Abstract
Chronic hepatitis C virus (HCV) infection is typically characterized by a lack of virus-specific CD4+ T-cell–proliferative responses, but strong responses have been described in a subset of persons with persistent viremia. One possible explanation for these responses is that they were primed by an earlier resolved infection and do not recognize the current circulating virus. We defined all targeted epitopes using overlapping peptides corresponding to a genotype 1a strain in 44 patients chronically infected with different HCV genotypes (GT). Surprisingly, more HCV-specific CD4+ T-cell responses were detected in patients with chronic non-GT1 infection compared with patients with chronic GT1 infection (P = .017). Notably, we found serologic evidence of a previous exposure to GT1 in 4 patients with non-GT1 infection, and these persons also demonstrated significantly more responses than non-GT1 patients in whom genotype and HCV serotype were identical (P < .001). Comparison of recognition of GT1-specific peptides to peptides representing autologous virus revealed the absence of cross-recognition of the autologous circulating virus. These data indicate that persisent HCV infection can occur in the presence of an HCV-specific T-cell response primed against a heterologous HCV strain, and suggest that clearance of 1 GT does not necessarily protect against subsequent exposure to a second GT.
Introduction
More than 170 million people are infected with the hepatitis C virus (HCV) worldwide. However, a significant proportion of acutely infected individuals are able to spontaneously clear the virus, indicating that acquisition of at least partial protective immunity against HCV persistence is possible.1-5 Moreover, spontaneous clearance of HCV is associated with a broad and durable HCV-specific proliferative CD4+ T-cell response.6-9 In contrast, HCV-specific CD4+ T-cell responses are rarely detected in chronically infected persons and are usually weak and narrow in breadth.9-11 The reasons for the diminished T helper response in chronic HCV infection are unclear, but multiple mechanisms have been suggested, including a failure to prime responses, an immunosuppressive effect of HCV proteins on T cells, and exhaustion of HCV-specific T cells by persistently high antigen levels.3,12-15 Elucidating the mechanisms of this diminished T helper response may be critical to the development of a therapeutic HCV vaccine
A total of 6 distinct HCV genotypes and multiple subtypes have been identified that demonstrate up to 50% cross genotype nucleotide divergence.16 Therefore, a major challenge in the development of an effective vaccine will be to render protective immunity to all of the different HCV genotypes.17 The degree of possible HCV cross-protective immunity against homologous or heterologous HCV strains has been addressed in a number of observational and experimental studies both in humans2,5,18-21 and chimpanzees.22-26 Some of these studies demonstrate a relative lack of protection against rechallenge with heterologous virus strains, indicating poor cross-genotype recognition.23
Multiple studies have shown that a subgroup of chronically infected patients, typically those infected with genotypes other than GT1, can display rather broad and vigorous T-helper cell responses targeting GT1-derived antigens.27,28 The role of these responses,27-29 and their contribution to viral control or the favorable outcome of antiviral therapy in certain genotypes,30 remain unclear. It is equally unclear whether the persistent CD4+ T-cell responses to HCV antigens in some chronically infected patients with non-GT1 infection is due to an altered pathobiology or immunogenicity of these HCV strains, and whether these responses actually target the current, persistent virus. Recent reports show serologic and cellular evidence of prior exposure to heterologous HCV strains in some chronically infected patients31,32 with vigorous CD4+ T-cell–proliferative responses against HCV GT1 proteins. Therefore, the detection of stronger proliferative responses against GT1a antigens observed in patients with chronic non-GT1 infection may indicate a previous exposure and clearance of GT1 infection in these persons.33 It has been suggested that these responses are not cross-reactive to the autologous persisting virus, but data supporting this hypothesis are limited.
In this study, we analyzed HCV-specific CD4+ T-cell responses at the single epitope level in a large cohort of patients chronically infected with different HCV genotypes, and examined these individuals for serologic evidence of previous infection with heterologous HCV genotypes. We also determined the ability of the HCV-specific CD4+ T-cell responses to recognize circulating autologous virus. The cellular, serologic, and HCV sequence data presented here suggest that the persistent HCV-specific CD4+ T-cell responses detected in a subset of chronically infected patients are unable to recognize the contemporary circulating viruses, but most likely represent immunologic scars from a previously resolved HCV infection. The data also highlight that HCV viremia per se is not immunosuppressive for HCV-specific CD4+ T cells,32 but rather that chronic viremia leads to selective elimination of adaptive CD4 responses to the epitopes presented in vivo.
Patients, materials, and methods
Study patients
This study was approved by the Institutional Review Board of the Massachusetts General Hospital or the Instituto Oswaldo Cruz/Fiocruz, Rio de Janeiro, Brazil. Informed consent was obtained in accordance with the Declaration of Helsinki from all patients upon enrollment. All patients, with the exception of C38 and C39, who were recruited in Brazil, were recruited in Boston, were naive for anti-HCV treatment, and had been confirmed anti-HCV+ by third-generation enzyme immunoassay. HCV RNA was measured by the Roche Amplicor Monitor assay (detection limit of 600 IU of HCV RNA per milliliter of plasma; Roche, Indianapolis, IN). All patients were HCV RNA+ for at least 24 months. HCV RNA+ patients were tested for HCV genotype by INNOLIPA (Innogenetics, Ghent, Belgium). In addition, a 1.2-kb fragment in NS5B of the HCV genome was polymerase-chain reaction (PCR) amplified, sequenced, and then compared with genotype reference sequences from the Los Alamos HCV database34 by phylogenetic analysis using the neighbor joining method. A total of 22 patients with spontaneously resolved HCV infection (previously published in detail10 ) and 10 unexposed, seronegative participants served as control groups.
HCV serotyping
HCV serotypes were determined using the Murex HCV Serotyping 1-6 assay (Abbott, Wiesbaden, Germany) according to the manufacturer's instructions and as previously published.10,31,32 Briefly, immunoplates were coated with synthetic peptides representing variable regions within NS4 from HCV types 1-6. Serum dilutions were then added in the presence of competing peptide in free solution to block cross-reactivity.
HCV peptides and recombinant HCV proteins
Peptides were synthesized corresponding to the amino acid sequences of the HCV-1a strain HCV-1 (GenBank accession number M62321). A total of 301 20mers overlapping by 10 amino acids (aa) and spanning the entire polyprotein were used for screening responses.11 Additional peptides were synthesized according to autologous viral sequences. Recombinant HCV proteins used in this study were expressed as carboxy-terminal fusion proteins with human superoxide dismutase in Saccharomyces cerevisiae or Escherichia coli and were kindly provided by Michael Houghton (Chiron, Emeryville, CA). These proteins were derived from the HCV-1 sequence and encoded core (C22-3 aa2 to 120), NS3 (C33C aa1192 to 1457), NS4 (C100.3 aa1569 to 1931), NS3/NS4 (C200 aa1192 to 1931), and NS5 (NS5 aa2054 to 2995).
Sequencing of autologous virus
Viral RNA was extracted from plasma using the vRNA extraction kit (QIAGEN, Santa Clarita, CA). Genotype-specific primers were designed based on alignments of all available sequences from public HCV databases (European HCV database,35 HCV Database Project36 ). Using the QIAGEN One-Step RT-PCR kit, reverse transcription (RT)–PCR cycling conditions were as follows: 50°C for 60 minutes and 95°C for 15 minutes, followed by 35 cycles of 30 seconds at 94°C, 30 seconds at 54°C, 1.5 minutes at 72°C, and a final extension of 68°C for 20 minutes. Nested PCR conditions were 35 cycles of 30 seconds at 94°C, 30 seconds at 62°C, 1 minute at 72°C, and a final extension of 68°C for 20 minutes using Titanium Taq DNA polymerase (CLONTECH Laboratories, Palo Alto, CA) as previously described.37 PCR fragments were gel- or PCR-purified (QIAGEN QIAquick Gel extraction/PCR purification kit), and the population was sequenced bidirectionally on an ABI 3100 PRISM automated sequencer (Applied Biosystems, Foster City, CA). Sequencher (Gene Codes, Ann Arbor, MI) and MacVector 4.1 (Oxford Molecular, Hunt Valley, MD) software programs were used to edit and align sequences.
HLA typing
DNA for HLA typing was extracted according to the manufacturer's instructions using the Puregene DNA Isolation Kit for blood (Gentra Systems, Minneapolis, MN). HLA class II typing was performed at the Massachusetts General Hospital (MGH) Tissue Typing Laboratory and by Dynal Biotech HLA Diagnostics (Bromborough, United Kingdom) using sequence-specific primer PCR.38,39
Lymphocyte proliferation assays
Lymphocyte proliferation assays were performed as described previously using c22.3, c33c, c100.3, C200, and NS5 HCV proteins (Chiron) at concentrations of 10 μg/mL.11
Bulk stimulation of PBMCs
A total of 20 million fresh or thawed peripheral blood mononuclear cells (PBMCs) were depleted of CD8+ cells with magnetic beads according to the manufacturer's instructions (111-47D CD8 Dynal beads; Invitrogen, Carlsbad, CA). The CD8+-depleted cells were stimulated with 1 μg/mL recombinant HCV antigens (either C22-3, C33C, C100.3, C200, or NS5) in 2 mL R10/50 medium (RPMI 1640 medium, 10 mM HEPES with 2 mM glutamine and antibiotics [penicillin-streptomycin, 50 U/ml] and 10% fetal calf serum [Sigma Aldrich, St Louis, MO] supplemented with recombinant IL-2 [rIL-2; 50 U/mL]).11 Alternatively, for the regions of the HCV genome for which recombinant antigens were not available (E1, E2, p7, and NS2), PBMCs were stimulated with overlapping 20mer peptides (1 μg/mL each) in pools of 10 peptides.11 After 11 to 13 days in culture, cells were assayed for IFN-γ production by enzyme-linked immunospot assay (ELISPOT) and intracellular cytokine staining in response to stimulation with HCV peptides.
ELISPOT assay
ELISPOT assays were performed as previously described.10,40 Responses were considered positive if the number of spots was 3 times greater than the background; phytohemagglutinin served as a positive control. All positive responses were reconfirmed by intracellular cytokine assay following stimulation with the specific peptide.
Intracellular IFN-γ staining by flow cytometry
Intracellular cytokine staining (ICS) was performed as previously described.10,40 Cells were analyzed on a FACS-Calibur flow cytometer using CELL Quest software (Becton Dickinson, San Jose, CA). Single peptides that elicited distinct responses of more than 3 times background IFN-γ production were considered positive.
Statistical analysis
Nonparametric Mann-Whitney U tests were performed using GraphPad Prism 4.0 software (GraphPad Software, San Diego, CA). To avoid overestimation of the total breadth of antiviral responses, responses to adjacent overlapping peptides were counted as responses to one epitopic region, with only the stronger response shown.
Results
CD4+ T-cell lines from chronically infected patients with non-GT1 infection show broadly directed responses against GT1a peptides
A total of 44 treatment-naive patients, 24 with chronic HCV GT1 infection and 20 with chronic HCV non-GT1 infection (Table 1), were recruited, and HCV-specific CD4+ T-cell responses were determined at the single-epitope level. There were no significant differences between patients with GT1 and without GT1 with respect to risk factor for the acquisition of infection (> 50% intravenous drug use [IVDU]) or other clinical parameters such as viral HCV RNA titers (Table 1).
Demographics and virologic and immunologic characteristics of the cohort
| Patient no. . | Age, y/sex . | Genotype . | HCV RNA, IU/mL . | CD4+ responses in short-term culture . | Risk factor . | Murex HCV serotype . | HCV proliferative response (SI >5) . |
|---|---|---|---|---|---|---|---|
| GT1 C1 | 48F | 1b | 300 000 | 1 | IDU | 1 | — |
| GT1 C2 | 43F | 1b | > 500 000 | 0 | IDU | 1 | — |
| GT1 C3 | 22F | 1a | 2 240 | 0 | IDU | 1 | — |
| GT1 C4 | 29F | 1a | > 500 000 | 0 | Unknown | ND | — |
| GT1 C5 | 31F | 1a | > 500 000 | 3 | IDU | 1 | — |
| GT1 C6 | 38M | 1a/1b | 450 000 | 0 | Transfusion | 1 | — |
| GT1 C7 | 40M | 1b | > 500 000 | 0 | Transfusion | 1 | — |
| GT1 C8 | 37M | 1a/1b | 420 000 | 0 | Transfusion | 1 | — |
| GT1 C9 | 59F | 1a | 39 600 | 0 | Transfusion | 1 | — |
| GT1 C10 | 47M | 1 | 100 000 | 0 | IDU | 1 | ND |
| GT1 C11 | 25M | 1 | > 500 000 | 1 | IDU | 1 | — |
| GT1 C12 | 64F | 1a | > 500 000 | 0 | Transfusion | 1 | — |
| GT1 C13 | 53M | 1 | > 500 000 | 0 | IDU | 1 | ND |
| GT1 C14 | 66F | 1 | 410 000 | 0 | Unknown | ND | ND |
| GT1 C15 | 21F | 1b | 160 000 | 0 | Transfusion | ND | — |
| GT1 C16 | 68M | 1a | > 500 000 | 0 | Transfusion | 1 | — |
| GT1 C17 | 37F | 1a | 330 000 | 0 | IDU | 1 | — |
| GT1 C18 | 49M | 1a | 420 000 | 0 | IDU | ND | — |
| GT1 C19 | 43M | 1a | > 500 000 | 0 | Multiple risk factors | 1 | — |
| GT1 C20 | 25M | 1a | > 500 000 | 0 | Unknown | ND | ND |
| GT1 C21 | 21M | 1a/1b | > 500 000 | 0 | IDU | 1 | ND |
| GT1 C22 | 56M | 1a | > 500 000 | 0 | IDU | 1 | ND |
| GT1 C23 | 75M | 1a | 474 000 | 0 | Transfusion | ND | — |
| GT1 C24 | 38M | 1a | > 500 000 | 0 | IDU | 1 | — |
| Non-GT1 C25 | 41F | 2a/2c | 38 600 | 2 | Needle stick | 2 | — |
| Non-GT1 C26 | 37F | 3a | 87 000 | 0 | IDU | ND | — |
| Non-GT1 C27 | 39M | 3a | > 500 000 | 0 | IDU | 3 | — |
| Non-GT1 C28 | 38M | 4c | 120 000 | 0 | Unknown | 4 | NS5 |
| Non-GT1 C29 | 45M | 4c | 230 000 | 5 | IDU | 1+4 | NS5 |
| Non-GT1 C30 | 47F | 4a/4b | > 500 000 | 1 | Unknown | 4 | NS4 |
| Non-GT1 C31 | 45F | 2 | 750 | 9 | Transfusion | 2 | — |
| Non-GT1 C32 | 49M | 2 | > 500 000 | 0 | IDU | 2 | — |
| Non-GT1 C33 | 27M | 3a | 100 000 | 0 | IDU | 3 | — |
| Non-GT1 C34 | 34M | 6a | 500 000 | 0 | Unknown | 6 | ND |
| Non-GT1 C35 | 50F | 2 | > 500 000 | 27 | IDU | 1+2 | NS4 + NS5 |
| Non-GT1 C36 | 52M | 3a | 57 000 | 12 | Unknown | 1+3 | — |
| Non-GT1 C37 | 47M | 3a | > 500 000 | 0 | IDU | 3 | — |
| Non-GT1 C38 | 43M | 3 | > 500 000 | 0 | Nosokomial | 3 | — |
| Non-GT1 C39 | 37M | 4 | > 500 000 | 0 | Sexual transmission | 4 | — |
| Non-GT1 C40 | 56F | 4a | 310 000 | 1 | IDU | 4 | — |
| Non-GT1 C41 | 57F | 3 | 396 000 | 16 | IDU | 2 | — |
| Non-GT1 C42 | 55M | 3a | 96 000 | 9 | IDU | 1 | NS4 + NS5 |
| Non-GT1 C43 | 50M | 2 | 500 000 | 6 | IDU | 2 | — |
| Non-GT1 C44 | 50M | 3a | 500 000 | 0 | IDU | 3 | NS4 |
| Patient no. . | Age, y/sex . | Genotype . | HCV RNA, IU/mL . | CD4+ responses in short-term culture . | Risk factor . | Murex HCV serotype . | HCV proliferative response (SI >5) . |
|---|---|---|---|---|---|---|---|
| GT1 C1 | 48F | 1b | 300 000 | 1 | IDU | 1 | — |
| GT1 C2 | 43F | 1b | > 500 000 | 0 | IDU | 1 | — |
| GT1 C3 | 22F | 1a | 2 240 | 0 | IDU | 1 | — |
| GT1 C4 | 29F | 1a | > 500 000 | 0 | Unknown | ND | — |
| GT1 C5 | 31F | 1a | > 500 000 | 3 | IDU | 1 | — |
| GT1 C6 | 38M | 1a/1b | 450 000 | 0 | Transfusion | 1 | — |
| GT1 C7 | 40M | 1b | > 500 000 | 0 | Transfusion | 1 | — |
| GT1 C8 | 37M | 1a/1b | 420 000 | 0 | Transfusion | 1 | — |
| GT1 C9 | 59F | 1a | 39 600 | 0 | Transfusion | 1 | — |
| GT1 C10 | 47M | 1 | 100 000 | 0 | IDU | 1 | ND |
| GT1 C11 | 25M | 1 | > 500 000 | 1 | IDU | 1 | — |
| GT1 C12 | 64F | 1a | > 500 000 | 0 | Transfusion | 1 | — |
| GT1 C13 | 53M | 1 | > 500 000 | 0 | IDU | 1 | ND |
| GT1 C14 | 66F | 1 | 410 000 | 0 | Unknown | ND | ND |
| GT1 C15 | 21F | 1b | 160 000 | 0 | Transfusion | ND | — |
| GT1 C16 | 68M | 1a | > 500 000 | 0 | Transfusion | 1 | — |
| GT1 C17 | 37F | 1a | 330 000 | 0 | IDU | 1 | — |
| GT1 C18 | 49M | 1a | 420 000 | 0 | IDU | ND | — |
| GT1 C19 | 43M | 1a | > 500 000 | 0 | Multiple risk factors | 1 | — |
| GT1 C20 | 25M | 1a | > 500 000 | 0 | Unknown | ND | ND |
| GT1 C21 | 21M | 1a/1b | > 500 000 | 0 | IDU | 1 | ND |
| GT1 C22 | 56M | 1a | > 500 000 | 0 | IDU | 1 | ND |
| GT1 C23 | 75M | 1a | 474 000 | 0 | Transfusion | ND | — |
| GT1 C24 | 38M | 1a | > 500 000 | 0 | IDU | 1 | — |
| Non-GT1 C25 | 41F | 2a/2c | 38 600 | 2 | Needle stick | 2 | — |
| Non-GT1 C26 | 37F | 3a | 87 000 | 0 | IDU | ND | — |
| Non-GT1 C27 | 39M | 3a | > 500 000 | 0 | IDU | 3 | — |
| Non-GT1 C28 | 38M | 4c | 120 000 | 0 | Unknown | 4 | NS5 |
| Non-GT1 C29 | 45M | 4c | 230 000 | 5 | IDU | 1+4 | NS5 |
| Non-GT1 C30 | 47F | 4a/4b | > 500 000 | 1 | Unknown | 4 | NS4 |
| Non-GT1 C31 | 45F | 2 | 750 | 9 | Transfusion | 2 | — |
| Non-GT1 C32 | 49M | 2 | > 500 000 | 0 | IDU | 2 | — |
| Non-GT1 C33 | 27M | 3a | 100 000 | 0 | IDU | 3 | — |
| Non-GT1 C34 | 34M | 6a | 500 000 | 0 | Unknown | 6 | ND |
| Non-GT1 C35 | 50F | 2 | > 500 000 | 27 | IDU | 1+2 | NS4 + NS5 |
| Non-GT1 C36 | 52M | 3a | 57 000 | 12 | Unknown | 1+3 | — |
| Non-GT1 C37 | 47M | 3a | > 500 000 | 0 | IDU | 3 | — |
| Non-GT1 C38 | 43M | 3 | > 500 000 | 0 | Nosokomial | 3 | — |
| Non-GT1 C39 | 37M | 4 | > 500 000 | 0 | Sexual transmission | 4 | — |
| Non-GT1 C40 | 56F | 4a | 310 000 | 1 | IDU | 4 | — |
| Non-GT1 C41 | 57F | 3 | 396 000 | 16 | IDU | 2 | — |
| Non-GT1 C42 | 55M | 3a | 96 000 | 9 | IDU | 1 | NS4 + NS5 |
| Non-GT1 C43 | 50M | 2 | 500 000 | 6 | IDU | 2 | — |
| Non-GT1 C44 | 50M | 3a | 500 000 | 0 | IDU | 3 | NS4 |
SI indicates stimulation index; —, no proliferation; and ND, not determined.
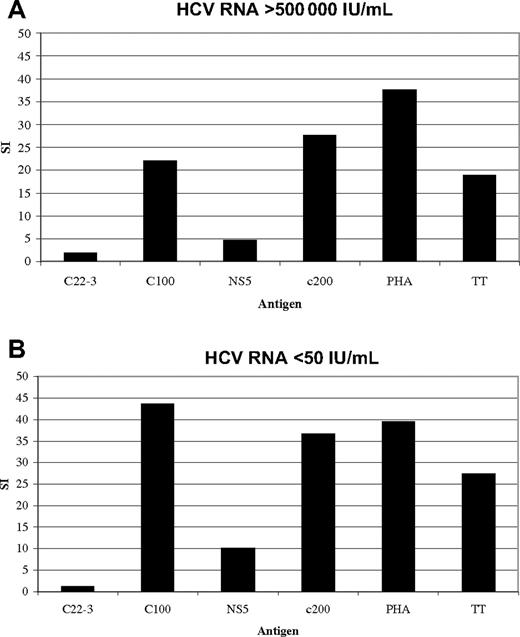
The HCV-specific lymphocyte proliferative responses against the HCV proteins were absent in most of the 37 patients with chronic infection (18 GT1/19 non-GT1) for whom lymphoproliferative assays with fresh PBMCs could be performed for each of the recombinant HCV proteins. Proliferative responses were more readily detected in persons with non-GT1 infection. None of the 18 patients with GT1 infection and 6 of 19 of the non-GT1 patients displayed significant proliferative responses (stimulation index [SI] higher than 5) to 1 or more of the nonstructural proteins (P < .03; Table 1). Our findings are in accordance with lymphoproliferative results published by other groups.27,28,32 A representative plot of a patient with chronic non-GT1 infection with strong proliferative HCV-specific responses is shown in Figure 1A. The responses in patient C35 were stable over 1 year of follow-up despite persistent GT2b viremia greater than 500 000 IU of HCV RNA per milliliter of plasma. In addition, there was no significant alteration in the response at later time points following a sustained virologic response after standard 24-week therapy with PEG IFN-α and ribavirin (Figure 1B).
Strong lymphoproliferative responses to recombinant HCV antigens are detectable in patient C35 despite high GT2b viremia. Lymphocyte proliferation assays (LPAs) with recombinant HCV antigens based on GT1 sequence: core (c22.3), NS4 (c100.3), NS3 + NS4 (c200), NS5, and tetanus toxoid (TT) and PHA as control were performed on fresh PBMCs of patient C35 (“Patients, materials, and methods”) and demonstrate (A) strong responses to multiple antigens despite high untreated viremia. These responses stayed stable over time (data not shown), and did not change (B) after successful antiviral treatment (sustained virologic response) with Peg-interferon and ribavirin for 24 weeks. Patient C35 showed serologic evidence of prior exposure to GT1 virus. Serotyping was performed with the Murex HCV serotyping 1-6 assay (Abbott; “Patients, materials, and methods”).
Strong lymphoproliferative responses to recombinant HCV antigens are detectable in patient C35 despite high GT2b viremia. Lymphocyte proliferation assays (LPAs) with recombinant HCV antigens based on GT1 sequence: core (c22.3), NS4 (c100.3), NS3 + NS4 (c200), NS5, and tetanus toxoid (TT) and PHA as control were performed on fresh PBMCs of patient C35 (“Patients, materials, and methods”) and demonstrate (A) strong responses to multiple antigens despite high untreated viremia. These responses stayed stable over time (data not shown), and did not change (B) after successful antiviral treatment (sustained virologic response) with Peg-interferon and ribavirin for 24 weeks. Patient C35 showed serologic evidence of prior exposure to GT1 virus. Serotyping was performed with the Murex HCV serotyping 1-6 assay (Abbott; “Patients, materials, and methods”).
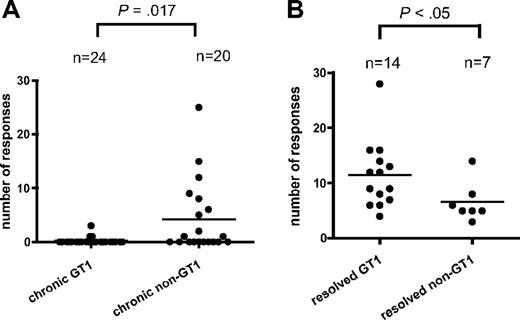
In order to characterize all of the epitopes targeted, a high-resolution screening for HCV-specific T-helper responses was conducted in all of the 44 patients with chronic infection by antigen-specific polyclonal expansion of CD4+ T cells in the presence of rIL-2.10,11 These lines were tested using IFN-γ ELISPOT with pools of HCV GT-1 peptides spanning the entire HCV polyprotein. Responses to single 20mer peptides were then confirmed by intracellular cytokine staining for IFN-γ production. Only in 3 of 24 patients with GT1 infection we could detect 1 or more CD4+ responses compared with 10 of 20 of the patients with non-GT1 infection (P < .01), despite the fact that we conducted these screens using GT1a peptides. Altogether, we detected significantly more HCV CD4+ T-cell responses in patients with chronic non-GT1 infection compared with patients with GT1 infection (patients with non-GT1 infection: mean, 4.2 responses; range, 0-25 responses; patients with GT1 infection: mean, 0.2 responses; range, 0-3 responses; P = .017; Figure 2A; Table 1). A maximum of 3 responses were identified in patients with chronic GT1 infection, whereas up to 25 responses were detected in 1 of the patients with non-GT1 infection (patient C35; Table 1). Overall, we detected 89 responses against 67 different peptide specificities/epitopes in 13 patients of our cohort of 44 chronically infected patients. These responses were broadly distributed across all of the HCV structural and nonstructural proteins (Figure 3). Most of these CD4+ specificities have previously been described in patients with spontaneously resolved infection and, therefore, are not unique to chronic HCV infection. These responses were stably maintained in several patients that were studied sequentially for up to 6 years of chronic untreated infection (data not shown).
Comparison of the breath of the HCV-specific CD4+ T-cell response in patients with chronic GT1 or non-GT1 infection or spontaneously resolved HCV. (A) PBMCs from 44 patients with chronic HCV infection were CD8+ depleted and stimulated with recombinant HCV proteins (or overlapping peptides) covering the entire polyprotein in the presence of recombinant IL-2. Expanded cell lines were tested for IFN-γ production upon stimulation with pools of 10 to 20 overlapping peptides by ELISPOT. All peptide pools eliciting a positive response by IFN-γ production were further deconvoluted to identify specific 20mer peptides targeted by the CD4+ T-cell response. Each single response detected in the ELISPOT assay was confirmed by ICS for IFN-γ production as well. Horizontal bars indicate the mean numbers of targeted HCV CD4+ T-cell responses against single peptides for each patient as tested by ICS. The number of responses is significantly higher (P = .017) in chronically infected patients with non-GT1 virus than in patients infected with GT1 virus when comparing the number of CD4+ T-cell responses detected with GT1-specific peptides in short-term lines across the entire polyprotein. (B) Genotype-specific subanalysis of previously published data10 reveals that within the group of patients with spontaneously resolved HCV, more responses are detected with GT1 peptide set in patients who are serologically GT1 than patients who are tested non-GT1 (P < .05). Serotyping was performed with the Murex HCV serotyping 1-6 assay (Abbott; “Patients, materials, and methods”).
Comparison of the breath of the HCV-specific CD4+ T-cell response in patients with chronic GT1 or non-GT1 infection or spontaneously resolved HCV. (A) PBMCs from 44 patients with chronic HCV infection were CD8+ depleted and stimulated with recombinant HCV proteins (or overlapping peptides) covering the entire polyprotein in the presence of recombinant IL-2. Expanded cell lines were tested for IFN-γ production upon stimulation with pools of 10 to 20 overlapping peptides by ELISPOT. All peptide pools eliciting a positive response by IFN-γ production were further deconvoluted to identify specific 20mer peptides targeted by the CD4+ T-cell response. Each single response detected in the ELISPOT assay was confirmed by ICS for IFN-γ production as well. Horizontal bars indicate the mean numbers of targeted HCV CD4+ T-cell responses against single peptides for each patient as tested by ICS. The number of responses is significantly higher (P = .017) in chronically infected patients with non-GT1 virus than in patients infected with GT1 virus when comparing the number of CD4+ T-cell responses detected with GT1-specific peptides in short-term lines across the entire polyprotein. (B) Genotype-specific subanalysis of previously published data10 reveals that within the group of patients with spontaneously resolved HCV, more responses are detected with GT1 peptide set in patients who are serologically GT1 than patients who are tested non-GT1 (P < .05). Serotyping was performed with the Murex HCV serotyping 1-6 assay (Abbott; “Patients, materials, and methods”).
Summary and location of all the specificities recognized by each individual with chronic infection. Shaded boxes indicate a positive CD4+ response. Peptides: p1 = aa11-30, p298 = aa2981-3000.
Summary and location of all the specificities recognized by each individual with chronic infection. Shaded boxes indicate a positive CD4+ response. Peptides: p1 = aa11-30, p298 = aa2981-3000.
Although the HCV-specific CD4+ T-cell responses detected in this cohort with chronic HCV infection have also been described in patients with resolved infection, we noticed the relative paucity of responses to a group of previously described, dominant, promiscuous, and highly conserved HCV-specific epitopes characteristic of patients with spontaneously resolved HCV.10,41,42 For example, peptide P177 (aa1771-1790), which is targeted by most patients with spontaneously resolved HCV infection,10,41,43 was not detected in a single patients with chronic infection regardless of the genotype. While these dominant epitopes in resolved infection are typically located in relatively conserved areas of the HCV genome, many of the responses detected in the patients with chronic non-GT1 infection were located in regions of high sequence variability among the different genotypes. For example, peptides P21 (aa211-230) and P26 (aa261-280), both located in the extremely variable region of E1, were each detected in 3 patients with chronic non-GT1 infection (Figure 3).
There is significant disparity between autologous virus sequence and GT1a-based peptides
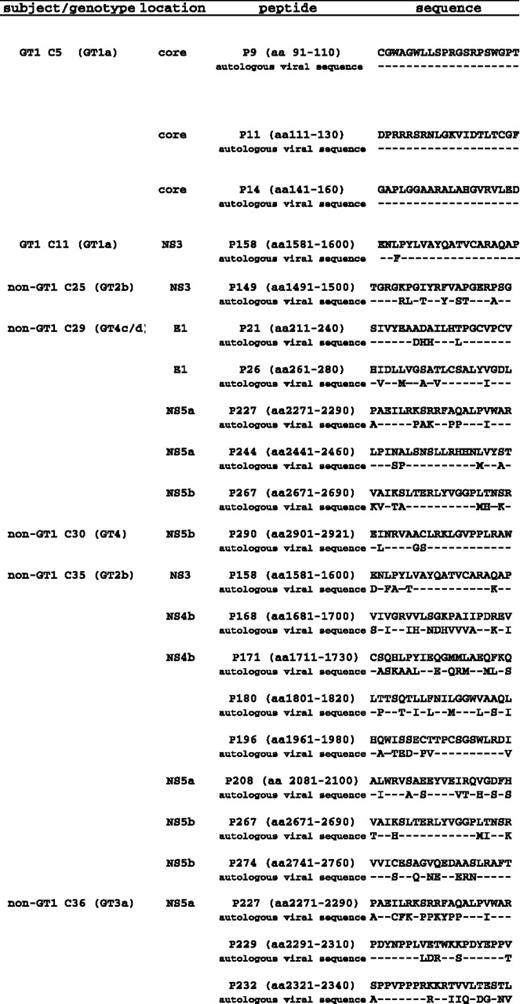
Persistent HCV infection is typically associated with a limited or absent HCV-specific CD4+ response. It was therefore surprising that in a subset of patients with chronic non-GT1 HCV infection we were able to detect up to 25 HCV CD4+ specificities despite high levels of viremia. Since the number of HCV CD4+ specificities detected in short-term cell lines did not correlate with lower viral load in the group of patients with non-GT1 infection (Table 1; chi-square χ = .05), we probed whether these responses targeted the circulating virus sequence as an indication of their ability to contribute to viral control. To investigate this relationship, we generated autologous sequence data by bulk sequencing and compared the results with the peptide sequence of GT1a peptides that had elicited responses in these patients. In the group of 24 patients with chronic GT1 infection, a total of only 5 different HCV-specific CD4+ T-cell responses were detected. Autologous sequence was available for 4 of 5 of these responses, and this sequence did not differ signifcantly from the reference GT1a peptide sequence (Figure 4). For 7 patients in the group with chronic GT1a infection, autologous sequences for the entire polyprotein were available. This enabled us to compare not only autologous virus in regions where we detected responses, but also to take a detailed look at regions that are commonly targeted in spontaneously resolved HCV infection that were not targeted in the GT1 patients in our cohort. Analysis of autologous sequences in these patients showed nearly complete conservation in the regions of the 3 most frequently detected epitopes in spontanously resolved infection,10 P124(aa1241-1260), P177(aa1771-1790), and P257(aa2571-2590) (Figure 5). None of the 7 patients showed evidence of escape mutations in these 3 epitopes at this chronic stage of infection.
Comparison of GT1a peptide sequence with autologous sequences from chronic subjects.
Comparison of GT1a peptide sequence with autologous sequences from chronic subjects.
Autologous sequence of GT1a-infected chronic patients in the region of three frequently detected epitopes in resolved HCV infection.
Autologous sequence of GT1a-infected chronic patients in the region of three frequently detected epitopes in resolved HCV infection.
In contrast to the 5 CD4+ responses detected in the group of patients with GT1 infection, we detected a total of 84 responses in our 20 patients with chronic non-GT1 infection. For 5 patients with non-GT1 infection, autologous sequence was generated using genotype-specific primers (GT2, GT3, or GT4, respectively). Figure 4 displays the sequences of all the epitopes for which autologous sequence was available. These sequences are aligned to the GT1a peptide sequence that was used to screen these individuals. In each instance, the GT1 sequences targeted by these CD4+ responses differed substantially from the autologous viral sequence. Importantly, these sequences were representative of the identified currently circulating genotype, and only minor sequence variations were detected when comparing the data with reference strains available in the Los Alamos database.34
In order to rule out possible mixed-genotype infections in the patients with non-GT1 infection, we attempted to amplify virus in the NS5 region using GT1-specific primers in 2 patients, C29 and C35. No amplification was noted in either of these patients, suggesting the absence of a possible mixed infection.
Broad HCV-specific CD4+ responses are detectable in patients with non-GT1 infection with serologic evidence of prior exposure to GT1 virus
In this study, we detected a greater number of GT1-specific responses in patients with chronic non-GT1 infection, which is a departure from our previous observations in patients with resolved HCV infection.10 In those studies, an average of approximately 10 epitopes targeted by CD4+ T cells were detectable in short-term cell lines isolated from patients with resolved HCV infection.10 Genotype-specific subanalysis of the data from that study10 showed that slightly fewer responses (8 vs 14; P < .05) were detected in the patients who had spontaneously resolved a non-GT1 HCV infection as compared with patients with serologic indication for clearance of a GT1 virus (Figure 2B). Since our screening is performed with GT1 peptides, these data suggest that in patients with resolved infection, more responses are missed in patients with resolved non-GT1 infection, likely due to the greater degree of HCV sequence variation between the autologous sequence and the prototype sequence of our reagents. However, in the patients with chronic infection studied here, we found the opposite to be the case; when HCV-specific CD4+ T-cell responses were detected, they were more likely to be found in patients with chronic non-GT1 virus infection.
The above data suggest that these patients with chronic infection, who are at high risk for repeated HCV exposure due to injection drug use, might have residual responses to a previously cleared infection with a hetreogenous genotype virus. Therefore, in order to determine whether a prior exposure to (and clearance of) GT1 HCV might explain the detection of durable CD4+ T-cell responses in patients with chronic non-GT1 infection, we tested for serologic evidence of prior exposure to different HCV genotypes by typing sera of the entire cohort with a commercially available enzyme-linked immunosorbent assay (ELISA; Murex, Cambridge, United Kingdom) for genotype 1-6–specific HCV antibodies31,32 (Table 1).
HCV serotypes were successfully defined in 39 of the 44 patients in our cohort. In agreement with recent results observed by other groups,32 we found serological evidence of exposure to GT1 in 4 of 19 patients with non-GT1 infection, but there was no evidence of previous exposure to a non-GT1 virus in the 19 patients with GT1 infection.
Serological detection of GT1 in the patients with non-GT1 infection significantly correlated (P < .001) with the number of CD4+ responses detected in our comprehensive expansion experiments. These patient had on average 11 responses (range, 5-25 responses), which was comparable with the number of responses detected in patients with spontaneously resolved GT1 infection (Figure 6). In addition, patient C41 (chronically infected with HCV GT3) had serologic evidence of a previous exposure to GT2. After exclusion of patients in whom we had detected serologic evidence of exposure to heterologous strains, analysis revealed that responses in the remaining 14 patients were not significantly greater than in patients with chronic GT1 infection (P = .1).
Patients with chronic non-GT1 infection who have serologic evidence of previous exposure to GT1 show significantly more responses than patients with non-GT1 infection in whom genotype and serotype correspond. Horizontal bars indicate the mean numbers of targeted HCV CD4+ T-cell responses against single peptides for each patient as tested by ICS. The number of responses is significantly higher (P < .001) in chronically infected patients with non-GT1 virus that serologically showed evidence for exposure to GT1 than patients chronically infected with a non-GT1 virus in whom genotype and serotype corresponded non-GT1. Serotyping test was performed with the Murex HCV serotyping 1-6 assay (Abbott; “Patients, materials, and methods”).
Patients with chronic non-GT1 infection who have serologic evidence of previous exposure to GT1 show significantly more responses than patients with non-GT1 infection in whom genotype and serotype correspond. Horizontal bars indicate the mean numbers of targeted HCV CD4+ T-cell responses against single peptides for each patient as tested by ICS. The number of responses is significantly higher (P < .001) in chronically infected patients with non-GT1 virus that serologically showed evidence for exposure to GT1 than patients chronically infected with a non-GT1 virus in whom genotype and serotype corresponded non-GT1. Serotyping test was performed with the Murex HCV serotyping 1-6 assay (Abbott; “Patients, materials, and methods”).
Further analysis of the pattern of cellular responses in the 4 patients with non-GT1 infection who had serologic evidence of previous exposure to GT1 virus revealed that while these patients mounted similar numbers of CD4+ responses as did patients with spontaneously resolved GT1 infection, there was a significant difference in the epitope repertoire. Previously, we described a group of 6 CD4+ epitopes that are highly conserved between the different HCV genotypes and were frequently targeted by patients with spontaneously resolved HCV infection.10 In contrast, those conserved epitopes were much less frequently targeted by patients with persistent non-GT1 infection which had serologic evidence for exposure to GT1. On average, more than 2 of these 6 epitopes were detectable in each patient of a previously published cohort of 22 patients with spontaneously resolved HCV (54 responses in total). In contrast, we detected an average of less than 1 of these epitopes (3 in total) in the 4 patients with non-GT1 infection mentioned previously (2.5 vs 0.8; P < .001). These data indicate that only CD4+ responses with a recognition pattern limited to GT1 infection, but not those responses that showed cross-reactivity with other genotypes (including the persistent autologous virus), were maintained in these 4 patients.
Memory HCV GT1-specific CD4+ T-cell responses are not cross-reactive with circulating non-GT1 virus
In order to elucidate whether the HCV-specific CD4+ responses that we detected using recombinant HCV proteins and peptides based on GT1a sequence would recognize autologous circulating virus, we generated peptides based on autologous sequence in 3 patients with non-GT1 infection (C29, C30, and C35) and compared the cellular responses to these peptides.
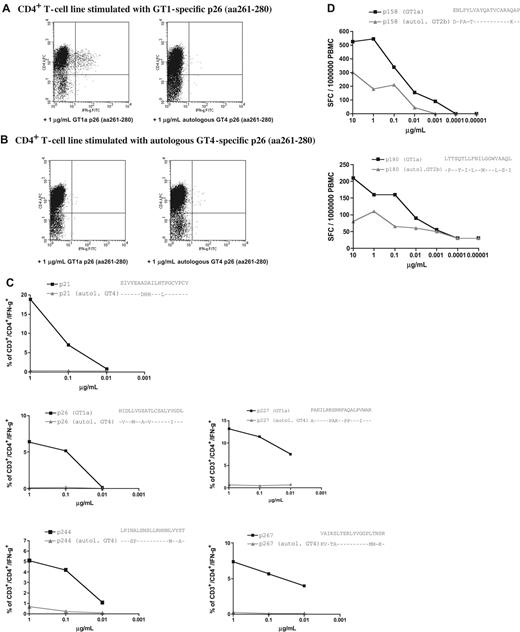
In patient C29, a 49-year-old man and former IVD user with chronic GT4 infection and serologic evidence of exposure to a GT1 strain, we compared the peptide sequence of all 5 of the different GT1a peptides that had reproducibly elicited strong responses at different time points in short-term lines (Figure 3). The GT1a peptides, but not peptides representing autologous viral sequences, lead to expansion of peptide-specific T cells in vitro (Figure 7A,B). In addition, none of the 5 T-cell lines that were established with the GT1-specific peptides cross-reacted with peptides representing the circulating autologous virus in an ICS assay (Figure 7C). Similar results were obtained for patient C35, who was infected with GT2b but in whom we also detected a serologic response to GT1. In ex vivo ELISPOT experiments, we observed strong ex vivo CD4+ responses against GT1a peptide P158 (aa1581-1600) and P180 (aa1881-1900) in the PBMCs of this patient, and we confirmed that these GT1-based peptides elicited a much stronger response than peptides based on autologous GT2b sequence (Figure 7D).
HCV GT1a-based peptides, but not peptides representing autologous sequence, are recognized in patients infected with non-GT1 strains. Strong and durable responses against 5 different GT1a-based HCV peptides were detected in patient C29 (Figure 3). This patient was infected with GT4 virus and showed a serologic response to GT1 virus. We were able to cultivate peptide-specific lines (A) by stimulating with GT1-based peptides, but not with autologous peptides (B) (“Patients, materials, and methods”). (C) dilution ICS assays performed on GT1-specific CD4+ T-cell lines for all specificities confirm the lack of recognition of peptides representing the autologous sequence of patient C29 (“Patients, materials, and methods”) (D) Ex vivo dilution ELISPOT data for 2 strong responses against p158 (1581-1600) and p180 (1801-1820) confirm that peptides based on GT1a sequence (■) elicited much higher responses than peptides based on autologous GT2b sequence (▲) in patient non-GT1 C35 (“Patients, materials, and methods”).
HCV GT1a-based peptides, but not peptides representing autologous sequence, are recognized in patients infected with non-GT1 strains. Strong and durable responses against 5 different GT1a-based HCV peptides were detected in patient C29 (Figure 3). This patient was infected with GT4 virus and showed a serologic response to GT1 virus. We were able to cultivate peptide-specific lines (A) by stimulating with GT1-based peptides, but not with autologous peptides (B) (“Patients, materials, and methods”). (C) dilution ICS assays performed on GT1-specific CD4+ T-cell lines for all specificities confirm the lack of recognition of peptides representing the autologous sequence of patient C29 (“Patients, materials, and methods”) (D) Ex vivo dilution ELISPOT data for 2 strong responses against p158 (1581-1600) and p180 (1801-1820) confirm that peptides based on GT1a sequence (■) elicited much higher responses than peptides based on autologous GT2b sequence (▲) in patient non-GT1 C35 (“Patients, materials, and methods”).
These results diverge from observations in patient C30, a female with no history of IVDU with chronic GT4 infection and no serologic evidence of a heterologous HCV serotype. Cellular assays showed that the autologous variant of the only HCV CD4+ epitope detected in this patient was equally well recognized as the GT1 peptide (data not shown). The reason for persistence of this single response that we could confirm ex vivo despite high viremia is not clear, but 1 possibilty is that this epitope cannot be processed due to selected mutations altering processing and presentation of the epitope.44
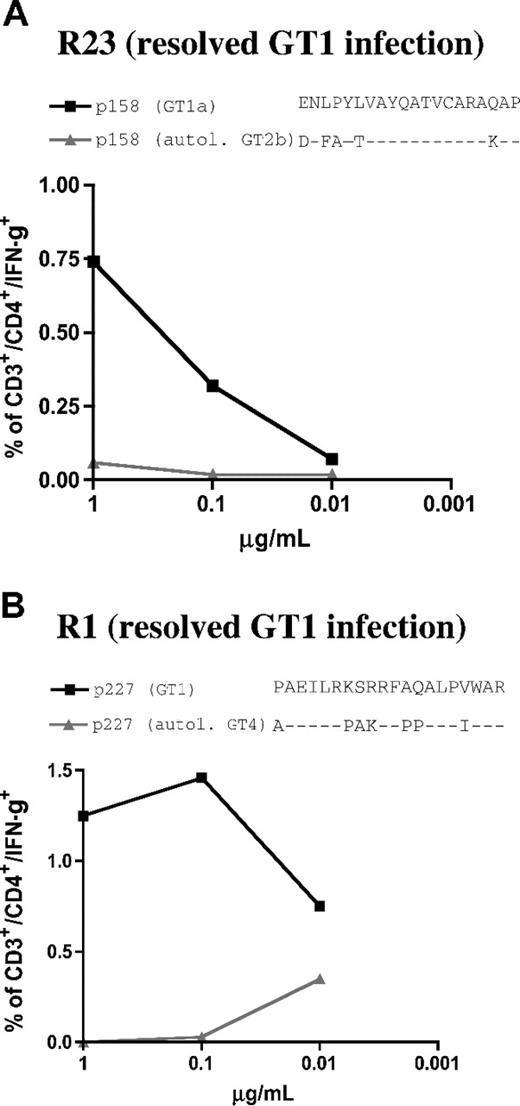
In order to confirm the lack of cross-genotype recognition of the autologous non-GT1 peptide in patients whose responses were primed with GT1 virus, we tested the cellular responses against 2 well-defined HCV-specific CD4+ cell epitopes, P227 (aa2271-2290) and P158 (aa1581-1600).10 Here, we used HCV-specific cell lines from 2 HLA-matched patients with spontaneously (and serologically defined) resolved GT1 infection, resolvers R1 and R23. Peptides representing GT1 sequence were recognized in both patients with resolved infection while the autologous non-GT1 peptide variants of C29 and C35, respectively, failed to induce a response (Figure 8). These data further support the hypothesis that the responses detected in patients C29 and C35 (with chronic non-GTI infection) were indeed primed by GT1 strains, and that these responses do not cross-react with the respective circulating virus.
GT4 and GT2-specific peptide variants representing autologous viral sequence of patients C29 and C35 are not cross-recognized in patients with resolved GT1 infection. HCV-specific cell lines of 2 patients with spontaneously resolved HCV infection (serologic GT1 according to Murex serotyping test: “Patients, materials, and methods”) were tested by dilution ICS for IFN-γ production for their responses against GT1 peptides (■) and autologous peptides (▲) of non-GT1 patients C29 and C35. (A) spontaneous resolver patient R23 shared the restricting HLA DR1*1 alleles for epitope p158 (aa1581-1600) that was detected in patient C35. The GT1-specific peptdie sequence but not the GT2b-specific autologous variant of the non-GT1 patient was recognized in dilution ICS experiments (“Patients, materials, and methods”). (B) Spontaneous resolver patient R1 shared the restricting HLA allele DRB1*11 for the previously described epitope P227 (aa2271-2290) that was detected in non-GT1 patient C29. Here also, the GT1 peptide sequence, but not the GT4-specific autologous variant of non-GT1 patient C29 was recognized.
GT4 and GT2-specific peptide variants representing autologous viral sequence of patients C29 and C35 are not cross-recognized in patients with resolved GT1 infection. HCV-specific cell lines of 2 patients with spontaneously resolved HCV infection (serologic GT1 according to Murex serotyping test: “Patients, materials, and methods”) were tested by dilution ICS for IFN-γ production for their responses against GT1 peptides (■) and autologous peptides (▲) of non-GT1 patients C29 and C35. (A) spontaneous resolver patient R23 shared the restricting HLA DR1*1 alleles for epitope p158 (aa1581-1600) that was detected in patient C35. The GT1-specific peptdie sequence but not the GT2b-specific autologous variant of the non-GT1 patient was recognized in dilution ICS experiments (“Patients, materials, and methods”). (B) Spontaneous resolver patient R1 shared the restricting HLA allele DRB1*11 for the previously described epitope P227 (aa2271-2290) that was detected in non-GT1 patient C29. Here also, the GT1 peptide sequence, but not the GT4-specific autologous variant of non-GT1 patient C29 was recognized.
Discussion
CD4+ T-helper-cell responses play a key role in the control of viral infections,45-53 and sustained HCV-specific CD4+ proliferative responses have consistently been demonstrated for spontaneous resolution of acute HCV infection.6,54-56 In contrast, HCV-specific CD4+ responses are usually narrowly directed or undetectable in patients with chronic infection.9,10,14,57 However, individual case reports and studies have described strong proliferative HCV-specific responses in individual patients with chronic HCV infection and persistently high viremia.27,31,32 Interestingly, this is most often observed in patients with chronic non-genotype 1 (non-GT1) infection. Previously, it was suggested that such responses were induced by previously resolved infections with other HCV strains, and the responses detected might not be cross-reactive to the persisting virus in those patients, but this hypothesis remained to be rigorously tested. The current study was specifically designed to analyze the CD4+ T-cell response against HCV in its full breadth at the single-epitope level in a larger cohort, thus allowing a detailed assessment of individual responses and their reactivity against autologous virus as well as the reference GT1 strain.
We observed significantly more HCV-specific CD4+ T-cell responses in patients with chronic non-GT1 infection than in patients with GT1 infection when comparing the number of CD4+ epitopes targeted in each patient. Further sequence analysis of the circulating HCV genomes in patients with non-GT1 infection who displayed significant responses against GT1-derived peptides revealed that the targeted epitopes were not present in vivo. Notably, we found autologous sequences that were distinct from the HCV GT1 prototype sequence of the targeted epitopes and autologous sequences that did not have a close phylogenetic relationship to the GT1 sequence (Figure 4). Rather, they were typical for the chronic HCV genotype as indicated by comparison with sequences published in the HCV database (data not shown), making viral escape an unlikely explanation for the variant sequences.
We observed a lack of cross-genotype reactivity between the T cells and the circulating virus in these patients, using HCV peptide–specific T-cell lines. Most importantly, 74% (66 of 89) of the responses detected in this cohort screened with GT1 peptides were detected in patients with non-GT1 infection in whom serologic evidence of previous exposure to a heterologous HCV strains could be documented (GT1 in 4 of 5 patients). In additional experiments, cell lines were generated using GT1-specific and autologous non-GT1 peptides. We confirmed the lack of cross-genotype recognition using these lines by demonstrating that the elicited responses were indeed type specific. Taken together, the results of these experiments argue against the suggestion of strain-specific suppression of responses in patients with chronic infection.32 However, they do provide further support for the hypothesis that responses detected in this subset of patients with non-GT1 infection were primed by prior exposure to a GT1 virus and are ineffective with regards to the non-GT1 virus persistently infecting those patients.
Animal models of HCV infection show that protective immunity to homologous virus challenge can be induced after spontaneous resolution of acute infection.2 In contrast, there is good evidence that this acquired immunity is not fully protective against heterologous genotypes.23,25 Here, we provide further evidence for relative lack of cross protective immunity in individuals chronically infected with GT2, GT3, or GT4 virus who each show serologic and cellular evidence of previous exposure to a heterologous strain. Together, these data demonstrate that reinfection by a heterologous HCV genotype is possible particularly in populations like IVDUs with a high risk for multiple exposures.33
In dengue virus infection, another flavivirus, the concept of antigenic sin is believed to be important in understanding viral pathogenesis and the severe clinical complications that arise upon secondary infection with a different-genotype dengue strain. This secondary viral encounter is believed to lead to the expansion of previously primed partly cross-reactive cells that have a suboptimal avidity for the current virus and thereby interfere with the kinetics and quality of the cellular antiviral response.58-61 In the case of reinfection with a heterologous HCV genotype, we hypothesize that any HCV cross-genotype–reactive HCV-specific CD4 response exhausts over time, while responses against subdominant GT1 epitopes with a high degree of cross-genotype variance are still detectable in these patients with non-GT1 viremia. The exact prerequisites of cross-protective immunity will only be understood by prospectively monitoring and studying the viral sequence and immune responses of sequential acute episodes in the chimp model or in high-risk populations. Only then will it be possible to determine whether the cellular responses primed by 1 HCV strain may be partially protective or detrimental to the clearance of a heterologous virus.31
Unfortunately, we cannot reconstruct the exact timeline of exposure to viruses of different genotypes in this study. Based upon the broad GT1-specific CD4+ T-cell responses we detected in these patients, and the known epidemiologic prevalence of HCV infection the United States, these data are most consistent with these patients clearing a prior GT1 infection and then facing subsequent exposure to a non-GT1 strain. We cannot rule out, however, the possibility that a subset of patients in this cohort were coinfected with different HCV genotypes during the acute event and have subsequently cleared only 1 of these strains.
Interestingly, 3 of the patients with non-GT1 infection, C29, C35, and C42, who have been described here in detail as having cellular and/or serologic responses specific to GT1, expressed the HLA molecules DR1 or DR11. Both of these HLA class II DR molecules have been correlated with spontaneous resolution of acute HCV GT1 infection.62 Future studies will be needed to determine whether this correlation with resolution is specific for only GT1 infection.
The majority of immunologic studies on HCV, including this report, are conducted with antigens and peptides based on the GT1 sequence. Our data indicate that future studies on HCV-specific CD4+ and CD8+ T-cell responses in acute, resolved, and chronic infection should take genotype into consideration. In particular, data might be more informative if genotype-specific reagents are used to define immunodominant epitopes in non-GT1 infection. Furthermore, a complete characterization of the cross-genotype reactivity of frequently detected genotype-specific CD4+ and CD8+ epitopes should be performed. Finally, our observation that none of the patients with chronic infection in this study (even those patients with non-GT1 infections who showed responses in our assays) targeted previously identified, well-conserved epitopes (eg, P177 [aa1771-1790]), frequently detected in patients with spontaneously resolved infection, is of significant importance with regards to future vaccine design.63
In summary, we provide evidence at the single-epitope level that a large percentage of HCV-specific CD4+ T-cell responses detected in patients with persistent HCV viremia are not directed against the circulating virus, but are most likely “immunologic scars” resulting from previous exposure to a heterologous virus strains. This study provides a comprehensive approach and analysis to extend previous findings based on case reports or studies using limited immunologic assays. Our data strongly indicate an inherent difficulty in generating cross-genotype protection during natural HCV infection in a subset of patients studied in this cohort. However, we have observed a lack of responses to a set of highly conserved CD4+ epitopes in chronic infection that may have significant implications for future vaccine development.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank foremost all patients who generously enrolled in this study and the dedicated clinical research staff and nurses who helped to recruit them. We thank Michael Houghton and Kevin Crawford at Chiron Corporation for generous provision of the recombinant antigens. We thank Mark Brockman for critical discussion of the manuscript.
This study was supported by the Howard Hughes Medical Institute (J.S.z.W., B.D.W.), Deutscher Akademischer Austauschdienst (J.S.z.W.), Deutsche Forschungsgemeinschaft (J.T.,T.K.), the National Institutes of Health grants U19-AI066345-02 (T.M.A., G.L., A.Y.K., L.L., and B.D.W.), RO1-AI067926-01 (T.M.A.), and RO1-AI031563-13 (B.D.W.), and the American Liver Foundation (G.L.).
National Institutes of Health
Authorship
Contribution: J.S.z.W. designed and performed experiments, analyzed data, enrolled patients, and wrote the first draft. G.L., A.Y.K., and A.W.L. designed experiments and revised the manuscript. B.D.W. gave financial support, designed experiments, and wrote and revised the manuscript. T.M.A., J.T., T.K., M.N., and A.B. performed and analyzed the sequencing experiments and revised the manuscript. A.J., B.E.N., S.A.L., and V.K. perfomed experiments and revised the manuscript. A.W., R.T.C., L.L., and C.McM. enrolled patients, analyzed data, and revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Bruce D. Walker, Partners AIDS Research Center, Howard Hughes Medical Institute, Massachusetts General Hospital, 149 13th St, Rm 5212D, Charlestown, MA 02129; e-mail: bwalker@partners.org; or Julian Schulze zur Wiesch, Medizinische Klinik 1, Universitätsklinikum Hamburg Eppendorf, Martinistr. 52, D-20246 Hamburg, Germany; email: julianszw@gmail.com.