Abstract
Ubiquitously acting chromatin opening elements (UCOEs) consist of methylation-free CpG islands encompassing dual divergently transcribed promoters of housekeeping genes that have been shown to confer resistance to transcriptional silencing and to produce consistent and stable transgene expression in tissue culture systems. To develop improved strategies for hematopoietic cell gene therapy, we have assessed the potential of the novel human HNRPA2B1-CBX3 UCOE (A2UCOE) within the context of a self-inactivating (SIN) lentiviral vector. Unlike viral promoters, the enhancer-less A2UCOE gave rise to populations of cells that expressed a reporter transgene at a highly reproducible level. The efficiency of expression per vector genome was also markedly increased in vivo compared with vectors incorporating either spleen focus-forming virus (SFFV) or cytomegalovirus (CMV) promoters, suggesting a relative resistance to silencing. Furthermore, an A2UCOE-IL2RG vector fully restored the IL-2 signaling pathway within IL2RG-deficient human cells in vitro and successfully rescued the X-linked severe combined immunodeficiency (SCID-X1) phenotype in a mouse model of this disease. These data indicate that the A2UCOE displays highly reliable transcriptional activity within a lentiviral vector, largely overcoming insertion-site position effects and giving rise to therapeutically relevant levels of gene expression. These properties are achieved in the absence of classic enhancer activity and therefore may confer a high safety profile.
Introduction
Retroviral vector–mediated gene transfer into hematopoietic stem cells (HSCs) has become a useful and promising tool for treatment of life-threatening inherited hematologic disorders.1-6 However, a significant risk of insertional mutagenesis has emerged as evidenced by 4 patients with X-linked severe combined immunodeficiency (SCID-X1) treated with a Moloney murine leukemia virus (MLV)–based vector developing clonal T-cell lymphoproliferation.6-10 In 2 patients, the cause appears to be at least in part due to MLV proviral vector integration either within or near the known T-cell proto-oncogene LMO2, which led to up-regulation of its expression, probably mediated via the enhancer elements within the viral long-terminal repeats (LTRs). In another gene therapy trial for chronic granulomatous disease (CGD), nonmalignant amplification of myeloid clones contributed to the efficacy but occurred due to similar spleen focus-forming virus (SFFV) LTR-mediated activation of MDS1-EVI1, PRDM16, or SETBP1 genes.6 In animal model systems, retroviral vector transgenes have additionally been susceptible to a substantial reduction and variegation in expression largely attributable to DNA methylation and histone deacetylation.11-21
The risk of enhancer-mediated mutagenesis may be partially reduced by the development of self-inactivating (SIN) retroviral vectors in which the U3 region of the 3′ LTR containing the viral enhancer sequence is deleted, leading to inactivation of both LTRs upon integration of the vector provirus into the target cell genome.22-25 Transcription of a therapeutic gene within SIN vectors is via an internal promoter. However, all enhancer elements associated with internal regulatory elements will have a potential for mutagenesis, which may be exacerbated by a preference of some vectors for integration either within or close to active transcriptional domains.26-28 The development of safer vectors incorporating enhancer-less regulatory elements that are capable of establishing and maintaining a transcriptionally competent chromatin domain, and which give rise to reproducible and stable transgene expression irrespective of tissue type or site of integration, is therefore of considerable interest. The recently described ubiquitously acting chromatin opening element (UCOE)21,29 appears to meet these requirements.
UCOEs consist of a methylation-free CpG island extending over closely spaced, dual divergently transcribed promoters derived from housekeeping gene loci.21,29 The UCOE from the human HNRPA2B1-CBX3 locus (A2UCOE) gives rise to completely stable transgene expression in stably transfected tissue culture cells in the absence of drug selection, even when integrated within centromeric heterochromatin29 rather than the typical rapid functional decline mediated by chromatin components.30 In addition, linking the A2UCOE upstream of CMV promoter–driven cassettes prevents transgene silencing and markedly increases median levels of expression in stably transfected cells, thereby substantially expediting the isolation of lines for the manufacture of therapeutic proteins.21 These data strongly suggest that the A2UCOE possesses a dominant chromatin remodeling or opening function and is therefore able to resist transcriptional silencing effects.
In the present study, we have assessed the efficiency and efficacy of the A2UCOE in regulating transgene expression in vitro and in vivo within lentiviral vectors for possible hematopoietic gene therapy applications. Our data show that the A2UCOE largely overcomes insertion-site position effects, giving rise to a reproducible, stable, and therapeutically relevant pattern of gene expression.
Materials and methods
Plasmid and lentiviral vector construction
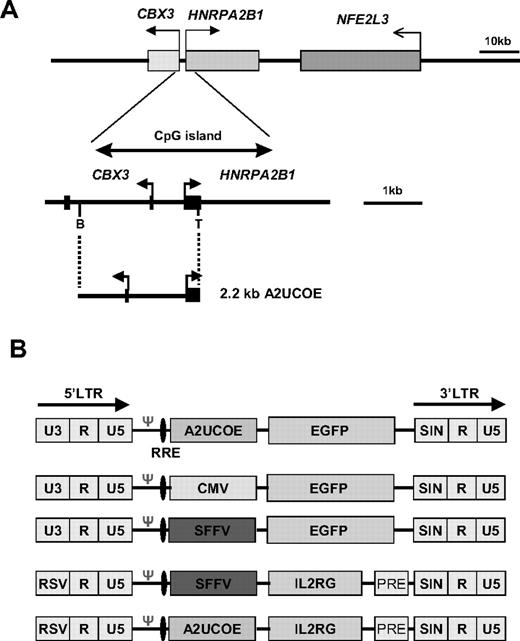
The pGL-2 UCOE construct: a 2.2-kb fragment extending from the BamHI site within the first intron of CBX3 to a (BamHI-linkered) TthIII I site just upstream of the ATG translational start codon within exon I of HNRPA2B1 (A2UCOE; Figure 1A) was inserted into the BamHI site of the pGL-2 promoter vector (Promega, Southampton, United Kingdom) downstream of the luciferase reporter gene in both forward and reverse orientations (Figure 2A; pGL-2 UCOE 5′, pGL-2 UCOE 3′). The pGL-2 control plasmid (Promega), which contains the SV40 enhancer in the same position as the A2UCOE, acted as a positive control. Lentiviral A2UCOE-EGFP: a 2.5-kb fragment extending from the EcoRI site within intron 2 of CBX3 to a SalI-linkered TthIII I site within exon I of HNRPA2B1 was inserted between the EcoRI and SalI polylinker sites upstream of the enhanced green fluorescent protein (EGFP) gene in pEGFP-N1 (Clontech Laboratories, Oxford, United Kingdom) to give the construct pA2UCOE-EGFP-N1. An EcoRI/NotI fragment of 3.2 kb containing the A2UCOE-EGFP cassette from pA2UCOE-EGFP-N1 was inserted between the EcoRI/XhoI sites of pHR'SINcPPT-CE,31 thereby replacing CMV-EGFP to give pHR'SINcPPT-UCOE-E. The pHR'SINcPPT-SE lentiviral vector31 containing an SFFV-EGFP combination was as previously described.31 Lentiviral A2UCOE-IL2RG: the SFFV promoter from the SINLV-SF-IL2RG vector (kindly provided by Christopher Baum, Hannover Medical School, Germany) was removed by digestion with XhoI/ClaI and the vector was religated after blunting of ends. The 2.2-kb A2UCOE BamH1 fragment was then inserted into a BamHI site upstream of the IL2RG cDNA of the SINLV-IL2RG vector to give A2UCOE-IL2RG (Figure 1B).
Illustration of the human HNRPA2B1-CBX3-NFE2L3 locus and derivation of the A2UCOE. (A, top panel) Gene organization of the HNRPA2B1-CBX3-NFE2L3 locus located at chromosome position 7p15.2 (GenBank accession nos. D28877, AC004520, and AC010677) is shown with gene positions indicated (shaded rectangles). (Bottom panel) Expanded view of the 3-kb methylation-free CpG island region encompassing the divergent CBX3 and HNRPA2B1 promoters. The region covered by the minimal 2.2-kb UCOE (A2UCOE; M.A. et al29 ) extending from the TthIII I (T) site within exon I of HNRPA2B1 to a BamHI (B) site within the first intron of CBX3 is shown. Black rectangles denote exons. Horizontal arrows denote the direction of transcription. (B) Illustration of the lentiviral vector constructs. LTR indicates long-terminal repeat; RRE, rev-response element; Ψ, packaging signal; RSV, Rous sarcoma virus U3; and PRE, mutated Woodchuck hepatitis posttranscriptional regulatory element.
Illustration of the human HNRPA2B1-CBX3-NFE2L3 locus and derivation of the A2UCOE. (A, top panel) Gene organization of the HNRPA2B1-CBX3-NFE2L3 locus located at chromosome position 7p15.2 (GenBank accession nos. D28877, AC004520, and AC010677) is shown with gene positions indicated (shaded rectangles). (Bottom panel) Expanded view of the 3-kb methylation-free CpG island region encompassing the divergent CBX3 and HNRPA2B1 promoters. The region covered by the minimal 2.2-kb UCOE (A2UCOE; M.A. et al29 ) extending from the TthIII I (T) site within exon I of HNRPA2B1 to a BamHI (B) site within the first intron of CBX3 is shown. Black rectangles denote exons. Horizontal arrows denote the direction of transcription. (B) Illustration of the lentiviral vector constructs. LTR indicates long-terminal repeat; RRE, rev-response element; Ψ, packaging signal; RSV, Rous sarcoma virus U3; and PRE, mutated Woodchuck hepatitis posttranscriptional regulatory element.
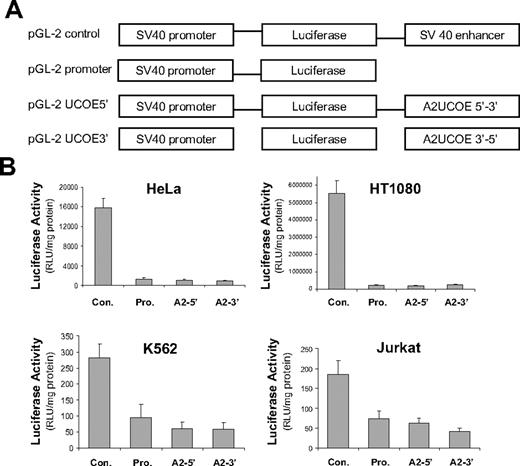
The A2UCOE lacks classic enhancer activity. (A) Illustration of the pGL-2 plasmid vector–based constructs. The pGL-2 promoter construct consists of a minimal, enhancer-less SV40 promoter driving a luciferase reporter gene. A 2.2-kb BamHI fragment consisting of a minimal A2UCOE (Figure 1A) was inserted downstream of the luciferase gene in pGL-2 promoter in both forward (pGL-2 UCOE5′) and reverse (pGL-2 UCOE3′) orientations. The pGL-2 control construct has the SV40 enhancer element inserted at the same position as the A2UCOE test fragment and acts as a positive enhancer control. (B) The 4 plasmids illustrated in panel A were used to conduct transient transfection assays in HeLa, HT1080, Jurkat, and K562 cells. Total protein cell lysates were analyzed for luciferase activity 24 hours after transfection. The mean and standard deviation of triplicate experiments for each cell line are shown. Con indicates pGL-2 control; Pro, pGL-2 promoter; A2-5′, pGL-2 A2UCOE5′; and A2-3′, pGL-2 A2UCOE3′. Note that the pGL-2 A2UCOE5′ and pGL-2 A2UCOE3′ test constructs give luciferase activities that are no higher than pGL-2 promoter in all 4 cell lines, indicating the absence of a classic enhancer function within this element.
The A2UCOE lacks classic enhancer activity. (A) Illustration of the pGL-2 plasmid vector–based constructs. The pGL-2 promoter construct consists of a minimal, enhancer-less SV40 promoter driving a luciferase reporter gene. A 2.2-kb BamHI fragment consisting of a minimal A2UCOE (Figure 1A) was inserted downstream of the luciferase gene in pGL-2 promoter in both forward (pGL-2 UCOE5′) and reverse (pGL-2 UCOE3′) orientations. The pGL-2 control construct has the SV40 enhancer element inserted at the same position as the A2UCOE test fragment and acts as a positive enhancer control. (B) The 4 plasmids illustrated in panel A were used to conduct transient transfection assays in HeLa, HT1080, Jurkat, and K562 cells. Total protein cell lysates were analyzed for luciferase activity 24 hours after transfection. The mean and standard deviation of triplicate experiments for each cell line are shown. Con indicates pGL-2 control; Pro, pGL-2 promoter; A2-5′, pGL-2 A2UCOE5′; and A2-3′, pGL-2 A2UCOE3′. Note that the pGL-2 A2UCOE5′ and pGL-2 A2UCOE3′ test constructs give luciferase activities that are no higher than pGL-2 promoter in all 4 cell lines, indicating the absence of a classic enhancer function within this element.
Maintenance of tissue culture cell lines and transient transfection assays with luciferase reporter gene constructs
Human HeLa, HEK293T, and HT1080 cells were maintained in DMEM medium (Invitrogen, Paisley, United Kingdom), whereas the Jurkat and K562 lines were cultured in RPMI 1640 (Invitrogen). All media were supplemented with 10% fetal calf serum (FCS; Sigma-Aldrich, Poole, United Kingdom). Transient transfections of HeLa and HT1080 cells were performed using SuperFect (Qiagen, Crawley, United Kingdom) and FuGENE 6 (Roche, Welwyn Garden City, United Kingdom) with Jurkat and K562 cells. Cell lysates were prepared 24 hours after transfection and protein was quantified using Burton Reagent (Bio-Rad, Hemel Hempstead, United Kingdom). Luciferase activities were assessed using the Luciferase Assay System (Promega), with light emission quantified using a FLUOstar OPTIMA luminometer (BMG LABTECH, Aylesburg, United Kingdom) and normalized per mg lysate protein. Statistical analysis was performed using a Student t test.
Lentiviral vector preparation and transduction of cell lines
Lentiviral vectors were produced by transient cotransfection of HEK293T cells with 3 plasmids (the lentiviral vector, pMD.G2 [envelop plasmid], and pCMVΔ8.91 [packaging plasmid, both produced by Plasmid Factory, Bielefeld, Germany]), employing polyethylenimine (PEI; Sigma-Aldrich) as previously described.31 Viral vector titer of EGFP-containing preparations was determined by transducing HeLa, Jurkat, and K562 cells with serial dilutions of virus and monitoring expression after 3 days by fluorescence-activated cell sorting (FACS) analysis. Titer of IL2RG transgene-containing vectors was by transduction of mouse fibroblast SC-1 cells followed by staining with anti–human CD132 (IL2RG) antibody (BD Biosciences, Oxford, United Kingdom) for FACS analysis of IL2RG expression. Lentiviral vectors containing EGFP under control of the A2UCOE, CMV, and SFFV promoters were used to transduce Jurkat, K562, and HeLa cells at a multiplicity of infection (MOI) of 1. Transduced cells were collected every 10 to 14 days for a total period of 78 days of continuous culture and EGFP gene expression analyzed by FACS.
Ex vivo transduction and analysis of murine hematopoietic stem cell transduction with EGFP-lentiviral vectors
All animal experiments were conducted under the Home Office project license number PPL 70/6146. Bone marrow lineage-negative (lin−) HSCs were isolated32 from the tibiae and femora of C57BL/6J mice at approximately 10 weeks of age. HSCs were seeded at 1 × 106/mL in StemSpan medium (StemCell Technologies, London, United Kingdom) and transduced with viral vector added at an MOI of 20 to 25. Transduced cells were cultured overnight and then injected into lethally irradiated recipient mice via the tail vein (2.5 × 105 to 5 × 105 cells/mouse).33 Peripheral blood and bone marrow cells were analyzed 3 months after transplantation by flow cytometry for EGFP transgene expression and lineage markers. Remaining cells were collected and stored at −20°C for subsequent isolation of genomic DNA. Statistical analysis was performed using a Wilcoxon sum rank test (Mann-Whitney test; http://elegans.swmed.edu/∼leon/stats/utest.cgi) to assess the significance of differences in expression between A2UCOE-, SFFV-, and CMV-EGFP transgenes.
Determination of EGFP and IL2RG lentiviral vector copy number by PCR
Genomic DNA was isolated from cells using the DNeasy kit (Qiagen). The primers employed for analysis of EGFP reporter gene vector–containing cells by standard (28 cycle) polymerase chain reaction (PCR) were as follows: forward, 5′AGCTGACCCTGAAGTTCATCTG3′; reverse, 5′GACGTTGTGGCTGATGTAGTTGTA3′. The mouse titin gene (Ttn)34 was used as an endogenous 2-copy gene control and was amplified with forward primer 5′AAAACGAGCAGTGACCTGAGG3′ and reverse primer 5′TTCAGTCATGCTGCTAGCGC3′ under the same conditions. PCR products were resolved by electrophoresis on agarose gels.
Real-time quantitative PCR (QPCR) to determine both EGFP and IL2RG transgene lentiviral vector copy number was performed using an ABI 7000 Sequence Detection System (ABI, Applied Biosystems, Warrington, United Kingdom). The primer sequences for EGFP were as follows: forward, 5′GCTACCCCGACCACATGAAG3′; reverse, 5′CGGGCATGGCGGACTT3′. The EGFP probe sequence was 5′-FAM-CAGCACGACTTCTTC-NFQ. Ttn primer sequences were as described previously in this section. The Ttn probe sequence was 5′-FAM-TGCACGGAATCTCGTCTCAGTC-TAMRA-3′. The IL2RG primer sequences were as follows: forward, 5′AGTAGACGGCATCGCAGCTT3′; reverse, 5′GGCTTCAACATGGCAGTCTAGAG3′. The IL2RG probe sequence was 5′ FAM-TCACGTGGGCGGCG-TAMRA-3′. The primer sequences for the human β-actin gene (ACTB) were as follows: forward, 5′TCACCCACACTGTGCCCATCTACGA3′; reverse, 5′CAGCGGAACCGCTCATTGCCAATGG3′. The ACTB probe sequence was 5′-FAM-ATGCCCTCCCCCATGCCA TCCTGCGT-TAMRA-3′.
STAT-5 phosphorylation assay in vitro
ED7R cells derived from an adult human T-cell leukemia line deficient in the IL2RG gene expression35 were cultured in RPMI 1640 plus 10% FCS. Cells were seeded into 12-well plates (105 cells/mL/well) and transduced with the SFFV-IL2RG and A2UCOE-IL2RG lentiviral vectors at different MOIs. On day 3 after transduction, the culture medium was replaced with serum-free medium and incubated overnight. Cells were then washed with PBS and resuspended in RPMI medium alone. Half of the cells were subsequently incubated with 600 ng interleukin-2 (IL-2; Peprotech, London, United Kingdom) at 37°C for 10 minutes followed by incubation with 2 mL prewarmed FACS Lyse/Fix buffer (BD Biosciences, Oxford, United Kingdom) for 30 minutes at 37°C. Cells were then washed in FACS buffer (0.5% BSA/PBS) and permeabilized in cold Perm Buffer III (BD Biosciences) for 30 minutes at 4°C. After washing in FACS buffer, cells were collected by centrifugation, resuspended in 100 μL FACS buffer containing 5 μL antiphosphorylated STAT-5 antibody (BD Biosciences) and 0.5 μL anti–human IL-2RG (BD Biosciences), and incubated for 30 minutes at 4°C. Cells were then washed with FACS buffer and fixed with 1% PFA/PBS for analysis by FACS (the protocol kindly provided by Kimberly Gilmour, Great Ormond Street Hospital, London, United Kingdom). ED7R cells stably transfected with an IL2RG transgene acted as the positive control.
SCID mouse model and ex vivo lentiviral vector–mediated IL2RG gene transfer
SCID mouse model: triple knock-out (3KO; Il2rg−/−Rag2−/−c5−/−) and 3KO X c57Il2rg−/− (Il2rg−/−Rag2+/+c5+/+) were generated by crossing Il2rg−/−Rag2−/−c5+/+ mice (obtained from Dr DiSanto, Hopital Necker-Enfants Malades, Paris, France) with A/J mice (Il2rg+/+Rag2+/+c5−/−; Harlan UK Limited, Bicester, United Kingdom). Il2rg−/−Rag2−/−c5−/− mice are deficient in T, B, and natural killer (NK) cells and provide a more complete immunologic defect more closely resembling the human phenotype of the disease. For ex vivo gene transfer, bone marrow was isolated from the tibiae and femora of 3KO X c57Il2rg−/− mice and lin− HSCs were purified by standard methods.32 HSCs were transduced at an MOI of 10 for A2UCOE-IL2RG and an MOI of 25 for SFFV-IL2RG. Transduced cells were cultured for 16 to 24 hours and injected into lethally irradiated recipient 3KO mice. At 3 months after transplantation, animals were culled and spleens were isolated and analyzed for immunoreconstitution. Splenocytes were cleared of red blood cells using red cell lysis buffer; stained with anti-CD8, -CD4, -CD3, and -NK1.1 antibodies for T cells, anti-IgM for mature B cells, and anti-B220 for immature B cells (BD Biosciences); and analyzed by FACS. T-cell proliferation assays were performed as previously described.33
Results
The A2UCOE is devoid of classic enhancer function
A minimal 2.2-kb A2UCOE fragment (Figure 1A) is sufficient to direct reproducible and stable EGFP transgene expression in transfected tissue culture cells.29 We assessed whether these properties were associated with classic enhancer activity by using a luciferase reporter gene system (Figure 2A) and transient transfection assays in a variety of human cell lines (carcinoma HeLa cells, fibrosarcoma HT1080 cells, leukemic lymphoblast Jurkat cells, and myelogenous leukemia K562 cells). In contrast to a pGL-2 control vector containing an SV40 enhancer, each configuration of the pGL-2 A2UCOE test constructs exhibited no statistically significant difference in activity compared with that of the enhancer-less pGL-2 promoter plasmid in any of the cell lines (Figure 2B; data not shown). To exclude the possibility that antisense transcripts from the divergent promoters within the A2UCOE were suppressing luciferase gene expression, reverse transcriptase–PCR (RT-PCR) was performed on RNA from transfected HeLa cells. No antisense transcripts were detectable, which may be due to the potential bidirectional nature of the SV40 polyadenylation signal downstream of the luciferase gene (data not shown). These data imply that the A2UCOE lacks classic enhancer activity, which in the context of gene therapy vectors is likely to reduce the risk of enhancer-mediated insertional mutagenesis.
The A2UCOE in a lentiviral vector context provides consistent gene expression in cell lines
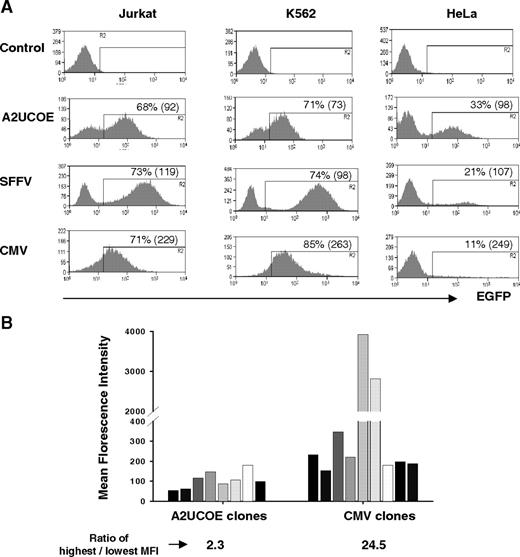
The A2UCOE can drive reproducible, stable, and long-term EGFP transgene expression in cell lines in the absence of drug-selective pressure, even when integrated within centromeric heterochromatin.29 We therefore assessed the efficiency and efficacy of the minimal 2.2-kb A2UCOE to regulate EGFP transgene expression within a SIN lentiviral vector system in comparison with the well-characterized CMV and SFFV viral promoters (Figure 1B). K562 (myeloid), Jurkat (lymphoid), and HeLa (carcinoma) cell lines were transduced at an MOI of 1 and analyzed at periodic intervals up to 78 days of continuous culture. The expression profile of polyclonal pools of cells transduced with the A2UCOE-EGFP vector was relatively reproducible, as indicated by a peak expression profile in all 3 cell lines (Figure 3A). We quantified the variation in the range of expression by reference to the coefficient of variation (CV; a parameter shown in histogram/plot statistics) by FACS for EGFP-expressing cells (Figure 3A). A2UCOE-EGFP–transduced cells consistently gave lower CV values than the SFFV and particularly CMV promoters, even though relative total transduction efficiencies as assessed by EGFP fluorescence were similar. This distinct difference in the expression profile between the A2UCOE and viral promoters was maintained over the entire 78-day period of cell culture (data not shown). We further explored these findings by assessing EGFP expression by FACS analysis of transduced HeLa cell clones (Figure 3B) carrying a singe copy of vector (confirmed by Southern-blot analysis; data not shown). In marked contrast to the CMV-EGFP vector, all A2UCOE-EGFP clones exhibited a very similar level of fluorescence intensity regardless of integration site. Furthermore, the CV values within individual A2UCOE-EGFP clones were markedly lower compared with those with CMV-EGFP (mean CV A2UCOE = 51, range 42-83, n = 8; CMV = 210, range 69-336, n = 7; Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). The consistent and stable transgene expression pattern between cells transduced with the A2UCOE-EGFP vector suggests that this element is less susceptible to insertion-site position effects.
An A2UCOE regulatory element within a lentiviral vector confers consistent transgene expression. (A) Lentiviral vectors containing an EGFP reporter gene under the control of the A2UCOE, SFFV, or CMV promoters (Figure 1B) were used to transduce Jurkat (T-cell), K562 (myeloid), and HeLa (carcinoma) human cell lines at an MOI of 1. Cells were analyzed by flow cytometry, and plots of percentage EGFP-positive cells (y-axis) verses fluorescence intensity (x-axis) are shown. Data shown are from 14 days of culture. The percentage of total EGFP-positive cells is shown in each plot. The coefficient of variation (CV) of EGFP-positive cells for each type cell is shown in the parentheses. Note that discrete peaks of EGFP-positive cells that are seen with A2UCOE-EGFP in all cells lines in conjunction with a lower CV suggest a more homogeneous transgene expression pattern compared with the SFFV and especially the CMV viral promoters. (B) Lentiviral vectors containing an EGFP reporter gene under control of either the A2UCOE or CMV promoters (Figure 1B) were used to transduce HeLa cells at an MOI of 0.3. Single EGFP-expressing cells were sorted by FACS and clonally expanded in culture. EGFP fluorescence intensity was analyzed by FACS in selected cell clones carrying a single copy of vector transgene. Note that expression from the A2UCOE construct between clones is highly reproducible and 10-fold lower in degree of variation than with the CMV promoter.
An A2UCOE regulatory element within a lentiviral vector confers consistent transgene expression. (A) Lentiviral vectors containing an EGFP reporter gene under the control of the A2UCOE, SFFV, or CMV promoters (Figure 1B) were used to transduce Jurkat (T-cell), K562 (myeloid), and HeLa (carcinoma) human cell lines at an MOI of 1. Cells were analyzed by flow cytometry, and plots of percentage EGFP-positive cells (y-axis) verses fluorescence intensity (x-axis) are shown. Data shown are from 14 days of culture. The percentage of total EGFP-positive cells is shown in each plot. The coefficient of variation (CV) of EGFP-positive cells for each type cell is shown in the parentheses. Note that discrete peaks of EGFP-positive cells that are seen with A2UCOE-EGFP in all cells lines in conjunction with a lower CV suggest a more homogeneous transgene expression pattern compared with the SFFV and especially the CMV viral promoters. (B) Lentiviral vectors containing an EGFP reporter gene under control of either the A2UCOE or CMV promoters (Figure 1B) were used to transduce HeLa cells at an MOI of 0.3. Single EGFP-expressing cells were sorted by FACS and clonally expanded in culture. EGFP fluorescence intensity was analyzed by FACS in selected cell clones carrying a single copy of vector transgene. Note that expression from the A2UCOE construct between clones is highly reproducible and 10-fold lower in degree of variation than with the CMV promoter.
The A2UCOE-EGFP vector gives rise to reproducible transgene expression in hematopoietic cells in vivo
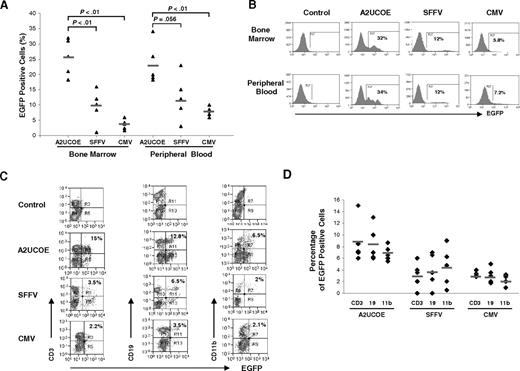
The A2UCOE-, SFFV-, or CMV-EGFP lentiviral vectors (Figure 1B) were used in an ex vivo protocol to transduce C57BL/6J murine bone marrow–derived HSCs that were then engrafted into lethally irradiated recipients. At 3 months after transplantation, bone marrow and peripheral blood cells were harvested and analyzed for EGFP expression and vector copy number. In experiment 1, HSCs were transduced at an MOI of 20 and engrafted into 6 recipient mice (2 animals per vector). In experiment 2, HSCs were transduced at an MOI of 25 and engrafted into 9 recipients (3 animals per vector). The initial transduction efficiency was assessed by FACS analysis 2 days following ex vivo infection and was found to be similar in all cases (data not shown). However, at 3 months after transplantation, a consistently higher percentage of EGFP-positive cells was found among the A2UCOE-EGFP vector recipient animals compared with those transduced by the SFFV- and CMV-regulated constructs, both in bone marrow and peripheral blood cells (Figure 4A). The mean percentage of EGFP-expressing cells transduced by the A2UCOE-EGFP vector from the 2 sets of experimental mice was 25.6%, which is 2.5-fold and 6.2-fold higher than the SFFV and CMV constructs, respectively, in bone marrow cells (Figure 4A left). Similarly, in peripheral blood from the same animals, the mean percentage of EGFP-positive cells transduced by the A2UCOE-EGFP vector was 23.9%, which is 2.1-fold and 2.8-fold higher than that observed in SFFV and CMV vector–transduced cells, respectively (Figure 4A right). Of greater significance in the context of this study, EGFP-expressing cells transduced by the A2UCOE-EGFP vector appeared as discrete peaks in the fluorescence intensity profiles both in bone marrow and peripheral blood (Figure 4B, UCOE panels).
The A2UCOE-EGFP vector gives rise to consistent transgene expression in bone marrow and peripheral blood hematopoietic cells in vivo. HSCs were isolated from the bone marrow of C57BL/6J mice and transduced with the A2UCOE-EGFP, SFFV-EGFP, and CMV-EGFP lentiviral vectors (Figure 1B) at an MOI of 20 to 25 and transplanted into lethally irradiated recipient animals. Total peripheral blood and bone marrow cells were obtained at 3 months after transplantation and analyzed for the presence of EGFP-positive cells by FACS. (A) Percentage of total EGFP-positive cells in bone marrow and peripheral blood from A2UCOE, SFFV, and CMV mice (n = 5 in each group). The horizontal bar shown in each column denotes the mean percentage value of EGFP-positive cells in each group. The P values were determined using the Wilcoxon rank sum test. (B) Expression profiles of a representative mouse that received a transplant in each group. The percentage of total EGFP-positive cells is shown in each plot. Note that the A2UCOE-EGFP vector generates a higher percentage of transgene-expressing cells (panel A) as well as discrete peaks of EGFP-positive cell populations (panel B), suggesting a negation of integration-site position effects leading to more consistent transgene expression compared with that achieved with SFFV and CMV viral promoters. (C) Peripheral blood cells from mice transduced with the A2UCOE-, SFFV-, and CMV-EGFP lentiviral vectors were incubated with antibodies against CD3 (T-cell), CD19 (B-cell), and CD11b (myeloid) lineage markers and scored against EGFP expression by FACS analysis. EGFP expression profile in different cell lineages in a representative mouse that received a transplant, corresponding to panel B (bottom row, peripheral blood), in each lentiviral vector group. (D) Total percentage of EGFP-positive cells in T-, B-, and myeloid cell lineages in peripheral blood from A2UCOE, SFFV, and CMV vector–transduced mice (n = 5 in each group). The horizontal bar shown in each column depicts the mean percentage value of EGFP-positive cells in each cell lineage.
The A2UCOE-EGFP vector gives rise to consistent transgene expression in bone marrow and peripheral blood hematopoietic cells in vivo. HSCs were isolated from the bone marrow of C57BL/6J mice and transduced with the A2UCOE-EGFP, SFFV-EGFP, and CMV-EGFP lentiviral vectors (Figure 1B) at an MOI of 20 to 25 and transplanted into lethally irradiated recipient animals. Total peripheral blood and bone marrow cells were obtained at 3 months after transplantation and analyzed for the presence of EGFP-positive cells by FACS. (A) Percentage of total EGFP-positive cells in bone marrow and peripheral blood from A2UCOE, SFFV, and CMV mice (n = 5 in each group). The horizontal bar shown in each column denotes the mean percentage value of EGFP-positive cells in each group. The P values were determined using the Wilcoxon rank sum test. (B) Expression profiles of a representative mouse that received a transplant in each group. The percentage of total EGFP-positive cells is shown in each plot. Note that the A2UCOE-EGFP vector generates a higher percentage of transgene-expressing cells (panel A) as well as discrete peaks of EGFP-positive cell populations (panel B), suggesting a negation of integration-site position effects leading to more consistent transgene expression compared with that achieved with SFFV and CMV viral promoters. (C) Peripheral blood cells from mice transduced with the A2UCOE-, SFFV-, and CMV-EGFP lentiviral vectors were incubated with antibodies against CD3 (T-cell), CD19 (B-cell), and CD11b (myeloid) lineage markers and scored against EGFP expression by FACS analysis. EGFP expression profile in different cell lineages in a representative mouse that received a transplant, corresponding to panel B (bottom row, peripheral blood), in each lentiviral vector group. (D) Total percentage of EGFP-positive cells in T-, B-, and myeloid cell lineages in peripheral blood from A2UCOE, SFFV, and CMV vector–transduced mice (n = 5 in each group). The horizontal bar shown in each column depicts the mean percentage value of EGFP-positive cells in each cell lineage.
The profile of EGFP transgene–expressing cells in different hematopoietic cell lineages was then determined in peripheral blood. Representative plots for each test group of animals are shown in Figure 4C and a compilation of data sets from all animals is summarized in Figure 4D. EGFP expression for all 3 vectors is distributed in all cell lineages (Figure 4D), suggesting that long-lived HSCs were successfully transduced. The average percentage of EGFP-positive cells given by the A2UCOE-EGFP vector was clearly higher in each lineage when compared with the other vectors.
Taken together, these data suggest that the SFFV and particularly CMV promoters are highly prone to position effects and silencing, resulting in a variable expression pattern. In contrast, the consistent peak transgene expression profiles conferred by the A2UCOE show that this element can largely overcome insertion-site position effects, giving rise to stable and reproducible transgene expression in HSCs and their progeny in vivo.
A2UCOE gives more reliable expression per vector copy than SFFV or CMV
We quantified the efficiency of A2UCOE-EGFP expression in comparison to the SFFV and CMV constructs by determining vector copy number and correlated this to the mean percentage of cells expressing EGFP within bone marrow of mice. Genomic DNA from bone marrow of all transplant recipients (Figure 4) was assayed by QPCR employing primers for the EGFP gene. Simultaneous amplification of murine Ttn sequences acted as an endogenous 2-copy gene control. Initial PCR analysis showed that the transgene is present in all recipients (Figure 5A). Following QPCR, comparison of the EGFP and Ttn amplification products showed that, in general, the average vector copy number in recipients of HSCs transduced with the A2UCOE-EGFP construct were significantly lower than those harboring the SFFV- and CMV-EGFP vectors (Figure 5B; Table 1). The average copy number across all samples for each vector was 0.25 per cell (range, 0.13 to 0.44 per cell) for the A2UCOE group, 0.96 per cell (range, 0.02 to 1.3 per cell) for SFFV, and 3.7 per cell (range, 1.2 to 6.5 per cell) for CMV vector recipients. A comparison of average vector copy number with average EFGP expression in bone marrow (Table 1) reveals a 1:1 correlation for the EGFP transgene regulated by the A2UCOE. In marked contrast, the ratio of expression to vector copy number is 1:9 for the SFFV and 1:90 for the CMV vectors (Table 1), which is indicative of either very low levels of expression per transgene or, more likely, extensive transgene silencing.17,18
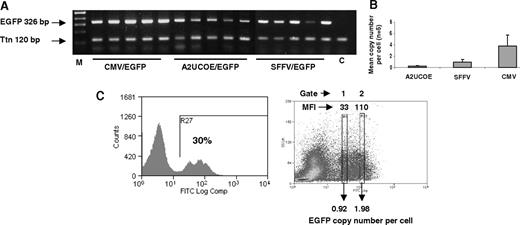
A2UCOE-EGFP gives stable, high efficiency expression per vector copy. (A) Genomic DNA derived from bone marrow of mice that received transplants ex vivo of HSCs transduced with the CMV-EGFP, SFFV-EGFP, and A2UCOE-EGFP vectors (Figure 4) was subjected to standard PCR for the presence of transgene (EGFP) and endogenous murine titin (Ttn) sequences and products were resolved by agarose gel electrophoresis. M indicates DNA size markers; C, mock control mouse bone marrow sample. (B) Summary of real-time quantitative PCR analysis of the same samples shown in panel A to determine lentiviral vector copy number. Error bars denote 1 standard deviation about the mean. (C) Determination of vector copy number in subpopulations of EGFP-expressing total bone marrow cells transduced with the A2UCOE-EGFP lentiviral vector. (Left) A representative sample of A2UCOE-EGFP vector–transduced bone marrow cells was sorted by FACS to isolate either low (gate 1) or high (gate 2) EGFP fluorescence intensity cells. DNA was then isolated from the sorted pools of cells and analyzed by QPCR as in panel B. (Right) Profile showing the sorting gates and corresponding mean fluorescence intensity (MFI). Average lentiviral vector copy number per cell is indicated. Note that the A2UCOE gives a higher number of EGFP-positive cells at a lower vector copy number than either the SFFV or CMV promoters (summarized in Table 1), with a clear trend toward copy number–dependent expression.
A2UCOE-EGFP gives stable, high efficiency expression per vector copy. (A) Genomic DNA derived from bone marrow of mice that received transplants ex vivo of HSCs transduced with the CMV-EGFP, SFFV-EGFP, and A2UCOE-EGFP vectors (Figure 4) was subjected to standard PCR for the presence of transgene (EGFP) and endogenous murine titin (Ttn) sequences and products were resolved by agarose gel electrophoresis. M indicates DNA size markers; C, mock control mouse bone marrow sample. (B) Summary of real-time quantitative PCR analysis of the same samples shown in panel A to determine lentiviral vector copy number. Error bars denote 1 standard deviation about the mean. (C) Determination of vector copy number in subpopulations of EGFP-expressing total bone marrow cells transduced with the A2UCOE-EGFP lentiviral vector. (Left) A representative sample of A2UCOE-EGFP vector–transduced bone marrow cells was sorted by FACS to isolate either low (gate 1) or high (gate 2) EGFP fluorescence intensity cells. DNA was then isolated from the sorted pools of cells and analyzed by QPCR as in panel B. (Right) Profile showing the sorting gates and corresponding mean fluorescence intensity (MFI). Average lentiviral vector copy number per cell is indicated. Note that the A2UCOE gives a higher number of EGFP-positive cells at a lower vector copy number than either the SFFV or CMV promoters (summarized in Table 1), with a clear trend toward copy number–dependent expression.
Correlation of lentiviral vector copy number with EGFP-positive cells in bone marrow of mice that received a transplant ex vivo
| . | A2UCOE . | SFFV . | CMV . |
|---|---|---|---|
| Mean % of EFGP-positive cells, n=5 | 25.6 | 10.2 | 4.1 |
| Mean vector copy number per cell, n=5 | 0.25 | 0.92 | 3.7 |
| Ratio of EGFP expression/copy number | 1/1 | 1/9 | 1/90 |
| . | A2UCOE . | SFFV . | CMV . |
|---|---|---|---|
| Mean % of EFGP-positive cells, n=5 | 25.6 | 10.2 | 4.1 |
| Mean vector copy number per cell, n=5 | 0.25 | 0.92 | 3.7 |
| Ratio of EGFP expression/copy number | 1/1 | 1/9 | 1/90 |
The average percentage of EGFP-positive ex vivo–manipulated bone marrow cells (Figure 4A) was correlated with the average A2UCOE-, SFFV-, and CMV-EGFP lentiviral vector copy number (Figure 5B). A ratio of average EGFP-positive cells to vector copy shows a 1:1 correlation in the case of A2UCOE-EGFP–transduced bone marrow but only 1:9 and 1:90 in the case of the SFFV- and CMV-EGFP vectors. This suggests that the A2UCOE negates transgene silencing to which the SFFV and especially the CMV viral promoters are highly prone.
As noted previously, the A2UCOE-EGFP expression profile frequently manifested as discrete peaks (Figure 5C left), which may consist of pools of cells harboring similar vector copy numbers and expression levels. We tested this possibility by sorting A2UCOE-EGFP–expressing cells according to fluorescence intensity and determining vector copy number by QPCR (Figure 5A,B). EGFP-expressing cells from the peak with lower fluorescence intensity (gate 1) carried a single copy of the vector, whereas the higher fluorescence intensity (gate 2) population contained an average of 2 copies per cell (Figure 5C). Overall, these data provide good evidence that the A2UCOE can prevent transgene silencing and give rise to stable and consistently reproducible expression levels when introduced into HSCs in vivo, which is reflective of its dominant chromatin opening function.21,29 Furthermore, the positive trend of transgene expression correlating with copy number is a property akin to that observed with locus control regions (LCRs).36
Functional reconstitution of IL-2R by A2UCOE-IL2RG
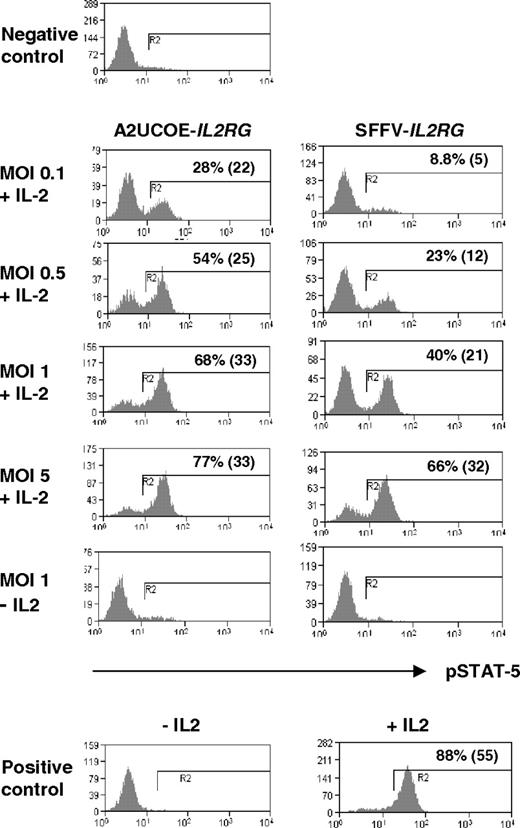
X-linked severe combined immunodeficiency is an immune disorder caused by mutations in the interleukin-2 receptor gamma gene (IL2RG), which encodes the common cytokine receptor gamma chain (γc, CD132). A key signaling pathway activated by IL-2 is phosphorylation of STAT-5 by JAK3. In order to determine whether the A2UCOE can direct a therapeutic level of IL2RG expression, human ED-7R cells, which are deficient in γc, were transduced with vectors containing either the A2UCOE-IL2RG or SFFV-IL2RG cDNA transcription unit (Figure 1B) at different MOIs. At 4 days after transduction, cells were stimulated with IL-2 and subsequently stained with antiphosphorylated STAT-5 (pSTAT-5) antibodies and analyzed by FACS. Expression of pSTAT-5 was readily detected at low MOIs and reached saturation at an MOI of 5 (Figure 6, A2UCOE-IL2RG panels). A parallel increase in γc levels was also demonstrated (Figure S2). The efficiency of rescue of pSTAT-5 function seemed higher with the A2UCOE-IL2RG than the SFFV-IL2RG vector (Figure 6, compare A2UCOE-IL2RG and SFFV-IL2RG panels). In order to quantify expression and determine whether this is indeed the case, vector copy number was assessed by QPCR and, as expected, increased steadily with increasing MOI (Table 2). At presaturation levels (MOI of 0.1-1), higher percentages of pSTAT-5–positive cells were achieved per copy with the A2UCOE-IL2RG–transduced populations compared with SFFV-IL2RG–transduced populations (Table 2). This suggests a higher functional efficiency of the A2UCOE compared with the SFFV promoter in regulating IL2RG expression in this in vitro assay, which is in agreement with the results obtained with the EGFP reporter gene constructs (Figures 4,5). The slight discrepancy between copy number and the percentage of pSTAT-5–expressing cells is most likely due to limitations in the accuracy of quantification by FACS and QPCR.
Rescue of JAK3-mediated STAT-5 tyrosine phosphorylation in human ED-7R cells. The A2UCOE-IL2RG and SFFV-IL2RG lentiviral vectors (Figure 1B) were used to transduce human ED-7R cells at MOIs of 0.1, 0.5, 1, and 5. At 4 days following transduction, cells were stimulated with IL-2, subsequently stained with antiphosphorylated STAT-5 (pSTAT-5) antibody, and assessed for the presence of pSTAT-5 expression by FACS. The percentage of pSTAT-5–positive cells and MFI (in parentheses) is shown. Negative control indicates untransduced ED-7R cells stimulated with IL-2; positive control, ED-7R cells stably transfected with an IL2RG transgene.
Rescue of JAK3-mediated STAT-5 tyrosine phosphorylation in human ED-7R cells. The A2UCOE-IL2RG and SFFV-IL2RG lentiviral vectors (Figure 1B) were used to transduce human ED-7R cells at MOIs of 0.1, 0.5, 1, and 5. At 4 days following transduction, cells were stimulated with IL-2, subsequently stained with antiphosphorylated STAT-5 (pSTAT-5) antibody, and assessed for the presence of pSTAT-5 expression by FACS. The percentage of pSTAT-5–positive cells and MFI (in parentheses) is shown. Negative control indicates untransduced ED-7R cells stimulated with IL-2; positive control, ED-7R cells stably transfected with an IL2RG transgene.
Correlation of IL2RG lentiviral vector copy number with restoration of pSTAT-5 in IL2RG-defficient human ED-7R cells
| MOI . | A2UCOE-IL2RG . | SFFV-IL2RG . | ||
|---|---|---|---|---|
| Copy number/cell . | % STAT-5 . | Copy number/cell . | % STAT-5 . | |
| 0.1 | 0.11 | 28 | 0.16 | 9 |
| 0.5 | 0.28 | 54 | 0.16 | 23 |
| 1 | 0.44 | 68 | 0.25 | 40 |
| 5 | 1.2 | 77 | 0.44 | 65 |
| MOI . | A2UCOE-IL2RG . | SFFV-IL2RG . | ||
|---|---|---|---|---|
| Copy number/cell . | % STAT-5 . | Copy number/cell . | % STAT-5 . | |
| 0.1 | 0.11 | 28 | 0.16 | 9 |
| 0.5 | 0.28 | 54 | 0.16 | 23 |
| 1 | 0.44 | 68 | 0.25 | 40 |
| 5 | 1.2 | 77 | 0.44 | 65 |
Percentage of pSTAT-5–positive cells after transduction with either the A2UCOE-IL2RG or SFFV-IL2RG lentiviral vectors and stimulation with IL-2 (Figure 6A) correlated with vector copy number determined by QPCR at different MOIs. Note that at low (0.1, 0.5) MOI A2UCOE-IL2RG gives a higher proportion of pSTAT-5–positive cells than the SFFV-IL2RG vector.
Immunologi reconstitution in a mouse model of SCID-X1
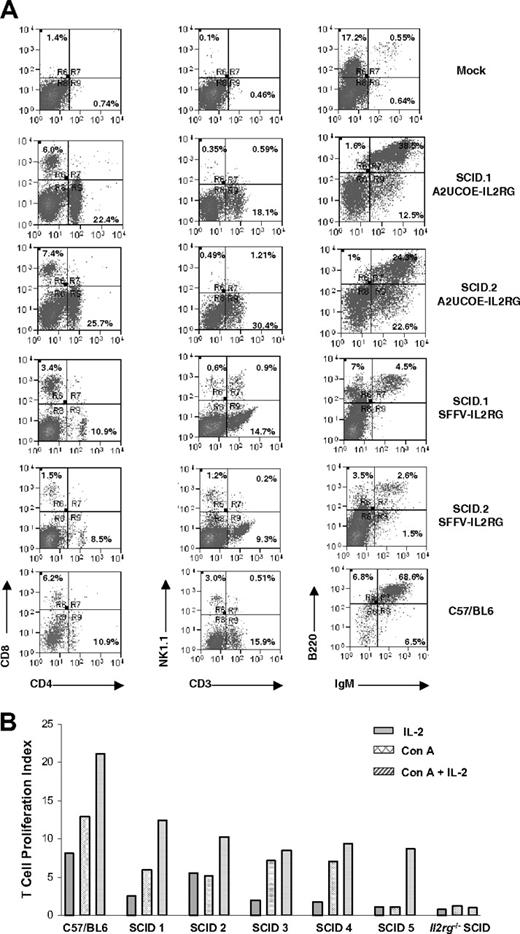
We next tested the efficacy of the A2UCOE in vivo in a mouse model of SCID-X1 gene therapy. HSCs isolated from SCID-X1 mice (3KO X c57Il2rg−/−) were transduced with the A2UCOE-IL2RG vector (Figure 1B) at an MOI of 10 and engrafted into 5 lethally irradiated recipients (Il2rg−/−, Rag2−/−, c5−/−; 2 in group 1 and 3 in group 2). In an independent experiment, HSCs were transduced with an SFFV-IL2RG vector (Figure 1B) at an MOI of 25. At 15 weeks after transplantation, animals were killed and the bone marrow, spleens, and thymi were removed and analyzed for immunologic reconstitution. Western-blot analysis of extracts from bone marrow revealed that IL2RG protein was present in all mice transduced with the A2UCOE-IL2RG vector (Figure S3). FACS analysis showed that in all animals treated with A2UCOE-IL2RG, the levels of T, B, and NK cells were substantially restored when compared with a wild-type control and with equivalent levels observed previously with therapeutic gammaretroviral vectors (Figure 7A; Table 3; S.I.T., A.S., S.J.H., and A.J.T., manuscript in preparation). T-cell proliferative responses to Concanavalin A (ConA) and IL-2 were also restored (Figure 7B). Reconstitution of SFFV-IL2RG–treated animals appeared less complete, but a statistical comparison was not possible due to small sample numbers. We also assessed transgene copy number in spleens recovered from treated SCID-X1 recipient animals by QPCR. The mean vector copy number in recipients transduced with A2UCOE-IL2RG (0.072 per cell; range, 0.03 to 0.19 per cell) was lower than in those transduced with SFFV-IL2RG (0.19 per cell; range, 0.12 to 0.26 per cell), which is in keeping with the different MOIs during initial transduction.
Efficient immunologic reconstitution in mice following ex vivo bone marrow HSC gene transfer. (A) HSCs from SCID-X1 mice were transduced with either the A2UCOE-IL2RG or SFFV-IL2RG lentiviral vectors (Figure 1B) and transplanted into lethally irradiated 3KO Il2rg−/−Rag2−/−c5−/− mice (see “Materials and methods, SCID mouse model and ex vivo lentiviral vector–mediated IL2RG gene transfer”). At 3 months following engraftment, spleens were analyzed for reconstitution of T-, B-, and NK-cell lineages by FACS. Cells were stained with anti-CD8, -CD4, -NK1.1, -IgM, and -B220 antibodies. Reconstitution of all cell lineages is observed in A2UCOE-IL2RG vector–transduced mice and SFFV-IL2RG vector–transduced mice (2 SCID-X1 mice transduced with the A2UCOE-IL2RG and 2 transduced with SFFV-IL2RG lentiviral vectors are shown). Mock indicates 3KO; Il2rg−/−Rag2−/−c5−/− SCID-X1 mouse that received only untransduced HSCs. Percentages within quadrants represent the percentage of the total cell population analyzed that is present in that quadrant. (B) T-cell proliferation assay. Splenocytes isolated from mice transduced with A2UCOE-IL2RG were stimulated with Concanavalin A (Con A), IL-2, and Con A plus IL-2. Proliferating cells were assessed by incorporation of 3H-thymidine and expressed as a proliferation index (the ratio of the stimulated cells to unstimulated cells). All 5 mice transduced with A2UCOE-IL2RG showed an increased cell proliferation index compared with an untreated animal.
Efficient immunologic reconstitution in mice following ex vivo bone marrow HSC gene transfer. (A) HSCs from SCID-X1 mice were transduced with either the A2UCOE-IL2RG or SFFV-IL2RG lentiviral vectors (Figure 1B) and transplanted into lethally irradiated 3KO Il2rg−/−Rag2−/−c5−/− mice (see “Materials and methods, SCID mouse model and ex vivo lentiviral vector–mediated IL2RG gene transfer”). At 3 months following engraftment, spleens were analyzed for reconstitution of T-, B-, and NK-cell lineages by FACS. Cells were stained with anti-CD8, -CD4, -NK1.1, -IgM, and -B220 antibodies. Reconstitution of all cell lineages is observed in A2UCOE-IL2RG vector–transduced mice and SFFV-IL2RG vector–transduced mice (2 SCID-X1 mice transduced with the A2UCOE-IL2RG and 2 transduced with SFFV-IL2RG lentiviral vectors are shown). Mock indicates 3KO; Il2rg−/−Rag2−/−c5−/− SCID-X1 mouse that received only untransduced HSCs. Percentages within quadrants represent the percentage of the total cell population analyzed that is present in that quadrant. (B) T-cell proliferation assay. Splenocytes isolated from mice transduced with A2UCOE-IL2RG were stimulated with Concanavalin A (Con A), IL-2, and Con A plus IL-2. Proliferating cells were assessed by incorporation of 3H-thymidine and expressed as a proliferation index (the ratio of the stimulated cells to unstimulated cells). All 5 mice transduced with A2UCOE-IL2RG showed an increased cell proliferation index compared with an untreated animal.
Reconstitution in spleens of mice following ex vivo bone marrow HSC gene transfer with the A2UCOE-IL2RG and SFFV-IL2RG lentiviral vector
| . | CD3, % . | CD4, % . | CD8, % . | B220/IgM, % . | IgM, % . |
|---|---|---|---|---|---|
| Mock | 0.46 | 0.74 | 1.4 | 0.55 | 0.64 |
| A2UCOE-γ c 1 | 18.1 | 22.4 | 6 | 38.5 | 12.5 |
| A2UCOE-γ c 2 | 30.4 | 25.7 | 7.4 | 24.3 | 22.5 |
| A2UCOE-γ c 3 | 34 | 19.2 | 7.4 | 9.5 | 24 |
| A2UCOE-γ c 4 | 37 | 29 | 9.3 | 18.1 | 22 |
| A2UCOE-γ c 5 | 23 | 17.4 | 7.7 | 5.1 | 9.4 |
| SFFV-γ c 1 | 14.7 | 10.9 | 3.4 | 4.5 | 0 |
| SFFV-γ c 2 | 9.3 | 8.5 | 1.5 | 2.6 | 1.5 |
| Control | 15.9 | 10.9 | 6.2 | 68.6 | 6.5 |
| . | CD3, % . | CD4, % . | CD8, % . | B220/IgM, % . | IgM, % . |
|---|---|---|---|---|---|
| Mock | 0.46 | 0.74 | 1.4 | 0.55 | 0.64 |
| A2UCOE-γ c 1 | 18.1 | 22.4 | 6 | 38.5 | 12.5 |
| A2UCOE-γ c 2 | 30.4 | 25.7 | 7.4 | 24.3 | 22.5 |
| A2UCOE-γ c 3 | 34 | 19.2 | 7.4 | 9.5 | 24 |
| A2UCOE-γ c 4 | 37 | 29 | 9.3 | 18.1 | 22 |
| A2UCOE-γ c 5 | 23 | 17.4 | 7.7 | 5.1 | 9.4 |
| SFFV-γ c 1 | 14.7 | 10.9 | 3.4 | 4.5 | 0 |
| SFFV-γ c 2 | 9.3 | 8.5 | 1.5 | 2.6 | 1.5 |
| Control | 15.9 | 10.9 | 6.2 | 68.6 | 6.5 |
SCID-X1 mice were subjected to an ex vivo procedure with HSCs transduced with the A2UCOE-IL2RG lentiviral vector (Figure 1B). At 3 months following engraftment, spleens were excised and analyzed for reconstitution of T-, B-, and NK cell lineages by FACS. Cells were stained with anti-CD8, -CD4, -NK1.1, -IgM, and -B220 antibodies. Efficient reconstitution of all cell lineages is observed in A2UCOE-IL2RG vector and SFFV-IL2RG–positive control transduced mice.
Discussion
Strategies that minimize the possibility of insertional mutagenesis by integrating viral vectors and yet retain functional levels of gene expression have recently become a major priority. The efficacy of gene transfer in vivo has in many cases been compromised by instability (high variability, silencing) of transgene expression from viral promoters.17,18 Furthermore, potentially mutagenic enhancer activity may be maintained even when linked promoters are silenced by DNA methylation (Wang et al37 ; and Manuel Grez, oral communication, at European Society for Gene Therapy conference, November 2006). We show for the first time that A2UCOE-regulated transgenes within lentiviral vectors produce a high, consistent, and homogeneous population of expressing cells that is not prone to gene silencing both in vitro (Figure 3) and more importantly within either HSCs or their progeny in vivo (Figure 4). A comparison of the average number of EGFP-positive cells with average vector copy number in bone marrow of recipient mice at 3 months after ex vivo transplantation indicates that almost all A2UCOE-EGFP transgenes are continuing to be successfully transcribed (Figure 5; Table 1). In marked contrast, the majority of EGFP transgenes under control of the SFFV and especially the CMV viral promoters appear to be subject to silencing under the same transduction and model conditions (Figure 5; Table 1).
We have found no classic enhancer function to be associated with the A2UCOE (Figure 2). This supports our previous suggestions that this element functions by virtue of its methylation-free CpG island status and divergent promoter activity generating a dominant transcriptional activating and chromatin remodeling function.21,29 This lack of enhancer function within the A2UCOE suggests that it should possess a far lower potential for inadvertent activation of genes in the vicinity of the vector integration site than enhancer-associated promoters. Formal evidence that this element is less mutagenic is under investigation. The divergent transcription from the A2UCOE through a nearby promoter could also potentially have either an activating or negative interference effect. However, our latest work shows that the placement of the A2UCOE upstream of tissue-specific promoter/enhancer combinations does not interfere with the specificity of these elements (Talbot, Waddington, M.A., Santilli, A.J.T., unpublished results). This suggests that the open chromatin environment established by the UCOE simply allows a linked, nearby promoter to function with its normal specificity and capacity. Nevertheless, potential problems of UCOE-mediated host promoter activation/inhibition can be avoided by placement of a transcriptional termination element 5′ of the A2UCOE. This would limit the divergent transcription-mediated chromatin opening function of the A2UCOE to the transgene region. These types of modifications are part of future long-term developments of this system.
The ability of the A2UCOE to largely negate position effects is further exemplified by the positive trend by this element to confer expression that is proportional to transgene copy number (Figure 5C; see Antoniou et al29 ). This property of the A2UCOE is akin to that possessed by LCRs, which are tissue-specific elements that participate in processes that establish a transcriptionally permissive chromatin structure, thereby protecting against position effects and repressive chromatin.35,38,39 Components of the human erythroid–specific HBB40,41 and T-cell specific CD242 LCRs have been incorporated into lentiviral vectors and have been found to partially overcome position effects resulting in more reproducible gene expression. However, the tissue-specific nature of LCRs and their general large size36 places significant restrictions on their utility, which has resulted in suboptimal functioning and only partial negation of insertion-site position effects.40,41 In addition, the potent enhancer-like properties possessed by LCR elements may cause insertional mutagenesis, compounded by the fact that LCRs have been shown to activate heterologous promoters (Greaves et al43 ; Collis et al44 ; M.A. and Grosveld45 ). The small size of the A2UCOE means that it is readily adaptable for incorporation into vectors with limiting capacity21,29 (Figure 1A) and, being derived from a housekeeping gene locus, it is capable of being active in all cell types (Figures 3-7; Williams et al21 ; M.A. et al29 ; and M.A., Edwards, Holdstock, Mountain and Crombie, unpublished results). Studies have also shown that even without enhancer activity, A2UCOE can be successfully employed in combination with heterologous promoters (Williams et al21 ; and Talbot, Waddington, and M.A., unpublished results). Therefore, apart from circumstances when exceptionally high levels of protein product are required to reach a therapeutic threshold (for example, HBB in thalassemia and sickle cell disease), which would necessitate the use of an LCR element,40,41 the versatility of the A2UCOE makes it highly attractive for generating standard vectors for a wide range of gene transfer applications.
The phosphoglycerate kinase (PGK1) and elongation factor 1α (EF1α) housekeeping gene promoters have also been used to regulate expression of various transgenes. However, based on our studies with A2UCOE-like dual divergently transcribed housekeeping gene promoters (Williams et al21 ; M.A. et al29 ), we would predict that CpG island–associated single promoters such as PGK1 and EF1α would be prone to insertion-site position effects and silencing, as has recently been found to be the case following in vivo transplantation of transduced cells.16,20,46 Flanking transgenes with insulator elements has also been attempted to minimize position effects and insertional mutagenesis.47,48 The insulator that has been most frequently employed within gammaretroviral and lentiviral vectors is the cHS4 element from the chicken β-globin (HBB_CHICK) LCR.48 This element has at best given only partial protection from position effects in a cell-type–dependent manner.48-50 Furthermore, no experimental data are as yet available to show whether 5′ and 3′ flanking insulators protect host gene promoters from activation by enhancer or LCR-type elements that may be regulating expression of the transgene. As insulators in lentiviral vectors have been found to provide only partial protection of the transgene from insertion-site position effects, these elements are unlikely to completely negate gene activation through insertional mutagenesis. The simple enhancer-less configuration of the A2UCOE and its ability to function as a single internal element avoids these potential problems.
Our data show that the A2UCOE is able to consistently drive therapeutic IL2RG cDNA expression, thereby restoring gene function efficiently both in vitro and in vivo, even at relatively low MOI and vector copy number and at an efficiency exceeding that obtained from the SFFV promoter in vitro and possibly in vivo. These findings are consistent with our observation that in marked contrast to the SFFV and especially the CMV promoters, the A2UCOE is not prone to transcriptional silencing and that the majority of vector integration events are stably productive, giving rise to an efficient rescue of IL2RG deficiency both in vitro (Figure 6) and in vivo (Figure 7; Table 3). Importantly, these data suggest that therapeutic efficacy may be achieved with a much lower vector dose using A2UCOE-regulated transgenes compared with conventional viral promoter/enhancer elements. This may have significant advantages in terms of mutagenic risk.
In summary, our data provide compelling evidence that the A2UCOE retains its dominant chromatin opening and transcriptional activating functions21,29 within a lentiviral vector context and is therefore able to largely overcome insertion-site position effects and give rise to reproducible and stable transgene expression. Furthermore, the enhancer-less nature of the A2UCOE implies that it should possess far lower insertional mutagenesis activation potential than commonly used enhancer-associated viral and nonviral promoters.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Assistance with the statistical analysis by Cathryn Lewis is gratefully acknowledged. Thanks to Luigi Naldini for the lentiviral packaging plasmids, which were produced by the Plasmid Factory. Thanks also to Ian Alexander for the gift of ED7R and ED7R-γc cell lines and Christopher Baum for the SINLV-SF-IL2RG vector.
This work was supported by the Wellcome Trust (A.J.T.), Medical Research Council United Kingdom (S.I.T.), the Department of Health (M.U., F.Z.), the Chronic Granulomatous Disorder Research Trust, and European Union Framework VI program CONSERT (LSHB-CT-2004-005242).
Wellcome Trust
Authorship
Contribution: F.Z., S.I.T., S.J.H., M.U., A.S., and J.S. conducted experimental work. H.B.G., C.K., M.A., and A.J.T. contributed to the design and supervision of the study. F.Z., M.A., and A.J.T. wrote the manuscript.
Conflict-of-interest disclosure: Author Michael Antoniou is an inventor on a patent for biotechnological application of UCOE. All other authors declare no competing financial interests.
Correspondence: Adrian J. Thrasher, Centre for Immunodeficiency, Molecular Immunology Unit, Institute of Child Health, University College London, 30 Guilford Street, London, WC1N 1EH, United Kingdom; e-mail: a.thrasher@ich.ucl.ac.uk, or Michael Antoniou, Nuclear Biology Group, Department of Medical and Molecular Genetics, Kings College London School of Medicine-Guy's Campus, 8th Floor Guy's Tower, Guy's Hospital, London, SE1 9RT, United Kingdom; e-mail: michael.antoniou@genetics.kcl.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal