Macrophages phagocytose particles to resolve infections and remove apoptotic cells. Phosphoinositide 3-kinase generates phosphatidylinositol-3,4,5-trisphosphate [PtdIns(3,4,5)P3] is restricted to the phagocytic cup, promoting phagocytosis. The PtdIns(3,4,5)P3 5-phosphatase (5-ptase) Src homology 2 (SH2) domain-containing inositol-5-phosphatase 1 (SHIP1) inhibits phagocytosis. We report here that another PtdIns(3,4,5)P3-5-ptase, the 72-kDa-5-phosphatase (72-5ptase), inhibits Fcγ receptor (FcγR)– but not complement receptor 3 (CR3)–mediated phagocytosis, affecting pseudopod extension and phagosome closure. In contrast, SHIP1 inhibited FcγR and CR3 phagocytosis with greater effects on CR3-stimulated phagocytosis. The 72-5ptase and SHIP1 were both dynamically recruited to FcγR-stimulated phagocytic cups, but only SHIP1 was recruited to CR3-stimulated phagocytic cups. To determine whether 5-ptases focally degrade PtdIns(3,4,5)P3 at the phagocytic cup after specific stimuli, time-lapse imaging of specific biosensors was performed. Transfection of dominant-negative 72-5ptase or 72-5ptase small interfering RNA (siRNA) resulted in amplified and prolonged PtdIns(3,4,5)P3 at the phagocytic cup in response to FcγR- but not CR3-stimulation. In contrast, macrophages from Ship1−/−/AktPH-GFP transgenic mice exhibited increased and sustained PtdIns(3,4,5)P3 at the cup in response to CR3 activation, with minimal changes to FcγR activation. Therefore, 72-5ptase and SHIP1 exhibit specificity in regulating FcγR- versus CR3-stimulated phagocytosis by controlling the amplitude and duration of PtdIns(3,4,5)P3 at the phagocytic cup.
Introduction
Macrophages engulf large particles (> 1.0 μm), including invading pathogens and apoptotic cells, by phagocytosis to resolve infections and regulate tissue remodeling. Phagocytosis is a dynamic process triggered by particle binding to cell surface receptors including Fc γ receptor (FcγR) and complement receptor 3 (CR3),1 which initiate a complex series of events including actin and membrane remodeling, leading to the extension of pseudopods, phagosome closure, and particle engulfment. A critical event in phagocytosis is the generation of phosphatidylinositol-3,4,5-trisphosphate [PtdIns(3,4,5)P3] at the phagocytic cup. PtdIns(3,4,5)P3 is synthesized by the class I phosphoinositide-3 (PI3)–kinase after clustering of FcγRs in response to immunoglobulin G (IgG)–opsonized particle binding or CR3 activation2,–4 and promotes pseudopod extension and phagosome closure. Macrophages with reduced PtdIns(3,4,5)P3, as consequence of targeted deletion of the p85 adapter subunit of the PI3-kinase, exhibit decreased phagocytic ability.5 PI3-kinase inhibition prevents the complete phagocytosis of erythrocytes coated with IgG (eIgG) or complement component C3bi (eC3bi),6,7 with this defect specific for large but not small particles.7 PtdIns(3,4,5)P3 recruits pleckstrin homology (PH)–domain-containing effectors including phospholipase C-γ and myosin X, which hydrolyze PtdIns(4,5)P2 and drive pseudopod extension, respectively.8,9
PtdIns(3,4,5)P3 is generated de novo at the base of the forming phagocytic cup, extending only to the sides of the cup but never diffusing beyond the lateral boundaries of the phagosome, disappearing following phagosome closure.2,3,10 PtdIns(3,4,5)P3 spatial distribution may be regulated by lipid phosphatases or active endocytosis at the edge of the cup and/or by the activation of tyrosine kinases and the recruitment of signaling complexes.3,11,12 Three lipid phosphatases have been identified that regulate phagocytosis, PTEN (phosphatase and tensin homologue deleted on chromosome 10), Src homology 2 (SH2) domain-containing inositol-5-phosphatase 1 (SHIP1), and SH2 domain-containing inositol-5-phosphatase 2 (SHIP2). PTEN, a PtdIns(3,4,5)P3 3-phosphatase, inhibits phagocytosis but its role in vivo in this process is unclear because it is not recruited to the phagocytic cup.3,10,13 SHIP1 and SHIP2 are inositol polyphosphate 5-phosphatases (5-ptases) that hydrolyze PtdIns(3,4,5)P3 forming PtdIns(3,4)P2, and both are recruited to the phagocytic cup and inhibit phagocytosis.14,15 Macrophages, which lack SHIP1 or SHIP2, exhibit enhanced phagocytic ability after FcγR activation; however, SHIP1−/− macrophages exhibit greater increases in phagocytosis in response to CR3 activation.14 Whether these or other 5-ptases regulate the spatial distribution of PtdIns(3,4,5)P3 at the phagocytic cup or rather limit the amplitude and/or duration of PtdIns(3,4,5)P3 at this site is unknown.
The 72-kDa 5-phosphatase (72-5ptase) (also called the type IV 5-phosphatase, or pharbin) shows the highest affinity toward PtdIns(3,4,5)P3 of any 5-ptase, greater than SHIP1 with a Michaelis-Menten-constant (Km) of 0.6 μM.16 The 72-5ptase hydrolyzes the 5-position phosphate from PtdIns(3,4,5)P3, forming PtdIns(3,4)P216,–18 and regulating PtdIns(3,4,5)P3-dependent Akt phosphorylation.19,–21 In this study we demonstrate the 72-5ptase is dynamically recruited to the phagocytic cup and inhibits phagocytosis in response to FcγR but not CR3 activation. In contrast, SHIP1 is dynamically recruited to both FcγR- and CR3-stimulated phagosomes to inhibit phagocytosis. Using time-lapse imaging of a PtdIns(3,4,5)P3-specific biosensor during phagocytosis, we demonstrate the 72-5ptase governs the duration and amplitude of PtdIns(3,4,5)P3 signals at the phagocytic cup in response to FcγR- but not CR3-mediated phagocytosis, whereas SHIP1 exhibits preference for hydrolyzing PtdIns(3,4,5)P3 signals in response to CR3 activation. These studies have identified that distinct 5-phosphatases regulate phagocytosis in response to specific agonists.
Materials and methods
Reagents
The RAW264.7 cell line was from Dr Julian Quinn (St Vincent's Institute, Melbourne, Australia), RPMI-1640 and Dulbecco modified Eagle medium (DMEM) were from Gibco (Mount Waverley, Australia), L-glutamine, penicillin/streptomycin, and fetal calf serum (FCS) were from JRH (Melbourne, Australia), sheep red blood cells (SRBC) and rabbit anti-SRBC IgG were from MP Biomedicals (Solon, OH), 6- and 10-μm latex beads were from Bangs Laboratories (Carmel, IN), monoclonal antibodies to hemagglutinin (HA) tag were from Babco (Richmond, CA), and Texas-Red, Alexa-Fluor 488, 647-labeled phalloidin, and secondary antibodies were from Molecular Probes (Eugene, OR). All other reagents were from Sigma-Aldrich (Castle Hill, Australia).
Constructs
pCGN (vector), pCGN-72 kDa-5ptase wild-type (HA-72-5ptase), and pCGN-72 kDa-5ptase catalytically inactive (HA-72-D480N5ptase) plasmids were generated as previously described.18 Green fluorescent protein (GFP)–VAMP3 was from Prof Jennifer Stow (Institute for Molecular Bioscience, University of Queensland, Brisbane, Australia). GFP-adenosine 5′-diphosphate-ribosylation factor nucleotide-binding-site opener (ARNO)/PH and GFP-PH-PLCδ plasmids were from Dr Tamas Balla (National Institute of Child Health and Human Development, Bethesda, MD). Yellow fluorescent protein (YFP)–TAPP1/PH was from Dr Dario Alessi (University of Dundee, Dundee, United Kingdom). His-HA-SHIP1 was from Prof Gerald Krystal (British Columbia Cancer Research Center, Vancouver, BC).
Adenoviral constructs
Recombinant adenoviral plasmids encoding wild-type or dominant-negative HA-72-5ptase were generated as described.21
AktPH-GFP transgenic mice
Cell culture/transfection/adenoviral transduction
RAW264.7 macrophages were cultured as described.23 For electroporation and adenoviral transduction, cells were seeded onto 10-cm Petri dishes in RPMI-1640 with 5% FCS or at 5 × 104 cells per mL and transduced with the adenoviral constructs (multiplicity of infection 40) in RPMI-1640 with 1% FCS overnight, respectively. Transfection of siRNA-transfected RAW264.7 macrophages with Lipofectamine LTX (Invitrogen, Eugene, OR) was performed according to manufacturers' instructions.
siRNA-mediated knock-down of 72-5ptase
Annealed RNA oligonucleotides (Dharmacon, Lafayette, CO) specific for murine 72-5ptase (GenBank accession no. NM_033134) were labeled using the Silencer siRNA labeling kit (Ambion, Austin, TX) according to the manufacturer's instructions. For transient transfection of siRNA, RAW264.7 macrophages or differentiated BMM, seeded at 105 cells per mL were transfected with 25 nM siRNA using 0.5 μL of Lipofectamine 2000. Seventy-two hours after transfection, cells were lysed [50 mM Tris, pH 7.4, 1% Triton-X-100, 0.2% sodium dodecyl sulfate (SDS), 0.2% sodium deoxycholate, 1 mM (ethylenedinitrilo)tetraacetic acid (EDTA)] for immunoblotting or stimulated for phagocytosis assays.
72-5ptase immunoprecipitation
RAW264.7 macrophages were lysed (50 mM Tris, pH 8.0, 125 mM NaCl, 1% Triton-X-100, 10 mM NaF, 10 mM Na3VO4, 10 μg/mL aprotinin and leupeptin) and fractionated by centrifugation at 320g for 20 minutes (4°C). Immunoprecipitation was performed overnight with protein-A-Sepharose and rabbit nonimmune serum or 72-5ptase-specific antibodies at 4°C. Immunoprecipitates were immunoblottted with 72-5ptase-specific antibodies.
Confocal microscopy
Cells were fixed with 3% paraformaldehyde (PFA; 20 minutes) or 4% formaldehyde (10 minutes) and stained as described.21 Cells were imaged using the Olympus FV1000 confocal microscope, 60×/1.4 NA oil objective. Images were collected using Olympus FV10 ASW version 1.6.01 software, saved as 16-bit greyscale images (Monash Micro Imaging, Clayton, Australia) and color added in Corel Photo-Paint version 9 (Ottawa, ON).
Phagocytosis assay
IgG opsonization of SRBC and latex beads and serum opsonization of zymosan were as described.2,25,26 RAW264.7 macrophages were incubated in low serum media (1% FCS) overnight, phagocytosis was stimulated as indicated, and cells were fixed/stained as described above. The phagocytic index was determined as the number of particles phagocytosed per 100 transfected cells and expressed as a percentage of vector or nonsilencing control siRNA (siCON)–transfected cells.
Opsonization of latex beads for phagocytosis by peritoneal macrophages
A 2.5% suspension of POLYBEAD carboxylate microspheres (Polysciences, Warrington, PA) was incubated in 50 mM 2-(N-morpholino)-ethanesulfonic acid buffer (pH 6.0) with 5 mg/mL bovine serum albumin (BSA) for 15 minutes at room temperature (RT) and added to 1-ethyl-3-(3-dimethylaminopropyl)–carbodiimide (10 mg) (Molecular Probes). The reaction was stopped by 100 mM glycine. BSA-coated beads were opsonized with rabbit anti–BSA IgG (Biogenesis, Bournemouth, United Kingdom) at RT for 4 hours. IgG-coated beads were collected by centrifugation at 3000g for 20 minutes and stored at 4°C until use.
GFP-VAMP3 recruitment to phagocytic cups and analysis of phagosome closure
RAW264.7 macrophages were cotransfected with excess vector, wild-type HA-72-5ptase, or HA-72-D480N5ptase relative to GFP-VAMP3. To observe phagocytosis, cells were transferred to a thermostatted (37°C) chamber containing phenol red- and bicarbonate-free DMEM buffered with 25 mM 4-(2-hydroxyethyl)–1-piperazineethanesulfonic acid (Hepes; pH 6.8) on an Olympus FV1000 confocal microscope (Monash Micro Imaging). IgG-opsonized SRBC (eIgG) or serum-opsonized zymosan (SOZ) were added to the chamber to initiate phagocytosis. Images were gathered every 12.5 seconds. Recruitment of GFP-VAMP3 was monitored, and duration of phagosome closure is expressed as the time (sec) following phagocytic cup formation.
Analysis of phosphoinositide dynamics
RAW264.7 macrophages transiently expressing GFP-ARNO/PH or YFP-TAPP1/PH were stimulated, and phagocytosis was monitored as described above. To characterize the temporal and spatial association of fluorescently labeled proteins with phagosomes, only phagosomes forming perpendicular to the coverslip were analyzed. To analyze the accumulation of lipid-biosensors during phagocytosis, the average phagosomal fluorescence was determined using ImageJ (version 1.34; National Institutes of Health, Bethesda, MD).27 To ensure that recruitment of lipid biosensors was specific, a lower threshold equivalent to the average cytosolic fluorescence plus 2 standard deviations (SD) was subtracted from the phagosomal fluorescence yielding relative fluorescence units (RFU). To allow for comparison between experiments, each RFU was divided by the maximum RFU obtained in each experiment (adapted from Scott et al26 ).
Time-lapse studies, in primary peritoneal macrophages, were performed at 37°C using a DM IRE2 microscope (Leica, Wetzlar, Germany) fitted with a confocal imaging system (Yokogawa, Musashino-shi, Japan) as described previously.22 Peritoneal macrophages (2.5 × 105 cells) isolated from wild-type (SHIP1+/+)–AktPH-GFP-Tg and Ship1−/−-AktPH-GFP-Tg were plated onto glass-bottom dishes for 1 hour at 37°C. Cells were rinsed and replaced with Krebs-Ringer buffer (130 mM NaCl, 4.7 mM KCl, 1.2 mM KH2PO4, 1.2 mM MgCl2, 1.5 mM CaCl2, 5 mM glucose, 0.2% BSA, and 10 mM Hepes-NaOH, pH 7.2) with or without 100 nM wortmannin. Phagocytosis was stimulated by SOZ, and images were captured every 10 seconds by a cooled charge-coupled-device camera (Orca-ER; Hamamatsu Photonics, Hamamatsu City, Japan) driven by IPLab software (Scanalytics, Fairfax, VA). The fluorescence intensity of AktPH-GFP at the phagocytic cup in the cytosol was analyzed by IPLab software.
Statistical analysis
Statistical analyses of data were performed using Student t test.
Results
72-5ptase regulates macrophage phagocytosis
We recently reported the 72-5ptase is expressed in adipocytes,21 which share a common cell lineage with macrophages.28 Antibodies to the 72-5ptase (Figure 1Ai)18 detected an immunoreactive 72-kDa polypeptide in RAW264.7 macrophage cytosol, Triton-X-100 soluble and insoluble-membrane fractions (Figure 1Aii). In addition, the 72-5ptase antibody immunoprecipitated the 72-5ptase (Figure 1Aiii).
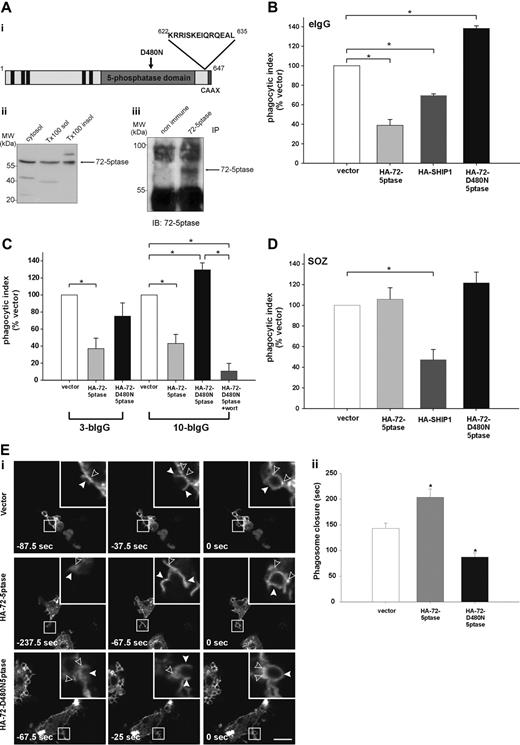
The 72-5ptase is expressed in RAW264.7 macrophages and inhibits FcγR-mediated phagocytosis. (A) (i) 72-5ptase domain structure, showing proline-rich (PxxP) motifs (■), catalytic domain (▒), D480N mutation which renders the enzyme inactive (↓) and peptide used to raise polyclonal antibodies. (ii) Cytosol, Triton-X-100-soluble (Tx100 sol) and -insoluble (Tx100 insol) membrane fractions from RAW264.7 macrophages were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblotted with 72-5ptase-specific antibodies. (iii) RAW264.7 Triton-X-100-soluble fraction was immunoprecipitated (IP) with nonimmune or immune 72-5ptase-specific antibodies and immunoblotted (IB) with anti-72-5ptase antibodies. MW, molecular weight. RAW264.7 macrophages ectopically expressing (B and D) vector, HA-72-5ptase, HA-SHIP1, or HA-72-D480N5ptase, or (C) vector, HA-72-5ptase, or HA-72-D480N5ptase were stimulated to phagocytose (B) eIgG, (C) 3-bIgG and 10-bIgG (−/+ wortmannin), or (D) SOZ. The phagocytic index represents the number of particles phagocytosed per 100 cells for at least 3 independent experiments, normalized to vector-expressing cells, designated 100%. Data are expressed as the mean (± SEM) of at least 3 independent experiments (*P < .05). (E) RAW264.7 macrophages were cotransfected with GFP-VAMP3 and vector, HA-72-5ptase, or HA-72-D480N5ptase, and phagocytosis was stimulated by the addition of eIgG. (i) Images represent a time course of phagocytosis in cells expressing vector, HA-72-5ptase, or HA-72-D480N5ptase imaged by confocal microscopy. Expression of GFP-VAMP3 is shown. Insets show enlargements of boxed areas. Closed arrowheads indicate forming phagocytic cups, and open arrowheads indicate GFP-VAMP3-positive vesicle recruitment. Scale bar represents 20 μm. In the middle panels closed arrowheads indicate distorted phagocytic cup in HA-72-5ptase-expressing cell. Time of phagocytic cup closure was designated 0 seconds. (ii) Time for phagosome closure was determined by scoring 16 to 20 phagocytic events over 4 independent experiments for each construct. Data represent mean (± SEM) time of phagosome closure from the commencement of phagocytic cup formation (sec) of 4 independent experiments. *P < .05.
The 72-5ptase is expressed in RAW264.7 macrophages and inhibits FcγR-mediated phagocytosis. (A) (i) 72-5ptase domain structure, showing proline-rich (PxxP) motifs (■), catalytic domain (▒), D480N mutation which renders the enzyme inactive (↓) and peptide used to raise polyclonal antibodies. (ii) Cytosol, Triton-X-100-soluble (Tx100 sol) and -insoluble (Tx100 insol) membrane fractions from RAW264.7 macrophages were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblotted with 72-5ptase-specific antibodies. (iii) RAW264.7 Triton-X-100-soluble fraction was immunoprecipitated (IP) with nonimmune or immune 72-5ptase-specific antibodies and immunoblotted (IB) with anti-72-5ptase antibodies. MW, molecular weight. RAW264.7 macrophages ectopically expressing (B and D) vector, HA-72-5ptase, HA-SHIP1, or HA-72-D480N5ptase, or (C) vector, HA-72-5ptase, or HA-72-D480N5ptase were stimulated to phagocytose (B) eIgG, (C) 3-bIgG and 10-bIgG (−/+ wortmannin), or (D) SOZ. The phagocytic index represents the number of particles phagocytosed per 100 cells for at least 3 independent experiments, normalized to vector-expressing cells, designated 100%. Data are expressed as the mean (± SEM) of at least 3 independent experiments (*P < .05). (E) RAW264.7 macrophages were cotransfected with GFP-VAMP3 and vector, HA-72-5ptase, or HA-72-D480N5ptase, and phagocytosis was stimulated by the addition of eIgG. (i) Images represent a time course of phagocytosis in cells expressing vector, HA-72-5ptase, or HA-72-D480N5ptase imaged by confocal microscopy. Expression of GFP-VAMP3 is shown. Insets show enlargements of boxed areas. Closed arrowheads indicate forming phagocytic cups, and open arrowheads indicate GFP-VAMP3-positive vesicle recruitment. Scale bar represents 20 μm. In the middle panels closed arrowheads indicate distorted phagocytic cup in HA-72-5ptase-expressing cell. Time of phagocytic cup closure was designated 0 seconds. (ii) Time for phagosome closure was determined by scoring 16 to 20 phagocytic events over 4 independent experiments for each construct. Data represent mean (± SEM) time of phagosome closure from the commencement of phagocytic cup formation (sec) of 4 independent experiments. *P < .05.
SHIP1 inhibits phagocytosis stimulated via FcγR and CR3 receptors.14,15 We investigated the role the 72-5ptase plays in FcγR-mediated phagocytosis by ectopically expressing wild-type HA-72-5ptase, a catalytically inactive HA-72-5ptase construct containing a mutation of a catalytic aspartic acid (HA-72-D480N5ptase),29 or HA-SHIP1 in RAW264.7 macrophages. Cells were stimulated to phagocytose eIgG (5-8 μm in size), which activates FcγR, and the number of completely phagocytosed particles were scored (Figure 1B). Ectopic expression of the 72-5ptase inhibited FcγR-mediated phagocytosis by 60% (Figure 1B), whereas under the same conditions, expression of HA-SHIP1 inhibited FcγR-mediated phagocytosis by 35%. Therefore the 72-5ptase exhibits greater activity in inhibiting FcγR-mediated phagocytosis than SHIP1, perhaps related to its reported higher affinity for PtdIns(3,4,5)P3.16 In contrast, HA-72-D480N5ptase increased phagocytosis by 1.4-fold (Figure 1B), suggesting the construct functions as a dominant-negative by binding PtdIns(3,4,5)P3 and preventing its hydrolysis by endogenous 5-ptases. In control studies immunoblot and immunofluorescence analysis using antibodies to the HA-tag demonstrated both HA-72-5ptase and HA-SHIP1 were expressed intact with similar transfection efficiencies (data not shown).
PtdIns(3,4,5)P3 is required for maximal pseudopod extension. PI3-kinase inhibition prevents phagocytosis of large particles (> 6 μm), which require increased pseudopod extension, but not small particles (< 3 μm).7 To examine the role the 72-5ptase plays in regulating pseudopod extension, we analyzed the phagocytosis of antibody-coated beads of various sizes. Phagocytosis of latex beads opsonized with IgG (bIgG) of both 3 μm (3-bIgG) and 10 μm (10-bIgG; Figure 1C) was inhibited by the HA-72-5ptase. However, HA-72-D480N5ptase showed dominant-negative activity in the phagocytosis of only 10-bIgG, suggesting the construct enhances PI3-kinase-dependent pseudopod extension (Figure 1C). In support of this contention, PI3-kinase inhibition by 100 nM wortmannin (30 minutes) treatment before challenge with 10-bIgG significantly inhibited HA-72-D480N5ptase-enhanced phagocytosis (Figure 1C).
CR3-mediated phagocytosis is mechanistically distinct from FcγR-mediated phagocytosis, although PtdIns(3,4,5)P3 has been implicated in both events.15,30,31 SHIP1 regulates both FcγR- and CR3-mediated phagocytosis, however, a greater increase in phagocytosis has been noted in response to CR3 relative to FcγR activation in SHIP1−/− macrophages.15 We investigated whether 72-5ptase regulates the phagocytosis of SOZ which activates CR325,32 (Figure 1D) relative to the effects of HA-SHIP1. No significant differences in phagocytosis were noted in cells expressing vector, HA-72-5ptase, or HA-72-D480N5ptase. In addition 72-5ptase ectopic expression also failed to inhibit phagocytosis of eC3bi over a prolonged time course (data not shown). In contrast HA-SHIP1 inhibited CR3-mediated phagocytosis of SOZ by approximately 50%. Thus 72-5ptase activity is specific for FcγR-mediated phagocytosis, whereas SHIP1 inhibits both FcγR- and CR3-mediated phagocytosis.
Pseudopod extension and phagosome closure are facilitated by the recruitment of intracellular vesicles to the phagocytic cup in a process called “focal exocytosis.”33 PI3-kinase inhibition partially inhibits the recruitment of GFP-VAMP3-positive vesicles from recycling endosomes to the plasma membrane during FcγR-stimulated phagocytosis.34 However, capacitance measurements of cell surface area during phagocytosis indicate PI3-kinase inhibition does not effect focal exocytosis.35 To determine the role 72-5ptase activity plays in focal exocytosis, we analyzed the recruitment of GFP-VAMP3 to the phagocytic cup. HA-72-5ptase or HA-72-D480N5ptase expression had no effect on GFP-VAMP3 distribution in quiescent macrophages (data not shown), GFP-VAMP3 recruitment to the forming phagocytic cup (Figure 1Ei), or the fusion of exofacially tagged VAMP3 with the plasma membrane (data not shown) during FcγR-stimulated phagocytosis. GFP-VAMP3 demarcates the membranes of the forming phagocytic cup facilitating the assessment of phagosome closure,34,36,37 a PI3-kinase-dependent event.6,8 We noted, as reported, that GFP-VAMP3 accumulated at the base of the phagocytic cup.37 After particle binding, phagosome closure proceeded over 140 to 160 seconds (Figure 1Ei,ii). GFP-VAMP3 outlined the forming phagocytic cup and remained associated with the phagosome after its closure for up to 5 minutes (data not shown). In cells expressing HA-72-5ptase, a small percentage (approximately 20%) of phagocytic cups formed but failed to close (data not shown); this was not detected in vector- or HA-72-D480N5ptase-expressing cells. In addition, the morphology of phagocytic cups in cells overexpressing HA-72-5ptase appeared distorted (Figure 1Di middle panels, see arrowheads). HA-72-5ptase prolonged phagosome closure, whereas HA-72-D480N5ptase increased closure rate (Figure 1Eii). In contrast, in response to CR3 activation, expression of the 72-5ptase constructs had no effect on the rate of phagosome closure (data not shown). Therefore 72-5ptase inhibits FcγR-stimulated PI3-kinasedependent phagocytosis, regulating pseudopod extension and phagosome closure.
siRNA-mediated depletion of the 72-5ptase increases FcγR- but not CR3-mediated phagocytosis
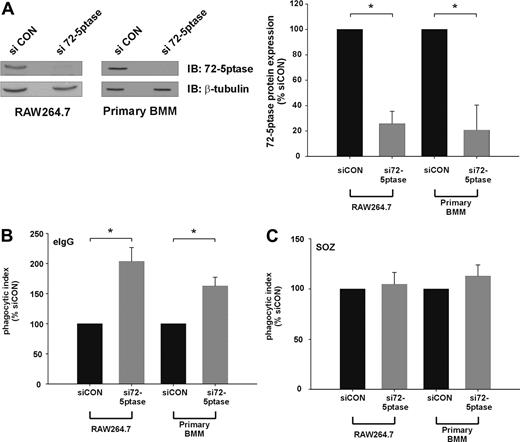
We next determined the effect of extinguishing endogenous 72-5ptase expression using siRNA technology. Transfection of siRNA oligonucleotides directed toward the GenBank entry NM_033134 (si72-5ptase) significantly decreased 72-5ptase protein levels in both RAW264.7 cells and murine bone marrow–derived macrophages (BMM) by approximately 80% relative to siCON, as assessed by 72-5ptase immunoblot and densitometry analysis (Figure 2A). Significantly, si72-5ptase increased FcγR-stimulated phagocytosis by 2-fold in RAW264.7 cells and 1.7-fold in primary mouse macrophages (Figure 2B), but in response to SOZ no change in phagocytosis was detected (Figure 2C). These results reveal the endogenous 72-5ptase specifically regulates FcγR- but not CR3-mediated phagocytosis.
siRNA mediated knock-down of the 72-5ptase enhances FcγR-mediated but not CR3-mediated phagocytosis in RAW264.7 macrophages and primary BMM. RAW264.7 macrophages or primary bone marrow macrophages (BMM) differentiated for 6 days were transiently transfected with siCON or si72-5ptase oligonucleotides. (A) After 72 hours cells were lysed and immunoblotted with 72-5ptase-specific antibodies or β-tubulin-specific antibodies, the latter to detect protein loading. The relative 72-5ptase protein expression in RAW264.7 macrophages and primary BMM was determined by densitometry and normalized to β-tubulin loading control and expressed as percentage of siCON, which was designated 100%. Data represent the mean (± SEM) of 3 (RAW264.7) or 2 (primary BMM) independent experiments. Cells transfected with siCON or si72-5ptase were stimulated with (B) eIgG or (C) SOZ for 10 minutes. Phagocytic index represents the number of particles phagocytosed per 100 cells and normalized to vector, which was designated 100%. Data are expressed as the mean (± SEM) of 3 independent experiments. *P ≤ .05.
siRNA mediated knock-down of the 72-5ptase enhances FcγR-mediated but not CR3-mediated phagocytosis in RAW264.7 macrophages and primary BMM. RAW264.7 macrophages or primary bone marrow macrophages (BMM) differentiated for 6 days were transiently transfected with siCON or si72-5ptase oligonucleotides. (A) After 72 hours cells were lysed and immunoblotted with 72-5ptase-specific antibodies or β-tubulin-specific antibodies, the latter to detect protein loading. The relative 72-5ptase protein expression in RAW264.7 macrophages and primary BMM was determined by densitometry and normalized to β-tubulin loading control and expressed as percentage of siCON, which was designated 100%. Data represent the mean (± SEM) of 3 (RAW264.7) or 2 (primary BMM) independent experiments. Cells transfected with siCON or si72-5ptase were stimulated with (B) eIgG or (C) SOZ for 10 minutes. Phagocytic index represents the number of particles phagocytosed per 100 cells and normalized to vector, which was designated 100%. Data are expressed as the mean (± SEM) of 3 independent experiments. *P ≤ .05.
The 72-5ptase is differentially recruited to the phagocytic cup in response to FcγR but not CR3 stimulation
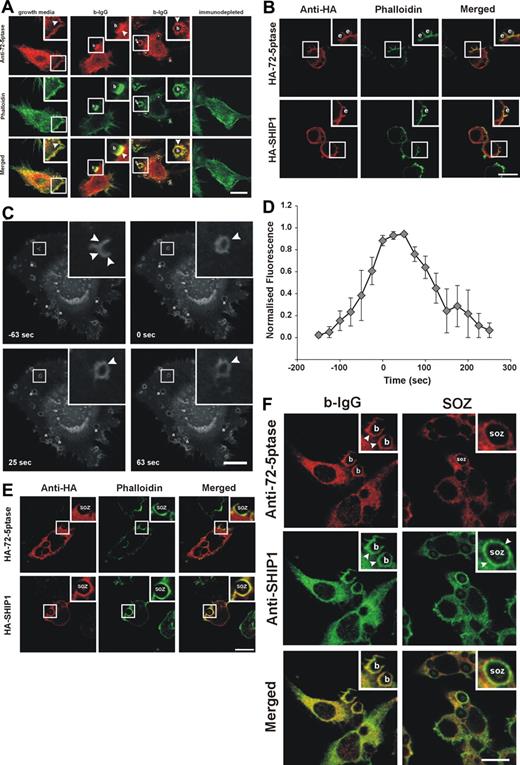
If the 72-5ptase regulates phagocytosis then it may localize to the phagocytic cup and exhibit differential recruitment in response to FcγR versus CR3 stimulation. In macrophages grown in serum endogenous 72-5ptase exhibited a perinuclear and cytoplasmic distribution, with enrichment at the plasma membrane colocalizing at this site with submembraneous polymerized actin as shown by costaining with phalloidin (Figure 3A yellow in merged images). Peptide-deplete serum was nonreactive. After the addition of 3-bIgG, endogenous 72-5ptase was enriched at the base and sides of the phagocytic cup and transiently on the internalized phagosome (Figure 3A).
Intracellular localization of 72-5ptase during phagocytosis. (A) RAW264.7 macrophages grown in serum or serum-starved and stimulated with 3-bIgG for 7 minutes were fixed and labeled with 72-5ptase (red) or immunodepleted-72-5ptase-specific antibodies and colocalized with phalloidin (green). Arrowheads indicate sites of 72-5ptase localization, “b” indicates 3-bIgG, and asterisks represent fully internalized beads. Scale bar represents 10 μm. (B) RAW264.7 cells transfected with HA-72-5ptase or HA-SHIP1 were stimulated with eIgG for 7 minutes, fixed, and labeled with HA antibodies (red) and phalloidin (green). Scale bar represents 10 μm; “e” indicates eIgG. (C) RAW264.7 transfected with GFP-72-D480N5ptase were stimulated with eIgG to initiate phagocytosis, and fluorescence was monitored using confocal microscopy. Images represent a time course of phagocytosis of eIgG. The time of closure was arbitrarily chosen as time 0 seconds. Arrowheads indicate sites of GFP-72-D480N5ptase accumulation, and asterisks indicate phagosomes forming parallel to the coverslip. Scale bar represents 20 μm. The complete sequence from which these images were taken is shown in Video S1. (D) To characterize the temporal and spatial association of GFP-72-D480N5ptase with phagosomes, the accumulation of fluorescence at the phagocytic cup normalized to the cytosolic level (RFU) was quantified. To allow for comparison between experiments the RFUs were normalized to the maximum RFU obtained for each experiment. Data are expressed as the mean (± SEM) of 3 independent experiments. (E) RAW264.7 cells transfected with HA-72-5ptase or HA-SHIP1 were stimulated with SOZ for 10 minutes, fixed, and labeled with HA antibodies (red) and phalloidin (green). “SOZ” indicates serum-opsonized zymosan. Scale bar represents 10 μm. (F) RAW264.7 macrophages stimulated with 6-bIgG or SOZ for 7 and 10 minutes, respectively, were fixed and labeled with 72-5ptase (red)– and SHIP1 (green)–specific antibodies. Arrowheads indicate protein localization, “b” indicates 6-bIgG, and “SOZ” indicates SOZ. Scale bar represents 10 μm. Insets show enlargements of boxed areas.
Intracellular localization of 72-5ptase during phagocytosis. (A) RAW264.7 macrophages grown in serum or serum-starved and stimulated with 3-bIgG for 7 minutes were fixed and labeled with 72-5ptase (red) or immunodepleted-72-5ptase-specific antibodies and colocalized with phalloidin (green). Arrowheads indicate sites of 72-5ptase localization, “b” indicates 3-bIgG, and asterisks represent fully internalized beads. Scale bar represents 10 μm. (B) RAW264.7 cells transfected with HA-72-5ptase or HA-SHIP1 were stimulated with eIgG for 7 minutes, fixed, and labeled with HA antibodies (red) and phalloidin (green). Scale bar represents 10 μm; “e” indicates eIgG. (C) RAW264.7 transfected with GFP-72-D480N5ptase were stimulated with eIgG to initiate phagocytosis, and fluorescence was monitored using confocal microscopy. Images represent a time course of phagocytosis of eIgG. The time of closure was arbitrarily chosen as time 0 seconds. Arrowheads indicate sites of GFP-72-D480N5ptase accumulation, and asterisks indicate phagosomes forming parallel to the coverslip. Scale bar represents 20 μm. The complete sequence from which these images were taken is shown in Video S1. (D) To characterize the temporal and spatial association of GFP-72-D480N5ptase with phagosomes, the accumulation of fluorescence at the phagocytic cup normalized to the cytosolic level (RFU) was quantified. To allow for comparison between experiments the RFUs were normalized to the maximum RFU obtained for each experiment. Data are expressed as the mean (± SEM) of 3 independent experiments. (E) RAW264.7 cells transfected with HA-72-5ptase or HA-SHIP1 were stimulated with SOZ for 10 minutes, fixed, and labeled with HA antibodies (red) and phalloidin (green). “SOZ” indicates serum-opsonized zymosan. Scale bar represents 10 μm. (F) RAW264.7 macrophages stimulated with 6-bIgG or SOZ for 7 and 10 minutes, respectively, were fixed and labeled with 72-5ptase (red)– and SHIP1 (green)–specific antibodies. Arrowheads indicate protein localization, “b” indicates 6-bIgG, and “SOZ” indicates SOZ. Scale bar represents 10 μm. Insets show enlargements of boxed areas.
The recruitment of HA-72-5ptase to the phagocytic cup was compared with HA-SHIP1 in response to FcγR activation. After stimulation of phagocytosis with eIgG, HA-72-5ptase and HA-SHIP1 were both recruited to the phagocytic cup, which was outlined by phalloidin staining of F-actin (Figure 3B). Live cell imaging of GFP-72-D480N5ptase was used for a detailed temporal analysis of 72-5ptase phagocytic cup recruitment. GFP-72-D480N5ptase was recruited from the cytosol to the base and extending pseudopods of the phagocytic cup after eIgG addition and persisted after phagosome closure (arbitrarily designated time 0 seconds) as a continuous ring surrounding the phagosome (Figure 3C; Video S1, available on the Blood website; see the Supplemental Materials link at the top of the online article) for up to 100 seconds after closure (Figure 3C,D) and subsequently dissociated symmetrically. In contrast, in response to CR3 activation, HA-72-5ptase remained cytosolic after SOZ addition, with little to no enrichment at the phagocytic cup. Under these conditions HA-SHIP1 was recruited to the phagocytic cup, exhibiting enhanced staining at the cup relative to its cytosolic distribution (Figure 3E). To confirm the differential recruitment of these 5-phosphatases in response to these distinct agonists, we colabeled the same RAW264.7 macrophages with polyclonal 72-5ptase and monoclonal SHIP1 antibodies.38 In response to 6-bIgG-stimulation, which activates FcγR-mediated phagocytosis, allowing spatial analysis of the relative 5-phosphatase localization, we noted both endogenous 72-5ptase and SHIP1 were enriched at the phagocytic cup (Figure 3F). However, after CR3 activation little to no 72-5ptase was detected at the phagocytic cup, relative to its cytosolic distribution whereas SHIP1 was enriched at this site. Collectively these studies are consistent with the contention that the 72-5ptase and SHIP1 exhibit distinct recruitment to the phagocytic cup in response to FcγR versus CR3 stimulation.
The 72-5ptase regulates the amplitude and duration of PtdIns(3,4,5)P3 on FcγR-containing phagosomes
Although the PtdIns(3,4,5)P3 5-ptases SHIP1,15 SHIP2,14 and, as shown here, the 72-5ptase negatively regulate phagocytosis, no studies have reported whether these enzymes focally degrade PtdIns(3,4,5)P3 at the phagocytic cup, regulating its spatial distribution, duration, and/or signal amplitude. PtdIns(3,4,5)P3 and the product of 5-ptase hydrolysis, PtdIns(3,4)P2, bind an overlapping subset of PH domains from Akt, ASAP-related protein (ARAP), adenosine-diphosphate ribosylation factor 6 guanine nucleotide exchange factor (ARF6 GEF), and phosphoinositide dependent kinase 1 (PDK1),39,–41 whereas the PH domain of the Arf-GEF ARNO only binds PtdIns(3,4,5)P3.42,–44 Green fluorescent protein (GFP) chimeras with the PH domain of ARNO can be used to monitor PtdIns(3,4,5)P3 dynamics in live cells.44,,–47 RAW264.7 macrophages coexpressing HA-72-5ptase or HA-72-D480N5ptase and GFP-ARNO/PH were examined live during phagocytosis to investigate the role the 72-ptase plays in regulating PtdIns(3,4,5)P3 dynamics at the phagocytic cup. Secondly, to assess SHIP1 activity at the cup we examined PtdIns(3,4,5)P3/PtdIns(3,4)P2 signals in macrophages derived from SHIP1−/− mice that had been crossed with AktPH-GFP transgenic mice (Ship1−/−/AktPH-GFP-Tg).22
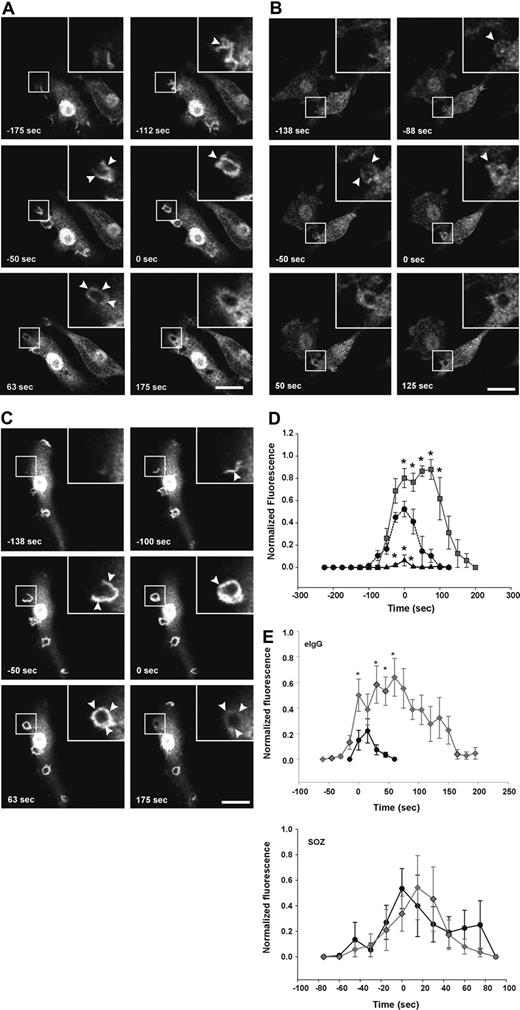
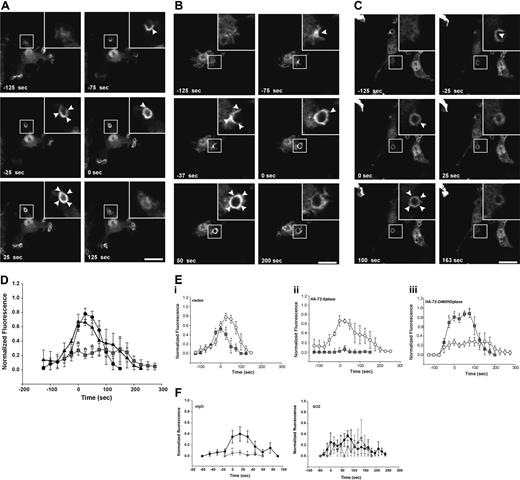
First, we determined whether the 72-5ptase focally degrades PtdIns(3,4,5)P3 at the macrophage phagocytic cup. After initiation of phagocytosis with eIgG, in vector coexpressing RAW264.7 cells, GFP-ARNO/PH accumulated at the base of the forming phagocytic cup and extended into the pseudopodia correlating with PtdIns(4,5)P2 disappearance from the phagocytic cup (data not shown). Upon phagosome closure, GFP-ARNO/PH appeared as a homogenous ring surrounding the recently engulfed particle and gradually disappeared after phagosome closure and was not detected after 175 seconds (Figure 4A; Video S2). HA-72-5ptase expression significantly decreased GFP-ARNO/PH association with the forming phagocytic cup and extending pseudopodia (Figure 4B; Video S3). GFP-ARNO/PH recruitment to phagocytic cups that formed perpendicular to the cell was analyzed and fluorescence was quantified (Figure 4D). An extremely low level of PtdIns(3,4,5)P3 was detected at the phagocytic cup before closure in HA-72-5ptase cells, followed by its more rapid disappearance soon after phagosome closure (< 50 seconds), relative to vector-expressing cells (Figure 4D). In contrast, HA-72-D480N5ptase expression enhanced fluorescence in the base and extending pseudopodia (Figure 4C; Video S4), and at the time of closure an intense homogeneous ring around the phagosome was detected that was increased greater than 1.5-fold relative to vector-expressing cells (Figure 4D). 3-dimensional visualization revealed GFP-ARNO/PH accumulated over the entire phagocytic cup surface but in no optical plane could fluorescence be detected outside the unclosed phagosome (Figure 4C). In cells expressing HA-72-D480N5ptase after phagosome closure, GFP-ARNO/PH persisted at high levels for a prolonged period (Figure 4D) but eventually dissociated symmetrically. Similar results were obtained for all 5-ptase constructs regardless of whether the GFP-ARNO/PH was expressed at moderate or low levels (data not shown). Therefore despite amplified and sustained PtdIns(3,4,5)P3 signals on the phagosome, this phosphoinositide remained confined to the phagosome.
FcγR-stimulated accumulation of PtdIns(3,4,5)P3 at forming phagosomes is regulated by 5-phosphatase activity. RAW264.7 macrophages cotransfected with GFP-ARNO/PH and (A) vector, (B) HA-72-5ptase, or (C) HA-72-D480N5ptase were stimulated with eIgG to initiate phagocytosis, and GFP fluorescence was monitored using confocal microscopy. (A-C) GFP-ARNO/PH expression is shown. Images represent a time course of phagocytosis of eIgG. Insets show enlargements of boxed areas. The time of closure was arbitrarily designated as time 0. Arrowheads indicate sites of GFP-ARNO/PH accumulation. Scale bar represents 20 μm. The complete sequences from which these images were taken are shown in Videos S2, S3, and S4. (D) Results shown represent the fluorescence intensity of GFP-ARNO/PH at phagosomes normalized to the cytosolic fluorescence. To allow for comparison between experiments, each RFU was normalized to the maximum RFU obtained for each experiment. ---●--- represents vector; ▴, HA-72-5ptase; and ▒, HA-72-D480N5ptase. Data represents mean (± SEM) of 3 to 5 independent determinations. *P < .05 relative to vector-expressing cells. (E) siCON or si72-5ptase-transfected RAW264.7 macrophages were further transfected with GFP-ARNO/PH 48 hours after transfection and stimulated with eIgG or SOZ, and phagocytosis was monitored using confocal microscopy. Results shown represent the fluorescence intensity of GFP-ARNO/PH at phagosomes normalized to the cytosolic fluorescence. To allow for comparison between experiments each RFU was normalized to the maximum RFU obtained for each experiment. • represent siCON, and ⧫ represent si72-5ptase. Data represents mean (± SEM) from 3 to 7 independent determinations. *P < .05 relative to siCON-expressing cells.
FcγR-stimulated accumulation of PtdIns(3,4,5)P3 at forming phagosomes is regulated by 5-phosphatase activity. RAW264.7 macrophages cotransfected with GFP-ARNO/PH and (A) vector, (B) HA-72-5ptase, or (C) HA-72-D480N5ptase were stimulated with eIgG to initiate phagocytosis, and GFP fluorescence was monitored using confocal microscopy. (A-C) GFP-ARNO/PH expression is shown. Images represent a time course of phagocytosis of eIgG. Insets show enlargements of boxed areas. The time of closure was arbitrarily designated as time 0. Arrowheads indicate sites of GFP-ARNO/PH accumulation. Scale bar represents 20 μm. The complete sequences from which these images were taken are shown in Videos S2, S3, and S4. (D) Results shown represent the fluorescence intensity of GFP-ARNO/PH at phagosomes normalized to the cytosolic fluorescence. To allow for comparison between experiments, each RFU was normalized to the maximum RFU obtained for each experiment. ---●--- represents vector; ▴, HA-72-5ptase; and ▒, HA-72-D480N5ptase. Data represents mean (± SEM) of 3 to 5 independent determinations. *P < .05 relative to vector-expressing cells. (E) siCON or si72-5ptase-transfected RAW264.7 macrophages were further transfected with GFP-ARNO/PH 48 hours after transfection and stimulated with eIgG or SOZ, and phagocytosis was monitored using confocal microscopy. Results shown represent the fluorescence intensity of GFP-ARNO/PH at phagosomes normalized to the cytosolic fluorescence. To allow for comparison between experiments each RFU was normalized to the maximum RFU obtained for each experiment. • represent siCON, and ⧫ represent si72-5ptase. Data represents mean (± SEM) from 3 to 7 independent determinations. *P < .05 relative to siCON-expressing cells.
PtdIns(3,4,5)P3 focal degradation at the phagocytic cup in response to FcγR versus CR3 activation was also examined under conditions of 72-5ptase siRNA-knock-down. Although the biosensor-signal was reduced in intensity, due to the use of different transfection techniques used for siRNA-transfected cells, a sustained and amplified PtdIns(3,4,5)P3 signal was detected in si72-5ptase-transfected cells only at the phagocytic cup in response to FcγR-activation, relative to siCON. In contrast, in response to CR3 activation si72-5ptase-transfected cells showed no change in the level or duration of PtdIns(3,4,5)P3 (Figure 4E). To confirm the 72-5ptase regulates PtdIns(3,4,5)P3 hydrolysis only in response to FcγR, we also examined the activation of the PtdIns(3,4,5)P3-dependent effector Akt using Ser473 phosphospecific antibodies. After FcγR activation Ser473Akt was enhanced approximately 1.5-fold in si72-5ptase-transfected cells relative to siCON-transfected cells, however there was no change in the relative Akt phosphorylation compared with nonsilencing siRNA controls in response to CR3 activation (data not shown).
PtdIns(3,4)P2 accumulates at the phagocytic cup during FcγR-mediated phagocytosis
Recently, in Dictyostelium PtdIns(3,4)P2 has been visualized at the phagosome during phagocytosis, however, its functional role in this process remains unexplored.10 We investigated the role of the 72-5ptase in PtdIns(3,4)P2 generation during phagocytosis using the PtdIns(3,4)P2-specific biosensor, YFP-TAPP1/PH.48 After the addition of eIgG to cells expressing control vector, YFP-TAPP1/PH accumulated at the phagocytic cup base and extending pseudopods almost simultaneously, indicating that PtdIns(3,4)P2 was generated after phagocytic cup formation. YFP-TAPP1/PH was maintained at the phagosome during pseudopod extension, peaking after closure, followed by its symmetrical dissociation (Figure 5A; Video S5). Comparison of PtdIns(3,4,5)P3 versus PtdIns(3,4)P2 spatial distribution revealed both signaling molecules were exclusively confined to the phagosome (Figures 4A and 5A), however, PtdIns(3,4)P2 maximum signals peaked after the maximum PtdIns(3,4,5)P3 (Figure 5Ei-iii). YFP-TAPP1/PH spatial distribution in HA-72-5ptase overexpressing cells was unchanged (Figure 5B; Video S6). However, under these conditions PtdIns(3,4)P2 appeared earlier and persisted for longer (Figure 5D) although signal amplitude was not increased. This may be due to the actions of inositol polyphosphate 4-phosphatases that hydrolyze PtdIns(3,4)P2 rapidly as it is formed.49 Comparison of PtdIns(3,4,5)P3 versus PtdIns(3,4)P2 in HA-72-5ptase-expressing cells revealed that low PtdIns(3,4,5)P3 correlated with early and prolonged PtdIns(3,4)P2 generation. This may result from sustained PtdIns(3,4,5)P3 hydrolysis by 72-5ptase generating its product, PtdIns(3,4)P2 (Figure 5Eii). In contrast, expression of HA-72-D480N5ptase was associated with a 4-fold reduction in the amplitude of YFP-TAPP1/PH fluorescence (Figure 5C,D; Video S7) across the entire phagosome after closure. Comparison of PtdIns(3,4,5)P3 versus PtdIns(3,4)P2 levels in cells expressing HA-72-D480N5ptase revealed prolonged duration of both signals, but the level of PtdIns(3,4,5)P3 was increased, whereas PtdIns(3,4)P2 levels were decreased (Figure 5Eiii).
5-phosphatases regulate PtdIns(3,4)P2-association with forming phagosomes. RAW264.7 macrophages cotransfected with YFP-TAPP1/PH and (A) vector, (B) HA-72-5ptase, or (C) HA-72-D480N5ptase were stimulated with eIgG to initiate phagocytosis, and fluorescence was monitored using confocal microscopy. (A-C) YFP-TAPP1/PH expression is shown. Images represent a time course of phagocytosis of eIgG. The time of closure was arbitrarily designated as time 0 seconds. Arrowheads indicate sites of YFP-TAPP1/PH accumulation. Scale bar represents 20 μm. Insets show enlargements of boxed areas. The complete sequences from which these images are taken are shown in Videos S5, S6, and S7. (D) Results shown represent the fluorescence intensity of YFP-TAPP1/PH with phagosomes, normalized to the cytosolic fluorescence. To allow for comparison between experiments, each RFU was normalized to the maximum RFU obtained for each experiment. ---●--- represents vector; ▴, HA-72-5ptase; and ▒, HA-72-D480N5ptase. Data represent mean (± SEM) of 3 to 5 independent determinations. *P < .05 compared with vector-expressing cells. (E) The normalized fluorescence intensity of GFP-ARNO/PH and YFP-TAPP1/PH at the phagosomal membrane in cells coexpressing (i) vector, (ii) HA-72-5ptase, or (iii) HA-72-D480N5ptase were plotted on the same graphs for direct comparison. ○ represents YFP-TAPP1/PH, and ▒ represents GFP-ARNO/PH. *P < .05 compared with YFP-TAPP1/PH RFU. (F) siCON- or si72-5ptase-transfected RAW264.7 macrophages were further transfected with YFP-TAPP1/PH 48 hours after transfection and stimulated with eIgG or SOZ, and phagocytosis was monitored using confocal microscopy. Results shown represent the fluorescence intensity of GFP-ARNO/PH at phagosomes normalized to the cytosolic fluorescence. To allow for comparison between experiments, each RFU was normalized to the maximum RFU obtained for each experiment. • represents siCON, and ⧫ represents si72-5ptase. Data represent mean (± SEM) from 3 to 7 independent determinations. *P < .05 relative to siCON-expressing cells.
5-phosphatases regulate PtdIns(3,4)P2-association with forming phagosomes. RAW264.7 macrophages cotransfected with YFP-TAPP1/PH and (A) vector, (B) HA-72-5ptase, or (C) HA-72-D480N5ptase were stimulated with eIgG to initiate phagocytosis, and fluorescence was monitored using confocal microscopy. (A-C) YFP-TAPP1/PH expression is shown. Images represent a time course of phagocytosis of eIgG. The time of closure was arbitrarily designated as time 0 seconds. Arrowheads indicate sites of YFP-TAPP1/PH accumulation. Scale bar represents 20 μm. Insets show enlargements of boxed areas. The complete sequences from which these images are taken are shown in Videos S5, S6, and S7. (D) Results shown represent the fluorescence intensity of YFP-TAPP1/PH with phagosomes, normalized to the cytosolic fluorescence. To allow for comparison between experiments, each RFU was normalized to the maximum RFU obtained for each experiment. ---●--- represents vector; ▴, HA-72-5ptase; and ▒, HA-72-D480N5ptase. Data represent mean (± SEM) of 3 to 5 independent determinations. *P < .05 compared with vector-expressing cells. (E) The normalized fluorescence intensity of GFP-ARNO/PH and YFP-TAPP1/PH at the phagosomal membrane in cells coexpressing (i) vector, (ii) HA-72-5ptase, or (iii) HA-72-D480N5ptase were plotted on the same graphs for direct comparison. ○ represents YFP-TAPP1/PH, and ▒ represents GFP-ARNO/PH. *P < .05 compared with YFP-TAPP1/PH RFU. (F) siCON- or si72-5ptase-transfected RAW264.7 macrophages were further transfected with YFP-TAPP1/PH 48 hours after transfection and stimulated with eIgG or SOZ, and phagocytosis was monitored using confocal microscopy. Results shown represent the fluorescence intensity of GFP-ARNO/PH at phagosomes normalized to the cytosolic fluorescence. To allow for comparison between experiments, each RFU was normalized to the maximum RFU obtained for each experiment. • represents siCON, and ⧫ represents si72-5ptase. Data represent mean (± SEM) from 3 to 7 independent determinations. *P < .05 relative to siCON-expressing cells.
We also determined the levels and duration of PtdIns(3,4)P2 signals in si72-5ptase macrophages in response to FcγR stimulation, revealing PtdIns(3,4)P2 signals were of significantly lower amplitude and duration and were virtually undetectable relative to siCON-transfected cells, consistent with the contention that this lipid signaling molecule is derived from 72-5ptase-mediated hydrolysis of PtdIns(3,4,5)P3 (Figure 5F). In contrast, after CR3 activation, little change in the levels or duration of PtdIns(3,4)P2 were noted (Figure 5F).
SHIP1 regulates the amplitude and duration of PtdIns(3,4,5)P3 during phagocytosis of zymosan
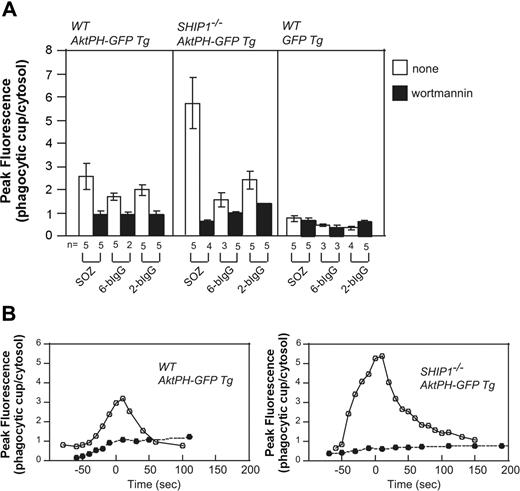
Macrophages from Ship1−/− mice exhibit significant defects in CR3-mediated phagocytosis,15 with less effects documented for FcγR-mediated phagocytosis. The PH domain of Akt (AktPH-GFP) binds both PtdIns(3,4,5)P3 and PtdIns(3,4)P2 with equal affinity. Transgenic (Tg) mice expressing Akt-PH-GFP were mated with Ship1−/− mice,50 generating Ship1−/−-AktPH-GF-Tg mice, and peritoneal macrophages were isolated. PtdIns(3,4,5)P3/PtdIns(3,4)P2 generation at the phagocytic cup was examined during the phagocytosis of SOZ, 2-bIgG, or 6-bIgG. AktPH-GFP accumulated at the phagosome of wild-type macrophages in response to all stimuli that were reduced after wortmannin treatment. This localization cannot be accounted for by cytosolic entrapment because in macrophages expressing only GFP, little fluorescence was associated specifically with the phagosome (Figure 6A). Significantly in Ship1−/−-AktPH-GFP-Tg macrophages a greater than 2-fold wortmannin-sensitive increase in fluorescence at the phagosome was noted, relative to that detected in wild-type (WT)-AktPH-GFP-Tg macrophages in response to SOZ. Interestingly, Ship1 deficiency did not result in enhanced AktPH-GFP association with the phagosome in response to IgG-opsonized particles, relative to wild type (Figure 6A).
Enhanced translocation of AktPH-GFP to the phagocytic cup of Ship1−/− macrophages in response to serum-opsonized zymosan. Peritoneal macrophages from wild-type (WT; Ship1+/+) AktPH-GFP Tg, Ship1−/−AktPH-GFP Tg, or WT GFP Tg mice that had been treated with (■) or without (□) wortmannin were incubated with IgG-coated beads or SOZ. The fluorescence intensity of AktPH-GFP or GFP at the phagocytic cup was normalized to the cytosolic fluorescence intensity. (A) Results are the peak values of the relative intensity in a cell (± SEM or range) from 2 to 5 independent experiments. (B) Time course analyses of the relative fluorescence intensity (phagocytic cup/cytosol) during phagocytosis of SOZ. The time of closure arbitrarily defines the 0 time point. • indicates 100 nM wortmannin treatment, and ○ represents no inhibitor treatment. Data shown are representative of 5 independent experiments. The time course from which these data were obtained is shown in Videos S8 and S9.
Enhanced translocation of AktPH-GFP to the phagocytic cup of Ship1−/− macrophages in response to serum-opsonized zymosan. Peritoneal macrophages from wild-type (WT; Ship1+/+) AktPH-GFP Tg, Ship1−/−AktPH-GFP Tg, or WT GFP Tg mice that had been treated with (■) or without (□) wortmannin were incubated with IgG-coated beads or SOZ. The fluorescence intensity of AktPH-GFP or GFP at the phagocytic cup was normalized to the cytosolic fluorescence intensity. (A) Results are the peak values of the relative intensity in a cell (± SEM or range) from 2 to 5 independent experiments. (B) Time course analyses of the relative fluorescence intensity (phagocytic cup/cytosol) during phagocytosis of SOZ. The time of closure arbitrarily defines the 0 time point. • indicates 100 nM wortmannin treatment, and ○ represents no inhibitor treatment. Data shown are representative of 5 independent experiments. The time course from which these data were obtained is shown in Videos S8 and S9.
After the addition of SOZ, AktPH-GFP accumulation in WT-AktPH-GFP-Tg macrophages began at the base of the phagocytic cup and as the phagosome formed fluorescence was associated with the extending pseudopods, with no loss of fluorescence at the base. Upon phagocytic cup closure, AktPH-GFP was observed as a homogenous ring surrounding the phagosome (Video S8). Significantly the spatial distribution of AktPH-GFP during phagocytosis was not altered in Ship1−/−-AktPH-GFP-Tg and at all times remained confined to the phagosome. However, the level and duration of phagosomal AktPH-GFP was enhanced in SHIP1−/− macrophages (Figure 6B; Video S9). Wortmannin treatment blocked the recruitment of AktPH-GFP to the phagocytic cup in WT-Akt-PH-GFP-Tg and Ship1−/−-AktPH-GFP-Tg macrophages (Figure 6B). Collectively these experiments confirm that SHIP1 enzyme activity regulates the amplitude and duration of PtdIns(3,4,5)P3/PtdIns(3,4)P2 during complement-mediated phagocytosis but not the spatial distribution of these phosphoinositides.
Discussion
This study has demonstrated that the 72-5ptase is expressed in macrophages and functions to negatively regulate FcγR- but not CR3-mediated phagocytosis. Interestingly the 72-5ptase was a more potent inhibitor of FcγR-induced phagocytosis than SHIP1, with the latter exhibiting greater activity in regulating CR3 phagocytosis. Whereas both the 72-5ptase and SHIP1 were recruited to phagocytic cups that formed in response to FcγR, only SHIP1 localized to the phagocytic cup in response to CR3 activation. We have noted for both SHIP1 and the 72-5ptase, inhibition of phagocytosis correlated with the ability to focally degrade PtdIns(3,4,5)P3 at the phagocytic cup. The 72-5ptase regulated both the amplitude and duration but not spatial distribution of PtdIns(3,4,5)P3 signals formed at the phagocytic cup in response to FcγR but not CR3 stimulation. In contrast SHIP1 governed CR3- but not FcγR-stimulated PtdIns(3,4,5)P3 dynamics at the cup. Therefore these studies have identified a novel lipid phosphatase, the 72-5ptase, in addition to SHIP1, SHIP2, and PTEN that inhibits phagocytosis, revealing agonist-specific regulation of PI3-kinase signals.
We have reported here that a relatively uncharacterized member of the inositol polyphosphate 5-phosphatase family, the 72-5ptase, negatively regulates FcγR-mediated phagocytosis. No mouse knock-out for this enzyme has been reported and there are few studies showing a functional role for this lipid phosphatase. As described here the 72-5ptase inhibited FcγR- but not CR3-stimulated phagocytosis correlating with its dynamic recruitment to FcγR- but not CR3-stimulated phagosomes. In addition, siRNA-mediated 72-5ptase knock-down in macrophages significantly increased phagocytosis in response to FcγR but not CR3 activation. These results suggest that this lipid phosphatase must gain access to PtdIns(3,4,5)P3 at the phagocytic cup to inhibit phagocytosis. Interestingly we noted siRNA-targeted depletion of the 72-5ptase led to a 2-fold increase in FcγR-stimulated PtdIns(3,4,5)P3 levels at the cup correlating with a 2-fold increase in phagocytosis, whereas 72-5ptase overexpression significantly inhibited both phagocytosis and PtdIns(3,4,5)P3 generation at the cup.
Why are so many 5-ptases required to negatively regulate phagocytosis? We have shown here the 72-5ptase does not regulate CR3-mediated phagocytosis but rather inhibits FcγR-mediated phagocytosis, indicating specificity of function. In addition our results support the contention that the 72-5ptase plays a more dominant role in regulating FcγR-mediated phagocytosis than SHIP1. We noted only a 35% decrease in FcγR-mediated phagocytosis after SHIP1 expression, compared with a greater than 60% inhibition by 72-5ptase under the same assay conditions. Cox et al15 have reported only a mild increase in phagocytosis (1.4-fold) in response to FcγR-mediated phagocytosis in SHIP1−/− macrophages. In support of this observation we did not detect increased or sustained PtdIns(3,4,5)P3 at the phagosome in SHIP1−/− macrophages in response to FcγR-stimulated phagocytosis. In contrast siRNA-mediated knock-down of the 72-5ptase was associated with amplified and sustained PtdIns(3,4,5)P3 signals at the phagocytic cup after FcγR activation. This may result from the different enzyme activity exhibited by 72-5ptase versus SHIP1. Kinetic analysis has revealed the 72-5ptase exhibits a 10-fold greater affinity for PtdIns(3,4,5)P3 than SHIP1.16 In addition although both the 72-5ptase and SHIP1 are recruited to FcγR-stimulated phagocytic cups, a recent detailed report examining SHIP1 recruitment to the cup has revealed it is only transiently and focally recruited to the lateral margins of the cup.51 These authors predicted SHIP1 would only transiently suppress PtdIns(3,4,5)P3, although this was not directly shown.51 This recruitment may preclude SHIP1 sufficient spatial or temporal access to its substrate, PtdIns(3,4,5)P3, thereby preventing significant inhibition of FcγR-mediated phagocytosis. In contrast SHIP1 appears to be the dominant 5-phosphatase in regulating CR3-mediated phagocytosis. We were unable to demonstrate any effects of the 72-5ptase in regulating this event and although SHIP2 inhibits FcγR-mediated phagocytosis, its effects on complement-mediated events are unknown.14 All 3 5-ptases, SHIP1, SHIP2, and the 72-5ptase, are dynamically recruited to the phagocytic cup during phagocytosis. Of note, unlike SHIP1 and SHIP2, the 72-5ptase lacks an SH2 domain and is therefore unlikely to associate with the phagocytic cup in the same manner as SHIP1 and SHIP2, which complex with FcγR via SH2-mediated mechanisms.52,53 SHIP1 is recruited to CR3-stimulated phagosomes via both SH2- and NPXY-mediated mechanisms.15,52
During phagocytosis the distribution of PtdIns(3,4,5)P3 remains confined to the phagocytic cup, however, the molecular mechanisms mediating this restriction remain unresolved. It has been proposed localized PtdIns(3,4,5)P3 generation at the phagocytic cup and its hydrolysis at the edge of the cup may result in its restricted spatial distribution.3,12 Recent elegant studies by Grinstein and colleagues11 have suggested that lipids binding to immobile signaling complexes, initiated by tyrosine kinase activation and the recruitment of signaling molecules, may restrict lipid diffusion. PtdIns(3,4,5)P3 is generated at the phagocytic cup by the dynamic recruitment of the class I PI3-kinase to the cup, via interaction with Src kinase-associated receptors.4 Its loss from the cup has been attributed to the activities of SHIP1 and/or SHIP2,14,15 but this has not been directly shown and analysis of the temporal-spatial regulation of PtdIns(3,4,5)P3 dynamics at the phagosome by any 5-ptase has not been previously reported. We have demonstrated here using multiple experimental models that 5-ptases focally degrade PtdIns(3,4,5)P3 at the phagocytic cup in response to distinct stimuli. Overexpression of the 72-5ptase reduced PtdIns(3,4,5)P3 to almost undetectable levels at the cup, correlating with significantly decreased phagocytosis. Under conditions where 5-ptase enzyme activity was significantly reduced by siRNA-mediated depletion, gene-targeted deletion or blocked by dominant-negative 5ptase, amplified and prolonged PtdIns(3,4,5)P3 at the cup was detected, but this was not associated with its diffusion at the edges of the phagocytic cup during either FcγR- or CR3-mediated phagocytosis. Although the AktPH-GFP reporter does not discriminate between PtdIns(3,4,5)P3 versus PtdIns(3,4)P2, the increased and prolonged association of AktPH-GFP with the phagosome in SHIP1−/− macrophages was not associated with lateral diffusion of this lipid biosensor.
As shown here PtdIns(3,4)P2 exhibits a similar spatial distribution to PtdIns(3,4,5)P3, forming at the base and extending pseudopodia of the nascent phagosome and surrounding the sealed phagosome, but at no time diffused laterally outside the phagocytic cup. PtdIns(3,4)P2 accumulation on the phagosome temporally closely followed PtdIns(3,4,5)P3, consistent with the contention that PtdIns(3,4)P2 is generated via 5-position hydrolysis of PtdIns(3,4,5)P3 rather than by direct phosphorylation of PtdIns(3)P or PtdIns(4)P. Furthermore, expression of dominant-negative 72-5ptase or its siRNA-mediated depletion blocked PtdIns(3,4)P2 formation on the phagosome. Interestingly we observed significantly enhanced phagocytosis even in the absence of PtdIns(3,4)P2 generation at the cup, suggesting PtdIns(3,4)P2 formation is not essential for FcγR-induced phagocytosis. In cells with low PtdIns(3,4)P2 levels at the phagosome but high PtdIns(3,4,5)P3, phagocytosis of large particles was increased and phagosome closure accelerated, indicating PtdIns(3,4,5)P3 can promote these events independent of PtdIns(3,4)P2. In contrast, 5ptase overexpression correlated with decreased PtdIns(3,4,5)P3 levels on the phagocytic cup, decreased phagocytosis, pseudopod extension, and phagosome closure, despite normal and sustained PtdIns(3,4)P2. Thus PtdIns(3,4)P2 levels do not correlate with phagocytic events, acting downstream of FcγR when PtdIns(3,4,5)P3 is present.
In summary, this study has identified that the relatively uncharacterized 72-5ptase is dynamically recruited to the phagocytic cup to negatively regulate FcγR- but not CR3-mediated phagocytosis by focally degrading PtdIns(3,4,5)P3 forming PtdIns(3,4)P2 only at the phagosome. In contrast SHIP1 exhibits greater activity in regulating CR3- rather than FcγR-mediated phagocytosis, revealing nonredundant functions for these distinct inositol polyphosphate 5-phosphatases.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from the National Health and Medical Research Council Australia and the Australian Research Council, Center of Excellence for Functional Microbial Genomics.
The authors thank Monash Micro Imaging and Mr Stephen Firth and Dr Ian Harper for technical assistance.
Kristy Horan was funded by the Cancer Council of Victoria postgraduate scholarship.
This work is dedicated to Amanda Horan.
Authorship
Contribution: K.A.H. and K.W. designed and performed experiments and analyzed data; A.M.K. performed experiments; C.G.B. and J.E.J.R. generated constructs and expression vectors and provided experimental design advice; T.S. designed and performed experiments and contributed to writing the manuscript; and C.A.M. designed experiments and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Christina A. Mitchell, Department of Biochemistry and Molecular Biology, Monash University, Wellington Rd, Clayton, Victoria 3800, Australia; e-mail:Christina.Mitchell@med.monash.edu.au.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal