Abstract
Chronic inflammatory diseases often have residual CD8+ T-cell infiltration despite treatment with systemic corticosteroids, which suggests divergent steroid responses between CD4+ and CD8+ cells. To examine steroid sensitivity, dexamethasone (DEX)–induced histone H4 lysine 5 (K5) acetylation and glucocorticoid receptor α (GCRα) translocation were evaluated. DEX treatment for 6 hours significantly induced histone H4 K5 acetylation in normal CD4+ cells (P = .001) but not in CD8+ cells. DEX responses were functionally impaired in CD8+ compared with CD4+ cells when using mitogen-activated protein kinase phosphatase (1 hour; P = .02) and interleukin 10 mRNA (24 hours; P = .004) induction as a readout of steroid-induced transactivation. Normal DEX-induced GCRα nuclear translocation and no significant difference in GCRα and GCRβ mRNA expression were observed in both T-cell types. In addition, no significant difference in SRC-1, p300, or TIP60 expression was found. However, activating transcription factor-2 (ATF2) expression was significantly lower in CD8+ compared with CD4+ cells (P = .009). Importantly, inhibition of ATF2 expression by small interfering RNA in CD4+ cells resulted in inhibition of DEX-induced transactivation in CD4+ cells. The data indicate refractory steroid-induced transactivation but similar steroid-induced transrepression of CD8+ cells compared with CD4+ cells caused by decreased levels of the histone acetyltransferase ATF2.
Introduction
Currently, glucocorticoids (GC)s are the most effective anti-inflammatory therapy used for treatment of chronic inflammatory and immune diseases.1,2 However, sensitivity to GCs varies considerably among immune cells.1,3,4 For instance, clinical data demonstrate residual CD8+ T-cell infiltration despite treatment with systemic GCs with more-severe disease outcomes. These cells, therefore, could be one of the key mediators for resistance to steroid therapy. It was found that in patients with multiple myeloma, a decrease in the CD4+/CD8+ ratio that results from an increased number of HLA-DR–expressing5 and cancer germline–specific CD8+ cells6 is usually a good indicator of poor steroid response. In GC-resistant cases of systemic lupus erythematosus, CD8+ T cells have been shown to be refractory to steroid-mediated apoptosis,7 and this is monitored as an indicator for the therapeutic efficacy of steroids. Relapses of multiple sclerosis are treated commonly with high-dose intravenous methylprednisolone.8 Several independent studies have reported that steroid treatment can significantly decrease the numbers of CD4+ T cells in these patients8 ; however, the same studies observed no change or even an increase in the number of CD8+ T cells after treatment in patients with poorly controlled disease.9 In patients with asthma, a decline in lung function as an asthma outcome has been shown to correlate with the number of lung-infiltrating CD8+ cells.10 In patients with chronic obstructive pulmonary disease, it has been shown that CD8+ and CD68+ cells and neutrophils are refractory to treatment with inhaled steroids, which highlights a need for understanding differential cell response to GCs.11,12
GCs exert their biologic effect through a specific receptor, GC receptor α (GCRα), which is expressed in virtually all cells. GCRα is a DNA-binding protein located in the cell cytoplasm. Its nuclear translocation is induced after ligand binding. GCs suppress production of multiple inflammatory proteins via transrepression and transactivation.13 GCRα directly interacts with proinflammatory transcription factors, which prevents transcription of inflammatory genes (transrepression).1,13,14 Activated GCRα also directly binds to its recognition sites in the promoters of certain genes to activate their transcription (transactivation), resulting in production of anti-inflammatory proteins such as mitogen-activated protein kinase phosphatase (MKP-1), which inhibits mitogen-activated protein kinase signaling pathways.15 Recently new insights were gained into the molecular mechanisms of how GCs suppress inflammation through transactivation and transrepression,2,13 as well as the importance of histone modification in steroid responsiveness.16-18
Several molecular mechanisms have been reported to be involved in GC resistance, including increased expression of GCRβ.1,19 GCRβ is the homologous isoform of GCRα in human cells, which is generated from alternative splicing of the human GCR gene.20,21 GCRβ differs from GCRα in its carboxyl terminus, where the last 50 amino acids of GCRα are replaced by a nonhomologous, 15 amino acid sequence. As a result of this difference, GCRβ may compete with GCRα for binding to glucocorticoid response element (GRE) sites22 or compete for the transcriptional coactivator molecules,23 inhibiting steroid responses.24,25 The expression level of GCRβ and GCRα in different cell types is the factor that determines variations in cellular responses to steroids.4,26-28 To gain information about responses of different cell types to steroids, we analyzed whether the responses of primary human CD8+ cells are refractory to steroids compared with CD4+ cells and the potential molecular mechanism(s) for this response.
Materials and methods
Reagents
Antihuman CD3 antibody (Ortho Biotech Products, Raritan, NJ) and dexamethasone (DEX) (Sigma Chemicals, St Louis, MO) were used for cell stimulation. Allophycocyanin (APC)–labeled CD4 (clone RPA-T4) and CD8 (clone RPA-T8) antibodies were purchased from BD Pharmingen (San Diego, CA). Human CD4+ T-cell isolation kit II and human CD8+ T-cell isolation kit II (Miltenyi Biotec, Auburn, CA) were used for the isolation of CD4+ and CD8+ T-cell populations, respectively. Primers and probes for MKP-1, interleukin 10 (IL-10), activating transcription factor 2 (ATF2), and glyceraldehyde-3-phosphate dehydrogenase (GADPH) were purchased from Applied Biosystems (Foster City, CA). GCRα and GCRβ primers were custom-ordered from Applied Biosystems on the basis of the sequences published by DeRijk et al.29
Subjects
This study was approved by the institutional review board at National Jewish Medical and Research Center. Eight healthy adult donors with no history of atopic or respiratory diseases were enrolled in the study. Informed consent in accordance with the Declaration of Helsinki was obtained during the time of blood draw.
Isolation of human peripheral blood mononuclear cells and magnetic cell sorting
Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll-Hypaque density-gradient centrifugation from heparinized venous blood of healthy donors as previously described.30 Cells were subsequently seeded to the slides to stain for acetylated histone H4 K5 or sorted into CD4+ and CD8+ T cells by negative depletion using magnetic cell sorting on the basis of the manufacture's instructions (Miltenyi Biotec). The purity of the enriched cells was evaluated by flow cytometry (FACSCalibur; Becton Dickinson, Franklin Lakes, NJ) and was always more than 90% positive.
Real-time polymerase chain reaction assay for GCR, MKP-1, and IL-10 mRNA
Total RNA from purified T cells was extracted using an RNeasy minikit (Qiagen, Valencia, CA) and reverse-transcribed into cDNA using reverse-transcription reagents (Invitrogen, Carlsbad, CA) following the manufacturer's instructions as described.31 To measure steroid-induced transactivation, DEX-induced MKP-1 and IL-10 mRNA were evaluated in CD4+ and CD8+ cells. For DEX-induced MKP-1 experiments, the cells were incubated with or without 10−7 M DEX for 1 hour before RNA extraction; for DEX-induced IL-10 experiments, the cells were incubated with or without 10−7 M DEX for 24 hours before RNA extraction. The quantitative real-time polymerase chain reaction (PCR) was performed as previously described.31 Briefly, the reactions were carried out using the dual-labeled fluorigenic-probe method. An ABI prism 7000 sequence detector (Applied Biosystems) was used to run real-time PCR and collect fluorescence data. Relative gene-expression levels were calculated and normalized to the corresponding levels of the housekeeping gene (GADPH). Standard curves for all targets were generated using the fluorescent data from twofold serial dilutions of 1000 ng of total RNA of the target sample. Standard curves for GCRα and GCRβ were generated from 10-fold serial dilutions of the GCRα and GCRβ plasmids as described by us earlier.26
GCRα nuclear translocation
GCRα intracellular translocation in response to 10−7 M DEX (Sigma) treatment was analyzed by immunofluorescent staining. In these experiments, freshly isolated PBMCs were seeded at 106 cells/mL on poly-D-lysine–coated cover slips. Cells then were treated with 10−7 M DEX or cultured in medium alone for 1 hour, fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS), and stained with anti-CD4 APC or anti-CD8 APC antibodies for 30 minutes on ice. After incubation, the cells were washed with PBS, permeabilized for 15 minutes at room temperature in permeabilization solution (PBS containing 0.1% [vol/vol] Tween 20, 0.1% [wt/vol] bovine serum albumin [Sigma], and 0.01% [wt/vol] saponin [Sigma]), and blocked with a commercial blocking solution (Superblock; Scytek, Logan, UT) for 15 minutes at room temperature. The cells then were incubated with an affinity-purified polyclonal antibody to GCRα (Affinity Bioreagents, Golden, CO) diluted in permeabilization solution (1:250) overnight at 4°C, washed, and then incubated with donkey antirabbit IgG, F(ab′)2-cy3–conjugated secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) (1:200), and the nucleus was counterstained with 300 nM of 4′,6′-diamidino-2-phenylindole (DAPI) (Sigma) for 1 hour at room temperature, washed, and mounted on slides. Purified nonimmune rabbit IgG (Southern Biotechnology Associates, Birmingham, AL) was used as an isotype control. The slides were analyzed by fluorescent microscopy (Leica, Wetzlar, Germany) with 63×/1.32-0.6 oil-immersion objective. The images were acquired using photometrics Coolsnap HQ Camera (Roper Scientific GmbH, Ottorunm, Germany) and analyzed by the imaging-analysis software Slidebook (Intelligent Imaging Innovations, Denver, CO). The data were presented as a ratio of mean fluorescence intensity (MFI) for nuclear and cytoplasmic cy3 (GCRα) staining as described by us earlier32 for CD4+ and CD8+ cells. Fifty CD4+ and CD8+ cells were analyzed per slide.
The TransAM GR transcription assay kit (Active Motive, Carlsbad, CA) was used to quantify GCRα cellular translocation of the bulk cell population. Freshly isolated CD4+ and CD8+ cells were cultured with 10−7 M DEX or medium alone for 1 hour. Then, nuclear and cytoplasmic cellular extracts were prepared from cells with an NE-PER nuclear and cytoplasmic extraction reagent (Pierce, Rockford, IL). Nuclear extracts (5 μg) from untreated and DEX-treated cells were added to the plate coated with GRE consensus sequence (5′-GGTACAnnnTGTTCT-3′), incubated for 1 hour at room temperature with mild agitation, and washed 3 times. GCR present in nuclear extracts bound specifically to the oligonucleotide and was detected with an antibody directed against GCR, recognized by secondary antirabbit horseradish peroxidase-conjugated antibody after addition of the developing solution. Absorbance was read on a spectrophotometer at 450 nm with a reference wavelength of 655 nm. Nuclear extract from DEX-treated HeLa cells was used as a positive control. Competitor GRE consensus sequence oligonucleotide (40 pmol/well) was added to the wells before the addition of nuclear extracts to prevent GCR binding to the probe immobilized to the plate to validate the specificity of the GCR-GRE interaction. GCR activation was expressed as an absorbance at 450 nm in all nuclear extracts.
Suppression of anti-CD3–induced CD4+ and CD8+ T-cell proliferation and cytokine production by DEX
Purified T cells (105/well) were stimulated with 0.5 μg/mL of soluble anti-CD3 in the absence or presence of 10−9 and 10−7 M DEX. Irradiated PBMCs (104/well) were used as antigen-presenting cells for the assay. The cultures were incubated for 72 hours at 37°C in a humidified 5% CO2 incubator. The cell proliferation was assessed on the basis of [3H]-thymidine incorporation as previously described.33
To examine DEX-induced suppression of cytokine production, cells were treated as described in “Materials and methods, GCRα nuclear translocation” in the 96-well plates for 24 hours. The supernatants were collected, frozen, and stored at −80°C until use. Commercial enzyme-linked immunosorbent assay kits were used to measure tumor necrosis factor α (TNF-α), interferon γ (IFN-γ), and IL-13 in cell culture supernatants (R&D Systems, Minneapolis, MN). The assays were performed according to the manufacturer's instructions. Absorbance readings were transformed to cytokine concentrations using standard curves. IC50, defined as the concentration of DEX that inhibits the proliferation or cytokine production by anti-CD3-stimulated lymphocytes to 50% of the level seen in the absence of DEX, was used as the parameter to measure the steroid sensitivity of the cells.
Assessment of histone H4 lysine 5 acetylation by immunostaining
Human PBMCs (106 cells/mL) were cultured in 4-well poly-D-lysine–coated Lab-Tek II chamber slides (Nalge Nunc International, Naperville, IL) in the presence or absence of 10−7 M DEX for 6 hours. Cells were washed with Hanks balanced salt solution and fixed in 4% paraformaldehyde in PBS for 5 minutes followed by blocking in a commercial blocking solution (Superblock) for 15 minutes at room temperature. Cells were incubated with either anti-CD4 APC or anti-CD8 APC antibodies for 1 hour at 4°C, permeabilized for another 15 minutes in permeabilization solution (PBS containing 0.1% [vol/vol], Tween 20, 0.1% [wt/vol] bovine serum albumin [Sigma], and 0.01% [wt/vol] saponin [Sigma]), and incubated with antiacetylated histone H4 lysine 5 (K5) antibody (Santa Cruz Biotechnology, Santa Cruz, CA) diluted in permeabilization solution (1:500) overnight at 4°C. Purified nonimmune rabbit IgG (Southern Biotechnology Associates) was used as an isotype control. After washing, the cells were incubated with a donkey antirabbit IgG, F(ab′)2-Cy3–conjugated secondary antibody (Jackson ImmunoResearch Laboratories; 1:200), and the nucleus was counterstained with 300 nM of DAPI (Sigma) for 45 minutes at room temperature and washed in PBS/0.1% Tween 20 for 15 minutes. Stained cells were observed by fluorescence microscopy (Leica) with an × 63 objective. One hundred cells were analyzed per slide after coding the slides to blind the slide reader. The MFI of cy3 staining (acetylated K5) in CD4+ and CD8+ cells was assessed by the analysis software within the computer-generated masks for nuclear (DAPI stained) regions of the cells. The data were presented as cy3 (acetylated K5) MFI before and after DEX treatment in CD4+ and CD8+ T cells.
Western blotting
Purified CD4+ and CD8+ T cells were treated with DEX (10−9 M, 10−7 M) before lysis in ice-cold radioimmunoprecipitation assay buffer containing protease inhibitor cocktail (Sigma). Total protein (30 μg) was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to the nitrocellulose membrane. MKP-1 was assayed using anti–MKP-1 antibody (Santa Cruz Biotechnology).
Nuclear extracts were prepared from freshly isolated CD4+ and CD8+ T cells with NE-PER nuclear and cytoplasmic extraction reagents (Pierce). Nuclear protein (10 μg per condition) was run on 4% to 15% gradient gel (Bio-Rad Laboratories, Hercules, CA) and transferred to the nitrocellulose membrane. The membranes were blotted with anti-steroid hormone receptor coactivator 1 (SRC-1), ATF2, and TIP60 (Santa Cruz Biotechnology) antibodies. For p300 detection, 60 μg of whole-cell extracts were run on 7.5% gel (Bio-Rad Laboratories) and transferred to the membrane for 4 hours at 20 V in the cold room. The membrane then was incubated with 2 μg/mL anti-p300 antibody (Upstate Biotechnology, Lake Placid, NY). To control the quality of nuclear-protein preparation, the membranes were stripped and reprobed with anti-C23 antibody (Santa Cruz Biotechnology) used as nuclear control protein.34 Developed X-ray films were scanned, and densitometry of the bands was quantified using National Institutes of Health Image 1.63 software (available on the Internet at http://rsb.info.nih.gov/nih-image).
Silencing of ATF2 expression by specific small interfering RNA
Small interfering RNAs (siRNAs) were used to inhibit ATF2 gene expression. ATF2 siRNA and nonspecific siRNA were purchased from Santa Cruz Biotechnology. A commercial Nucleofector human T-cell kit (Amaxa, Gaithersburg, MD) and a special transfection program for human T cells on the Nucleofector device were used. In brief, 5 × 106 purified CD4+ T cells were suspended in 100 μL of transfection solution and transfected with 1 μg of ATF2 siRNA or nonsilencing control siRNA using the V014 program (an approach previously described by Goleva et al19 ). Transfected cells were diluted immediately with prewarmed T-cell growth medium (Amaxa) and cultured in 24-well plates or on poly-D-lysine–coated coverslips. To confirm inhibition of ATF2 mRNA and protein expression by ATF2 siRNA, transfected cells were serially assayed by real-time PCR and microscopy 24 to 72 hours after transfection for ATF2 expression using the methods described above. To determine the effect of siRNA delivery on cell viability, the trypan blue dye exclusion test was used. Cell viability was usually 70% to 80% at 48 hours and 50% to 60% at 72 hours after siRNA transfection.
Statistical analysis
Data were expressed as mean plus or minus the standard error of the mean (SEM). The paired t test was used to compare functional responses of CD4+ and CD8+ cells obtained from the same donors (hence, paired). The test was also used to compare pre-DEX and post-DEX responses within CD4+ and CD8+ cells types. Before testing, paired difference distributions were examined for outliers, which can indicate violation to the normality assumption of the parametric t test. No outliers were apparent. A P value of less than .05 was considered statistically significant. All reported P values were based on 2-sided tests.
Results
Differential gene activation by GCs in human CD4+ vs CD8+ T cells
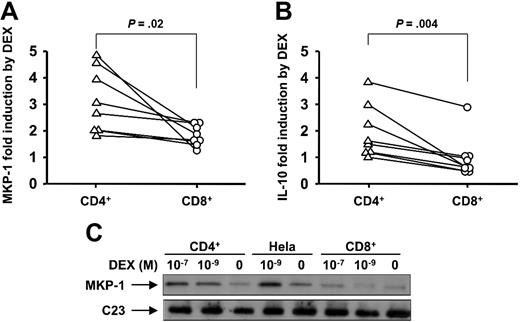
To characterize GC-mediated transactivation in human CD4+ compared with CD8+ T cells, we investigated the effect of DEX on MKP-1 gene induction by real-time PCR. MKP-1 is an important GC-induced anti-inflammatory gene that reflects early GCR transactivation response.26,35 There was no significant difference in MKP-1 mRNA levels in resting CD4+ and CD8+ cells (3.05 ng ± 0.99 ng vs 4.48 ng ± 1.12 ng of MKP-1 per ng of GADPH in CD4+ vs CD8+ cells, respectively [n = 8]). In human T cells, DEX rapidly induced MKP-1 mRNA gene expression. However, we found significantly different levels of MKP-1 induction by DEX in CD4+ and CD8+ T cells (Figure 1A). CD4+ T cells showed significantly greater induction of MKP-1 expression after short-term DEX exposure than did CD8+ T cells (3.13 ± 0.42-fold in CD4+ T cells vs 1.83 ± 0.14-fold in CD8+ T cells [n = 8]; P = .02) (Figure 1A).
Divergent effects of DEX on anti-inflammatory gene induction in human CD4+ and CD8+ cells. Shown is DEX-induced activation of (A) MKP-1 and (B) IL-10 gene expression by purified human CD4+ and CD8+ cells. MKP-1 and IL-10 mRNA induction by DEX- compared with medium-treated cells were analyzed by real-time PCR. Purified human CD4+ and CD8+ T cells were collected 1 hour after DEX treatment to analyze MKP-1 induction and 24 hours after DEX treatment to analyze IL-10 induction. (C) Differential induction of MKP-1 protein expression by DEX in CD4+ vs CD8+ cells. The cells had been treated by DEX or cultured in medium only for 9 hours. A representative Western blot of 3 independent experiments performed is shown.
Divergent effects of DEX on anti-inflammatory gene induction in human CD4+ and CD8+ cells. Shown is DEX-induced activation of (A) MKP-1 and (B) IL-10 gene expression by purified human CD4+ and CD8+ cells. MKP-1 and IL-10 mRNA induction by DEX- compared with medium-treated cells were analyzed by real-time PCR. Purified human CD4+ and CD8+ T cells were collected 1 hour after DEX treatment to analyze MKP-1 induction and 24 hours after DEX treatment to analyze IL-10 induction. (C) Differential induction of MKP-1 protein expression by DEX in CD4+ vs CD8+ cells. The cells had been treated by DEX or cultured in medium only for 9 hours. A representative Western blot of 3 independent experiments performed is shown.
To provide more definitive evidence regarding the divergent GCR transactivation activity in CD4+ vs CD8+ T cells, we measured the effects of DEX on another GC-induced anti-inflammatory gene, IL-10, known to be induced by GCR transactivation. The basal levels of IL-10 mRNA in CD4+ and CD8+ cells were not significantly different (0.43 ng ± 0.11 ng vs 0.71 ng ± 0.29 ng of IL-10 per ng of GADPH [n = 8]). It was found that DEX treatment for 24 hours significantly increased IL-10 gene expression in CD4+ T cells, whereas nearly no effect on IL-10 gene expression in CD8+ T cells was seen (IL-10 induction by DEX was 1.95 ± 0.35-fold in CD4+ T cells and 0.95 ± 0.28-fold in CD8+ T cells [n = 8]; P = .004) (Figure 1B).
Along with mRNA, significantly lower MKP-1 protein induction by DEX was found in CD8+ cells compared with CD4+ cells. The cells were treated with 10−7 M and 10−9 M DEX for 9 hours or cultured in medium alone. The DEX-mediated increase in MKP-1 protein level was dose dependent in both CD4+ and CD8+ T cells; however, a much stronger induction of MKP-1 was observed in CD4+ than that in CD8+ T cells (Figure 1C).
DEX induces similar GCRα nuclear translocation in CD4+ and CD8+ T cells
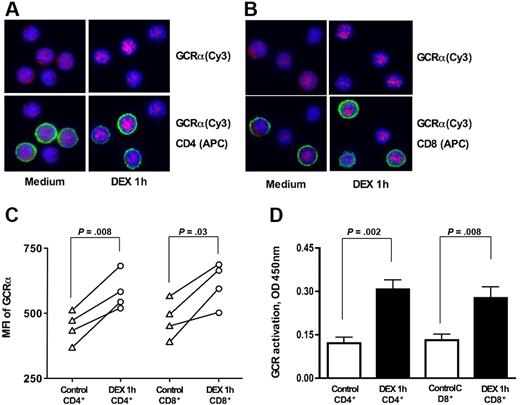
To determine whether the differences in GCR transactivation properties of CD4+ vs CD8+ T cells were caused by the difference in GCRα nuclear translocation in response to steroids, we measured the GCRα cellular translocation in response to DEX in normal CD4+ vs CD8+ T cells. GCRα nuclear translocation was assessed in response to 10−7 M DEX by immunostaining. In the absence of DEX, GCRα was localized uniformly in T cells; DEX induced GCRα nuclear translocation within 1 hour. Increasing the time of DEX treatment for greater than 1 hour had no further effect on GCRα nuclear translocation; therefore, we used this time point to assay additional subjects. As shown in Figure 2A-C, DEX induced similar levels of GCRα nuclear translocation in both CD4+ and CD8+ T cells. Cy3 MFIs for the nuclear region of CD4+ were 447.1 (± 52.9) and 585.7 (± 62.0) before and after DEX treatment, respectively (P = .008; n = 4) and for CD8+ were 476.7 (± 64.0) vs 615.2 (± 72.0), respectively (P = .03; n = 4).
GCRα cellular translocation in human CD4+ and CD8+ lymphocytes in response to 10−7 M DEX treatment in vitro. Freshly isolated PBMCs were stimulated for 1 hour with 10−7 M DEX or remained untreated. Representative images of GCRα cellular translocation in (A) CD4+ and (B) CD8+ T cells in response to DEX treatment are shown (original magnification × 630; blue, DAPI, nuclear staining; red, cy3, GCRα; green, APC, CD4+ or CD8+ T-cell surface staining) (the cells from 4 different donors were evaluated in this assay). Note cytoplasmic localization of GCRα before DEX treatment and increase in nuclear localization of the GCRα after DEX treatment in both CD4+ and CD8+ T cells. See “Materials and methods, GCRα nuclear translocation” for more detailed image acquisition information. (C) GCRα (Cy3) MFI for the nuclear region of CD4+ and CD8+ before and after DEX treatment. (D) Addition of DEX resulted in significant increase in the amount of nuclear GCR in both purified CD4+ and CD8+ cells. GCR was measured in nuclear extracts from CD4+ vs CD8+ T cells plus or minus DEX (1 hour) on the basis of its interaction with GRE consensus motive immobilized to the plate (TransAM GR transcription factor assay). Data are expressed as mean plus or minus SEM (n = 6).
GCRα cellular translocation in human CD4+ and CD8+ lymphocytes in response to 10−7 M DEX treatment in vitro. Freshly isolated PBMCs were stimulated for 1 hour with 10−7 M DEX or remained untreated. Representative images of GCRα cellular translocation in (A) CD4+ and (B) CD8+ T cells in response to DEX treatment are shown (original magnification × 630; blue, DAPI, nuclear staining; red, cy3, GCRα; green, APC, CD4+ or CD8+ T-cell surface staining) (the cells from 4 different donors were evaluated in this assay). Note cytoplasmic localization of GCRα before DEX treatment and increase in nuclear localization of the GCRα after DEX treatment in both CD4+ and CD8+ T cells. See “Materials and methods, GCRα nuclear translocation” for more detailed image acquisition information. (C) GCRα (Cy3) MFI for the nuclear region of CD4+ and CD8+ before and after DEX treatment. (D) Addition of DEX resulted in significant increase in the amount of nuclear GCR in both purified CD4+ and CD8+ cells. GCR was measured in nuclear extracts from CD4+ vs CD8+ T cells plus or minus DEX (1 hour) on the basis of its interaction with GRE consensus motive immobilized to the plate (TransAM GR transcription factor assay). Data are expressed as mean plus or minus SEM (n = 6).
An alternative technique was applied to evaluate GCR cellular translocation in bulk CD4+ and CD8+ T cells. GCR was measured in nuclear extracts from these cells on the basis of its interaction with GRE consensus motive immobilized to the plate (TransAM GR transcription factor assay). No difference in the amount of nuclear GCR was found in freshly isolated CD4+ and CD8+ T cells. Addition of DEX to the cells resulted in a significant increase in the amount of nuclear GCR in both CD4+ and CD8+ T cells (Figure 2D). Both cell types showed similar amounts of nuclear GCR after DEX treatment.
Because GC anti-inflammatory response is mediated through GCRα, and an alternatively spliced isoform, GCRβ, can interfere with GCRα by either forming heterodimers with GCRα33 or competing for steroid receptor coactivator23 and GRE,25 displaying a dominant negative activity, we analyzed the expression of GCRα and GCRβ isoforms in CD4+ vs CD8+ T cells. A quantitative real-time PCR assay was used to compare GCRα and GCRβ mRNA amounts between purified CD4+ and CD8+ T cells. Primers and probes were designed to be complementary to the sequence within the region of exon 9α for GCRα or exon 9β for GCRβ (as previously described29 ). Our results demonstrate that there is no significant difference in GCRα mRNA and GCRβ mRNA expression (Table 1) between human CD4+ and CD8+ T cells. The molar ratios of GCRα mRNA to GCRβ in CD4+ and CD8+ were 2183 (± 506) vs 1834 (± 424), respectively (P = .53; n = 8). In addition, microscopy results also showed that GCRα and GCRβ protein expression in CD4+ T cells were similar to CD8+ T cells (data not shown).
GCRα and GCRβ mRNA expression between human CD4+ and CD8+ T cells
| Subjects . | GCRα mRNA expression . | GCRβ mRNA expression . | ||||||
|---|---|---|---|---|---|---|---|---|
| Ct values . | GCRα concentration, pg/mL* . | Ct values . | GCRβ concentration, fg/mL* . | |||||
| CD4+ . | CD8+ . | CD4+ . | CD8+ . | CD4+ . | CD8+ . | CD4+ . | CD8+ . | |
| 1 | 27.96 | 27.15 | 0.008 | 0.015 | 37.06 | 36.98 | 0.011 | 0.011 |
| 2 | 27.76 | 27.66 | 0.009 | 0.010 | 37.08 | 36.77 | 0.011 | 0.013 |
| 3 | 26.89 | 27.05 | 0.018 | 0.016 | 37.47 | 37.92 | 0.008 | 0.006 |
| 4 | 27.67 | 27.82 | 0.010 | 0.009 | 38.30 | 37.21 | 0.004 | 0.010 |
| 5 | 27.15 | 27.05 | 0.015 | 0.016 | 36.59 | 36.13 | 0.015 | 0.020 |
| 6 | 28.02 | 27.66 | 0.008 | 0.010 | 35.75 | 36.32 | 0.027 | 0.020 |
| 7 | 26.73 | 27.08 | 0.020 | 0.015 | 35.27 | 36.54 | 0.037 | 0.015 |
| 8 | 26.46 | 27.68 | 0.024 | 0.010 | 37.08 | 36.31 | 0.011 | 0.018 |
| Mean plus and minus SEM | 27.33 ± 0.21 | 27.39 ± 0.12 | 0.014 ± 0.002 | 0.013 ± 0.001 | 36.83 ± 0.34 | 36.77 ± 0.21 | 0.016 ± 0.004 | 0.014 ± 0.002 |
| Subjects . | GCRα mRNA expression . | GCRβ mRNA expression . | ||||||
|---|---|---|---|---|---|---|---|---|
| Ct values . | GCRα concentration, pg/mL* . | Ct values . | GCRβ concentration, fg/mL* . | |||||
| CD4+ . | CD8+ . | CD4+ . | CD8+ . | CD4+ . | CD8+ . | CD4+ . | CD8+ . | |
| 1 | 27.96 | 27.15 | 0.008 | 0.015 | 37.06 | 36.98 | 0.011 | 0.011 |
| 2 | 27.76 | 27.66 | 0.009 | 0.010 | 37.08 | 36.77 | 0.011 | 0.013 |
| 3 | 26.89 | 27.05 | 0.018 | 0.016 | 37.47 | 37.92 | 0.008 | 0.006 |
| 4 | 27.67 | 27.82 | 0.010 | 0.009 | 38.30 | 37.21 | 0.004 | 0.010 |
| 5 | 27.15 | 27.05 | 0.015 | 0.016 | 36.59 | 36.13 | 0.015 | 0.020 |
| 6 | 28.02 | 27.66 | 0.008 | 0.010 | 35.75 | 36.32 | 0.027 | 0.020 |
| 7 | 26.73 | 27.08 | 0.020 | 0.015 | 35.27 | 36.54 | 0.037 | 0.015 |
| 8 | 26.46 | 27.68 | 0.024 | 0.010 | 37.08 | 36.31 | 0.011 | 0.018 |
| Mean plus and minus SEM | 27.33 ± 0.21 | 27.39 ± 0.12 | 0.014 ± 0.002 | 0.013 ± 0.001 | 36.83 ± 0.34 | 36.77 ± 0.21 | 0.016 ± 0.004 | 0.014 ± 0.002 |
Ct indicates threshold cycle.
According to the standard curves measured from GCRα and GCRβ plasmids, respectively.
Similar gene suppression by DEX in CD4+ vs CD8+ T cells
The anti-inflammatory effects of GCs rely on both the direct activation of genes (eg, the induction of MKP-1 and IL-10 [transactivation]) and their indirect effects on inflammatory gene suppression via interaction with other transcription factors (transrepression).1 The results described above showed a significant difference in steroid-induced transactivation responses in CD4+ vs CD8+ T cells; therefore, it was of importance to compare steroid-induced transrepression responses in these 2 T-cell populations.
Purified CD4+ or CD8+ T cells were stimulated by anti-CD3 in the presence or absence of DEX for 72 hours. Irradiated PBMCs were used as antigen-presenting cells for the assay. The effect of DEX on T-cell proliferation induced by anti-CD3 was assessed on the basis of tritiated thymidine incorporation. As shown in Figure 3, DEX significantly inhibited CD4+ and CD8+ T-cell proliferation with a DEX IC50 of 5.73 nM (± 1.34 nM) for CD4+ cells and 7.60 nM (± 0.87 nM) for CD8+ cells. At the same time, TNF-α and IFN-γ were measured in cell culture supernatants from 24-hour–stimulated anti-CD3–stimulated cells. Production of TNF-α and IFN-γ by anti-CD3–stimulated CD4+ and CD8+ cells were comparable. A similar degree of DEX-induced suppression of cytokine production was observed in CD4+ and CD8+ T cells (the DEX IC50 for TNF-α production was 6.33 nM ± 2.18 nM for CD4+ cells and 6.37 nM ± 2.45 nM for CD8+ cells; the DEX IC50 for IFN-γ production was 0.59 nM ± 0.04 nM for CD4+ cells and 0.51 nM ± 0.07 nM for CD8+ cells).
DEX IC50 required for inhibition of anti-CD3–stimulated proliferation and cytokine secretion by purified human CD4+ and CD8+ T cells from normal donors. No significant difference was observed between CD4+ and CD8+ T cells. Data are expressed as mean plus or minus SEM (n = 3).
DEX IC50 required for inhibition of anti-CD3–stimulated proliferation and cytokine secretion by purified human CD4+ and CD8+ T cells from normal donors. No significant difference was observed between CD4+ and CD8+ T cells. Data are expressed as mean plus or minus SEM (n = 3).
We attempted to measure T-helper type 2 cytokine IL-13 in 24-hour cell culture supernatants from CD4+ and CD8+ cells stimulated with anti-CD3 in the presence of irradiated APC. However, IL-13 was below the detection limit of the assay. This is not surprising, because normal (not allergic) donors were used in this study.
Differential effect of DEX on histone acetylation in CD4+ vs CD8+ T cells
Histone acetylation status is one of the key factors in regulation of inflammatory gene transcription.36 Because histone H4 K5 acetylation is a marker of GC-mediated transactivation,17 we examined the ability of GCs to induce histone H4 acetylation at residue K5. Using immunostaining, we found (Figure 4) that DEX treatment for 6 hours significantly induced histone H4 K5 acetylation in normal CD4+ T cells (MFI, 397.0 ± 20.4 and 518.0 ± 31.0 before and after DEX, respectively [n = 8]; P = .001). There was a marginal change in histone H4 K5 acetylation detected in CD8+ T cells (MFI, 392.1 ± 18.3 vs 421.8 ± 22.7 [n = 8]; P = .06).
DEX-induced histone H4 K5 acetylation in human CD4+ and CD8+ T cells. Representative images of acetylated histone H4 K5 in CD4+ and CD8+ cells in response to 6 hours of 10−7 M DEX treatment in vitro are shown (original magnification × 630; blue, DAPI, nuclear staining; red, cy3, acetylated histone H4 K5; green, APC, surface staining with (A) anti-CD4 or (C) anti-CD8 APC antibodies). The MFI of cy3 staining (acetylated histone H4 K5) in CD4+ (B) and CD8+ (D) cells was assessed by analysis software within the computer-generated masks for the cell nuclei. Cells (50–100) were analyzed for each donor studied (n = 8). See “Materials and methods, GCRα nuclear translocation” for more detailed image acquisition information.
DEX-induced histone H4 K5 acetylation in human CD4+ and CD8+ T cells. Representative images of acetylated histone H4 K5 in CD4+ and CD8+ cells in response to 6 hours of 10−7 M DEX treatment in vitro are shown (original magnification × 630; blue, DAPI, nuclear staining; red, cy3, acetylated histone H4 K5; green, APC, surface staining with (A) anti-CD4 or (C) anti-CD8 APC antibodies). The MFI of cy3 staining (acetylated histone H4 K5) in CD4+ (B) and CD8+ (D) cells was assessed by analysis software within the computer-generated masks for the cell nuclei. Cells (50–100) were analyzed for each donor studied (n = 8). See “Materials and methods, GCRα nuclear translocation” for more detailed image acquisition information.
Expression of histone acetyltransferases that acetylate histone H4 K5 in CD4+ vs CD8+ T cells
Histone acetyltransferases (HATs) regulate gene transcription by histone acetylation. Acetylation of the ϵ group on lysine residues reduces the charge of histones and subsequently releases the tightly wound DNA, which allows the recruitment of additional large protein complexes.36,37 We hypothesized that the difference in DEX-induced acetylation and, hence, transactivation in CD8+ cells vs CD4+ cells could be a consequence of deficient HAT expression by CD8+ cells.
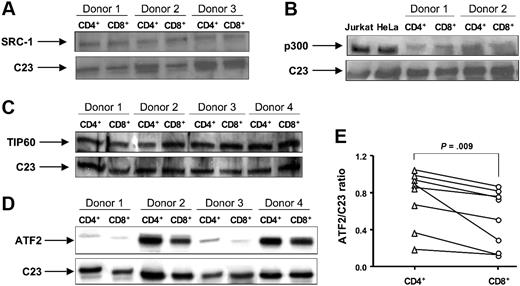
To test this hypothesis, nuclear protein extracts were prepared from CD4+ and CD8+ cells isolated by negative selection. SRC-1, ATF2, TIP60, and p300 have been reported to have HAT activity and are known to acetylate K5 of histone H4.38,39 Using Western blot analysis, we measured expression of these proteins in CD4+ and CD8+ cells. No difference in SRC-1 and TIP60 expression between these 2 cell populations was found (Figure 5A,C). We were unable to consistently detect p300 in T-cell nuclear extracts from all donors, probably because of the insufficient cell amount used (6 × 106 CD4+ or CD8+ was the maximal cell number we could recover). While using whole-cell extracts from CD4+ and CD8+ cells we did not detect a difference in p300 expression between these 2 cell types (Figure 5B). The immunostaining and real-time PCR also revealed that there was no difference in p300 expression in CD4+ vs CD8+ cells (data not shown). The amounts of ATF2 varied within 8 donors studied. Importantly, a significant decrease in ATF2 expression in CD8+ cells was found compared with CD4+ cells (Figure 5D,E). The densitometry ratio of ATF2 to C23 was 0.53 (± 0.11) in CD8+ cells compared with 0.75 (± 0.11) in CD4+ cells (P = .009; n = 8).
The expression of HATs SRC-1, p300, TIP60, and ATF2 in normal CD4+ and CD8+ lymphocytes. Nuclear or whole-cell proteins were extracted from purified CD4+ and CD8+ T cells. The expression of (A) SRC-1, (B) p300, (C) TIP60, and (D) ATF2 was evaluated by Western blot. C23 was used as a loading control for nuclear proteins. Nuclear extracts from Jurkat T cells and HeLa cells were used as positive controls for the anti-p300 antibody. Vertical lines have been inserted to indicate where a gel lane was cut. These gels came from 2 different experiments. (E) The ratio of AFT2 to C23 was calculated as a ratio of density for AFT2 and C23 bands measured with National Institutes of Health (Bethesda, MD) Image 1.63 software.
The expression of HATs SRC-1, p300, TIP60, and ATF2 in normal CD4+ and CD8+ lymphocytes. Nuclear or whole-cell proteins were extracted from purified CD4+ and CD8+ T cells. The expression of (A) SRC-1, (B) p300, (C) TIP60, and (D) ATF2 was evaluated by Western blot. C23 was used as a loading control for nuclear proteins. Nuclear extracts from Jurkat T cells and HeLa cells were used as positive controls for the anti-p300 antibody. Vertical lines have been inserted to indicate where a gel lane was cut. These gels came from 2 different experiments. (E) The ratio of AFT2 to C23 was calculated as a ratio of density for AFT2 and C23 bands measured with National Institutes of Health (Bethesda, MD) Image 1.63 software.
The effect of ATF2 gene silencing on DEX-induced gene transactivation in human CD4+ T cells
To evaluate whether different levels of ATF2 expression can modulate steroid-induced transactivation, we silenced the ATF2 gene expression in primary human CD4+ T cells and examined the effects of DEX on MKP-1 and IL-10 gene induction. Purified CD4+ T cells from normal donors were transfected by electroporation with ATF2 siRNA or nonsilencing control siRNA. Real-time PCR (Figure 6A) and microscopy (Figure 6B) results indicated that introduction of ATF2 siRNA 48 hours after transfection specifically inhibited ATF2 expression in the CD4+ T cells. The estimated silencing efficiency was 60.41% (± 7.22%, n = 4). Therefore, we selected this time point to compare MKP-1 induction followed by incubation with DEX for 1 hour and IL-10 induction for 24 hours. Figure 6C shows that silencing of ATF2 resulted in a significant decrease of DEX-induced MKP-1 production (MKP-1 mRNA induction was 3.42 ± 0.42-fold [n = 4]; P = .006 compared with the nonsilencing siRNA with values of 5.58 ± 0.70-fold). In addition, significantly reduced IL-10 gene induction was observed in ATF2-silenced CD4+ T cells compared with those in the nonsilencing RNA group (1.05 ± 0.06 vs 2.44 ± 0.38, respectively [n = 4]; P = .03) (Figure 6D).
Inhibition of ATF2 gene expression using specific siRNA in human CD4+ decreases the transactivation activity of steroids. Introduction of ATF2 siRNA into CD4+ T cells resulted in specific inhibition of ATF2 expression as shown by (A) real-time PCR and (B) immunostaining (original magnification ×630; blue, DAPI, nuclear staining; red, cy3, ATF2; see “Materials and methods, GCRα nuclear translocation” for more detailed image acquisition information). The pictures are representatives of 4 independent experiments. Silencing of ATF2 resulted in a significant decrease of DEX-induced (C) MKP-1 and (D) IL-10 production by human CD4+ T cells (n = 4) compared with a nonspecific siRNA control group.
Inhibition of ATF2 gene expression using specific siRNA in human CD4+ decreases the transactivation activity of steroids. Introduction of ATF2 siRNA into CD4+ T cells resulted in specific inhibition of ATF2 expression as shown by (A) real-time PCR and (B) immunostaining (original magnification ×630; blue, DAPI, nuclear staining; red, cy3, ATF2; see “Materials and methods, GCRα nuclear translocation” for more detailed image acquisition information). The pictures are representatives of 4 independent experiments. Silencing of ATF2 resulted in a significant decrease of DEX-induced (C) MKP-1 and (D) IL-10 production by human CD4+ T cells (n = 4) compared with a nonspecific siRNA control group.
These data support the concept that ATF2, via acetylation of the histone H4 K5 residue, controls GC-mediated gene transactivation in T lymphocytes and accounts for differences in steroid responsiveness of CD8+ vs CD4+ cells.
Discussion
We assessed the steroid responsiveness of human peripheral blood CD8+ T cells compared with CD4+ T cells. Functional assays were performed to evaluate steroid-induced transactivation and transrepression, because the anti-inflammatory and immunosuppressive effects of GCs rely on these 2 molecular mechanisms, respectively.1,2,14 Steroid-induced transactivation assays, including IL-10 and MKP-1 gene induction, demonstrated significantly reduced GCR-mediated transactivation in CD8+ T cells compared with CD4+ cells. To assess DEX-induced transrepression, purified CD4+ or CD8+ T cells were stimulated in the presence and absence of DEX. Our results show an inhibitory effect of DEX on cytokine secretion and cell proliferation in both T-cell populations, which suggests that DEX has similar transrepression activity in these 2 cell types. No difference in the maximal response (Emax) of DEX was seen on proliferation or cytokine release between CD4+ and CD8+ cells. In contrast, when we measured the transactivation effects of DEX on resting T cells, significantly different responses to GCs between CD4+ and CD8+ cells were found. The difference in DEX-induced transactivation between CD4+ and CD8+ cells was also detected with 10−6 M DEX (the highest physiologic dose) (data not shown), which suggests that both the DEX effect on the sensitivity of cells (EC50) and Emax were significantly altered in CD8+ cells compared with CD4+ cells. These data suggest that CD8+ T cells are refractory to steroids because of a failure of steroids to switch on anti-inflammatory gene expression rather than a failure to switch off inflammatory genes. Our data support the idea that steroid responses can be regulated not only by GCR transrepression but also by GCR transactivation.
Because reduced GCRα cellular translocation in response to steroids and divergent expression of GCRα vs the dominant negative isoform, GCRβ, by cells can modify cellular response to steroids,19,26,27,33 we evaluated these parameters of steroid response in our system. There was no difference in DEX-induced GCRα nuclear translocation of CD4+ vs CD8+ T cells. In addition, similar levels of GCRα and GCRβ mRNA were found in both cell types.
Histone acetylation has been found to differentially regulate gene transcription, and acetylation of histone H4 itself serves as an important mechanism for signaling-mediated chromatin remodeling. It has been demonstrated that activated GCR forms complexes with cAMP response element-binding protein (CREB) and its homologue p300, which has intrinsic HAT activity, and induces histone acetylation of K5 and K16. The effect of GCs on K5 has been established to be a target for steroid-induced transactivation.17,38 In our studies, we found reduced DEX-induced transactivation in CD8+ T cells compared with CD4+ cells, and this attenuated effect was related to histone H4 acetylation on K5 in response to DEX. This study was designed to test the hypothesis that differential expression of HATs could be associated with decreased GC responses in CD8+ compared with CD4+ T cells. Four enzymes (SRC-1, p300, ATF2, and TIP60) have been reported to regulate histone H4 K5 acetylation.38,39 By using Western-blotting difference in SRC-1, p300, and TIP60, expression between these 2 T-cell populations was detected. However, a significantly lower level of ATF2 protein was found in CD8+ T cells than in CD4+ T cells.
ATF2-activating transcription factor 2 (CREB2), is a member of the leucine zipper family of DNA-binding proteins. This protein binds to the cAMP-responsive element (CRE), an octameric palindrome. The protein forms a homodimer or heterodimer with c-Jun and stimulates CRE-dependent transcription. This protein is also an HAT that specifically acetylates histones H2B and H4 in vitro; thus, it may represent a class of sequence-specific factors that activate transcription by direct effects on chromatin components.40 To demonstrate that ATF2 expression has an important regulatory role in cellular steroid responses, we used siRNA transfection technology to silence ATF2 mRNA and examined the consequence of ATF2 elimination on GCR-mediated gene transcription. Indeed, we found that specific ATF2 siRNA, but not control siRNA, could significantly inhibit ATF2 protein and mRNA expression in human CD4+ T cells after transfection. In ATF2-silenced, but not control siRNA-silenced, CD4+ T cells, MKP-1 and IL-10 mRNA induction by steroids was decreased significantly. These data, for the first time, provide direct evidence supporting that ATF2 expression level is directly related to cellular steroid responses.
Our present studies demonstrate that peripheral blood CD8+ T cells are less sensitive to DEX compared with CD4+ T cells, which accounts for the clinical observation that CD8+ T cells often remain in persistently inflamed tissues of patients despite systemic steroid therapy.5-11 Moreover, we investigated the molecular mechanism of relative steroid insensitivity of CD8+ vs CD4+ T cells. We demonstrate for the first time that CD8+ T cells have a lower level of HAT ATF2 expression than CD4+ T cells. The decreased ATF2 in CD8+ seems to be able to functionally reduce the response of this cell type to steroids, because RNA silencing of ATF2 inhibited the steroid-responsive CD4+ T cells as well. These data suggest that molecular manipulation of ATF2 expression and activity may offer a novel approach for modifying cellular steroid responsiveness.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Maureen Sandoval for help in preparing this manuscript.
This work was supported in part by National Institutes of Health (NIH) grants AR41256, 5R21AR051634, AI070140, and HL37260, NIH/National Institute of Allergy and Infectious Diseases contracts N01 AI40029 and N01 AI40030, General Clinical Research Center grant MO1 RR00051 from the Division of Research Resources, the Edelstein Family Chair in Pediatric Allergy and Immunology, and the University of Colorado Cancer Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases, National Heart, Lung, and Blood Institute, or the National Institutes of Health.
National Institutes of Health
Authorship
Contribution: L.-b.L. performed experiments, analyzed the data, and wrote the manuscript; D.Y.M.L. helped with data interpretation and discussion; M.J.S. attested to the soundness of the experimental design, analysis, and conclusion of the manuscript; and E.G. designed experiments, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Elena Goleva, National Jewish Medical Research Center, 1400 Jackson St, Rm K1020, Denver, CO 80206; e-mail: golevae@njc.org.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal