Hematopoietic cell transplantation (HCT) from an HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 allele–matched unrelated donor is a well-recognized life-saving treatment modality for patients with hematologic disorders. The morbidity and mortality from clinically significant acute graft-versus-host disease (aGVHD) remains a limitation. The extent to which transplantation outcome may be improved with donor matching for HLA-DP is not well defined. The risks of aGVHD, relapse, and mortality associated with HLA-DPB1 allele mismatching were determined in 5929 patients who received a myeloablative HCT from an HLA-A–, HLA-B–, HLA-C–, HLA-DRB1–, and HLA-DQB1–matched or –mismatched donor. There was a statistically significantly higher risk of both grades 2 to 4 aGVHD (odds ratio [OR] = 1.33; P < .001) and grades 3 to 4 aGVHD (OR = 1.26; P < .001) after HCT from an HLA-DPB1–mismatched donor compared with a matched donor. The increased risk of aGVHD was accompanied by a statistically significantly decrease in disease relapse (hazard ratio [HR] = 0.82; P = .01). HLA-DPB1 functions as a classical transplantation antigen. The increased risk of GVHD associated with HLA-DPB1 mismatching is accompanied by a lower risk of relapse. Knowledge of the DPB1 matching status prior to transplantation will aid in more precise risk stratification for the individual patient.
Introduction
Patients with hematologic disorders who lack a human leukocyte antigen (HLA)–identical sibling donor can be successfully treated with a hematopoietic cell transplantation (HCT) from an HLA-matched unrelated donor. However, these patients are at a higher risk for posttransplantation immunologic sequelae, including graft-versus-host disease (GVHD) and delayed immune reconstitution, resulting from the greater degree of genetic disparity between such individuals.1,2
Historically, the selection of potential donors for HCT relied on serologic tests for donor-recipient identity for classical HLA-A, HLA-B, and HLA-DR antigens.3 Later, as molecular methods for typing HLA genes became available, the degree of diversity of this region became apparent.4 When such typing methods were applied to the analysis of HC transplant recipients and donors, many serologically identical unrelated donor–recipients were found to harbor mismatches between 2 unique alleles of the same antigen.5,6 Clinical transplantation studies have demonstrated that such allelic differences are clinically relevant, increasing the risks of graft failure, acute GVHD (aGVHD), and mortality.7,,,–11 These data have lead to the current gold standard of a “fully matched” unrelated donor being one who is allele-matched for HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1.
HLA-DPB1 is the sixth classic HLA molecule, located centromeric to HLA-DR and HLA-DQ in the class II region of chromosome 6p21.3. HLA-DP shares structural similarity with the other class II molecules, and is highly polymorphic (IMGT/HLA database12 ). Although donor-recipient HLA-DP typing and matching is not routinely performed prior to transplantation, we hypothesized that the risks of GVHD and mortality after HLA-matched unrelated donor HCT could be contributed to by disparity at the HLA-DP locus. Although HLA-DP has been reported to provoke GVHD after transplantation,13,,,–17 the balance of graft-versus-host and host-versus-graft allorecognition and its overall effect on survival are not well defined, and the potential additive effects of HLA-DP to mismatches at other class I and II loci are not known. We analyzed the risks associated with donor-recipient mismatching for HLA-DPB1 alleles in a large well-characterized international dataset. Information on the clinical importance of HLA-DP may aid in the selection of donors, in the assessment of individual patient risks, and in planning optimal transplantation regimens.
Patients, materials, and methods
Patients and data collection
All available donor-recipient pairs for whom retrospective high-resolution typing of HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DQB1, and HLA-DPB1 alleles and complete clinical data (aGVHD, relapse, survival) were available were eligible for inclusion in the study. A total of 5929 recipient-donor pairs met these criteria and were submitted to the HCT component of the International Histocompatibility Working Group (IHWG). Donors were unrelated volunteers provided by donor registries (Document S1; Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Clinical data were contributed by participating transplantation centers, transplantation registries, and donor registries detailed in Table S1.
Ethical approval
All research samples and data were collected according to institutional review board–approved guidelines and protocols of each participating institution. For samples contributed by the National Marrow Donor Program (NMDP), all surviving recipients included in this analysis were retrospectively contacted, and informed consent was obtained in accordance with the Declaration of Helsinki for participation in the research program. Informed consent was waived by the NMDP Institutional Review Board for all deceased recipients. To address potential bias introduced by inclusion of only a proportion of surviving patients (those who consented) but all deceased recipients, a sample of deceased patients was selected using a weighted randomized scheme that adjusts for overrepresentation of deceased patients in the cohort.
Tissue typing
HLA typing was performed by the following methods: sequencing-based typing (81.2%), sequence-specific priming (10.9%), reference strand conformation analysis (3.6%), sequence-specific oligonucleotide probing (3.3%), and reverse dot blot 1%. These methods are well characterized and described in full elsewhere.18 The choice of typing methods was made by participating laboratories and registries. A description of typing methods used for the NMDP samples can be found at http://bioinformatics.nmdp.org/HLA/hla_typing_idx.
Donor-recipient HLA disparity was defined on the basis of HLA alleles encoded in the donor and recipient as determined by DNA-based typing methods; analysis of serologically defined HLA-A, HLA-B, HLA-C, HLA-DR, or HLA-DQ antigens or primed lymphocyte test-defined HLA-DP alloantigens was not undertaken in the current analysis.
Statistical analysis
HLA-DPB1 mismatching for death and relapse endpoints denotes mismatching in either vector (graft-versus-host [GVH] or host-versus-graft [HVG]); mismatching for GVHD denotes mismatching in the GVH vector. Survival estimates were obtained using the Kaplan-Meier method. The probability of relapse was summarized using cumulative incidence estimates, where death without relapse was regarded as a competing risk. Multivariable Cox regression models were used to examine the association between DPB1 disparity and mortality and relapse outcomes; logistic regression was used for GVHD. Factors other than mismatching at DPB1 that were included in these models were severity of disease (categorized as low, intermediate, or high); patient age at transplantation; number of mismatched alleles at HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 (modeled as a continuous variable); patient/donor sex, patient/donor cytomegalovirus (CMV) status; source of stem cells; and use of T-cell depletion. All “adjusted models” have taken these factors into account. Tests for interaction between DPB1 disparity and each of these factors were made by including the appropriate term in a regression model. Two-sided P values were obtained from the Wald test, and no adjustments were made for multiple comparisons.
Results
Patient demographics
Transplantations were performed between 1984 and 2005. Patient and donor demographics and transplantation characteristics are summarized in Table 1. Patient diagnoses included both malignant and nonmalignant disorders.
Demographics of the study population
| Characteristic . | 0 DPB1 mismatch, n = 848 . | 1 DPB1 mismatch, n = 3205 . | 2 DPB1 mismatches, n = 1876 . |
|---|---|---|---|
| CMV status, p/d, no (%) | |||
| Positive/positive | 147 (17) | 544 (17) | 371 (20%) |
| Positive/negative | 220 (26) | 791 (25) | 485 (26%) |
| Negative/positive | 138 (16) | 516 (16) | 265 (14%) |
| Negative/negative | 261 (31) | 1045 (33) | 584 (31%) |
| Unknown | 82 (10) | 309 (10) | 171 (9%) |
| Sex, p/d | |||
| Female/female | 178 (21) | 614 (19) | 358 (19) |
| Female/male | 191 (23) | 698 (22) | 402 (21) |
| Male/female | 147 (17) | 620 (19) | 381 (20) |
| Male/male | 305 (36) | 1154 (36) | 658 (35) |
| Unknown | 27 (3) | 119 (4) | 77 (4) |
| Diagnosis* | |||
| ALL | 172 (20) | 643 (21) | 428 (23) |
| AML | 210 (25) | 775 (24) | 474 (25) |
| CML | 332 (39) | 1260 (39) | 694 (37) |
| MDS | 84 (10) | 350 (11) | 171 (9) |
| Other | 33 (4) | 130 (4) | 70 (4) |
| Unknown | 18 (2) | 48 (1) | 39 (2) |
| Disease severity† | |||
| High | 158 (19) | 672 (21) | 374 (20) |
| Intermediate | 380 (45) | 1458 (45) | 887 (47) |
| Low | 252 (30) | 849 (26) | 488 (26) |
| Unknown | 58 (7) | 226 (7) | 127 (7) |
| T-cell depletion | |||
| Yes | 163 (19) | 604 (19) | 386 (21) |
| No | 647 (76) | 2446 (76) | 1402 (75) |
| Unknown | 38 (4) | 155 (5) | 88 (5) |
| HLA matching‡ | |||
| Matched | 470 (55) | 1461 (46) | 773 (41) |
| Single mismatch | 180 (21) | 866 (27) | 515 (27) |
| Two or more mismatches | 198 (23) | 878 (27) | 588 (31) |
| Source of stem cells | |||
| Bone marrow | 734 (87) | 2775 (87) | 1657 (88) |
| PBMCs | 94 (11) | 347 (11) | 166 (9) |
| Unknown | 20 (2) | 83 (3) | 53 (3) |
| Patient age | |||
| 0th percentile | <1 | <1 | <1 |
| 25th percentile | 18 | 19 | 17 |
| 50th percentile | 34 | 34 | 32 |
| 75th percentile | 45 | 44 | 43 |
| 100th percentile | 68 | 69 | 70 |
| Characteristic . | 0 DPB1 mismatch, n = 848 . | 1 DPB1 mismatch, n = 3205 . | 2 DPB1 mismatches, n = 1876 . |
|---|---|---|---|
| CMV status, p/d, no (%) | |||
| Positive/positive | 147 (17) | 544 (17) | 371 (20%) |
| Positive/negative | 220 (26) | 791 (25) | 485 (26%) |
| Negative/positive | 138 (16) | 516 (16) | 265 (14%) |
| Negative/negative | 261 (31) | 1045 (33) | 584 (31%) |
| Unknown | 82 (10) | 309 (10) | 171 (9%) |
| Sex, p/d | |||
| Female/female | 178 (21) | 614 (19) | 358 (19) |
| Female/male | 191 (23) | 698 (22) | 402 (21) |
| Male/female | 147 (17) | 620 (19) | 381 (20) |
| Male/male | 305 (36) | 1154 (36) | 658 (35) |
| Unknown | 27 (3) | 119 (4) | 77 (4) |
| Diagnosis* | |||
| ALL | 172 (20) | 643 (21) | 428 (23) |
| AML | 210 (25) | 775 (24) | 474 (25) |
| CML | 332 (39) | 1260 (39) | 694 (37) |
| MDS | 84 (10) | 350 (11) | 171 (9) |
| Other | 33 (4) | 130 (4) | 70 (4) |
| Unknown | 18 (2) | 48 (1) | 39 (2) |
| Disease severity† | |||
| High | 158 (19) | 672 (21) | 374 (20) |
| Intermediate | 380 (45) | 1458 (45) | 887 (47) |
| Low | 252 (30) | 849 (26) | 488 (26) |
| Unknown | 58 (7) | 226 (7) | 127 (7) |
| T-cell depletion | |||
| Yes | 163 (19) | 604 (19) | 386 (21) |
| No | 647 (76) | 2446 (76) | 1402 (75) |
| Unknown | 38 (4) | 155 (5) | 88 (5) |
| HLA matching‡ | |||
| Matched | 470 (55) | 1461 (46) | 773 (41) |
| Single mismatch | 180 (21) | 866 (27) | 515 (27) |
| Two or more mismatches | 198 (23) | 878 (27) | 588 (31) |
| Source of stem cells | |||
| Bone marrow | 734 (87) | 2775 (87) | 1657 (88) |
| PBMCs | 94 (11) | 347 (11) | 166 (9) |
| Unknown | 20 (2) | 83 (3) | 53 (3) |
| Patient age | |||
| 0th percentile | <1 | <1 | <1 |
| 25th percentile | 18 | 19 | 17 |
| 50th percentile | 34 | 34 | 32 |
| 75th percentile | 45 | 44 | 43 |
| 100th percentile | 68 | 69 | 70 |
Data are numbers (%). The self-described patient and donor racial backgrounds were as follows: North American and European, 80.6% and 64.3%; Hispanic, 4.1% and 3.5%; black, 2.8% and 2.3%; Asian, 2.3% and 2.3%; Native American, 0.2% and 0.6%; and other or unknown, 9.9% and 26.9%.
p/d indicates patient/donor; and PBMCs, peripheral blood mononuclear cells.
The ″other″ category included both malignant and nonmalignant diagnoses: aplastic anemia, acute leukemia undifferentiated, chronic leukemia, chronic lymphocytic leukemia, chronic myelomonocytic leukemia, Fanconi anemia, hemoglobinopathy, histiocytic disorder, Hodgkin lymphoma, inherited disorder of metabolism, malignant other, multiple myeloma, myeloproliferative syndrome, non-Hodgkin lymphoma, nonmalignant other, other acute leukemia, other leukemia, plasma cell disorder, severe combined immunodeficiency and other immune system disorders, secondary acute leukemia, and thalassemia.
Low-risk indicates CML chronic phase; intermediate-risk, CML accelerated phase or blast crisis/remission, acute leukemia with transplantation in remission, and MDS–refractory anemia; high-risk, blast-crisis CML, acute leukemia with transplantation in relapse, and MDS other than refractory anemia.
HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1.
HLA-DPB1 allele frequencies and matching
Of the 126 known HLA-DPB1 alleles, 65 were seen in this population (Table 2). The frequency of HLA-DPB1 mismatching among HLA-A–, HLA-B–, HLA-C–, HLA-DRB1–, and HLA-DQB1–matched and –mismatched pairs is provided in Table 1.
Frequency of HLA-DPB1 alleles in the study population
| Allele . | No. (%) . |
|---|---|
| DPB1*0401 | 9943 (41.925) |
| DPB1*0201 | 2967 (12.511) |
| DPB1*0402 | 2825 (11.912) |
| DPB1*0301 | 2265 (9.551) |
| DPB1*0101 | 1404 (5.920) |
| DPB1*0501 | 635 (2.678) |
| DPB1*1101 | 506 (2.134) |
| DPB1*1301 | 458 (1.931) |
| DPB1*1701 | 397 (1.674) |
| DPB1*0601 | 375 (1.581) |
| DPB1*1001 | 360 (1.518) |
| DPB1*1401 | 301 (1.269) |
| DPB1*0202 | 223 (0.940) |
| DPB1*0901 | 187 (0.788) |
| DPB1*1501 | 158 (0.666) |
| DPB1*1901 | 125 (0.527) |
| DPB1*2301 | 122 (0.514) |
| DPB1*1601 | 120 (0.506) |
| DPB1*2001 | 120 (0.506) |
| DPB1*1801 | 47 (0.198) |
| DPB1*2701 | 20 (0.084) |
| DPB1*2101 | 14 (0.059) |
| DPB1*3401 | 10 (0.042) |
| DPB1*5101 | 10 (0.042) |
| DPB1*4001 | 9 (0.038) |
| DPB1*0801 | 8 (0.034) |
| DPB1*2901 | 8 (0.034) |
| DPB1*3901 | 7 (0.030) |
| DPB1*4501 | 7 (0.030) |
| DPB1*2801 | 6 (0.025) |
| DPB1*3501 | 6 (0.025) |
| DPB1*3801 | 6 (0.025) |
| DPB1*4601 | 6 (0.025) |
| DPB1*2601 | 5 (0.021) |
| DPB1*3001 | 5 (0.021) |
| DPB1*8501 | 5 (0.021) |
| DPB1*3301 | 3 (0.013) |
| DPB1*6501 | 3 (0.013) |
| DPB1*0602 | 2 (0.008) |
| DPB1*0802 | 2 (0.008) |
| DPB1*3101 | 2 (0.008) |
| DPB1*3601 | 2 (0.008) |
| DPB1*3701 | 2 (0.008) |
| DPB1*5201 | 2 (0.008) |
| DPB1*5401 | 2 (0.008) |
| DPB1*5701 | 2 (0.008) |
| DPB1*6301 | 2 (0.008) |
| DPB1*7201 | 2 (0.008) |
| DPB1*7301 | 2 (0.008) |
| DPB1*8101 | 2 (0.008) |
| DPB1*0305 | 1 (0.004) |
| DPB1*1302 | 1 (0.004) |
| DPB1*1402 | 1 (0.004) |
| DPB1*2201 | 1 (0.004) |
| DPB1*2401 | 1 (0.004) |
| DPB1*2501 | 1 (0.004) |
| DPB1*4101 | 1 (0.004) |
| DPB1*4801 | 1 (0.004) |
| DPB1*5001 | 1 (0.004) |
| DPB1*6101N | 1 (0.004) |
| DPB1*6601 | 1 (0.004) |
| DPB1*6801 | 1 (0.004) |
| DPB1*7001 | 1 (0.004) |
| DPB1*8601 | 1 (0.004) |
| DPB1*8701 | 1 (0.004) |
| Allele . | No. (%) . |
|---|---|
| DPB1*0401 | 9943 (41.925) |
| DPB1*0201 | 2967 (12.511) |
| DPB1*0402 | 2825 (11.912) |
| DPB1*0301 | 2265 (9.551) |
| DPB1*0101 | 1404 (5.920) |
| DPB1*0501 | 635 (2.678) |
| DPB1*1101 | 506 (2.134) |
| DPB1*1301 | 458 (1.931) |
| DPB1*1701 | 397 (1.674) |
| DPB1*0601 | 375 (1.581) |
| DPB1*1001 | 360 (1.518) |
| DPB1*1401 | 301 (1.269) |
| DPB1*0202 | 223 (0.940) |
| DPB1*0901 | 187 (0.788) |
| DPB1*1501 | 158 (0.666) |
| DPB1*1901 | 125 (0.527) |
| DPB1*2301 | 122 (0.514) |
| DPB1*1601 | 120 (0.506) |
| DPB1*2001 | 120 (0.506) |
| DPB1*1801 | 47 (0.198) |
| DPB1*2701 | 20 (0.084) |
| DPB1*2101 | 14 (0.059) |
| DPB1*3401 | 10 (0.042) |
| DPB1*5101 | 10 (0.042) |
| DPB1*4001 | 9 (0.038) |
| DPB1*0801 | 8 (0.034) |
| DPB1*2901 | 8 (0.034) |
| DPB1*3901 | 7 (0.030) |
| DPB1*4501 | 7 (0.030) |
| DPB1*2801 | 6 (0.025) |
| DPB1*3501 | 6 (0.025) |
| DPB1*3801 | 6 (0.025) |
| DPB1*4601 | 6 (0.025) |
| DPB1*2601 | 5 (0.021) |
| DPB1*3001 | 5 (0.021) |
| DPB1*8501 | 5 (0.021) |
| DPB1*3301 | 3 (0.013) |
| DPB1*6501 | 3 (0.013) |
| DPB1*0602 | 2 (0.008) |
| DPB1*0802 | 2 (0.008) |
| DPB1*3101 | 2 (0.008) |
| DPB1*3601 | 2 (0.008) |
| DPB1*3701 | 2 (0.008) |
| DPB1*5201 | 2 (0.008) |
| DPB1*5401 | 2 (0.008) |
| DPB1*5701 | 2 (0.008) |
| DPB1*6301 | 2 (0.008) |
| DPB1*7201 | 2 (0.008) |
| DPB1*7301 | 2 (0.008) |
| DPB1*8101 | 2 (0.008) |
| DPB1*0305 | 1 (0.004) |
| DPB1*1302 | 1 (0.004) |
| DPB1*1402 | 1 (0.004) |
| DPB1*2201 | 1 (0.004) |
| DPB1*2401 | 1 (0.004) |
| DPB1*2501 | 1 (0.004) |
| DPB1*4101 | 1 (0.004) |
| DPB1*4801 | 1 (0.004) |
| DPB1*5001 | 1 (0.004) |
| DPB1*6101N | 1 (0.004) |
| DPB1*6601 | 1 (0.004) |
| DPB1*6801 | 1 (0.004) |
| DPB1*7001 | 1 (0.004) |
| DPB1*8601 | 1 (0.004) |
| DPB1*8701 | 1 (0.004) |
The proportion of HLA-DPB1–matched pairs was statistically significantly higher among those who were also matched at HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 (matched for 10 of 10 alleles) compared with those mismatched for at least 1 of 10 alleles (P < .001). A total of 470 donor-recipient pairs were matched for all 6 HLA genes (12 of 12 matches at HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DQB1, and HLA-DPB1).
Clinical outcome
aGVHD.
The observed probabilities of grades 2 to 4 and grades 3 to 4 aGVHD in the group overall were 2918 (52%) of 5621 and 1738 (31%) of 5621, respectively (GVHD data were not available for 308 patients). Among 1384 patients who were matched at HLA-DPB1, 645 (47%) developed grades 2 to 4 aGVHD, while 1303 (53%) of 2456 patients with a single HLA-DPB1 mismatch and 970 (54%) of 1781 with 2 HLA-DPB1 mismatches developed grades 2 to 4 aGVHD. The observed probability of grades 3 to 4 aGVHD was 27% (377 of 1384), 31% (765 of 2456), and 33% (596 of 1781) in patients with 0, 1, or 2 mismatches at HLA-DPB1, respectively.
These proportions led to a statistically significantly higher odds of both grades 2 ot 4 and grades 3 to 4 aGVHD in mismatched compared with matched pairs, both in unadjusted and adjusted models. The increased risk of aGVHD among the HLA-DPB1–mismatched pairs was attributable to both single and double mismatches (Table 3).
Logistic regression models for grades II-IV and grades III-IV acute GVHD and Cox regression models for relapse and overall mortality
| . | Unadjusted models . | Adjusted models . | ||||||
|---|---|---|---|---|---|---|---|---|
| Grades 2-4 aGVHD . | Grades 3-4 aGVHD . | Relapse . | Overall mortality . | Grades 2-4 aGVHD . | Grades 3-4 aGVHD . | Relapse . | Overall mortality . | |
| Matched at HLA-DPB1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Mismatched at HLA-DPB1 | 1.33 (1.17-1.50; P < .001) | 1.26 (1.10-1.45; P < .001) | 0.82 (0.70-0.96; P = .01) | 1.15 (1.05-1.27; P = .003) | 1.33 (1.18-1.51; P < .001) | 1.22 (1.06-1.40; P = .005) | 0.78 (0.67-0.92; P = .002) | 1.09 (0.99-1.19; P = .07) |
| 1 allele mismatched | 1.30 (1.14-1.48; P = .001) | 1.21 (1.04-1.40; P = .01) | 0.82 (0.70-0.96; P = .01) | 1.14 (1.03-1.25; P = .01) | 1.31 (1.14-1.49; P < .001) | 1.18 (1.02-1.37; P = .03) | 0.79 (0.67-0.93; P = .005) | 1.08 (0.98-1.19; P = .13) |
| 2 alleles mismatched | 1.37 (1.19-1.58; P < .001) | 1.34 (1.15-1.57; P < .001) | 0.82 (0.69-0.98; P = .03) | 1.18 (1.07-1.31; P = .002) | 1.36 (1.18-1.58; P < .001) | 1.27 (1.09-1.49; P = .003) | 0.76 (0.64-0.91; P = .003) | 1.09 (0.98-1.21; P = .11) |
| . | Unadjusted models . | Adjusted models . | ||||||
|---|---|---|---|---|---|---|---|---|
| Grades 2-4 aGVHD . | Grades 3-4 aGVHD . | Relapse . | Overall mortality . | Grades 2-4 aGVHD . | Grades 3-4 aGVHD . | Relapse . | Overall mortality . | |
| Matched at HLA-DPB1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Mismatched at HLA-DPB1 | 1.33 (1.17-1.50; P < .001) | 1.26 (1.10-1.45; P < .001) | 0.82 (0.70-0.96; P = .01) | 1.15 (1.05-1.27; P = .003) | 1.33 (1.18-1.51; P < .001) | 1.22 (1.06-1.40; P = .005) | 0.78 (0.67-0.92; P = .002) | 1.09 (0.99-1.19; P = .07) |
| 1 allele mismatched | 1.30 (1.14-1.48; P = .001) | 1.21 (1.04-1.40; P = .01) | 0.82 (0.70-0.96; P = .01) | 1.14 (1.03-1.25; P = .01) | 1.31 (1.14-1.49; P < .001) | 1.18 (1.02-1.37; P = .03) | 0.79 (0.67-0.93; P = .005) | 1.08 (0.98-1.19; P = .13) |
| 2 alleles mismatched | 1.37 (1.19-1.58; P < .001) | 1.34 (1.15-1.57; P < .001) | 0.82 (0.69-0.98; P = .03) | 1.18 (1.07-1.31; P = .002) | 1.36 (1.18-1.58; P < .001) | 1.27 (1.09-1.49; P = .003) | 0.76 (0.64-0.91; P = .003) | 1.09 (0.98-1.21; P = .11) |
Adjusted models include severity of disease; patient age at transplantation; number of mismatched alleles at HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1; patient/donor sex; patient/donor CMV status; source of stem cells; and use of T-cell depletion.
Ranges in parentheses are 95% CI.
The number of mismatched alleles at HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 was categorized as 0, 1, 2, and 3 or more mismatches. The impact of an HLA-DPB1 mismatch on grades 2 to 4 (but not 3 to 4) aGVHD was reduced when there were 3 or more mismatches elsewhere (Table 4). A formal test of interaction between mismatching at HLA-DPB1 and 0 to 2 versus 3 or more mismatches at HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 yields a P value of .04.
Impact of number of mismatched alleles at HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 in the GVH vector on the association of mismatching at HLA-DPB1 with grades 2-4 aGVHD
| No. mismatched alleles at HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DQB1 . | Matched at HLA-DPB1, no. developing aGVHD (%) . | Mismatched at HLA-DPB1, no. developing aGVHD (%) . | Adjusted odds ratio . |
|---|---|---|---|
| 0 | 309/725 (43) | 967/1907 (51) | 1.38 |
| 1 | 155/326 (48) | 620/1152 (54) | 1.34 |
| 2 | 106/213 (50) | 382/654 (58) | 1.51 |
| 3 or more | 75/120 (63) | 304/524 (58) | 0.88 |
| No. mismatched alleles at HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DQB1 . | Matched at HLA-DPB1, no. developing aGVHD (%) . | Mismatched at HLA-DPB1, no. developing aGVHD (%) . | Adjusted odds ratio . |
|---|---|---|---|
| 0 | 309/725 (43) | 967/1907 (51) | 1.38 |
| 1 | 155/326 (48) | 620/1152 (54) | 1.34 |
| 2 | 106/213 (50) | 382/654 (58) | 1.51 |
| 3 or more | 75/120 (63) | 304/524 (58) | 0.88 |
Odds ratios adjusted for severity of disease, patient age at transplantation, patient/donor sex, source of stem cells, and use of T-cell depletion.
The impact of mismatching at HLA-DPB1 did not appear to depend on severity of disease, use of T-cell depletion, age, patient/donor sex, or source of stem cells (data not shown).
Relapse.
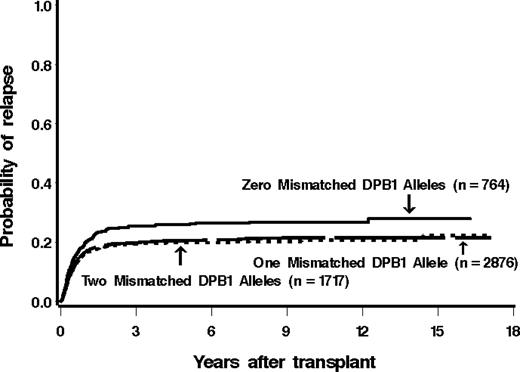
Among 790 patients matched at HLA-DPB1 in both the GVH and HVG vector, 221 (28%) relapsed. Of patients mismatched for 1 or 2 HLA-DPB1 alleles, 23% (679 of 2973) and 21% (363 of 1745) relapsed (421 patients lacked information for relapse). The estimated probability of relapse at 5 years among these groups was 26%, 20%, and 20%, respectively (Figure 1).
Cumulative incidence estimates of the probability of relapse among patients with 0, 1, or 2 mismatched alleles at HLA-DPB1. There was a significantly higher incidence of relapse in patients receiving HLA-DPB1–matched grafts.
Cumulative incidence estimates of the probability of relapse among patients with 0, 1, or 2 mismatched alleles at HLA-DPB1. There was a significantly higher incidence of relapse in patients receiving HLA-DPB1–matched grafts.
There was a statistically significantly lower hazard of disease relapse in those pairs that were mismatched for HLA-DPB1 compared with the pairs that were matched, both before (hazard ratio [HR] = 0.82; P = .01) and after (HR = 0.78; P = .001) adjustment. This decreased risk of relapse was attributable to both single and double mismatches (Table 3).
There was evidence to suggest that the impact of mismatching at HLA-DPB1 was influenced by the presence of mismatching atHLA-A, HLA-B, HLA-C, HLA-DRB1, or HLA-DQB1 (P = .04). In particular, among patients matched for 10 of 10 alleles, the hazard of relapse among patients mismatched for HLA-DPB1 was statistically significantly less than that among patients matched at HLA-DPB1 (adjusted HR = 0.70 [95% confidence interval (CI) 0.57-0.85[; P < .001). Conversely, among patients mismatched for at least 1 allele at HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1, the hazards of relapse were similar among those mismatched and matched at HLA-DPB1 (HR = 0.97 [95% CI 0.75-1.26]; P = .84).
The impact of HLA-DPB1 mismatching on relapse was similar for homozygous donors (GVH vector of disparity) and heterozygous donors (both GVH and HVG vectors), and there were no obvious influences on the impact of mismatching as related to age, severity of disease, source of stem cells, patient/donor sex, or use of T-cell depletion (data not shown).
Mortality.
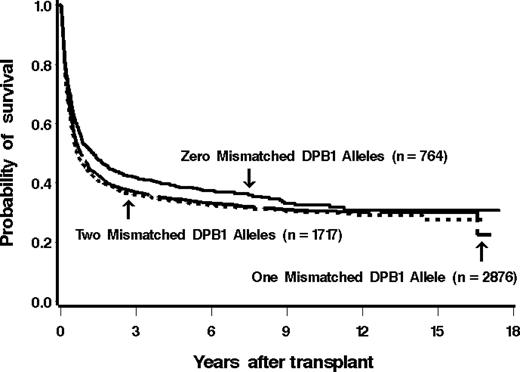
Among 825 patients matched at HLA-DPB1 in both the GVH and HVG vectors, 507 (61%) have died as of last contact. Of patients mismatched for 1 or 2 HLA-DPB1 alleles, 65% (2034 of 3120) and 66% (1209 of 1825) died (159 patients did not have information available for mortality). The estimated probability of overall survival at 5 years among these groups was 38%, 34%, and 33%, respectively (Figure 2).
Kaplan-Meier estimates of the probability of overall survival among patients with 0, 1, or 2 mismatched alleles at HLA-DPB1. The overall survival did not differ significantly between the groups.
Kaplan-Meier estimates of the probability of overall survival among patients with 0, 1, or 2 mismatched alleles at HLA-DPB1. The overall survival did not differ significantly between the groups.
Before adjusting for the factors listed, the hazard of death among patients mismatched for HLA-DPB1 was statistically significantly higher than that among patients matched for HLA-DPB1. After adjustment, however, this difference was no longer statistically significant (Table 3).
The hazard of transplantation-related mortality (TRM) was suggestively higher in DPB1-mismatched compared with -matched transplantations (HR = 1.12 [95% CI 1.00-1.26]; P = .06). This occurred in both 1 (HR = 1.15 [ 95% CI 1.01-1.31]; P = .03) and 2 (HR = 1.10 [95% CI 0.97-1.24]; P = .12) allele-mismatched transplantations.
The impact of a mismatch at HLA-DPB1 on overall mortality did not appear to depend on the level of matching at HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1, disease severity, diagnosis, age, source of stem cells, patient/donor sex, or use of T-cell depletion (data not shown).
Discussion
Hematopoietic cell transplantation from an HLA-DPB1–mismatched unrelated donor is associated with an increased risk of aGVHD, irrespective of the matching status for the other HLA class I and II genes. The increased risk of GVHD is accompanied by a decrease in disease recurrence, particularly in patients who receive a transplant from an HLA-A–, HLA-B–, HLA-C–, HLA-DRB1–, and HLA-DQB1–matched donor. These data support an immunologic role for HLA-DP in transplantation.
The DP series of antigens were defined in 1980 following characterization of HLA-DRB1 and HLA-DQB1. Like the other class II molecules, DPB1 was shown to function as an alloantigen. Disease associations were noted,19,–21 and HLA-DP was shown to present a range of peptides from different sources,22,–24 both allogeneic or autologous,25,26 including those derived from other HLA molecules,22,27 to CD4+ T cells in a major histocompatibility complex (MHC)–restricted fashion.
Despite these data supporting the role of HLA-DP in the immune response, the application of DPB1 typing in the transplantation setting has lagged behind that of the other classic HLA molecules for a number of reasons. Traditional methods for typing HLA-DP gene products include the primed lymphocyte test, a modified mixed lymphocyte culture method in which 6 alloantigens could be distinguished.28,29 The extensive sequence polymorphism of this locus mandated the use of modern DNA-based methods to uncover undetected donor-recipient disparity (http://www.ebi.ac.uk/imgt/hla/stats.html).12 In addition, it was shown that while much of the MHC displays strong positive linkage disequilibrium (LD; where certain alleles are seen together at a higher frequency than would be predicted by chance), LD weakens between HLA-DQB1 and HLA-DPB1, resulting in a high incidence of mismatching for DPB15,14,30 and a low chance of identifying DPB1-matched donors even when there is matching for other HLA genes.31
Since the development of DNA typing methods, the study of HLA-DP in clinical populations has become easier. Data from renal transplantation have suggested a role for DPB1 in graft rejection,32 and several small studies in HCT found an increase in GVHD associated with DPB1 mismatching in both unrelated donor5,14,15,17 and sibling donor transplantations.33 These findings are supportedby functional data showing that HLA-DP–specific T cells can be isolated from skin biopsies at the onset of GVHD in HLA-DP–mismatched (but otherwise HLA-matched) transplantations.34
Acute GVHD results in an increased morbidity and mortality in transplantation patients, particularly in those with grades 3 to 4 disease. In the current study, there was an increase in mortality in the DPB1-mismatched patients, but this was not statistically significant despite the significant increase in clinically severe GVHD. A possible explanation for this is that the increased risk of death in the DPB1-mismatched group coincides with an increased risk of relapse observed in the DPB1-matched cohort.
It is quite possible that HLA-DP could function as a target and facilitate the GVL effect. It has been shown that DPB1 is expressed on most leukemic cells, which are able to be killed by allogeneic HLA-DP–specific T cells.35 An increase in disease relapse in DPB1-matched pairs has previously been reported in a cohort of recipients of T cell–depleted (TCD) HCT.17 The current results confirm and extend these findings in a much larger study group. Interestingly, we did not see a demonstrable difference in the effect of DPB1 mismatching between TCD and T-cell–replete transplantations, suggesting that the absolute number of infused T cells may not be the critical determining factor. That the effect of HLA-DP on GVL was marked in 10 of 10–matched pairs suggests that when matching for all 6 HLA genes can be achieved, there may be insufficient allodisparity to maintain a GVL effect. The ability of HLA-DP to present peptides derived from other HLA molecules may be important in this context, as a mismatch for any of the other HLA loci diminished or abrogated the loss of GVL seen in 12 of 12–matched pairs. As this study was not designed to uncover the basic cellular mechanisms of GVL, it is not known whether the lower relapse was indeed due to the higher GVHD, or possibly to GVHD-independent mechanisms.
The observed increase in mortality in DPB1-mismatched pairs, although not statistically significant in the whole cohort, was suggestive. The analysis was not disease specific, although adjusted models did not suggest differences in the impact of mismatching across the diagnoses of acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), chronic myelogenous leukemia (CML), or myelodysplastic syndrome (MDS). It has previously been reported that patients receiving transplants from HLA-DPB1–matched donors for the treatment of ALL have lower overall survival due to an increase in posttransplantation relapse.17 It is well recognized that salvage therapy for patients whose ALL has recurred after transplantation is often unsuccessful.36 This clinical experience, together with the results of the current study, provides the impetus for exploring the disease-specific effects of HLA-DP in the future.
An accurate prediction of the likelihood of posttransplantation complications is not always straightforward in clinical practice, due to many factors that have to be taken into account. Therefore, a simple test that can aid in prediction is welcomed. Our results suggest that pretransplantation assessment of recipient-donor HLA-DPB1 type may aid in risk assessment, especially for certain high-risk patients who would not tolerate GVHD, or patients in whom disease recurrence represents the major limitation of antileukemic therapy. For patients with low-risk disease, common HLA haplotypes, and many potential 10 of 10–matched donors, further donor selection based on DPB1 matching status could provide a means to lower GVHD. If prospective DPB1 typing is initiated in this clinical setting, careful attention to the length of time required to identify DP-matched donors is required because of the low frequency of DP matching, and may be most useful for patients whose disease status affords sufficient time for typing and matching. For patients at high risk of relapse because of advanced or aggressive malignancy, the consequences of a prolonged search for an HLA-DPB1–matched donor on disease progression must be carefully weighed. When expediency of transplantation is desirable, the risks of acute GVHD associated with the use of an HLA-DPB1–mismatched donor must be balanced against the potential benefits of lowered disease recurrence. Patients who have only a single available donor can benefit from a more accurate estimate of the risk of relapse and GVHD, and the clinician may elect to alter the posttransplantation immunosuppression (and/or conditioning regimen) based on this knowledge.
The current study was not designed to measure risks associated with donor-recipient mismatching for the HLA-DPA1 gene. The LD between DPA1 and DPB1 is strong, and therefore not only is the DPA1 allele predictable for a given DPB1 genotype, but DPB1 identity may predict DPA1 identity in many cases. Indeed, among HLA-DPA1–typed pairs, more than 95% were matched at both DPA1 and DPB1, while 70% of pairs mismatched for DPB1 were matched for DPA1. Among pairs who were DPB1 matched, only 28 were DPA1 mismatched. Hence, investigation of the clinical significance of DPA1 disparity will require a much larger transplantation experience. Nevertheless, since the HLA-DP molecule is comprised of the products of both genes, such a study would be interest.
Unlike the other classic HLA genes, HLA-DPB1 displays marked skewing of allele frequencies in human population with a few commonly observed alleles (Table 2). Among our study population, 75% of patients encoded 2 common DPB1 alleles, 23% encoded 1 common DPB1 allele, and 2% encoded no common DPB1 allele, defined as an allele frequency greater than 2% (Table 2). Of the 10 of 10 allele–matched pairs, 76% encoded 2, 22% encoded 1, and 2% encoded no common DPB1 allele. These results suggest that any given patient has an equally likely probability of prospectively identifying an HLA-DPB1–matched donor.
Among patients with 0, 1, or 2 common DPB1 alleles, 42%, 45%, and 46% were 10 of 10 matched, respectively. These data indicate that the number of common DPB1 alleles does not predict matching for other classical HLA genes, and is likely a consequence of the relatively weak LD between the DP locus and the other classical HLA molecules.31 We observed an increased probability of DPB1 identity in the pairs who were 10 of 10 allele–matched compared with pairs with HLA-A, HLA-B, HLA-C, HLA-DRB1, or HLA-DQB1 mismatches. These data suggest that in some cases LD may extend across the entire MHC to include the DP locus, a phenomenon that may be confined to a selected set of extended HLA haplotypes.31 In the future, knowledge of extended haplotypes may allow for greater accuracy in predicting the likelihood of a donor being found who is matched at all 6 loci.
The existence of permissive mismatches at DPB1 has been reported.37,–39 Within the HLA-DPB1 chain there are 6 hypervariable regions (HVRs) in exon 2, which encodes for the β1 domain and forms the peptide-binding groove (PBG). Each DPB1 allele is characterized by a specific combination of the sequences of these 6 regions.40 Based on functional data, the relative immunogenicity of different DPB1 mismatches has been calculated by the group of Fleischhauer37,38,41 to develop an algorithm. Applying this algorithm in retrospective analyses of transplantation outcome, in 2 separate cohorts, they have shown that certain mismatches (predicted to have low immunogenicity) are associated with significantly fewer transplantation complications. Studies to confirm these findings in large numbers of patients are required.
In conclusion, this large analysis provides new information on the role of HLA-DP as a classic transplantation determinant. Our results support the inclusion of pretransplantation typing of HLA-DPB1 in current practice in order to assess risks of GVHD, relapse, and mortality in individual patients. The challenge remains in balancing acceptable levels of GVHD without compromising the GVL effect.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The members of the IHWG HCT Component who contributed data to this manuscript are listed in Document S1, available on the Blood website.
Authorship
Contribution: B.E.S., M.M., E.W.P., J.A.M., and S.G.E.M. contributed to the design of the study. B.E.S., T.G., M.M., S.G.E.M., and E.W.P. contributed to data generation, analysis, interpretation, and presentation. A.B.B., M.M.H., A.G., and O.R. contributed to data interpretation. All authors contributed to the writing of the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Bronwen E. Shaw, Anthony Nolan Research Institute, Royal Free Hospital, Pond St, Hampstead NW3 2QG, United Kingdom; e-mail:bshaw@doctors.org.uk.