Abstract
All-trans retinoic acid (ATRA) has been widely used in differentiation therapy for acute promyelocytic leukemia (APL). ATRA binds to retinoic acid receptor (RAR) and triggers the formation of the transcription coactivator complex, which leads to changes in gene expression, APL cell-cycle arrest and differentiation, and clinical remission. The mechanisms responsible for ATRA's beneficial effects are still ill-defined. Here, we conducted a large-scale, unbiased short hairpin RNA (shRNA) screen aiming to identify mediators of ATRA-induced differentiation and growth arrest of APL cells. Twenty-six proteins were identified. They cover a wide range of cellular functions, including gene expression, intracellular signaling, cell death control, stress responses, and metabolic regulation, indicating the complexity of ATRA-induced cell growth control and differentiation in APL. One of these proteins, the ubiquitin-conjugating enzyme UBE2D3, is up-regulated in ATRA-treated acute promyelocytic NB4 cells. UBE2D3 is physically associated with cyclin D1 and mediates ATRA-induced cyclin D1 degradation. Knocking down UBE2D3 by RNA interference (RNAi) leads to blockage of ATRA-induced cyclin D1 degradation and cell-cycle arrest. Thus, our results highlight the involvement of the ubiquitin-mediated proteolysis pathway in ATRA-induced cell-cycle arrest and provide a novel strategy for modulating ATRA-elicited cellular effects.
Introduction
Acute promyelocytic leukemia (APL) represents 10% to 15% of acute myeloid leukemias in adults.1-4 It is caused by a variety of chromosomal translocations into the retinoic acid receptor-α (RARα) gene. These chromosomal rearrangements lead to the generation of fusion proteins containing a fixed C-terminal segment originating from RARα protein (X-RARα) and a variable N-terminal segment derived from the PML, Numa, PLZF, nucleophosmin, or STAT5b proteins.4-8 That the RARα gene is consistently altered in APL suggests an essential role of RARα protein in the pathogenesis of APL.8 All-trans retinoic acid (ATRA), a derivative of vitamin A and the physiologic ligand of RARα, is able to elicit complete remission of APL and has been successfully used in clinical treatment of APL.2,3,8-10
RARα belongs to a family of hormone nuclear receptors that act as ligand-dependent transcription factors.5,11 RARα forms a heterodimeric complex with retinoid X receptors (RXRs). In the absence of ATRA, the RAR/RXR heterodimer recruits a multiprotein repression complex containing nuclear receptor corepressors N-CoR or SMRT, mSins, and histone deacetylases. Deacetylation of histone leads to chromatin condensation and transcriptional repression on crucial myeloid differentiation genes. ATRA can bind to RARα, induce the dissociation of the repression complex, and promote the formation of a coactivator complex containing nuclear receptor coactivator p160 family members and histone acetyltransferase CBP/p300. Histone acetylation results in chromatin decompactation, promoter clearance, and transcriptional activation of crucial myeloid differentiation genes.5,11,12 The X-RARα fusion protein associated with APL causes abnormal dimerization with RXR and aberrant transcription repression. The X-RARα fusion protein associates with the corepressor complex under physiologic (low) concentrations of ATRA, thus leading to transcriptional repression and promyelocytic differentiation blockage. Only pharmacologic (high) concentrations of ATRA trigger the formation of the coactivator complex and lead to changes in gene expression, APL cell differentiation, and clinical remission.7,12,13
Although ATRA has been widely used in differentiation therapy for APL and a variety of ATRA targets have been reported, the mechanisms responsible for its beneficial effects are still ill-defined.5,12,13 RARα modulates gene transcription by binding to specific retinoic acid response elements (RAREs) present in the promoters of the target genes. RAREs are composed of 2 direct repeats of a core hexameric motif, PuG(G/T)TCA, which are usually separated by a 5-bp (or 1- or 2-bp) sequence. RAREs have been identified in the promoters of a variety of genes such as Hoxa-1, Hoxd-4, CYP26, and CRBPII.5,12 Several other downstream factors, such as p21WAF, c-myc, C/EBPε, and C/EBPβ, have also been well characterized.5,13,14 Recently, gene-expression patterns in the APL cell line NB4 before and after ATRA treatment were analyzed using DNA microarrays.15,16 A comparative proteomic analysis was also conducted to reveal systematic posttranscriptional control mechanisms in ATRA-treated APL.17 However, whether any of these alterations in mRNA or protein levels are responsible for the clinical beneficial effects of ATRA has not been investigated.
To further investigate the molecular basis of ATRA-induced differentiation and growth arrest of APL cells, we conducted a large-scale RNAi screen and identified 26 proteins that are essential for ATRA-induced growth inhibition and differentiation. One of the proteins identified is the ubiquitin-conjugating enzyme UBE2D3. We further show that UBE2D3 level is up-regulated in ATRA-treated acute promyelocytic NB4 cells. UBE2D3 is physically associated with cyclin D1 and mediates ATRA-induced cyclin D1 degradation. Knocking down UBE2D3 by RNAi leads to blockage of ATRA-induced cyclin D1 degradation and cell-cycle arrest.
Materials and methods
Cell culture and chemicals
NB4 cells, a human promyelocytic leukemia cell line, were maintained in RPMI 1640 medium with 2 mM l-glutamine, 25 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), 10% heat-inactivated fetal bovine serum (FBS), and 100 units/mL of penicillin-streptomycin in a humidified atmosphere with 5% CO2 at 37°C.18 The ATRA-resistant NB4-R2 cell line was a generous gift from Dr Lanotte (Institut National de la Santé et de la Recherche Médicale [INSERM], Créteil, France) and was cultured under the same conditions. Both NB4 and NB4-R2 cells were grown and maintained at a density of 105 to 5 × 105 cells/mL. Staurosporine-elicited NB4 cell growth arrest was induced by addition of 0.5 μM staurosporine. Cell death was assessed as previously described.19 HEK-293 FT cells (Invitrogen, Carlsbad, CA), a kidney tumor cell line, were maintained in Dulbecco modified Eagle medium (DMEM) with 2 mM l-glutamine, 10% heat-inactivated FBS, and 100 units/mL of penicillin-streptomycin. Molecular cloning reagents were purchased from New England Biolabs (Beverly, MA) or Promega (Madison, WI). Protein A/G agarose was from Calbiochem (La Jolla, CA). MG-132 was purchased from Calbiochem. All other reagents, including ATRA, were purchased from Sigma (St Louis, MO), except as indicated.
Soft-agar colony-formation assay
Soft agar was prepared by autoclaving Bacto agar in distilled water before use. Infected NB4 cells were spun down, resuspended in cooled 0.2% agar in RPMI medium supplemented with 10% FBS and 0.5 μM ATRA at a density of 5 × 105 cells/100-mm plate, and then seeded onto a solidified base layer containing 0.5% agar and 0.5 μM ATRA in culture medium. Plates were kept at room temperature for 30 minutes and then transferred to a CO2 incubator. Cultures were incubated for 7 to 9 days at 37°C in a 5% CO2 atmosphere. Colonies were recognized and counted under a phase-contrast microscope.
Retroviral infection for high-throughput shRNA screening
The GeneNet Human 8.5K siRNA Library was purchased from System Biosciences (Mountain View, CA). The library was cloned into a feline immunodeficiency virus (FIV)-based pFIV-H1-copGFP shRNA expression vector.20 Viruses were used at a multiplicity of infection (MOI) of 0.5 to infect cultured NB4 cells, and 5% infection efficiency was obtained routinely. For the screening, we used 2 million NB4 cells that were infected by 1 million shRNA viral particles. The infection was conducted following a protocol provided by the manufacturer (System Biosciences). The cells were recovered for 1 to 2 days before being plated in soft-agar medium.
Generation of shRNA viral particles
The amplified polymerase chain reaction (PCR) products from each ATRA-resistant colony were digested with EcoR1 and BamH1 (New England BioLabs). The shRNA-containing fragments were then subcloned into the pFIV-H1-copGFP shRNA expression vector (System Biosciences). The shRNA expression plasmid and packaging plasmid mix (System Biosciences) were cotransfected into the HEK-293 FT packaging cell line using a lipo2000 transfection Kit (Invitrogen). Viral particles released in culture medium were collected 48 hours after transfection and concentrated using polyethylene glycol 8000 (PEG 8000) precipitation. Viral particles were incubated with 10% PEG 8000 (in 0.75 × RPMI) for 48 hours and then spun down at 1000g for 15 minutes. Viral titer was determined by a plaque assay on 293 cells based on a standard protocol provided by System Biosciences. We routinely obtained 5 × 106 viral particles from a transfection conducted in a 100-mm culture dish.
Confirmation of the identified ATRA-resistant colonies by a liquid culturing assay
NB4 cells were infected with the concentrated shRNA viruses at an MOI of 2.5 in the presence of 6 μg/mL polybrene (Sigma). We obtained a 10% to 15% infection efficiency under this condition. The percentages of transduced cells were analyzed for green fluorescent protein (GFP) expression by flow cytometry.
Coimmunoprecipitation
To test the physiologic association between UBE2D3 and cyclin D1, 7 × 106 ATRA-treated or untreated cells were lysed with 1 mL immunoprecipitation (IP) buffer21 and incubated at 4°C for 30 minutes. The lysate was then centrifuged at 14 000g for 10 minutes to remove insoluble materials. Equal aliquots of the supernatant were incubated with 5 μg first antibody (anti-cyclin D1 antibody [Ab] or mouse IgG) at 4°C overnight. Protein G Plus/protein A agarose suspension (20 μL) was then added to the lysate. The bound proteins were analyzed by Western immunoblotting with indicated antibody as previously described.21
Microarray analysis
Total RNA was isolated from undifferentiated or ATRA-differentiated NB4 cells (4 days) using Tri Reagent LS (Molecular Research Center, Cincinnati, OH), digested with DNase I, and purified by elution from a MegaClear Filter Cartridge (Ambion, Austin, TX). Genome-wide mRNA profiling was performed using high-density oligonucleotide arrays (GeneChip HG_U133 set; Affymetrix, Santa Clara, CA). Isolation of cRNA from purified total RNA samples was conducted according to the manufacturer's protocols (Affymetrix). Scan outputs of hybridization signals were modeled and normalized as summarized gene-expression values using DNA-Chip Analyzer (dChip) software (Affymetrix). The confirmation of microarray data by real-time PCR is described in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Cell-cycle analysis
The proportion of cells in each cell-cycle phase was determined by DNA content. Cells (106) were resuspended in 1 mL of hypotonic solution (0.1% sodium citrate) containing 0.05 mg/mL propidium iodide (PI) and incubated at 4°C for 10 minutes. The cellular DNA content, represented by PI staining, was analyzed by fluorescence-activated cell sorter (FACS) using the CellQuest ProTM software (Becton Dickinson, Mountain View, CA). The percentage of each population was measured using ModFIT software (Becton Dickinson). At least 10 000 cells were analyzed for each data point.
Statistical analysis
Values shown in each figure represent the mean plus and minus the standard deviation (SD). Statistical significances were calculated with the Student t test. Differences were considered significant for P values less than .05 (n = 3) or .01 (n = 6).
Results
shRNA screen for mediators of ATRA signaling in acute promyelocytic leukemia
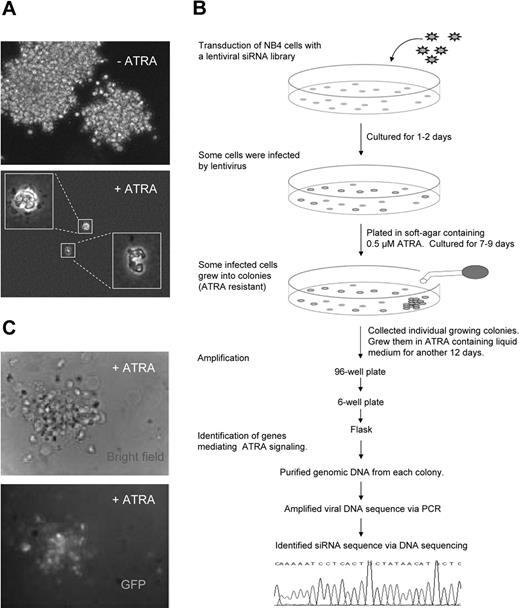
To further investigate the molecular basis of ATRA-induced differentiation and growth arrest of APL cells, we used a large-scale RNAi approach to identify genes that are essential for ATRA-induced growth inhibition and differentiation of acute promyelocytic NB4 cells. NB4 cells are maturation inducible by ATRA and bear the diagnostic chromosomal translocation t(15;17), which fuses RARα with the PML gene, and thus have been widely used and well accepted as a model system for studying ATRA signaling in APL.18,22 In the absence of ATRA, wild-type NB4 cells grew into colonies in soft-agar medium (Figure 1A top). In ATRA-containing soft-agar medium, cell proliferation was inhibited and cells differentiated into mature neutrophils with characteristic segmented and donut-shaped nuclei (Figure 1A bottom).
Identification of suppressors of ATRA-mediated cell growth arrest using an shRNA screening approach. (A) Soft-agar colony-formation assay. NB4 cells grew into colonies in RPMI/0.2% in the absence of ATRA. In the presence of 0.5 μM ATRA, NB4 cells stopped proliferation in soft agar and differentiated into mature neutrophils in 7 days. Cells were plated at a density of 5 × 105 cells/100-mm plate. Images were captured using a 20× dry objective (NIA 0.5) on an Olympus IX-71 microscope (Melville, NY). Images were acquired using a CCD camera (SensiCam, Cooke, Romulus, MI) controlled by IPLab imaging software (IPLab, Rockville, MD). The figure shows the results of a representative experiment. (B) The experimental flow chart of the functional shRNA screening. Details are provided in “Retroviral infection for high-throughput shRNA screening.” (C) An ATRA-resistant positive colony identified from the initial screening. This colony is GFP positive and thus contains the inserted viral sequence. The images were acquired as described in panel A.
Identification of suppressors of ATRA-mediated cell growth arrest using an shRNA screening approach. (A) Soft-agar colony-formation assay. NB4 cells grew into colonies in RPMI/0.2% in the absence of ATRA. In the presence of 0.5 μM ATRA, NB4 cells stopped proliferation in soft agar and differentiated into mature neutrophils in 7 days. Cells were plated at a density of 5 × 105 cells/100-mm plate. Images were captured using a 20× dry objective (NIA 0.5) on an Olympus IX-71 microscope (Melville, NY). Images were acquired using a CCD camera (SensiCam, Cooke, Romulus, MI) controlled by IPLab imaging software (IPLab, Rockville, MD). The figure shows the results of a representative experiment. (B) The experimental flow chart of the functional shRNA screening. Details are provided in “Retroviral infection for high-throughput shRNA screening.” (C) An ATRA-resistant positive colony identified from the initial screening. This colony is GFP positive and thus contains the inserted viral sequence. The images were acquired as described in panel A.
We infected NB4 cells with a FIV-based lentiviral shRNA library. This library contains 43 800 siRNA templates targeted to 8500 well-characterized human genes listed in the NCBI RefSeq database.23 The library was cloned into an FIV-based pFIV-H1-copGFP shRNA expression vector.20 This single-promoter siRNA vector expresses cloned shRNA from a single H1 RNA polymerase III promoter. Viral transduced cells could be recognized by their GFP expression. To obtain the maximal number of infected cells and deliver only one viral particle into each cell, we used a MOI of 1:2 (viral particle-cell ratio). Under these conditions, transduction efficiencies of approximately 5% were routinely obtained. Most infected cells were healthy, and less than 10% dead cells were detected using trypan blue staining. After 1 to 2 days of culture, the cells (including both the viral-infected GFP-positive and uninfected GFP-negative cells) were plated in soft-agar clonogenic culture medium containing 0.5 μM ATRA. Starting with a library containing 2 million viral particles, we obtained 960 positive colonies that could grow in ATRA-containing soft-agar medium (Figure 1B-C). Many of these colonies were relatively small and might not have been authentic ATRA-resistant colonies; thus we regrew all colonies in ATRA-containing liquid medium for another 15 to 18 days. At the end, we obtained 122 authentic ATRA-resistant colonies (Table S1).
The ATRA-resistant positive colonies from our initial screening can be divided into 2 major categories. The first one includes those from which no PCR products were recovered. Their ATRA resistance is probably caused by somatic mutations occurring on genes involved in ATRA signaling and thus has nothing to do with the viral infection. The majority of these colonies came from the uninfected cells that count for approximately 95% of the initial cell population (the infection efficiency is approximately 5%). These colonies were GFP negative and no viral fragment could be recovered from them. Seventy-four such colonies were obtained from our screening and the calculated frequency is approximately 35 per million cells. The other type of positive colonies includes those infected by virus. The PCR amplification of viral DNA from these colonies should be positive. We were able to recover the inserted shRNA templates via PCR from 48 colonies, and 33 of them were successfully sequenced (Table 1; Table S2). All of the sequenced shRNA templates could be matched to a specific gene (Table 1; Table S2); of these, 3 represent the UBE2D3 gene and 2 identified the BDH2 gene. Fifteen of the PCR products could not be clearly sequenced, which might be due to the mixed colonies (Table S1).
The list of positive hits from the shRNA screen sorted by their known cellular functions
| Gene name . | Symbol . | Activities . | Processes involved, known . | Positive hits . | Confirmation . |
|---|---|---|---|---|---|
| Gene expression regulation | |||||
| Splicing factor proline/glutamine-rich | SFPQ | Associated with polypyrimidine tract-binding protein | Transcription and pre-mRNA processing | 1 | N/A |
| Chromodomain helicase DNA-binding protein 1 | CHD1 | Binds to DNA helicase | Chromatin assembly or disassembly, transcription | 1 | Confirmed |
| SPANX-b, c, and d | SPANX | Unknown | Regulates transcription and translation, spermatogenesis | 1 | N/A |
| Cellular repressor of adenovirus E1A protein-stimulated genes 1 | CREG1 | Antagonizes transcriptional activation of E1A protein | Transcriptional control of cell growth and differentiation | 1 | N/A |
| Metabolic regulation | |||||
| Asparagine-linked glycosylation 5 homolog | ALG5 | Dolichyl-phosphate beta-glucosyltransferase activity | N-glycan biosynthesis | 1 | N/A |
| 3-hydroxybutyrate dehydrogenase, type 2 | BDH2 | Dehydrogenase/reductase enzyme | Fatty acid beta-oxidation | 2 | Confirmed |
| Cathepsin K | CTSK | Cysteine proteinase | Proteolysis, tumor invasiveness, bone remodeling | 1 | Confirmed |
| Peroxiredoxin 3 | PRDX3 | Peroxidase activity, alkyl hydroperoxide reductase activity | Activation of NF-kappaB transcription factor, antioxidant | 1 | Confirmed |
| Protein kinases | |||||
| Serine threonine kinase 39 | STK39 | Protein serine/threonine kinase activity | Mediates stress-activated signals | 1 | N/A |
| Protein kinase C, nu | PRKCN | Protein serine/threonine kinase activity | Intracellular signaling cascade | 1 | N/A |
| Serine/threonine protein kinase MST4 | MASK | Protein serine/threonine kinase activity | Regulation of apoptosis, can be cleaved by caspase-3 | 1 | N/A |
| Cell signaling related | |||||
| NCK adaptor protein 1 | NCK1 | Contains SH2 and SH3 domains, cytoskeletal adaptor | Intracellular signaling cascade, positive regulation of actin filament polymerization | 1 | Confirmed |
| NCK-associated protein 1 | NCKAP1 | Interacts with Nck | Lamellipodium biogenesis, apoptosis | 1 | Confirmed |
| Transient receptor potential cation channel, subfamily C, member 6 | TRPC6 | Calcium channel activity | Calcium ion transport, calcium signaling | 1 | Confirmed |
| Interleukin 1 receptor antagonist | IL1RN | Inhibits the activities of interleukin 1 | Regulates immune and inflammatory responses | 1 | N/A |
| Cell death/stress related | |||||
| Hypoxia-inducible factor 1, alpha subunit | HIF1A | Transcription factor | Cellular and systemic homeostatic responses to hypoxia | 1 | Confirmed |
| DED containing | DEDD | Contains DED, regulation of apoptosis | Cell death | 1 | N/A |
| Transmembrane BAX inhibitor motif containing 1 | TMBIM1 | Contains BAX inhibitor motif | Cell death | 1 | N/A |
| Others | |||||
| Ubiquitin-conjugated enzyme, UBE2D3 | UBE2D3 | Ubiquitin-protein ligase activity | Ubiquitin-mediated proteolysis | 3 | Confirmed |
| Nidogen 2/osteonidogen | NID2 | A basement membrane protein | Adhesion, migration, differentiation, gene expression, and apoptosis | 1 | N/A |
| Mutated in colorectal cancers | MCC | Interacts with NFKBIB | Putative colorectal tumor suppressor gene | 1 | Confirmed |
| Complement component 1, q subcomponent-binding protein | C1QBP | Inhibit C1 activation | Play a role in serum complement system | N/A | |
| Ancient ubiquitous protein 1 | AUP1 | Unknown | Play a crucial role in the integrin alpha(IIb)beta(3) inside-out signaling | 1 | N/A |
| Stromal interaction molecule 1 | STIM1 | Unknown | Located in an important tumor-suppressor gene region, calcium signaling | 1 | N/A |
| Spastic ataxia of Charlevoix-Saguenay, sacsin | SACS | Unknown | Mutated in ARSACS | 1 | N/A |
| Similar to interferon-induced transmembrane protein 3, LOC144383 | N/A | Unknown | Unknown | 1 | N/A |
| Gene name . | Symbol . | Activities . | Processes involved, known . | Positive hits . | Confirmation . |
|---|---|---|---|---|---|
| Gene expression regulation | |||||
| Splicing factor proline/glutamine-rich | SFPQ | Associated with polypyrimidine tract-binding protein | Transcription and pre-mRNA processing | 1 | N/A |
| Chromodomain helicase DNA-binding protein 1 | CHD1 | Binds to DNA helicase | Chromatin assembly or disassembly, transcription | 1 | Confirmed |
| SPANX-b, c, and d | SPANX | Unknown | Regulates transcription and translation, spermatogenesis | 1 | N/A |
| Cellular repressor of adenovirus E1A protein-stimulated genes 1 | CREG1 | Antagonizes transcriptional activation of E1A protein | Transcriptional control of cell growth and differentiation | 1 | N/A |
| Metabolic regulation | |||||
| Asparagine-linked glycosylation 5 homolog | ALG5 | Dolichyl-phosphate beta-glucosyltransferase activity | N-glycan biosynthesis | 1 | N/A |
| 3-hydroxybutyrate dehydrogenase, type 2 | BDH2 | Dehydrogenase/reductase enzyme | Fatty acid beta-oxidation | 2 | Confirmed |
| Cathepsin K | CTSK | Cysteine proteinase | Proteolysis, tumor invasiveness, bone remodeling | 1 | Confirmed |
| Peroxiredoxin 3 | PRDX3 | Peroxidase activity, alkyl hydroperoxide reductase activity | Activation of NF-kappaB transcription factor, antioxidant | 1 | Confirmed |
| Protein kinases | |||||
| Serine threonine kinase 39 | STK39 | Protein serine/threonine kinase activity | Mediates stress-activated signals | 1 | N/A |
| Protein kinase C, nu | PRKCN | Protein serine/threonine kinase activity | Intracellular signaling cascade | 1 | N/A |
| Serine/threonine protein kinase MST4 | MASK | Protein serine/threonine kinase activity | Regulation of apoptosis, can be cleaved by caspase-3 | 1 | N/A |
| Cell signaling related | |||||
| NCK adaptor protein 1 | NCK1 | Contains SH2 and SH3 domains, cytoskeletal adaptor | Intracellular signaling cascade, positive regulation of actin filament polymerization | 1 | Confirmed |
| NCK-associated protein 1 | NCKAP1 | Interacts with Nck | Lamellipodium biogenesis, apoptosis | 1 | Confirmed |
| Transient receptor potential cation channel, subfamily C, member 6 | TRPC6 | Calcium channel activity | Calcium ion transport, calcium signaling | 1 | Confirmed |
| Interleukin 1 receptor antagonist | IL1RN | Inhibits the activities of interleukin 1 | Regulates immune and inflammatory responses | 1 | N/A |
| Cell death/stress related | |||||
| Hypoxia-inducible factor 1, alpha subunit | HIF1A | Transcription factor | Cellular and systemic homeostatic responses to hypoxia | 1 | Confirmed |
| DED containing | DEDD | Contains DED, regulation of apoptosis | Cell death | 1 | N/A |
| Transmembrane BAX inhibitor motif containing 1 | TMBIM1 | Contains BAX inhibitor motif | Cell death | 1 | N/A |
| Others | |||||
| Ubiquitin-conjugated enzyme, UBE2D3 | UBE2D3 | Ubiquitin-protein ligase activity | Ubiquitin-mediated proteolysis | 3 | Confirmed |
| Nidogen 2/osteonidogen | NID2 | A basement membrane protein | Adhesion, migration, differentiation, gene expression, and apoptosis | 1 | N/A |
| Mutated in colorectal cancers | MCC | Interacts with NFKBIB | Putative colorectal tumor suppressor gene | 1 | Confirmed |
| Complement component 1, q subcomponent-binding protein | C1QBP | Inhibit C1 activation | Play a role in serum complement system | N/A | |
| Ancient ubiquitous protein 1 | AUP1 | Unknown | Play a crucial role in the integrin alpha(IIb)beta(3) inside-out signaling | 1 | N/A |
| Stromal interaction molecule 1 | STIM1 | Unknown | Located in an important tumor-suppressor gene region, calcium signaling | 1 | N/A |
| Spastic ataxia of Charlevoix-Saguenay, sacsin | SACS | Unknown | Mutated in ARSACS | 1 | N/A |
| Similar to interferon-induced transmembrane protein 3, LOC144383 | N/A | Unknown | Unknown | 1 | N/A |
The positive colonies that have been confirmed by the “liquid culturing assay” (details in “Materials and methods” and Table S2) are indicated.
Positive hits indicates the number of positive colonies matching the same gene; N/A, not available (has not been tested); DED, death effector domain; and ARSACS, autosomal recessive spastic ataxia of Charlevoix-Saguenay.
Confirmation of identified positive hits
The insertion of viral DNA into a host chromosome often leads to disruption of host genes. Therefore, to demonstrate that the ATRA-resistant phenotype of the positive cells is indeed caused directly by the shRNA knockdown of the targeted genes instead of nonspecific defects induced by random viral DNA insertion, we reinfected the wild-type NB4 cells with each positive lentivirus. We used a colony assay in our primary screening (Figure 1). For the confirmation, we used an alternative liquid culture method to demonstrate that the phenotypes we detected were not assay specific. Using our established viral infection protocol, we routinely obtained a transduction efficiency of 5%. If viral infection does not alter the ATRA resistance, the infected cell will not gain growth advantage in ATRA-containing medium, and thus the percentage of GFP-positive cells should remain at 5%. On the contrary, if NB4 cells do acquire ATRA resistance after viral infection, the percentage of GFP-positive cells should keep increasing in ATRA-containing culture medium. Eventually, the GFP-positive cells will constitute the majority of the cell population. So far we have produced lentiviruses corresponding to 14 of the positive colonies (Table 2). We confirmed that the ATRA resistance of 10 of them was indeed caused by specific knockdown of the siRNA-targeted genes; whereas the other 4 positive colonies might be a result of either random viral insertion or viral infection-independent somatic mutations (Table S1). Based on the calculated somatic mutation rate, among the 48 viral-infected positive colonies, there should be 3 to 4 colonies (3.5 of 0.1 million infected cells) resulting from somatic mutations.
ATRA treatment induces differentiation of NB4 cells into mature neutrophils. Thus, we examined whether the ATRA-resistance NB4 cells can be prevented from differentiating into neutrophils. We used CD11b as a marker for neutrophil maturation. ATRA treatment dramatically increased CD11b expression in uninfected or control virus-infected NB4 cells, indicating differentiation toward mature neutrophils. CD11b expression was completely inhibited in cells infected with the positive siRNAs, suggesting a blockage of differentiation (Figure S1).
Overall, 26 genes were identified as involved in ATRA-induced cell growth arrest and differentiation of NB4 cells. These genes covered a wide range of cellular functions such as gene expression, intracellular signaling, cell death control, stress responses, and metabolic regulation, indicating that ATRA-induced cell growth control and differentiation in APL is a complex process.
RNAi knockdown of UBE2D3 suppresses ATRA-induced cell growth arrest
Three of the positive colonies from our screening contained an shRNA template targeting the UBE2D3 gene, which has been implicated in protein ubiquitination. Ubiquitination involves at least 3 classes of enzymes: ubiquitin-activating enzymes (E1s); ubiquitin-conjugating enzymes (E2s); and ubiquitin-protein ligases (E3s).24,25 The UBE2D3 gene encodes a member of the E2 ubiquitin-conjugating enzyme family.26,27 Before studying its function in more detail, we first sought to confirm the screening result by reassaying the UBE2D3 shRNA. As expected, NB4 cells infected with the UBE2D3 shRNA viral construct grew into colonies in ATRA-containing soft-agar medium, whereas uninfected or control shRNA viral construct-infected NB4 cells failed to grow into recognizable colonies. The infection of viral construct was confirmed by GFP expression in the transduced cells (Figure 2A). When the infected cells were grown in ATRA-containing liquid medium, UBE2D3 shRNA-infected cells proliferated much faster than the control cells, which is consistent with their acquired ATRA resistance (Figure 2B). We also confirmed the ATRA resistance of UBE2D3 shRNA-infected cells using the “liquid culturing” strategy described in Table S2. When control shRNA-infected cells (GFP positive) grew together with uninfected cells (GFP negative), their percentage remained the same during the continuous culturing. On the contrary, when UBE2D3 shRNA-infected cells (GFP positive) grew together with uninfected cells (GFP negative), the percentage of GFP-positive cells increased from approximately 10% at day 0 to more than 70% at day 3, indicating an acquired growth advantage in these cells (Figure 2C). The effect of UBE2D3 shRNA appeared to be specific to ATRA-treated cells. In ATRA-free medium, knocking down UBE2D3 by shRNA did not promote cell proliferation. In fact, instead of promoting NB4 cell proliferation, UBE2D3 shRNA-infected cells displayed a slightly lower growth rate in ATRA-free medium compared with the control cells (Figure 2D). In addition, transduction with UBE2D3 shRNA only inhibited NB4 cell proliferation arrest induced by ATRA, but not by other cell proliferation inhibitors such as staurosporine (Figure 2E), suggesting that UBE2D3 specifically mediates ATRA-induced cellular processes.
shRNA knockdown of UBE2D3 suppresses ATRA-induced cell growth arrest. (A) Colony-forming assay in soft agar. Uninfected, control shRNA-infected, and UBE2D3 shRNA-infected NB4 cells were seeded in soft-agar plates in the presence (0.5 μM) or absence of ATRA. The expression of shRNA by the infected cells was confirmed by their coexpression of GFP. The figure shows the results of a representative experiment. Pictures at day 3 and day 7 are presented. Images were viewed using an Olympus 20×/0.5 NA dry objective microscope (Olympus, Melville, NY) and SensiCam CD camera (Cooke, Romulus, MI) and processed using IPlab imaging software (IPlab, Rockville, MD). (B) shRNA knockdown of UBE2D3 suppresses ATRA-induced cell growth arrest in liquid cultures. Control shRNA and UBE2D3 shRNA-infected NB4 cells were cultured in liquid medium containing 0.5 μM ATRA. At each indicated time point, the total number of cells was counted by hemocytometer. The percentages of transduced cells (n %) were analyzed for GFP expression by flow cytometry. The total number of infected cells equals the total amount of cells times n %. Bars indicate 1 standard deviation (SD) about the mean (n = 6, * P < .01). (C) Confirmation of the ATRA resistance by a liquid culturing assay. NB4 cells were infected with UBE2D3 shRNA viruses. The infection efficiency is 10% to 15% under our experimental condition. The infected cells (including 10%-15% infected and 85%-90% uninfected cells) were continuously cultured in liquid medium with or without 0.5 μM ATRA. The percentages of transduced (GFP positive) cells were analyzed by flow cytometry every day. The increase of this number indicates a growth advantage of these cells over the uninfected cells. Data presented are the means (± SD) of 6 independent experiments. (D) shRNA knockdown of UBE2D3 does not promote NB4 cell growth in the absences of ATRA. Cell proliferation was analyzed as described in (B). Data presented are the means (± SD) of 6 independent experiments. (E) shRNA knockdown of UBE2D3 does not suppress staurosporine (STS)-induced cell growth arrest. Control shRNA and UBE2D3 shRNA-infected NB4 cells were cultured in the presence of 0.5 μM staurosporine and the cell growth was assessed as described in (B). Data presented are the means (± SD) of 3 independent experiments.
shRNA knockdown of UBE2D3 suppresses ATRA-induced cell growth arrest. (A) Colony-forming assay in soft agar. Uninfected, control shRNA-infected, and UBE2D3 shRNA-infected NB4 cells were seeded in soft-agar plates in the presence (0.5 μM) or absence of ATRA. The expression of shRNA by the infected cells was confirmed by their coexpression of GFP. The figure shows the results of a representative experiment. Pictures at day 3 and day 7 are presented. Images were viewed using an Olympus 20×/0.5 NA dry objective microscope (Olympus, Melville, NY) and SensiCam CD camera (Cooke, Romulus, MI) and processed using IPlab imaging software (IPlab, Rockville, MD). (B) shRNA knockdown of UBE2D3 suppresses ATRA-induced cell growth arrest in liquid cultures. Control shRNA and UBE2D3 shRNA-infected NB4 cells were cultured in liquid medium containing 0.5 μM ATRA. At each indicated time point, the total number of cells was counted by hemocytometer. The percentages of transduced cells (n %) were analyzed for GFP expression by flow cytometry. The total number of infected cells equals the total amount of cells times n %. Bars indicate 1 standard deviation (SD) about the mean (n = 6, * P < .01). (C) Confirmation of the ATRA resistance by a liquid culturing assay. NB4 cells were infected with UBE2D3 shRNA viruses. The infection efficiency is 10% to 15% under our experimental condition. The infected cells (including 10%-15% infected and 85%-90% uninfected cells) were continuously cultured in liquid medium with or without 0.5 μM ATRA. The percentages of transduced (GFP positive) cells were analyzed by flow cytometry every day. The increase of this number indicates a growth advantage of these cells over the uninfected cells. Data presented are the means (± SD) of 6 independent experiments. (D) shRNA knockdown of UBE2D3 does not promote NB4 cell growth in the absences of ATRA. Cell proliferation was analyzed as described in (B). Data presented are the means (± SD) of 6 independent experiments. (E) shRNA knockdown of UBE2D3 does not suppress staurosporine (STS)-induced cell growth arrest. Control shRNA and UBE2D3 shRNA-infected NB4 cells were cultured in the presence of 0.5 μM staurosporine and the cell growth was assessed as described in (B). Data presented are the means (± SD) of 3 independent experiments.
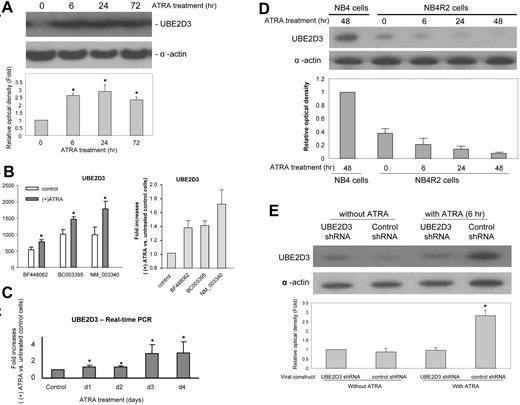
UBE2D3 expression is up-regulated in ATRA-treated NB4 cells
ATRA treatment in APL regulates the expression of a variety of genes. Because UBE2D3 was implicated in ATRA-mediated cell arrest and differentiation, we investigated whether the expression of UBE2D3 could also be regulated by ATRA treatment. We first measured the protein level via Western blotting (Figure 3A). Level of UBE2D3 protein increased approximately 2.5-fold in 6 hours and reached 3-fold above control level after 24 hours of ATRA treatment. The slight decline at 72 hours is likely to be the result of ATRA-induced apoptosis. The ATRA-induced increase of UBE2D3 protein level was at least partially due to the augmented gene transcription. Microarray analysis revealed a significant and consistent increase of the UBE2D3 mRNA in ATRA-treated NB4 cells. The magnitude of the increase varied between 1.4- and 2-fold, depending on the oligomeric probes used (Figure 3B). This result was further confirmed by real-time quantitative PCR analysis, which revealed significant increases in UBE2D3 mRNA level starting the second day after ATRA treatment (Figure 3C). However, the ATRA-induced elevation of UBE2D3 mRNA levels is somewhat slower than the increase of the protein levels, suggesting that the large early elevation in UBE2D3 protein level might be a result of up-regulated translation or/and enhanced protein stability.
UBE2D3 expression is up-regulated in ATRA-treated NB4 cells. (A) NB4 cells were cultured in the presence of 0.5 μM ATRA for the indicated time. Protein extracts were resolved on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). UBE2D3 and actin were detected by Western blotting analysis. The relative amounts of UBE2D3 were quantified using NIH ImageJ software (http://rsb.info.nih.gov/ij/). The UBE2D3 signals were normalized to the amount of actin in each sample. All samples were compared with the signal detected in untreated (time point 0) NB4 cells. Data presented are the means (± SD) of 3 independent experiments. (B) ATRA-induced up-regulation of UBE2D3 mRNA expression identified by RNA microarray. The mRNA was prepared from untreated or ATRA-treated (1 μM for 4 days) NB4 cells. Both absolute values (left) and fold increases (right) were presented. The IDs of each oligomeric probe are indicated. All probes showed a significant ATRA-induced augmentation of UBE2D3 mRNA level. Data presented are the means (± SD) of 3 independent experiments. (C) ATRA-induced up-regulation of UBE2D3 mRNA expression identified by real-time quantitative PCR. NB4 cells were cultured in the presence of 1 μM of ATRA for the indicated number of days. The 2-step quantitative reverse transcription-polymerase chain reactions (RT-PCRs) were conducted using purified total RNA as described in Document S1. The expression of alpha tubulin (k-alpha-1) was used as internal control gene. Shown are fold increases over untreated cells. Data presented are the means (± SD) of 3 independent experiments. *P < .01 versus control. (D) ATRA-induced UBE2D3 up-regulation could not be detected in the ATRA-resistant NB4-R2 cells. ATRA treatment and quantification of UBE2D3 protein level were conducted exactly as described in panel A. (E) UBE2D3 shRNA completely abolished ATRA-induced up-regulation of UBE2D3 in NB4 cells. Control shRNA and UBE2D3 shRNA-infected NB4 cells were incubated with or without 0.5 μM ATRA for 6 hours. The GFP-positive shRNA-expressing cells were sorted on a MoFlo High-Performance cell sorter (Dako, Carpinteria, CA). Protein extracts were resolved on SDS-PAGE and the amounts of UBE2D3 protein were measured as described in panel A. Data presented are the means (± SD) of 3 independent experiments. *P < .001
UBE2D3 expression is up-regulated in ATRA-treated NB4 cells. (A) NB4 cells were cultured in the presence of 0.5 μM ATRA for the indicated time. Protein extracts were resolved on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). UBE2D3 and actin were detected by Western blotting analysis. The relative amounts of UBE2D3 were quantified using NIH ImageJ software (http://rsb.info.nih.gov/ij/). The UBE2D3 signals were normalized to the amount of actin in each sample. All samples were compared with the signal detected in untreated (time point 0) NB4 cells. Data presented are the means (± SD) of 3 independent experiments. (B) ATRA-induced up-regulation of UBE2D3 mRNA expression identified by RNA microarray. The mRNA was prepared from untreated or ATRA-treated (1 μM for 4 days) NB4 cells. Both absolute values (left) and fold increases (right) were presented. The IDs of each oligomeric probe are indicated. All probes showed a significant ATRA-induced augmentation of UBE2D3 mRNA level. Data presented are the means (± SD) of 3 independent experiments. (C) ATRA-induced up-regulation of UBE2D3 mRNA expression identified by real-time quantitative PCR. NB4 cells were cultured in the presence of 1 μM of ATRA for the indicated number of days. The 2-step quantitative reverse transcription-polymerase chain reactions (RT-PCRs) were conducted using purified total RNA as described in Document S1. The expression of alpha tubulin (k-alpha-1) was used as internal control gene. Shown are fold increases over untreated cells. Data presented are the means (± SD) of 3 independent experiments. *P < .01 versus control. (D) ATRA-induced UBE2D3 up-regulation could not be detected in the ATRA-resistant NB4-R2 cells. ATRA treatment and quantification of UBE2D3 protein level were conducted exactly as described in panel A. (E) UBE2D3 shRNA completely abolished ATRA-induced up-regulation of UBE2D3 in NB4 cells. Control shRNA and UBE2D3 shRNA-infected NB4 cells were incubated with or without 0.5 μM ATRA for 6 hours. The GFP-positive shRNA-expressing cells were sorted on a MoFlo High-Performance cell sorter (Dako, Carpinteria, CA). Protein extracts were resolved on SDS-PAGE and the amounts of UBE2D3 protein were measured as described in panel A. Data presented are the means (± SD) of 3 independent experiments. *P < .001
The ATRA-elicited increase of UBE2D3 level was detected in wild-type NB4 cells but not in an ATRA-resistant NB4 cell line, NB4-R2. This result further suggests that up-regulation of UBE2D3 is involved in ATRA-mediated signaling and subsequent cellular events (Figure 3D). Interestingly, contrary to what was observed in wild-type NB4 cells, we detected a slight decrease of UBE2D3 protein level in ATRA-treated NB4-R2 cells. Whether this difference has a physiologic meaning is currently unknown.
We have shown that transduction of NB4 cells with the UBE2D3 shRNA viral construct suppressed ATRA-induced cell growth arrest and differentiation. To confirm that this effect is indeed due to the specific knockdown of UBE2D3 expression, we measured UBE2D3 protein levels in viral-infected cells. As expected, infection with the UBE2D3 shRNA viral construct almost completely abolished ATRA-induced up-regulation of UBE2D3, whereas infection with the control shRNA viral construct did not have any effect (Figure 3E).
UBE2D3 is required for ATRA-induced breakdown of cyclin D1
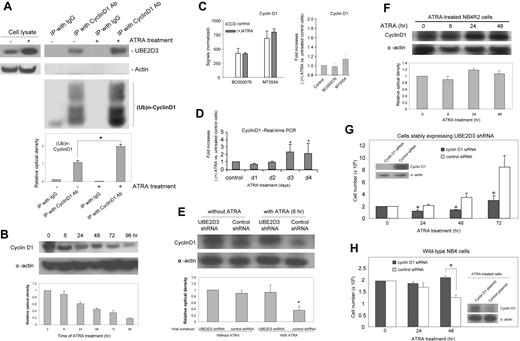
ATRA-induced cell growth arrest and terminal differentiation of APL involves the sequential regulation of cell-cycle regulatory proteins, such as D-type cyclins (cyclins D1, D2, and D3), which promote G1-S progression and are aberrantly expressed in various cancers.28-30 UBE2D3 is an E2 ubiquitin-conjugating enzyme and its in vitro targets include cyclin D1,31 raising a possibility that the function of UBE2D3 in ATRA signaling might be mediated by this cell-cycle protein. To test this hypothesis, we first explored the physical association of these 2 proteins in NB4 cells. We immunoprecipitated NB4 cell lysate with antibodies against cyclin D1 and observed selective coimmunoprecipitation of UBE2D3 and cyclin D1 in both ATRA-treated and untreated cells (Figure 4A), indicating an ATRA-independent robust interaction of the 2 proteins.
Cyclin D1 is a target of UBE2D3. (A) UBE2D3 physically associates with cyclin D1 in both ATRA-treated (48 hour) and untreated NB4 cells, and ATRA treatment leads to increased ubiquitination of cyclin D1. Cell lysates were immunoprecipitated with cyclin D1 antiserum or IgG. The precipitates were blotted with monoclonal anti-UBE2D3, antiubiquitin, as well as antiactin antibodies. The left two lanes contain small aliquots of the whole-cell lysates used for coimmunoprecipitation. The experiments were repeated multiple times. The figure shows the results of a representative experiment. The levels of ubiquitinated cyclin D1 were quantified using NIH ImageJ software. All samples were compared with the signal detected in untreated NB4 cell lysate immunoprecipitated with anti-cyclin D1 antiserum (lane 3). Data presented are the means (±SD) of 3 independent experiments. *P < .001. (B) Cyclin D1 protein level is down-regulated in ATRA-treated NB4 cells. NB4 cells were cultured in the presence of 0.5 μM ATRA for the indicated time. Cyclin D1 and actin were detected by Western-blotting analysis. The relative amounts of cyclin D1 protein were quantified using NIH ImageJ software. The cyclin D1 signals were normalized to the amount of actin in each sample. All samples were compared with the signal detected in untreated (time point 0) NB4 cells. Data are presented as mean values from 3 independent experiments whose results varied less than 5%. (C) Cyclin D1 mRNA level in ATRA-treated NB4 cells analyzed by RNA microarray. Cyclin D1 mRNA expression was analyzed using the same microarray data as described in Figure 3B. Both absolute values (left) and fold increases (right) were presented. The IDs of each oligomeric probe are indicated. Data presented are the means (± SD) of 3 independent experiments. (D) Cyclin D1 mRNA level in ATRA-treated NB4 cells analyzed by real-time quantitative PCR. The experiment was conducted as described in Figure 3C. Shown are fold increases over untreated cells. Data presented are the means (±SD) of 3 independent experiments. *P < .01. (E) UBE2D3 shRNA completely abolished ATRA-induced cyclin D1 degradation in NB4 cells. Control shRNA and UBE2D3 shRNA-infected NB4 cells were incubated with or without 0.5 μM ATRA for 6 hours. The GFP-positive shRNA-expressing cells were sorted on a MoFlo High-Performance cell sorter. Protein extracts were resolved on SDS-PAGE and the amounts of cyclin D1 protein were measured as described in panel B. Data presented are the means (± SD) of 3 independent experiments. *P < .001. (F) ATRA-induced cyclin D1 degradation could not be detected in the ATRA-resistant NB4-R2 cells. ATRA treatment and quantification of cyclin D1 protein level by Western blotting were conducted exactly as described in panel B. (G) Knocking down cyclin D1 protein by siRNA inhibits the growth of UBE2D3-shRNA-expressing NB4 cells in the presence of ATRA (abrogates the effect of the UBE2D3 shRNA). NB4 cells stably expressing UBE2D3 shRNA (2 million) were transfected with human cyclin D1 siRNA (100 pmol) (Santa Cruz Biotechnology, Santa Cruz, CA) or control siRNA (100 pmol) using a Nucleofector Kit (Amaxa, Koeln, Germany) and a protocol provided by the manufacturer. Using this electroporation-based method, transfection efficiencies (for siRNA) of greater than 80% are routinely obtained. Transfected NB4 cells were cultured in the presence of 0.5 μM ATRA and the cell growth was assessed as described in Figure 2B. Data presented are the means (± SD) of 3 independent experiments. *P < .01 versus cells transfected with control siRNA. (Inset) Western blot result showing the specific knockdown of cyclin D1 protein. (H) Overexpression of cyclin D1 suppresses ATRA-induced cell growth arrest in NB4 cells (mimics the effect of the UBE2D3 shRNA). NB4 cells (2 ×106) were transfected with a cyclin D1 expression plasmid (6 μg; Open Biosystems, Huntsville, AL) or a pEGFP expression plasmid (control) using the Nucleofector Kit. We routinely obtain transfection efficiencies (for plasmid) of greater than 20% using this method. Transfected NB4 cells were cultured in the presence of 0.5 μM ATRA and the cell growth was assessed as described in Figure 2B. The ATRA-resistant cells gain growth advantage in ATRA-containing medium. Data presented are the means (± SD) of 3 independent experiments. *P < .01 versus cells transfected with control plasmid. (Inset) Western blot result showing the overexpression of cyclin D1 protein in ATRA-treated (3 days) NB4 cells.
Cyclin D1 is a target of UBE2D3. (A) UBE2D3 physically associates with cyclin D1 in both ATRA-treated (48 hour) and untreated NB4 cells, and ATRA treatment leads to increased ubiquitination of cyclin D1. Cell lysates were immunoprecipitated with cyclin D1 antiserum or IgG. The precipitates were blotted with monoclonal anti-UBE2D3, antiubiquitin, as well as antiactin antibodies. The left two lanes contain small aliquots of the whole-cell lysates used for coimmunoprecipitation. The experiments were repeated multiple times. The figure shows the results of a representative experiment. The levels of ubiquitinated cyclin D1 were quantified using NIH ImageJ software. All samples were compared with the signal detected in untreated NB4 cell lysate immunoprecipitated with anti-cyclin D1 antiserum (lane 3). Data presented are the means (±SD) of 3 independent experiments. *P < .001. (B) Cyclin D1 protein level is down-regulated in ATRA-treated NB4 cells. NB4 cells were cultured in the presence of 0.5 μM ATRA for the indicated time. Cyclin D1 and actin were detected by Western-blotting analysis. The relative amounts of cyclin D1 protein were quantified using NIH ImageJ software. The cyclin D1 signals were normalized to the amount of actin in each sample. All samples were compared with the signal detected in untreated (time point 0) NB4 cells. Data are presented as mean values from 3 independent experiments whose results varied less than 5%. (C) Cyclin D1 mRNA level in ATRA-treated NB4 cells analyzed by RNA microarray. Cyclin D1 mRNA expression was analyzed using the same microarray data as described in Figure 3B. Both absolute values (left) and fold increases (right) were presented. The IDs of each oligomeric probe are indicated. Data presented are the means (± SD) of 3 independent experiments. (D) Cyclin D1 mRNA level in ATRA-treated NB4 cells analyzed by real-time quantitative PCR. The experiment was conducted as described in Figure 3C. Shown are fold increases over untreated cells. Data presented are the means (±SD) of 3 independent experiments. *P < .01. (E) UBE2D3 shRNA completely abolished ATRA-induced cyclin D1 degradation in NB4 cells. Control shRNA and UBE2D3 shRNA-infected NB4 cells were incubated with or without 0.5 μM ATRA for 6 hours. The GFP-positive shRNA-expressing cells were sorted on a MoFlo High-Performance cell sorter. Protein extracts were resolved on SDS-PAGE and the amounts of cyclin D1 protein were measured as described in panel B. Data presented are the means (± SD) of 3 independent experiments. *P < .001. (F) ATRA-induced cyclin D1 degradation could not be detected in the ATRA-resistant NB4-R2 cells. ATRA treatment and quantification of cyclin D1 protein level by Western blotting were conducted exactly as described in panel B. (G) Knocking down cyclin D1 protein by siRNA inhibits the growth of UBE2D3-shRNA-expressing NB4 cells in the presence of ATRA (abrogates the effect of the UBE2D3 shRNA). NB4 cells stably expressing UBE2D3 shRNA (2 million) were transfected with human cyclin D1 siRNA (100 pmol) (Santa Cruz Biotechnology, Santa Cruz, CA) or control siRNA (100 pmol) using a Nucleofector Kit (Amaxa, Koeln, Germany) and a protocol provided by the manufacturer. Using this electroporation-based method, transfection efficiencies (for siRNA) of greater than 80% are routinely obtained. Transfected NB4 cells were cultured in the presence of 0.5 μM ATRA and the cell growth was assessed as described in Figure 2B. Data presented are the means (± SD) of 3 independent experiments. *P < .01 versus cells transfected with control siRNA. (Inset) Western blot result showing the specific knockdown of cyclin D1 protein. (H) Overexpression of cyclin D1 suppresses ATRA-induced cell growth arrest in NB4 cells (mimics the effect of the UBE2D3 shRNA). NB4 cells (2 ×106) were transfected with a cyclin D1 expression plasmid (6 μg; Open Biosystems, Huntsville, AL) or a pEGFP expression plasmid (control) using the Nucleofector Kit. We routinely obtain transfection efficiencies (for plasmid) of greater than 20% using this method. Transfected NB4 cells were cultured in the presence of 0.5 μM ATRA and the cell growth was assessed as described in Figure 2B. The ATRA-resistant cells gain growth advantage in ATRA-containing medium. Data presented are the means (± SD) of 3 independent experiments. *P < .01 versus cells transfected with control plasmid. (Inset) Western blot result showing the overexpression of cyclin D1 protein in ATRA-treated (3 days) NB4 cells.
To investigate the role of cyclin D1 in ATRA signaling, we further examined whether ATRA treatment can regulate its protein expression. In ATRA-treated NB4 cells, the level of cyclin D1 protein was drastically decreased (Figure 4B). Interestingly, the microarray data displayed no obvious differences between its mRNA levels (Figure 4C) and the real-time PCR even displayed a slight increase in cyclin D1 mRNA levels in cells treated with ATRA for more than 3 days (Figure 4D), suggesting that the UBE2D3-mediated proteasomal degradation of cyclin D1 protein might be the predominant mechanism for the ATRA-induced down-regulation of cyclin D1. Supporting this interpretation, we detected dramatically increased ubiquitination of cyclin D1 after ATRA treatment (Figure 4A). In addition, ATRA-induced down-regulation of cyclin D1 was almost completely abolished in NB4 cells in which UBE2D3 expression was inhibited by its specific shRNA (Figure 4E). Consistent with the low expression of UBE2D3 in ATRA-resistant NB4-R2 cells (even after ATRA treatment), the level of cyclin D1 protein in these cells was not decreased after the ATRA treatment (Figure 4F). Lastly, knocking down cyclin D1 by siRNA abrogates the effect of UBE2D3 shRNA in ATRA-induced cell growth arrest (Figure 4G) and overexpression of cyclin D1 mimics the effect of the UBE2D3 shRNA (Figure 4H), suggesting that cyclin D1 is an essential component downstream of UBE2D3. In summary, our results demonstrate that UBE2D3 exerts its function in ATRA-mediated cell growth arrest of NB4 cells by promoting proteasomal degradation of cyclin D1 protein.
RNAi knockdown of UBE2D3 suppresses ATRA-induced cell-cycle arrest
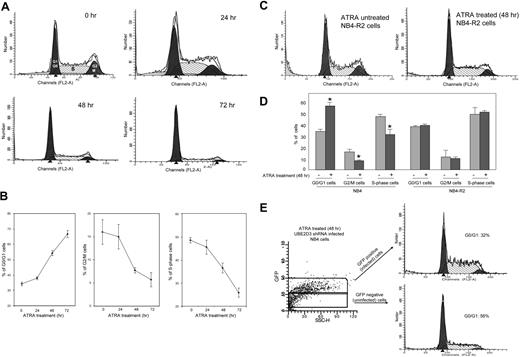
One of the hallmarks of terminal neutrophilic differentiation of NB4 cells is an irreversible arrest in the G0/G1 phase of the cell cycle.8,9,14 This arrest involves the coordinate regulation of signals that negatively control the cell-cycle machinery and inhibit the G1/S transition. Nevertheless, the molecular and cellular mechanisms by which ATRA induces cell-cycle arrest are still not completely understood. Cyclin D1 is a cell-cycle regulatory protein that functions early in the G1 phase and is essential for cell-cycle progression during the G1/S transition. Our results show that its level can be down-regulated by UBE2D3-mediated proteasomal degradation, thus providing a potential cellular mechanism of ATRA-induced cell-cycle arrest. To test this pathway directly, we analyzed cell-cycle distribution using PI staining as a measure of DNA content.
As expected, ATRA-induced cell growth arrest and differentiation in NB4 cells was accompanied by G0/G1 arrest (Figure 5A-B). The percentage of cells arrested in the G0/G1 phase increased with time, reaching 70% in 72 hours. Correspondingly, the percentage of cells in the S and G2/M phases drastically declined after ATRA treatment. As shown in Figure 5C-D, ATRA-induced G0/G1 arrest was completely abolished in ATRA-resistant NB4-R2 cells in which UBE2D3 expression was not up-regulated (Figure 3C) and cyclin D1 protein was not degraded (Figure 4E), consistent with the idea that UBE2D3-mediated proteasomal degradation of cyclin D1 is essential for ATRA-induced G0/G1 arrest. To examine directly whether UBE2D3 is required for ATRA-induced G0/G1 arrest, we compared cell-cycle distribution in UBE2D3 shRNA-infected (GFP positive) and uninfected (GFP negative) NB4 cells (Figure 5E). Knocking down UBE2D3 with shRNA resulted in a major redistribution of the cell cycle in ATRA-treated NB4 cells. After a 48-hour treatment with ATRA, the percentage of cells arrested in the G0-G1 phase reached 56% in uninfected cells. However, with the same ATRA treatment, only approximately 32% of UBE2D3 shRNA-infected cells were arrested in the G0-G1 phase. This level is almost as low as that observed in untreated NB4 cells (Figure 5E). Collectively, these results are consistent with the previous result that ATRA-induced cyclin D1 degradation was abolished in the UBE2D3 shRNA-infected cells and indicate that UBE2D3-mediated degradation of cyclin D1 is an essential mediating step in ATRA-induced G0/G1 arrest.
shRNA knockdown of UBE2D3 suppresses ATRA-induced cell-cycle arrest. (A) Cell-cycle analysis of ATRA-treated NB4 cells. NB4 cells were cultured in the presence of 0.5 μM ATRA for the indicated time. Cells were stained with propidium iodide and the cellular DNA content, which was reflected by PI staining, was analyzed by FACS using the Cell Quest software (Becton Dickinson). The percentage of cells in each cell-cycle phase was determined using a ModFIT software (Becton Dickinson), which deconvolves the flow-cytometer data. At least 10 000 cells were analyzed for each data point. (B) The cell percentages obtained from panel A were plotted against times of ATRA treatment. Data presented are the means (± SD) of 3 independent experiments. (C) ATRA treatment does not alter cell-cycle distribution in ATRA-resistant NB4-R2 cells. ATRA treatment and cell-cycle analysis were conducted exactly as described in panel A. (D) The proportions of cells that are in each cell-cycle phase were compared between ATRA-treated NB4 cells and NB4-R2 cells. Data presented are the means (± SD) of 3 independent experiments P < .01 versus untreated cells. (E) shRNA knockdown of UBE2D3 suppresses ATRA-induced G0/G1 arrest in NB4 cells. UBE2D3 shRNA-infected NB4 cells were incubated in the presence of 0.5 μM ATRA for 48 hours. Viral-infected and uninfected cells were gated by their high and low fluorescent intensity, respectively. Cell-cycle analysis was carried out as described in panel A. Data are presented as mean values from 3 independent experiments whose results varied less than 5%.
shRNA knockdown of UBE2D3 suppresses ATRA-induced cell-cycle arrest. (A) Cell-cycle analysis of ATRA-treated NB4 cells. NB4 cells were cultured in the presence of 0.5 μM ATRA for the indicated time. Cells were stained with propidium iodide and the cellular DNA content, which was reflected by PI staining, was analyzed by FACS using the Cell Quest software (Becton Dickinson). The percentage of cells in each cell-cycle phase was determined using a ModFIT software (Becton Dickinson), which deconvolves the flow-cytometer data. At least 10 000 cells were analyzed for each data point. (B) The cell percentages obtained from panel A were plotted against times of ATRA treatment. Data presented are the means (± SD) of 3 independent experiments. (C) ATRA treatment does not alter cell-cycle distribution in ATRA-resistant NB4-R2 cells. ATRA treatment and cell-cycle analysis were conducted exactly as described in panel A. (D) The proportions of cells that are in each cell-cycle phase were compared between ATRA-treated NB4 cells and NB4-R2 cells. Data presented are the means (± SD) of 3 independent experiments P < .01 versus untreated cells. (E) shRNA knockdown of UBE2D3 suppresses ATRA-induced G0/G1 arrest in NB4 cells. UBE2D3 shRNA-infected NB4 cells were incubated in the presence of 0.5 μM ATRA for 48 hours. Viral-infected and uninfected cells were gated by their high and low fluorescent intensity, respectively. Cell-cycle analysis was carried out as described in panel A. Data are presented as mean values from 3 independent experiments whose results varied less than 5%.
Proteosome inhibition suppresses ATRA-induced cell-cycle arrest
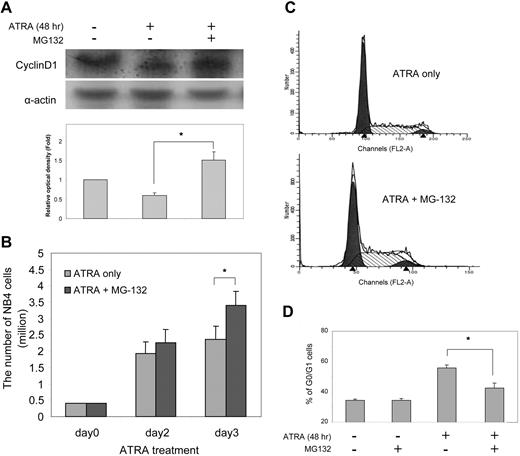
To demonstrate directly that UBE2D3-mediated down-regulation of cyclin D1 is mediated by proteasomal degradation, we examined whether cyclin D1 down-regulation and the resultant cell-cycle arrest can be inhibited by the proteosome inhibitor MG-132. This peptide aldehyde reversibly inhibits ubiquitin-mediated proteolysis by binding to and inactivating 20S and 26S proteasomes. Treatment with this inhibitor significantly reduced the ATRA-elicited cyclin D1 degradation in NB4 cells (Figure 6A). In fact, the level of cyclin D1 in cells treated with both MG-132 and ATRA was even higher than that in the untreated cells, suggesting that there might be basal cyclin D1 degradation in ATRA-untreated NB4 cells. Consistent with its effect on cyclin D1 degradation, MG-132 was able to reverse the cell growth arrest induced by ATRA treatment (Figure 6B). Treatment with MG-132 increased the number of NB4 cells grown in ATRA-containing medium by more than 40% in 3 days. This increase is probably due to the inhibition of ATRA-induced cell-cycle arrest, because the percentage of cells arrested in the G0/G1 phase was reduced to 40% in the cell population treated with both ATRA and MG-132 compared with 55% in the population treated with ATRA alone (Figure 6C-D). These results further highlight the involvement of UBE2D3-mediated proteasomal degradation of cyclin D1 in ATRA-induced cell proliferation and growth arrest.
MG-132, a proteosome inhibitor, suppresses ATRA-induced cyclin D1 degradation and cell-cycle arrest. (A) NB4 cells were cultured in the presence of 0.5 μM ATRA for 24 hours and then treated with or without 5 μM MG-132 for 30 minutes. After being incubated in ATRA for another 24 hours, the cells were lysed and cellular cyclin D1 protein level was analyzed using Western-blot analysis as described in Figure 4B. The degradation of cyclin D1 in ATRA-treated cells is significantly suppressed by MG-132 incubation (n = 3, *P < .05). (B) Treatment with MG-132 promotes NB4 cell proliferation in the presence of ATRA. NB4 cells were cultured in liquid medium containing 0.5 μM ATRA and treated with MG-132 as described in Figure 6A. Cell proliferation was assessed as described in Figure 2B. Bars indicate mean (± SD, n = 3, *P < .05). (C) MG-132 suppresses ATRA-induced G0/G1 cell-cycle arrest. NB4 cells were treated with ATRA and MG-132 as described in panel A. Cell-cycle analysis was carried out as described in Figure 5A. The figure shows the results of a representative experiment. (D) The percentage of cells arrested in G0/G1 phase was quantified using a ModFIT software as described in Figure 5B. Data presented are the means (± SD) of 3 independent experiments. P < .01 versus MG132-untreated cells.
MG-132, a proteosome inhibitor, suppresses ATRA-induced cyclin D1 degradation and cell-cycle arrest. (A) NB4 cells were cultured in the presence of 0.5 μM ATRA for 24 hours and then treated with or without 5 μM MG-132 for 30 minutes. After being incubated in ATRA for another 24 hours, the cells were lysed and cellular cyclin D1 protein level was analyzed using Western-blot analysis as described in Figure 4B. The degradation of cyclin D1 in ATRA-treated cells is significantly suppressed by MG-132 incubation (n = 3, *P < .05). (B) Treatment with MG-132 promotes NB4 cell proliferation in the presence of ATRA. NB4 cells were cultured in liquid medium containing 0.5 μM ATRA and treated with MG-132 as described in Figure 6A. Cell proliferation was assessed as described in Figure 2B. Bars indicate mean (± SD, n = 3, *P < .05). (C) MG-132 suppresses ATRA-induced G0/G1 cell-cycle arrest. NB4 cells were treated with ATRA and MG-132 as described in panel A. Cell-cycle analysis was carried out as described in Figure 5A. The figure shows the results of a representative experiment. (D) The percentage of cells arrested in G0/G1 phase was quantified using a ModFIT software as described in Figure 5B. Data presented are the means (± SD) of 3 independent experiments. P < .01 versus MG132-untreated cells.
Discussion
The biologic effects of ATRA have been well characterized; nevertheless, the molecular mechanisms regulating these processes are still largely unknown.1,9,12,14 ATRA-induced alterations in gene transcription and translation in APL have been analyzed using DNA microarray and comparative proteomics techniques.15-17 However, whether any of these alterations are responsible for ATRA-induced differentiation and growth arrest of APL cells has not been investigated. In addition, some ATRA-elicited cellular effects might not depend on alterations of gene expression. To systematically explore the functional mediators of the ATRA effect in APL, we conducted an unbiased, large-scale RNAi screen and identified 26 proteins that are essential for ATRA-induced growth inhibition and differentiation in acute promyelocytic NB4 cells. Most positive hits from this functional screen were confirmed as authentic mediators in the ATRA pathway. ATRA is not only used in differentiation therapy for APL but also plays an important role in cell differential, proliferation, reproduction, and development in many other systems such as stem cells, neurons, leukemias other than APL, and various solid cancers.32-37 Thus, the results presented here may also illuminate the mechanisms of action of ATRA in other systems.
ATRA treatment directly modulates the transcription of its target genes.12-14 Whether any of the identified genes are direct targets of ATRA has not been investigated. Future studies need to be conducted to connect our functional screening data directly to ATRA-induced gene-expression changes, as measured by microarray technology, and protein expression changes, as measured by quantitative proteomics. The 26 genes identified in our screening covered a wide range of cellular mechanisms such as gene expression, intracellular signaling, cell death control, stress responses, and metabolic regulation, indicating that ATRA-induced cell growth arrest and differentiation is a complex process. Integrated with other large-scale data sets (eg, microarray and proteomics), functional mediators identified in this study should provide an interesting foundation for a system-wide modeling of ATRA-induced cell signaling events leading to cell growth arrest and differentiation.
One of the proteins identified from our screening is the ubiquitin-conjugating enzyme UBE2D3. The modification of proteins with ubiquitin is an important cellular mechanism for targeting abnormal or short-lived proteins for degradation.24,25,38 Ubiquitination-mediated degradation of cell-cycle-related proteins, such as cyclins, p53, and anaphase-promoting complex, has been recognized as one of the mechanisms for cell-cycle control.39-43 However, its involvement in ATRA-induced G0/G1 arrest in APL has not been fully investigated. In this study, we showed that UBE2D3 level is up-regulated in ATRA-treated acute promyelocytic NB4 cells. In addition, we identified cyclin D1 as one of the substrates of UBE2D3. UBE2D3 is physically associated with cyclin D1 and mediates ATRA-induced cyclin D1 degradation. Knocking down UBE2D3 by RNAi leads to blockage of ATRA-induced cyclin D1 degradation and cell-cycle arrest. Forced cyclin D1 degradation by specific cyclin D1 siRNA abrogates the effect of UBE2D3 shRNA, whereas overexpression of cyclin D1 mimics the effect of the UBE2D3 shRNA, further confirming that cyclin D1 is an essential component downstream of UBE2D3 (Figure 4). Although UBE2D3 is critical for ATRA-induced cell-growth arrest, overexpression of UBE2D3 alone cannot induce cyclin D1 degradation or cell-growth arrest, nor can it make ATRA-resistant R2 cells regain ATRA sensitivity (data not shown). These findings suggest that other mediators or pathways besides UBE2D3 are also required for ATRA-induced cyclin D1 degradation and cell-growth arrest (eg, unidentified E3 ligase for cyclin D1).
In eukaryotes, there is only one E1, about 60 E2s (in mammals), and a much larger numbers of E3s.24,25,38 The specificity of ubiquitination is largely determined by E3 ligases, which recognize the protein substrate and facilitate ubiquitin transfer from the E2 ubiquitin-conjugating enzyme onto substrate.24,25,38,44 A single E2 such as UBE2D3 can usually interact with several E3 ubiquitin ligases and thereby affect multiple targets.45 Thus, proteins other than cyclin D1 could also act as downstream targets of UBE2D3. Although p53 is another known target of UBE2D3,26 we could not detect any change in p53 protein level in ATRA-treated NB4 cells. Consistent with this result, p53 does not associate with UBE2D3 in ATRA-treated or untreated NB4 cells. Thus, p53 might not be a downstream target of UBE2D3 in NB4 cells (Figure S2), perhaps due to the absence of other factors required for p53 ubiquitination and degradation in these cells. However, we cannot rule out the possibility that other unidentified UBE2D3 targets are also involved in ATRA-induced cell growth arrest and differentiation. Collectively, our results highlight the involvement of the ubiquitin-mediated proteolysis pathway in ATRA-induced cell-cycle arrest and thus provide a novel strategy for modulating ATRA-elicited cellular effects.
Another finding of this study is that MG-132, a specific proteasome inhibitor, suppresses ATRA-induced cell-cycle arrest in NB4 cells. It has been known that proteasomes control the half-life of many short-lived cell-cycle-related regulatory proteins. Proteasome malfunction leads to abnormal regulation of the cell cycle and uncontrolled cell proliferation. Inhibition of proteasome function can cause cell-cycle arrest and cell death in various cancers. Several proteasome inhibitors have been approved or proposed for use in the treatment of cancers.46,47 However, our observation that MG-132 suppresses ATRA-induced cell-cycle arrest in NB4 cells suggests that this proteasome inhibitor might not be a good choice for treating APL in combination with ATRA, because it antagonizes the ATRA effect.
The online version of this article contains a data supplement.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Leslie Silberstein, John Manis, James Campbell, and Li Cai for helpful discussions. We also thank Dr Lanotte for providing the ATRA-resistant NB4-R2 cell line. H.R.L. is supported by National Institutes of Health (NIH) grants NS052200, HL085100, and GM076084 and a Research Scholar Grant from American Cancer Society. P.E.N. is supported by NIH grant NIH DK054369.
National Institutes of Health
Authorship
Contribution: H.H., X.Z., P.E.N., and H.R.L. designed research and analyzed data. H.H., X.Z., Y.J., K.K.S., H.J., and F.L. performed research. H.H., P.E.N. and H.R.L. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hongbo R. Luo, Department of Pathology, Joint Program in Transfusion Medicine, Harvard Medical School, Department of Lab Medicine, Children's Hospital Boston, Karp Family Research Building, Room 10214, Boston, MA 02115; e-mail: hongbo.luo@childrens.harvard.edu.