, and CIN85-HA (D). Total-cell lysates and proteins precipitated with GST-pTα were analyzed by immunoblotting using either an anti-Flag mAb to detect CD2BP3 and CMS (top 2 panels) or an anti-HA mAb to detect CIN85 (bottom 2 panels). *Nonspecific band recognized by the anti-HA antibody. Data are representative of at least 3 independent experiments. Vertical lines have been inserted to indicate a repositioned gel lane.
Multiprotein complexes involving CMS, CIN85, and CD2BP3 can be recruited to the cytoplasmic domain of human pTα. Total lysates from COS7 cells transfected with the indicated amounts of CD2BP3-Flag (A) or CMS-Flag and CD2BP3-Flag (B) were subjected to pull-down with GST-pTα. Precipitated proteins were analyzed by Western blotting with an anti-Flag mAb. COS7 cells were transiently cotransfected with the indicated amounts of CD2BP3-Flag and CIN85-HA (C) or CD2BP3-Flag, CMS-Flag, and CIN85-HA (D). Total-cell lysates and proteins precipitated with GST-pTα were analyzed by immunoblotting using either an anti-Flag mAb to detect CD2BP3 and CMS (top 2 panels) or an anti-HA mAb to detect CIN85 (bottom 2 panels). *Nonspecific band recognized by the anti-HA antibody. Data are representative of at least 3 independent experiments. Vertical lines have been inserted to indicate a repositioned gel lane.
Identification of the specific CIN85/CMS SH3 binding motif in the human pTα cytoplasmic tail
To confirm the identity of the polyproline sequence/s in the human pTα tail responsible for the interaction with CIN85/CMS SH3 domains, pull-downs were next performed using GST-fused pTα tail mutant forms (pTαΔPro1-3) that lacked each of the 3 polyproline sequences shown in Figure 1A. We found that neither the distal C-term (Pro3) nor the CD2-like (Pro2) proline-rich pTα motifs were required for CIN85/CMS binding (Figure 2C,D). In contrast, deletion of the polyproline-arginine (Pro1) motif completely impaired the association of pTα to CIN85 and CMS (Figure 2C,D), indicating that Pro1 is a bona fide motif for CIN85 and CMS SH3 binding. As a whole, these data provide evidence that interaction of CIN85/CMS SH3 domains with the human pTα cytoplasmic tail specifically involves the Pro1 polyproline-arginine motif, and identify human pTα as a novel CIN85/CMS interacting partner.
Recruitment of multiprotein complexes involving CMS, CIN85, and CD2BP3 adaptors to the cytoplasmic domain of the human pTα chain
According to lack of SH3A, CD2BP3 was hardly precipitated with GST-pTα, even under overexpression conditions (Figure 3A). Since CMS and CD2BP3 can heterodimerize through their coiled-coil domains,27 it is possible that CMS-CD2BP3 heterodimers could be recruited to pTα. Confirming this possibility, CD2BP3 was coprecipitated with CMS in a dose-dependent manner from COS7 cells transfected with CMS and increasing amounts of CD2BP3 (Figure 3B). Similar experiments performed using HA-tagged CIN85 and Flag-tagged CD2BP3 showed that CD2BP3 was also coprecipitated with CIN85 bound to pTα (Figure 3C). Therefore, both CMS and CIN85 can mediate the recruitment of CD2BP3 to the pTα CT domain. Finally, GST-pTα pull-downs performed in COS7 cells transfected with CMS, CD2BP3, and increasing amounts of CIN85 revealed that CIN85 did not impair binding of CMS to pTα, nor CD2BP3 recruitment (Figure 3D). In contrast, amounts of CMS and CD2BP3 coprecipitated with pTα increased in the presence of CIN85 in a dose-dependent manner (Figure 3D). As a whole, we can conclude that multiprotein complexes involving CMS, CIN85 and CD2BP3 adaptors can be recruited to the CT domain of the human pTα chain, likely owing to hetero-oligomerization mediated by their coiled-coil domains.
Differential regulation of CIN85/CMS gene expression during human thymic development
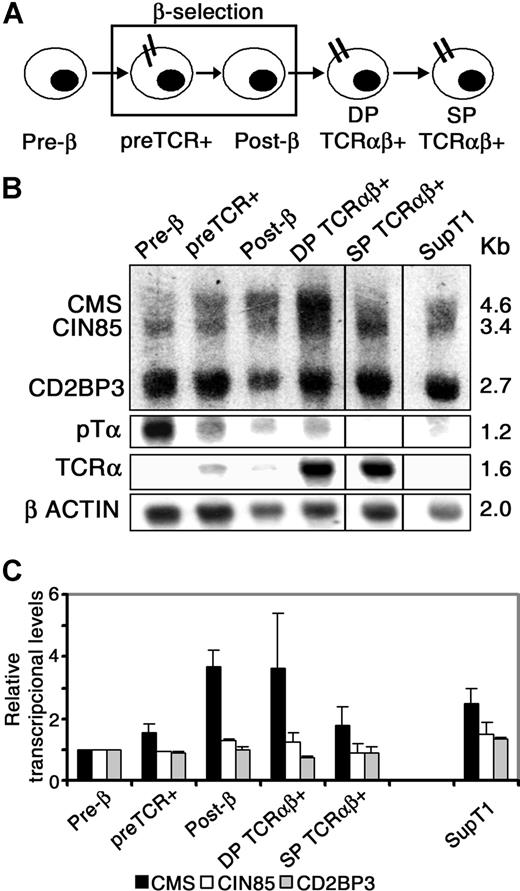
It has been reported that some CIN85/CMS adaptors are transcriptionally regulated during the development of particular tissues and organs in which they are functionally involved.35 To assess the regulation of CIN85/CMS transcription in vivo during human thymocyte development, gene expression was analyzed by Northern blotting at discrete intrathymic stages surrounding pre-TCR–mediated β-selection (Figure 4A).33 As shown in Figure 4B, transcription of the 3 members of the CIN85/CMS family was differentially regulated along human thymopoiesis. Measurements of the relative mRNA amounts normalized to β-actin transcripts from 2 independent experiments (Figure 4C) indicated that transcription of CIN85 and CD2BP3 splicing isoforms remained essentially constant throughout development. In contrast, CMS transcription was transiently up-regulated following pre-TCR expression: it increased up to 4.0-fold from pre-β to post-β selected pre-T cells, and thereafter it returned to basal levels at the SP stage. SupT1 pre-T cells included as a pre-TCR+ control also displayed relatively increased expression of CMS (Figure 4C). These results indicate that CMS transcription is specifically up-regulated in pre-T cells following pre-TCR expression, suggesting a role for CMS in pre-TCR function. Alternatively, since up-regulated CMS was also observed in DP thymocytes, CMS could be involved in attenuation of pre-TCR signaling in post-β selected thymocytes.
Differential regulation of CIN85/CMS transcription during human thymic development. (A) Scheme of the developmental progression of human intrathymic stages. (B) Total RNA from the indicated cell subsets was analyzed for CIN85/CMS/CD2BP3 transcription by Northern blotting. Blots were simultaneously hybridized with CIN85 and CMS probes, and sequentially rehybridized with pTα, TCRα, and β-actin probes. Purity of cell subsets was greater than 95% upon flow cytometric reanalysis, except for the pre-TCR+ subset, which included up to 10% of DP TCRαβ+ thymocytes (faint TCRα band). (C) Amounts of CMS, CIN85, and CD2BP3 transcripts were normalized to β-actin mRNA values. Relative transcriptional levels displayed in arbitrary units are referred to the pre-β cell stage. Data represent the means plus or minus SEM of 2 experiments corresponding to independent thymus samples. Vertical lines have been inserted to indicate a repositioned gel lane.
Differential regulation of CIN85/CMS transcription during human thymic development. (A) Scheme of the developmental progression of human intrathymic stages. (B) Total RNA from the indicated cell subsets was analyzed for CIN85/CMS/CD2BP3 transcription by Northern blotting. Blots were simultaneously hybridized with CIN85 and CMS probes, and sequentially rehybridized with pTα, TCRα, and β-actin probes. Purity of cell subsets was greater than 95% upon flow cytometric reanalysis, except for the pre-TCR+ subset, which included up to 10% of DP TCRαβ+ thymocytes (faint TCRα band). (C) Amounts of CMS, CIN85, and CD2BP3 transcripts were normalized to β-actin mRNA values. Relative transcriptional levels displayed in arbitrary units are referred to the pre-β cell stage. Data represent the means plus or minus SEM of 2 experiments corresponding to independent thymus samples. Vertical lines have been inserted to indicate a repositioned gel lane.
CMS colocalizes with the actin cytoskeleton to the pTα endocytic pathway
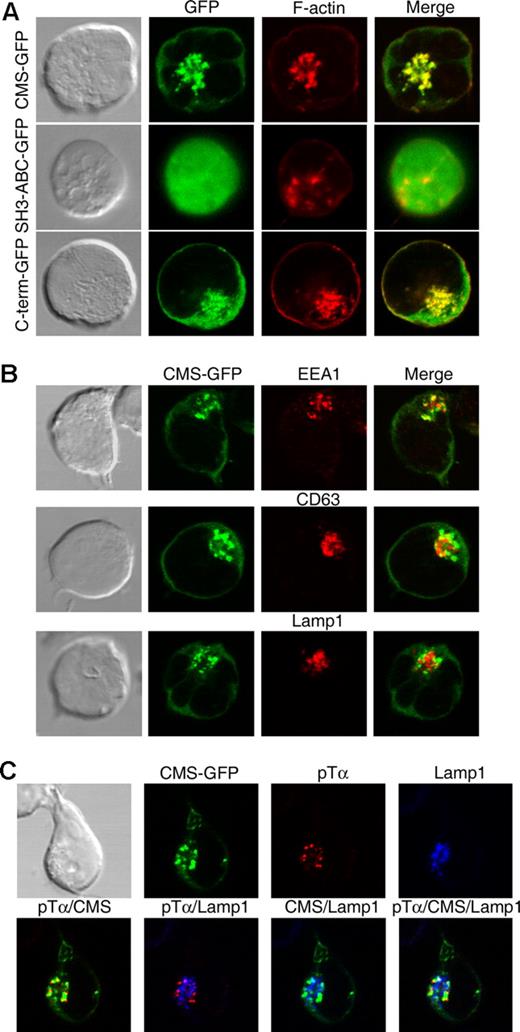
To get some insights into the function of CMS, we first assessed its subcellular distribution using SupT1 pre-T cells expressing GFP-tagged CMS. Confocal microscopy analysis showed that CMS-GFP expression was mostly confined to cytosolic vesicular structures that fully colocalized with polymerized actin (Figure 5A). Actin-rich vesicles were present as well in vivo in primary human pre-TCR+ thymocytes (Figure S1A), but not in mature T cells, in which CMS showed a diffuse expression throughout the cytosol (Figure S1B), as reported for murine T cells.36 These data suggest that formation of CMS/actin-rich vesicles is constitutive and pre-T cell specific. Since CMS contains 4 acting-binding domains in its C-term region,27 we next assessed whether the C-term is responsible for the association of CMS to the actin cytoskeleton in the vesicles found in pre-T cells. We found that overexpression of a GFP-tagged truncated form of CMS involving the 3 SH3 domains (SH3ABC) altered the vesicular pattern and prevented the colocalization of CMS to the actin cytoskeleton, this resulting in a diffuse intracellular distribution of GFP. In contrast, formation of CMS/actin-rich vesicles was conserved when a C-term CMS mutant lacking the SH3ABC domains was overexpressed (Figure 5A). Therefore, the C-term domain is responsible for the vesicular distribution and the colocalization of CMS with polymerized actin found in pre-T cells. Inmunofluorescence analyses performed to assess the identity of the constitutive CMS/actin-rich vesicles formed in pre-T cells (Figure 5B) revealed that GFP-CMS partly colocalized with the early endosomal compartment marker EEA1 and with CD63, which marks late endosomes and lysosomes,37 but colocalization with the lysosomal marker Lamp1 was hardly found (mean colocalization percentage ± SEM: 26.2 ± 1.3, 30.5 ± 2.2, and 15.9 ± 2.1, respectively). These data indicate that CMS localizes constitutively to the endocytic compartment, but is mostly excluded from the lysosomal compartment in pre-T cells.
CMS colocalizes with the actin cytoskeleton to the pTα endocytic pathway. (A) SupT1 pre-T cells were transfected with GFP-tagged versions of either full-length CMS, a truncated CMS form encompassing the SH3ABC domains, or a C-term CMS mutant lacking the SH3 domains. Transfected cells were labeled with phalloidin-TRITC and analyzed by confocal microscopy for the coexpression of polymerized actin (F-actin) and GFP. (B) SupT1 CMS-GFP+ transfectants were labeled with anti-EEA1, anti-CD63, or anti-Lamp1 mAbs followed by Alexa555–anti-mouse IgGs. Coexpression of CMS-GFP with EEA1, CD63, or Lamp1 was analyzed by confocal microsopy. (C) SupT1 CMS-GFP+ transfectants were incubated with an anti-pTα mAb for 15 at 37°C, washed, and fixed. After fixation, internalized pTα was detected by labeling with Alexa555–anti-mouse IgGs. Colocalization to the lysosomal compartment was determined by confocal microscopy after labeling with anti-Lamp1 plus Alexa647–anti-IgG1. Data are representative of at least 3 independent experiments.
CMS colocalizes with the actin cytoskeleton to the pTα endocytic pathway. (A) SupT1 pre-T cells were transfected with GFP-tagged versions of either full-length CMS, a truncated CMS form encompassing the SH3ABC domains, or a C-term CMS mutant lacking the SH3 domains. Transfected cells were labeled with phalloidin-TRITC and analyzed by confocal microscopy for the coexpression of polymerized actin (F-actin) and GFP. (B) SupT1 CMS-GFP+ transfectants were labeled with anti-EEA1, anti-CD63, or anti-Lamp1 mAbs followed by Alexa555–anti-mouse IgGs. Coexpression of CMS-GFP with EEA1, CD63, or Lamp1 was analyzed by confocal microsopy. (C) SupT1 CMS-GFP+ transfectants were incubated with an anti-pTα mAb for 15 at 37°C, washed, and fixed. After fixation, internalized pTα was detected by labeling with Alexa555–anti-mouse IgGs. Colocalization to the lysosomal compartment was determined by confocal microscopy after labeling with anti-Lamp1 plus Alexa647–anti-IgG1. Data are representative of at least 3 independent experiments.
Since pre-T cells are able to internalize and degrade the pre-TCR in an autonomous manner,16,17 it is possible that CMS/actin-rich vesicles participate in constitutive endocytosis of the pre-TCR. To investigate whether CMS+ vesicles do actually contain internalized pre-TCR proteins, the constitutive fate of surface pre-TCR complexes was monitored over time in nonstimulated CMS-GFP–expressing Sup-T1 cells fixed at various times after labeling with an anti-pTα mAb.10 Confocal microscopy analysis showed that the pTα chain was efficiently internalized within the first 15 minutes (Figure 5C). By this time, a major fraction of labeled pTα (up to 55%) colocalized to CMS-rich vesicles, and a minor proportion (less than 30%) colocalized with Lamp1+ lysosomes, from which CMS was excluded. Over time, increasing proportions of pTα localized to lysosomes (up to 70% within 30 minutes; Figure S2), but CMS and Lamp1 colocalization was difficult to observe (data not shown), confirming that CMS and pTα colocalize exclusively at the endocytic compartment. Notably, neither CMS nor polymerized actin colocalized with the conventional TCRαβ complex internalized upon receptor engagement on mature T cells (Figure S3), supporting a selective role for the CMS endocytic pathway in pre-T cells. As a whole, these results indicate that CMS colocalizes with polymerized actin to the pTα endocytic pathway, and marks pTα trafficking to lysosome degradation. Accordingly, disruption of the actin cytoskeleton by cytochalasin D treatment abrogated pTα transport to lysosomes in pre-T cells and resulted in the accumulation of pTα in early endosomes (Figure S2).
CMS-rich clusters colocalize with polymerized actin at the site of pre-TCR activation
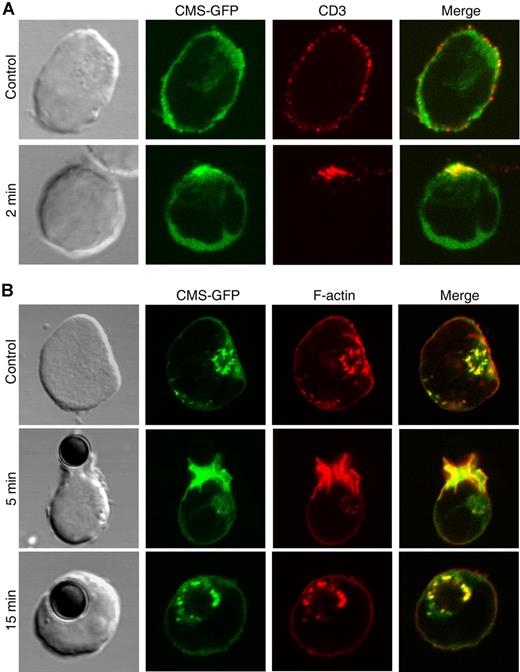
In mature T cells, interaction of CMS with the CD2 coreceptor is involved in CD2 clustering and cytoskeletal reorganization following CD2 ligand attachment.23 We thus assessed whether CMS participates in pre-TCR patterning following pre-T cell activation. As shown in Figure 6A, CD3ϵ engagement of CMS-GFP–transfected SupT1 cells resulted in the redistribution and clustering of the pre-TCR on the cell surface. Notably, CMS-rich clusters appeared at the area of pre-TCR engagement and colocalized with the pre-TCR. To confirm that CMS is recruited to the site of pre-TCR activation, the fate of CMS was then monitored over time in SupT1 cells fixed at various times after pre-TCR stimulation with anti-CD3ϵ–coated magnetic beads (Figure 6B). Recruitment of CMS-GFP to the zone of pre-TCR engagement was efficiently induced within the first 5 minutes of stimulation. Phalloidin staining revealed the redistribution and clustering of the actin cytoskeleton to the same area and its colocalization with CMS. In addition, CMS and polymerized actin fully colocalized in cytosolic vesicles that accumulate over time at the zone of tight contact of the pre-T cell with the stimulatory bead (35% vs 57% and 27% vs 48% at 5 vs 15 minutes, respectively, in 2 different experiments; Figure 6B). As shown for nonstimulated pre-T cells, these CMS-rich vesicles expressed early and/or late endosomal markers, but not lysosomal markers (data not shown). As a whole, these results indicate that signaling through the pre-TCR induces the simultaneous recruitment of CMS and polymerized actin to the site of pre-TCR clustering, and their colocalization to the pre-TCR endocytic pathway.
CMS and the actin cytoskeleton are simultaneously recruited to the site of pre-TCR activation. Confocal microscopy analysis of (A) CMS-GFP–transfected SupT1 cells stained with an anti-CD3ϵ mAb plus Alexa555–anti-mouse and left at 4°C (top row), or incubated at 37°C for 2 minutes to induce pre-TCR aggregation (bottom row); or (B) CMS-GFP transfectants either untreated (top row) or stimulated with anti-CD3ϵ–coated beads for 5 and 15 minutes (middle and bottom rows) and stained with phalloidin-TRITC. Data are representative of at least 3 independent experiments.
CMS and the actin cytoskeleton are simultaneously recruited to the site of pre-TCR activation. Confocal microscopy analysis of (A) CMS-GFP–transfected SupT1 cells stained with an anti-CD3ϵ mAb plus Alexa555–anti-mouse and left at 4°C (top row), or incubated at 37°C for 2 minutes to induce pre-TCR aggregation (bottom row); or (B) CMS-GFP transfectants either untreated (top row) or stimulated with anti-CD3ϵ–coated beads for 5 and 15 minutes (middle and bottom rows) and stained with phalloidin-TRITC. Data are representative of at least 3 independent experiments.
Binding of CMS to the proline-arginine motif in the human pTα cytoplasmic tail is involved in pre-TCR signaling
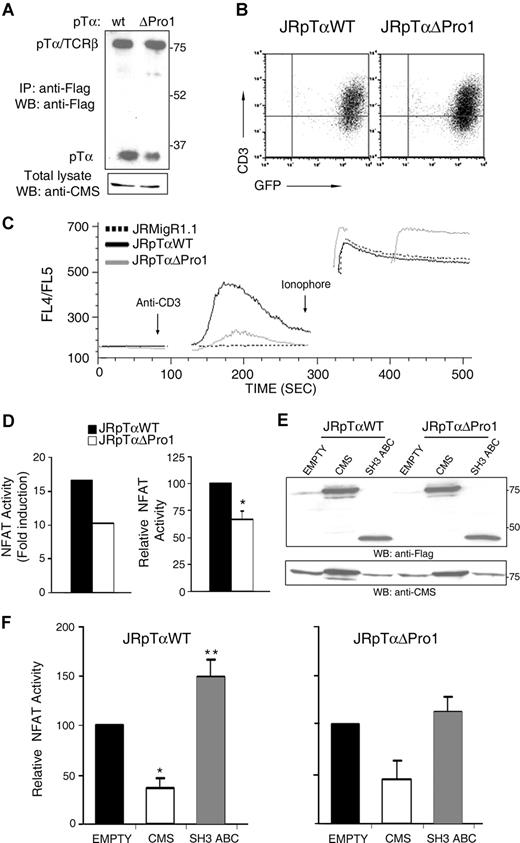
It has been shown that activation through the murine pre-TCR promotes Ca2+-dependent signals.8 To investigate whether the CIN85/CMS binding motif of human pTα is involved in pre-TCR signaling, Ca2+ mobilization was assessed after pre-TCR cross-linking in TCRα-deficient Jurkat cells (JR3.11)28 in which Flag-tagged versions of either a wild-type or a ΔPro1 mutant pTα chain were coexpressed with GFP by retroviral transduction. Anti-Flag immunoprecipitations of GFP+ sorted cells showed comparable levels of expression of the wild-type and mutant pre-TCRs (Figure 7A), and equivalent surface expression was observed as well by flow cytometry (Figure 7B). Also, western immunoblotting with anti-CMS revealed similar expression levels of endogenous CMS (Figure 7A). As shown previously in pre-T cells,8,10 CD3ϵ crosslinking of JR3.11 cells that expressed wild-type pTα induced a [Ca2+]i rise, which consisted of an initial rapid rise followed by a slow decrease. However, Ca2+ mobilization was markedly defective in JR.311 cells bearing the mutant pTαΔPro1 chain (Figure 7C), indicating that the proline-arginine CIN85/CMS binding motif is required for proper pre-TCR signaling.
Binding of CMS to the pTα cytoplasmic tail is involved in pre-TCR signaling. (A) Expression levels of pre-TCR (TCRβ-pTα) heterodimers and pTα monomers in JR3.11 cells transduced with Flag-tagged versions of either pTαWT or pTαΔPro1 were detected by anti-Flag immunoprecipitation, followed by nonreducing SDS-PAGE, and Western blotting with an anti-Flag mAb. Endogenous CMS expression was detected by inmmunoblotting with anti-CMS. (B) Surface expression levels of pre-TCR complexes on pTαWT and pTαΔPro1 JR3.11 GFP+-transduced cells were analyzed by flow cytometry with an anti-CD3ϵ mAb. (C) Time-course response in calcium mobilization obtained from the indicated cell lines after stimulation with an anti-CD3ϵ mAb. Ionophore treatment was used to control for cell viability and intact calcium stores. Data are represented as the FL4/FL5 ratio of emission analyzed by flow cytometry at the indicated times. Data are representative of at least 4 independent experiments. (D) Deletion of the Pro1 motif (ΔPro1) of the human pTα tail impairs pre-TCR–induced NFAT transcriptional activity. pTαWT and pTαΔPro1 JR3.11 cells transiently transfected with an NFAT–luciferase-reporter plasmid were stimulated with an anti-pTα mAb or left unstimulated. NFAT transcriptional activity is expressed as the fold increase of luciferase activity above levels obtained from the nonstimulated control in a representative experiment (left), or as the mean (± SEM) of relative NFAT activity of 6 independent experiments (right); *P < .005. (E) Western blot analysis of CMS expression in pTαWT and pTαΔPro1 JR3.11 cells. Total lysates of cells transfected with an empty vector, or with a vector encoding either full-length CMS or a CMS truncated form encompassing the SH3ABC CMS domains, were analyzed by immnubloting with anti-CMS or anti-Flag. (F) CMS modulates preTCR-induced NFAT transcriptional activity. pΤαWT and pTαΔPro1 JR3.11 cells were transfected with either an empty vector, or a vector encoding either full-length CMS or a CMS truncated form encompassing the SH3ABC CMS domains. NFAT transcriptional activity was analyzed upon stimulation with anti-pTα as the mean (± SEM) of relative NFAT activity of 4 independent experiments. *P < .001 and **P < .05.
Binding of CMS to the pTα cytoplasmic tail is involved in pre-TCR signaling. (A) Expression levels of pre-TCR (TCRβ-pTα) heterodimers and pTα monomers in JR3.11 cells transduced with Flag-tagged versions of either pTαWT or pTαΔPro1 were detected by anti-Flag immunoprecipitation, followed by nonreducing SDS-PAGE, and Western blotting with an anti-Flag mAb. Endogenous CMS expression was detected by inmmunoblotting with anti-CMS. (B) Surface expression levels of pre-TCR complexes on pTαWT and pTαΔPro1 JR3.11 GFP+-transduced cells were analyzed by flow cytometry with an anti-CD3ϵ mAb. (C) Time-course response in calcium mobilization obtained from the indicated cell lines after stimulation with an anti-CD3ϵ mAb. Ionophore treatment was used to control for cell viability and intact calcium stores. Data are represented as the FL4/FL5 ratio of emission analyzed by flow cytometry at the indicated times. Data are representative of at least 4 independent experiments. (D) Deletion of the Pro1 motif (ΔPro1) of the human pTα tail impairs pre-TCR–induced NFAT transcriptional activity. pTαWT and pTαΔPro1 JR3.11 cells transiently transfected with an NFAT–luciferase-reporter plasmid were stimulated with an anti-pTα mAb or left unstimulated. NFAT transcriptional activity is expressed as the fold increase of luciferase activity above levels obtained from the nonstimulated control in a representative experiment (left), or as the mean (± SEM) of relative NFAT activity of 6 independent experiments (right); *P < .005. (E) Western blot analysis of CMS expression in pTαWT and pTαΔPro1 JR3.11 cells. Total lysates of cells transfected with an empty vector, or with a vector encoding either full-length CMS or a CMS truncated form encompassing the SH3ABC CMS domains, were analyzed by immnubloting with anti-CMS or anti-Flag. (F) CMS modulates preTCR-induced NFAT transcriptional activity. pΤαWT and pTαΔPro1 JR3.11 cells were transfected with either an empty vector, or a vector encoding either full-length CMS or a CMS truncated form encompassing the SH3ABC CMS domains. NFAT transcriptional activity was analyzed upon stimulation with anti-pTα as the mean (± SEM) of relative NFAT activity of 4 independent experiments. *P < .001 and **P < .05.
Since intracellular Ca2+ rise is necessary for pre-TCR–induced activation of the NFAT transcription factor,8 we next assessed the functionality of the NFAT transcriptional pathway in JRpTαWT and JRpTαΔPro1 cells using a luciferase reporter assay.34 As shown in Figure 7D, pre-TCR crosslinking with an anti-pTα mAb promoted a marked luciferase increase in JRpTαWT cells, while NFAT activity was significantly lower in JRpTαΔPro1 mutants. Notably, overexpression of a truncated CMS form encompassing exclusively the SH3ABC domains (Figure 7E) significantly enhanced pre-TCR–mediated NFAT transcriptional activity in JRpTαWT cells, while this effect was prevented in pTαΔPro1 mutants (Figure 7F), supporting the theory that SH3ABC-promoted NFAT activation depends on pTα binding. In contrast, overexpression of either the entire CMS molecule (Figure 7E) or a CMS C-term form lacking the SH3 domains (not shown) markedly reduced NFAT transcriptional activity in both JRpTαWT and pTαΔPro1 cells (Figure 7F), thus revealing a dominant-negative role for CMS on NFAT activation independent of pTα binding. However, overexpressed CMS did not significantly affect Ca2+ mobilization in JRpTαWT cells (Figure S4), thus precluding a dominant negative role of CMS on Ca2+ mobilization. As a whole, our results showing that binding of CMS SH3 domains to the proline-arginine motif of pTα is in fact relevant for pre-TCR signaling provide the first molecular evidence for a cytosolic adaptor of the pTα chain involved in pre-TCR function.
Discussion
Current views support that the exclusive capability of the pre-TCR complex in promoting T-cell development maps to the CT domain of its pTα chain.7,18 It is thus expected that pTα interacts with intracellular adaptors that propagate specific pre-TCR signals; therefore, identification of such regulators is key for understanding the unique signaling properties of the pre-TCR. However, identification of pTα interacting partners has remained elusive. Here, we provide biochemical evidence that members of the CIN85/CMS family of adaptors are cytosolic interactors of human pTα. Binding of CIN85/CMS to the pTα CT domain did not involve the reported CD2-like proline-rich sequence conserved in mouse pTα.4,18 Rather, we identified a novel polyproline-arginine (SPRPQPR) sequence, which fits the atypical recognition consensus for binding of CIN85/CMS adaptors,25,26 and show that this sequence is indeed a bona fide binding site for CIN85/CMS SH3 domains in human pTα. Supporting the functional relevance of this interaction, we show that pre-TCR activation results in the simultaneous recruitment of CMS and the actin cytoskeleton to the site of pre-TCR clustering. In addition, CMS was found to associate with polymerized actin via its C-term region, colocalizing in the endocytic pathway with pTα molecules in traffic to the lysosomal compartment. These observations, together with the finding that deletion of the pTα CIN85/CMS-binding motif impaired pre-TCR–mediated Ca2+ mobilization and NFAT transcriptional activity, and precluded enhanced activation induced by overexpression of a CMS-SH3 N-term form, provide compelling evidence that CMS acts downstream of the pre-TCR to promote appropriate pre-TCR signaling. Biochemical studies addressing CMS-pTα interaction could not be performed in a more physiological condition due to the scarce numbers of primary pre-T cells, thus limiting the extension of our results to thymocyte development. However, we do provide evidence that the pTα polyproline-arginine domain identified here endows the human pre-TCR with a specific SH3-binding site, critical for interaction with intracellular adaptors involved in pre-TCR function.
Despite high sequence identity of CIN85 and CMS SH3 domains, CMS bound to pTα more efficiently than CIN85, which concurs with the observation that distinct SH3 domains of CIN85 and CMS were involved in pTα interaction. Since CIN85 but not CMS SH3 domains interact with proline-rich sequences within the same molecule,24 it was expected that full-length CIN85 interacted with pTα less efficiently than its SH3ABC and SH3AB forms. Besides this form of biochemical regulation, our data suggest that transcriptional regulation is an additional mechanism operating during T-cell development to favor binding of CMS to pTα in pre-TCR+ cells, and support a preferential function for CMS versus CIN85 and CD2BP3 in pre-T cells.
In mature T cells, CMS and CIN85 play a pivotal role in the regulation of cytoskeletal rearrangements and T-cell polarization.23,24 Binding of CMS and its mouse ortholog CD2AP to CD2 facilitates membrane receptor clustering and cytoskeletal polarization, which enable organization of the immunologic synapse (IS) and TCR-mediated signaling.23,24,38 Attempts to analyze similar morphologic and molecular events associated to pre-TCR activation have been hampered by the fact that the pre-TCR signals constitutively in a ligand-independent manner. By modeling pre-TCR activation with anti-CD3ϵ–coated beads,10,39 we show here that pre-TCR clustering and recruitment of polymerized actin and CMS to the site of pre-TCR activation are downstream events of pre-TCR activation. CMS recruitment was apparently independent of the pTα CMS-binding site (not shown), but proper pre-TCR signaling, as measured by Ca2+ mobilization, was contingent on CMS binding to an intact pTα polyproline-arginine domain. Similarly, TCR clustering and IS formation can occur in CMS/CD2AP-deficient T cells, or upon CD2-independent T-cell activation, while TCRαβ-mediated signal transduction is defective in both situations, mainly due to profound alterations in large-scale molecular segregation at the IS.23,36,40 Therefore, as proposed for the CMS/CD2AP-CD2 interaction,23 the function of the CMS-pTα interaction could be to promote a central cluster of pre-TCR complexes that could maintain a stable domain required for sustained pre-TCR signaling. In this scenario, either pTα multimerization induced by interaction of the 3 SH3 domains of a single CMS molecule to distinct pTα chains, or CMS multimerization mediated by a single pTα molecule,41 could also be predicted. An additional level of organization could significantly contribute to pre-TCR clustering, since hetero-oligomers involving CMS, CIN85, and CD2BP3 can be synergistically associated to pTα. Thus, it can be proposed that CMS acts as a scaffolding molecule that enables pre-TCR clustering required for propagation of autonomous cell signaling.
Besides the role of SH3 domains in mediating CMS-pTα interactions, the C-term region of CMS was shown to mediate CMS association with the actin cytoskeleton and CMS localization to the pTα endocytic pathway. It is thus possible that CMS participates in the regulation of endosomal vesicle formation in pre-T cells, as proposed in other cells.42 Our results also suggest a functional role for CMS in pTα trafficking to lysosomal degradation, which could finally result in attenuation of pre-TCR signaling. This possibility is strongly supported by the finding that CMS is up-regulated during thymocyte development in post-β–selected thymocytes. However, so far we have failed to provide evidence that the CMS-pTα interaction directly participates in pre-TCR internalization and/or degradation, although such a role could depend on an indirect association of CMS mediated through c-Cbl, as has been reported for other signaling receptors, including EGFR and TCR.40,43 It has recently been shown that the impaired ability of CD2AP/CMS-deficient T cells to degrade the activated TCR indirectly resulted from an impaired receptor activation, suggesting that CMS-mediated receptor clustering may finally balance TCR signaling and degradation.40 A similar CMS-dependent mechanism could finally regulate pre-TCR signaling and degradation, but an additional level of complexity can be expected considering that the proline-rich region of CIN85/CMS acts as an interaction module for SH3-containing signaling proteins, such as the p85α PI3K subunit, Grb2, or Src kinases,27 involved in T cell (and likely, pre-T cell) signaling. As a result, distinct parts of the CMS molecule could elicit divergent functional activities, whose interplay could finally determine the pre-TCR signaling output. According to this view, the SH3ABC CMS domains behave as constitutively active CMS forms upon binding to the proline-arginine motif of pTα, while full-length CMS markedly impaired SH3ABC-induced NFAT activation. Since the inhibitory effect of CMS was independent of pTα interaction, and involved the C-term portion of the molecule (data not shown), it could result from the specific binding and sequestration of molecular effectors, downstream of Ca2+ mobilization, involved in NFAT transactivation and pre-TCR signaling. The dominant-negative effect of CMS could also reflect an active role of the molecule in promoting and unbalance toward pTα degradation. Therefore, it is possible that distinct parts of the CMS molecule may differentially affect particular functions whose interplay and coordination finally determine pre-TCR signaling output.
While unraveling the molecular basis of CMS-mediated pre-TCR signaling will require further studies, our results provide the first molecular evidence that the CT domain of pTα provides the pre-TCR with the unique capacity to recruit CIN85/CMS adaptors crucial for propagation of downstream signals. Therefore, as previously proposed, the cytoplasmic tail of pTα could confer on the pre-TCR the capacity to “costimulate,”4 while self-oligomerization of pTα would be the mechanism responsible for initiation of ligand-independent pre-TCR signaling.12 Both signals, however, are delivered separately via engagement of the TCR and the costimulatory molecule CD2 in mature T cells.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs J. C. Aster for the MigR1.1-GFP retroviral vector, J. M. Redondo for the luciferase-NFAT plasmid, B. Rubin for JR3.11 cells, J. Bravo for helpful discussions, and V.G. de Yébenes for help with the Ca2+ mobilization assay, and the Pediatric Cardiosurgery Units from the C. E. Ramón y Cajal and Hospital La Paz (Madrid, Spain) for the thymus samples.
This work was supported in part by grants from the Plan Nacional de Biomedicina (SAF2004-01122), Comunidad de Madrid (GR/SAL/0143/2004, S-SAL0304-2006), Fundación la Caixa (ON03/109-00), and Fundación Rodríguez Pascual, and by an institutional grant from the Fundación Ramón Areces. G.N. and P.F. were supported by the Plan Nacional de Biomedicina SAF2001-1269 and SAF2004-01122 grants, respectively; M.N.N. was supported by a Formación Personal Investigador (FPI) fellowship.
Authorship
Contribution: M.N.N., G.N., and P.F. designed and performed the research and analyzed and interpreted data; S.G.-G. and J.A. performed the research; and M.L.T. designed the research and drafted the manuscript.
M.N.N. and G.N. contributed equally to this work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: María L. Toribio, Centro de Biología Molecular Severo Ochoa, Facultad de Ciencias, Universidad Autónoma de Madrid, Cantoblanco, 28049 Madrid, Spain; e-mail:mtoribio@cbm.uam.es.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal