Abstract
The incidence of respiratory virus infection after hematopoietic cell transplantation (HCT) has probably been underestimated with conventional testing methods in symptomatic patients. This prospective study assessed viral infection episodes by testing weekly respiratory samples collected from HCT recipients, with and without symptoms reported by questionnaire, for 100 days after HCT. Samples were tested by culture and direct fluorescent antibody testing for respiratory syncytial virus (RSV), parainfluenza virus (PIV), and influenza A and B, and by quantitative reverse transcription–polymerase chain reaction for RSV, PIV, influenza A and B, and metapneumovirus (MPV). Of 122 patients, 30 (25%) had 32 infection episodes caused by RSV (5), PIV (17), MPV (6), influenza (3), RSV, or influenza (1). PIV, with a cumulative incidence estimate of 17.9%, was the only virus for which asymptomatic infection was detected. Lower virus copy number in patients with no or one symptom compared with 2 or more symptoms was found for all viruses in all patients (P < .001), with PIV infection having a similar virus-specific comparison (P = .004). Subclinical infection with PIV may help explain why infection-control programs that emphasize symptoms are effective against RSV and influenza but often not against PIV.
Introduction
Respiratory virus infections after hematopoietic cell transplantation (HCT) have the potential to result in serious respiratory disease. Estimates of the incidence of respiratory virus infections vary widely over time depending on the patient population, transplantation regimen, and type of surveillance instituted, but the most common viruses described in the HCT population are respiratory syncytial virus (RSV), parainfluenza virus (PIV), and influenza virus.1-14 Recent studies have described overall virus-specific rates of 5%, 7%, and 1% to 2% for RSV, PIV, and influenza, respectively, during the first 100 days after transplantation.1-4 These viral infections have been reported to progress from upper respiratory tract infection (URI) to pneumonia in 18% to 44% of cases, with mortality ranging from 25% to 45% within 30 days after the diagnosis of pneumonia.1-4 Among long-term survivors, history of respiratory virus infection during the initial 100 days after HCT confers a significant risk for late airflow obstruction, which is also associated with increased overall mortality.15,16
Previous studies have probably underestimated the true incidence of respiratory virus infections in HCT recipients, because conventional diagnostic testing strategies lack sensitivity and testing has been traditionally applied only to symptomatic inpatients. Real-time quantitative reverse transcription–polymerase chain reaction (RT-PCR) assays now permit rapid, sensitive, and specific detection of PIV, RSV, and influenza virus infections, as well as detection of new viruses not demonstrated by culture-based methods.17 For example, newly described viruses such as human metapneumovirus (MPV) are best detected by RT-PCR.18 MPV has been reported to be associated with respiratory disease and to cause fatal pneumonia among HCT recipients.9,19-22 Sensitive molecular methods also allow for quantitation of viral load in respiratory specimens.
Although asymptomatic viral shedding in the nasopharynx among immunocompetent hosts is relatively common,23,24 previous data are inconclusive as to whether HCT recipients also exhibit asymptomatic respiratory virus infection, and it is unknown whether these infections are transient or progress to clinical infection.6,25-29 If subclinical respiratory virus infection is transmissible and leads to progression of symptomatic disease, asymptomatic shedding among HCT recipients could potentially play an important role in propagating outbreaks of respiratory disease. Detection of asymptomatic infection among HCT recipients might provide an opportunity for early therapeutic intervention before progression to clinical illness. In addition, even intervention for asymptomatic infection alone might be useful, because it has been shown that lower respiratory tract infection (LRI) with PIV, RSV, and influenza, and even PIV URI that does not progress to pneumonia, are associated with late pulmonary sequelae after HCT.15,16 Asymptomatic shedding of PIV in the respiratory tract may contribute to sustained inflammation or activation of an inflammatory process that leads to irreversible airway damage.16,30
To our knowledge, this prospective surveillance study is the first to describe quantitative RT-PCR detection of respiratory viruses after HCT in recipients both with and without reported symptoms by using a standardized symptom survey. This novel approach provides information regarding the full spectrum of respiratory virus infections in the HCT population, including the incidence of asymptomatic respiratory virus infections after HCT. We analyzed detailed symptom data and quantitative viral load patterns to determine the associations between viruses, symptoms, and viral quantity as determined by RT-PCR.
Patients, materials, and methods
Patients
This study was a prospective, longitudinal surveillance study of patients for 100 days after HCT and was approved by the Fred Hutchinson Cancer Research Center Institutional Review Board. Informed consent was obtained in accordance with the Declaration of Helsinki. The study period began in December 2000, and patients were followed during 4 winter/spring seasons (between the months of November and June) until June 2004. A denominator of 814 patients received transplants during the study period, and HCT recipients of all transplant types and all ages were eligible. Patients received information regarding the study after arrival to the center and were enrolled on the basis of willingness to provide informed consent. Patients were followed weekly while hospitalized on the oncology/HCT unit and after hospital discharge in the outpatient clinic. Primary end points included respiratory virus URI or LRI or asymptomatic shedding. Study subjects completed a baseline information form and a standardized weekly symptom survey that included 12 symptoms (Figure 2). Clinical chart review was performed for subjects with asymptomatic infection to determine whether symptoms occurred outside of the study visit.
Virologic methods
Weekly nasopharyngeal wash (or nasopharyngeal swab) and oropharyngeal swab samples were collected from each subject. Nasopharyngeal washes were collected with a total of 10 mL of saline (5 mL/nostril) for adults and 5 mL (2.5 mL/nostril) for children and combined into one sterile vial with the oropharyngeal swab. For one 1-year-old child, a range of 2 to 5 mL was used for each nasal wash. After collection, nasal-wash recovery volumes averaged 60% of the instilled volume. Bronchoalveolar (BAL) and other clinical respiratory specimens were collected when available. Lung biopsy and autopsy specimens were reviewed, and a representative block with features most suggestive of a viral etiology was tested by RT-PCR. Respiratory samples were tested for RSV, PIV, and influenza by conventional tube culture or shell vial centrifugation culture with rhesus monkey kidney, Buffalo green monkey kidney, HL (transformed HeLa), human foreskin fibroblasts, and A-549 cell lines. Specimens were also tested by type-specific respiratory direct fluorescent antibody (DFA) testing for RSV, influenza virus types A and B, and PIV types 1 through 3 using commercially available, type-specific antiserum (Chemicon, Temecula, CA).3,31
To prepare respiratory samples for RT-PCR assays, total nucleic acids were isolated from 200 μL of each respiratory specimen.17,32 To ensure that negative results were not a result of poor nucleic acid extraction or assay inhibition, a known quantity of a 262-base RNA transcript derived from jellyfish DNA (EXO) was added to the lysis buffer, and a corresponding primer was added to the reaction.17,32 All negative samples had evidence of amplification of the EXO control. One low positive control that contained 200 to 1000 copies/RT-PCR reaction of each respiratory virus harvested from cell culture and one negative control that consisted of cultured, uninfected human epithelial cells were processed with each batch of specimens. Separate previously described quantitative real-time RT-PCR assays were used to detect 7 RNA viruses, including RSV, influenza virus types A and B, PIV types 1 through 3, and MPV.17,32,33 Each assay reliably detected 10 viral copies per reaction, which provided a sensitivity of 1100 copies/mL (10 μL of specimen added per reaction).17,32,33 Specimens with positive results of less than 10 copies/reaction were repeated to confirm positivity. Interassay standard deviations and coefficients of variation (standard deviation divided by the mean × 100) were calculated within and between virus-specific RT-PCR runs to evaluate assay reproducibility. The coefficient of variation for viral quantification was less than 10% for each respiratory virus RT-PCR assay when estimated by using the results of the low positive control.32,33 The investigators who performed RT-PCR testing were blinded to the patients' culture and DFA results. Both the DNA and RT-PCR methods were performed according to College of American Pathologist standards, and the laboratories participated in and passed proficiency testing in viral diagnostics.
Criteria for analysis and definitions
Subjects who had submitted at least 4 serial samples within the first 100 days after transplantation were included in analysis. Patients who died before 100 days after transplantation but had submitted a sample within 14 days of death were also eligible. An infection episode was defined as detection of respiratory virus in a subject's respiratory sample by any laboratory method. Total duration of virus shedding was calculated from the first positive test result to the first negative test result after the final positive sample, provided that the patient did not become asymptomatic for more than 1 week. If 2 different respiratory viruses were detected and separated in time by at least 1 month, a subject was defined as having 2 infection episodes, whereas a subject infected with 2 viruses simultaneously had a coinfection (one infection episode). Asymptomatic infection was defined as detectable virus in the patient's upper respiratory sample without documented symptoms of respiratory illness on the symptom questionnaire. URI was defined as the detection of a respiratory virus from an upper respiratory sample in conjunction with any respiratory symptoms. LRI was defined as respiratory virus detection from a BAL specimen collected from patients with radiographic signs of LRI.
Underlying disease risk (risk of relapse for malignancies and survival for nonmalignant diseases) was categorized as low, intermediate, or high according to disease status at time of HCT by using previously published criteria.16 Acute graft-versus-host disease (GvHD) was graded by using conventional criteria and categorized as acute GvHD grades 0 to 1 and grades 2 through 4.34
Statistical methods
The probability of acquiring at least one infection was estimated by using a cumulative incidence curve for each type of infection and for any type. Patient records were censored at 14 days past the time of last eligible respiratory sample, death, or day 100 after transplantation, whichever occurred first. Death before day 100 and within 14 days of a patient's last sample was treated as a competing risk for infection.
Linear regression was used to analyze the virus copies per milliliter in secretion associated with the presence of symptoms. Logistic regression was used to analyze the occurrence of each symptom type as a function of the presence of seasonal allergies and the survey day relative to the date of transplantation. Measurements that were based on multiple respiratory specimens per patient were entered into the analysis as repeated measures, with adjustment for possible correlation between values within a subject by using generalized estimating equations.35
Cox regression models were used to estimate the associations of potential risk factors for respiratory virus acquisition. Lymphopenia within the previous 14 days and acute GvHD were included as time-dependent covariates. Lymphopenia was assessed as a predictor of respiratory virus infection only among patients with lymphocyte engraftment (defined as at least 3 consecutive days with an absolute lymphocyte count > 300). Adjusted hazard ratios were obtained by using a multivariable Cox regression model including any covariate found to be significant in univariate analysis and factors with biologic plausibility to contribute to respiratory virus acquisition.
P values from regression models were obtained from the Wald test, and no adjustments were made for multiple comparisons. Two-sided P values of less than .05 were considered to be statistically significant. Statistical analyses were performed by using Stata 9.0 (StataCorp, College Station, TX) and SAS 8.0 (SAS Institute, Cary, NC).
Results
Respiratory virus incidence
From December 2000 to June 2004, during the winter/spring seasons, 157 patients submitted at least one qualified sample, and 122 study subjects had at least 4 serial samples available for analysis. These 122 subjects completed a median number of 9 visits (range, 2-18 visits). The clinical characteristics of the study population are shown in Table 1.
Characteristics of the HCT recipient cohort at the Fred Hutchinson Cancer Research Center, winter/spring seasons of December 2000-June 2004 (N = 122)
| Characteristic . | No. . |
|---|---|
| Median age, y (range) | 47 (1-71) |
| Sex, male, no. (%) | 73 (60) |
| HCT donor type, no. (%) | |
| Autologous/syngeneic | 22 (18) |
| Matched sibling | 42 (34) |
| Mismatched or unrelated | 58 (48) |
| Underlying disease, no. (%) | |
| Acute leukemia | 34 (28) |
| Chronic leukemia | 17 (14) |
| Myelodysplastic syndrome | 20 (16) |
| Non-Hodgkin lymphoma | 23 (19) |
| Multiple myeloma | 14 (11) |
| Other | 14 (11) |
| Underlying disease risk, no. (%)* | |
| Low | 19 (16) |
| Intermediate | 42 (34) |
| High | 61 (50) |
| Nonmyeloablative conditioning regimen† | 35/100 (35) |
| Stem cell source, no. (%) | |
| Bone marrow | 18 (15) |
| Peripheral blood | 103 (84) |
| Bone marrow and peripheral blood | 1 (1) |
| CMV serostatus, no. (%) | |
| Recipient seropositivity | 60 (49) |
| Donor seropositivity† | 40/100 (40) |
| Acute GvHD†, no. (%) | |
| Grade 0 or 1 | 38/100 (38) |
| Grade 2-4 | 62/100 (62) |
| Underlying seasonal allergies‡ | 35/111 (32) |
| Characteristic . | No. . |
|---|---|
| Median age, y (range) | 47 (1-71) |
| Sex, male, no. (%) | 73 (60) |
| HCT donor type, no. (%) | |
| Autologous/syngeneic | 22 (18) |
| Matched sibling | 42 (34) |
| Mismatched or unrelated | 58 (48) |
| Underlying disease, no. (%) | |
| Acute leukemia | 34 (28) |
| Chronic leukemia | 17 (14) |
| Myelodysplastic syndrome | 20 (16) |
| Non-Hodgkin lymphoma | 23 (19) |
| Multiple myeloma | 14 (11) |
| Other | 14 (11) |
| Underlying disease risk, no. (%)* | |
| Low | 19 (16) |
| Intermediate | 42 (34) |
| High | 61 (50) |
| Nonmyeloablative conditioning regimen† | 35/100 (35) |
| Stem cell source, no. (%) | |
| Bone marrow | 18 (15) |
| Peripheral blood | 103 (84) |
| Bone marrow and peripheral blood | 1 (1) |
| CMV serostatus, no. (%) | |
| Recipient seropositivity | 60 (49) |
| Donor seropositivity† | 40/100 (40) |
| Acute GvHD†, no. (%) | |
| Grade 0 or 1 | 38/100 (38) |
| Grade 2-4 | 62/100 (62) |
| Underlying seasonal allergies‡ | 35/111 (32) |
Low-risk diseases included chronic myeloid leukemia (CML) in chronic phase, refractory anemia, and aplastic anemia. Intermediate-risk diseases included CML in accelerated phase or in chronic phase after blast phase, acute leukemia or lymphoma in remission, refractory anemia with excess blasts, chronic lymphocytic leukemia, and paroxysmal nocturnal hemoglobinuria. High-risk diseases included CML in blast phase, juvenile CML, acute leukemia or lymphoma in relapse, newly diagnosed acute leukemia, refractory anemia with excess blasts in transformation, myeloma, immunodeficiency syndromes, renal cell carcinoma, and testicular carcinoma.
Characteristic applies to allogeneic transplantations only.
Excluding food and drug allergies. Allergy data were not available for 11 subjects.
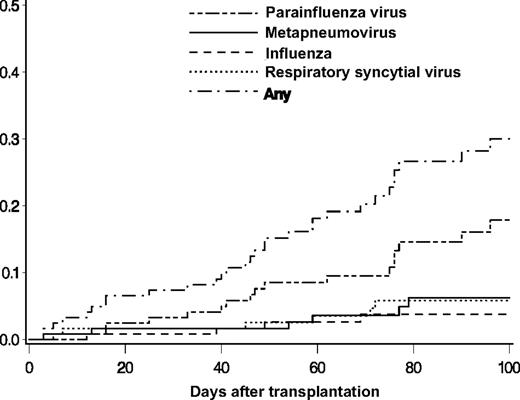
Respiratory samples from 30 (25%) of 122 subjects had a respiratory virus detected (Table 2). Of the 30 subjects, 2 were children (aged 1 and 6 years). There were 32 incident infection episodes in the 30 subjects; 2 patients had 2 separate infection episodes, one with RSV followed by MPV and the second with PIV followed by MPV. PIV was the most frequent infection, with 17 episodes and a cumulative incidence estimate of 17.9% (95% confidence interval [CI], 9.8-25.9) at day 100 (Figure 1). The cumulative incidence estimates of MPV, RSV, and influenza at day 100 were 6.2% (95% CI, 1.3-11.2), 5.8% (95% CI, 1.2-10.4), and 3.7% (95% CI, 0.1-7.3), respectively (Figure 1). One influenza infection episode was caused by influenza A, 2 were caused by influenza B, and one subject had a coinfection with RSV and influenza B. The probability of an infection episode with one of the 4 viruses in the first 100 days after HCT was 30% (95% CI, 20.5-39.5).
Characteristics of 32 primary infection episodes (in 30 subjects) attributable to respiratory viruses after HCT
| Virus . | No. of infection episodes . | Days to positive test, median (range) . | Days of viral shedding, median (range)* . | No. of infection episodes by diagnostic method . | No. of infection episodes with symptoms . | ||||
|---|---|---|---|---|---|---|---|---|---|
| RT-PCR . | RT-PCR + DFA/culture . | DFA/culture . | Asymptomatic . | LRI . | URI . | ||||
| RSV | 6 | 52 (5-72) | 16 (5-39) | 1 | 5 | 0 | 0 | 1 | 5 |
| PIV | 17 | 47 (12-96) | 11 (6-42) | 8 | 6 | 3† | 6 | 2 | 9 |
| MPV | 6 | 57 (3-79) | 16 (7-24) | 6 | — | — | 0 | 0 | 6 |
| Influenza | 4 | 44 (3-69) | 14 (7-24) | 0 | 4 | 0 | 0 | 0 | 4 |
| Total | 32‡ | 49 (3-96) | 14 (5-42) | 15 | 15 | 3 | 6 | 3 | 23 |
| Virus . | No. of infection episodes . | Days to positive test, median (range) . | Days of viral shedding, median (range)* . | No. of infection episodes by diagnostic method . | No. of infection episodes with symptoms . | ||||
|---|---|---|---|---|---|---|---|---|---|
| RT-PCR . | RT-PCR + DFA/culture . | DFA/culture . | Asymptomatic . | LRI . | URI . | ||||
| RSV | 6 | 52 (5-72) | 16 (5-39) | 1 | 5 | 0 | 0 | 1 | 5 |
| PIV | 17 | 47 (12-96) | 11 (6-42) | 8 | 6 | 3† | 6 | 2 | 9 |
| MPV | 6 | 57 (3-79) | 16 (7-24) | 6 | — | — | 0 | 0 | 6 |
| Influenza | 4 | 44 (3-69) | 14 (7-24) | 0 | 4 | 0 | 0 | 0 | 4 |
| Total | 32‡ | 49 (3-96) | 14 (5-42) | 15 | 15 | 3 | 6 | 3 | 23 |
— indicates not done.
Follow-up specimens were available for analysis in 19 of the 32 total infection episodes.
Three PIV infections were detected only by conventional testing; 2 of 3 subjects had samples sent for clinical testing that were unavailable to be tested by RT-PCR. The third subject's respiratory sample was tested by RT-PCR and culture and found to be positive by culture alone.
There was one influenza B/RSV coinfection; therefore, the column total is 32 instead of 33.
Cumulative incidences of first infection episodes of PIV, MPV, influenza, and RSV after transplantation in 122 HCT recipients.
Cumulative incidences of first infection episodes of PIV, MPV, influenza, and RSV after transplantation in 122 HCT recipients.
The 32 infection episodes were detected a median of 49 days (range, 3-96 days) after HCT (Table 2). Twenty-two (69%) of the episodes were diagnosed in the outpatient department. The number of infection episodes detected varied according to different testing methods. For RSV, 5 infection episodes were detected by both RT-PCR and conventional testing (DFA and/or culture), and 1 additional episode was detected by RT-PCR alone. All 4 influenza infections were detected by both RT-PCR and conventional testing, whereas the 6 MPV infections were detected by RT-PCR alone because DFA and culture were not available. RT-PCR testing gave the highest yield for PIV, for which 8 of 17 infection episodes were detected by RT-PCR alone (Table 2). Specimens were available for complete follow-up in 19 of the 32 infection episodes; the median length of virus shedding during these episodes was 14 days (range, 5-42 days; Table 2).
Respiratory specimens from 3 subjects with PIV3 exhibited prolonged and intermittent viral positivity. In all 3, PIV3 was initially undetectable after 7 to 9 days followed by an additional positive test result 16 to 37 days after the first test. Symptom surveys and RT-PCR testing of specimens were performed weekly during the negative interval, and respiratory symptoms were reported on all but one survey. Specimens from only one other patient with RSV, MPV, or influenza infections had a similar negative interval with symptoms present throughout the infection episode: a patient with influenza B initially had a respiratory specimen test result that was negative after 7 days followed by an additional positive test result 17 days after the first.
Asymptomatic infection
Six subjects had asymptomatic infection episodes, all associated with the detection of PIV. By contrast, none of the patients with RSV, MPV, or influenza had subclinical infection identified in the absence of symptoms. Of the 6 asymptomatic infection episodes, PIV1 and PIV3 were each detected in 3 episodes. Subject ages ranged from 26 to 57 years; 5 (83%) were male and had received allogeneic transplantation (Table 3). All were diagnosed in the outpatient department on days 33 to 96 after HCT. Patient 6 had a follow-up specimen that was cultured 6 days later that remained positive, and then no further testing was performed. Only patient 1 had a follow-up test result that was negative 2 weeks after the positive test result. The other 4 patients did not have a follow-up test or symptom survey; 3 were near the end of the study period. No symptoms were noted in clinic-visit notes at the time PIV was detected. Of 6 subjects, 5 were alive at the 6-month follow-up; 1 subject died 7 weeks after transplantation as a result of recurrent acute myeloid leukemia.
Characteristics of 6 patients with PIV asymptomatic respiratory virus infection episodes after HCT
| Patient no. . | No. of positive respiratory samples (virus) . | Age, y . | Sex . | Primary diagnosis . | Primary disease status . | HCT month/year . | HCT donor type . | Cell source . | Transplant no. . | Myeloablative . | Days to neutrophil engraftment after HCT . | Days to viral detection after HCT . | Detection method . | Symptom follow-up (chart review) . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 (PIV1) | 38 | M | ALL | Remission | April 2001 | Syn | PBSCs | 1 | No | 11 | 41 | PCR | Runny nose noted 1 wk before, no symptoms at or after PIV-positive test |
| 2 | 1 (PIV1) | 57 | M | AML | Relapse | May 2003 | Allo | PBSCs | 1 | No | 17 | 33 | PCR | No symptoms noted |
| 3 | 1 (PIV3) | 48 | M | NHL | Remission | May 2003 | Allo | PBSCs | 2 | Yes | 16 | 96 | PCR | No symptoms noted |
| 4 | 1 (PIV3) | 50 | F | AML | Remission | April 2003 | Allo | PBSCs | 1 | No | 17 | 90 | PCR and culture | 6 d after positive PIV test: runny nose, sneezing, cough, wheeze (noted as allergies in chart) |
| 5 | 1 (PIV3) | 55 | M | CML | Blast crisis | May 2004 | Allo | PBSCs | 1 | No | 13 | 76 | PCR | No symptoms noted |
| 6 | 2 (PIV1) | 26 | M | CML | Accelerated phase | May–June 2003 | Allo | PBSCs | 1 | No | 16 | 40 | Culture | No symptoms noted |
| Patient no. . | No. of positive respiratory samples (virus) . | Age, y . | Sex . | Primary diagnosis . | Primary disease status . | HCT month/year . | HCT donor type . | Cell source . | Transplant no. . | Myeloablative . | Days to neutrophil engraftment after HCT . | Days to viral detection after HCT . | Detection method . | Symptom follow-up (chart review) . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 (PIV1) | 38 | M | ALL | Remission | April 2001 | Syn | PBSCs | 1 | No | 11 | 41 | PCR | Runny nose noted 1 wk before, no symptoms at or after PIV-positive test |
| 2 | 1 (PIV1) | 57 | M | AML | Relapse | May 2003 | Allo | PBSCs | 1 | No | 17 | 33 | PCR | No symptoms noted |
| 3 | 1 (PIV3) | 48 | M | NHL | Remission | May 2003 | Allo | PBSCs | 2 | Yes | 16 | 96 | PCR | No symptoms noted |
| 4 | 1 (PIV3) | 50 | F | AML | Remission | April 2003 | Allo | PBSCs | 1 | No | 17 | 90 | PCR and culture | 6 d after positive PIV test: runny nose, sneezing, cough, wheeze (noted as allergies in chart) |
| 5 | 1 (PIV3) | 55 | M | CML | Blast crisis | May 2004 | Allo | PBSCs | 1 | No | 13 | 76 | PCR | No symptoms noted |
| 6 | 2 (PIV1) | 26 | M | CML | Accelerated phase | May–June 2003 | Allo | PBSCs | 1 | No | 16 | 40 | Culture | No symptoms noted |
ALL indicates acute lymphocytic leukemia; AML, acute myeloid leukemia; NHL, non-Hodgkin lymphoma; CML, chronic myeloid leukemia; Syn, Syngeneic; Allo, Allogeneic; PBSCs, peripheral blood stem cells.
Symptoms and respiratory virus infection episodes
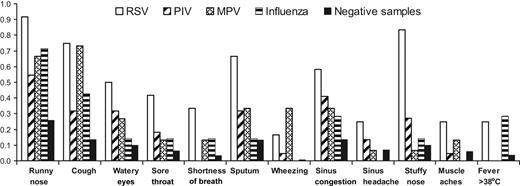
Twenty-three URI and 6 asymptomatic infection episodes were detected in 29 HCT recipients. These patients provided a total of 55 respiratory samples accompanied by symptom surveys in which RSV (n = 11), PIV (n = 22), MPV (n = 15), influenza (n = 6), or RSV/influenza B (n = 1) were detected by RT-PCR and 284 samples that tested negative for these viruses by any testing method. The mean number of symptoms (and range per positive sample) for RSV, PIV, MPV, and influenza were 5 (0-12), 2 (0-8), 3 (0-6), and 2 (0-5), respectively. In general, patients reported symptoms more frequently on the survey form when their respiratory samples had RSV, PIV, MPV, or influenza detected than when their samples tested negative (Figure 2). There was no association with preexisting seasonal allergies and symptoms, but symptoms were increased during the first 30 days after HCT, the period during which mucositis was most likely to be present (data not shown).
Symptoms reported on symptom surveys simultaneous to respiratory samples collected from HCT recipients with infection episodes caused by respiratory viruses after HCT. Proportion of surveys with symptoms reported are shown for weeks in which respiratory samples tested positive for RSV, PIV, MPV, or influenza vs weeks during which samples tested negative for all 4 viruses.
Symptoms reported on symptom surveys simultaneous to respiratory samples collected from HCT recipients with infection episodes caused by respiratory viruses after HCT. Proportion of surveys with symptoms reported are shown for weeks in which respiratory samples tested positive for RSV, PIV, MPV, or influenza vs weeks during which samples tested negative for all 4 viruses.
Nearly all symptoms were reported in the highest proportion of surveys when RSV was detected compared with other virus detections or when samples tested negative (Figure 2). The proportion of symptoms present during weeks in which samples tested positive for PIV were lower than for RSV, probably because of the inclusion of the 6 asymptomatic persons with PIV infection. Influenza was associated with symptoms similar to those with the other viruses, except no patients reported wheezing, sinus headache, or myalgias with influenza infection, and 2 (29%) of the 7 samples that tested positive for influenza were accompanied by fever (Figure 2). Fever was present with the first specimen collected in one patient and with the second specimen collected in the other patient. All patients with influenza were treated with oseltamivir, but the effect of treatment on symptom burden could not be analyzed because of small numbers.
Infection episodes resulted in LRI in 3 patients: one secondary to RSV on day 7 after transplantation and 2 attributable to PIV3 on days 16 and 77 after transplantation. Death of the patient with preengraftment PIV3 infection occurred on day 19; this patient also had cytomegalovirus (CMV) detected concurrently on BAL and a history of pretransplantation pulmonary aspergillus. Lung biopsy and autopsy specimens from 5 study subjects tested negative by RT-PCR. None of the tissue specimens were obtained at a time concurrent to when the patient had a documented infection episode.
Quantitative respiratory virus load
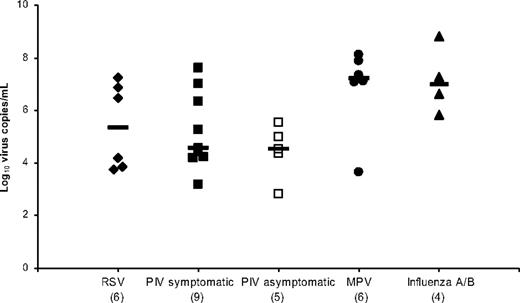
There were 27 patients with at least one positive RT-PCR value with a total of 57 positive RT-PCR measurements. Including only the maximum RT-PCR value per infection episode, the median virus-specific viral loads were highest for the samples that were positive for MPV (1.7 × 107 copies/mL) and influenza (1.2 × 107 copies/mL), with no significant difference between symptomatic and asymptomatic PIV episodes (Figure 3). Analysis of uncorrected viral loads was compared with viral loads corrected by volume of nasal-wash sample, and we found minimal differences that were within the range of assay reproducibility (less than one half of the log10 viral load; data not shown).
Respiratory virus–specific median viral loads calculated by using the maximum value per infection episode. The x-axis shows the virus responsible (number of infection episodes), and the horizontal bars show the median values.
Respiratory virus–specific median viral loads calculated by using the maximum value per infection episode. The x-axis shows the virus responsible (number of infection episodes), and the horizontal bars show the median values.
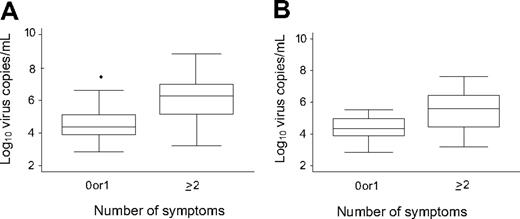
When all RT-PCR values were analyzed, the median viral load levels corresponding to one or more symptoms (8.6 × 105 copies/mL [range, 1.6 × 103 to 6.9 × 108 copies/mL]) were significantly higher than the median viral load that corresponded to no symptoms (2.33 × 105 copies/mL [range, 7.0 × 102 to 4.4 × 106 copies/mL]; P < .001). After exploratory analysis, we found that the same was true for viral load levels that corresponded to 2 or more symptoms (1.8 × 106 copies/mL [range, 1.6 × 103 to 6.9 × 108 copies/mL]) vs no or one symptom (2.4 × 104 copies/mL [range, 7.0 × 102 to 3.0 × 107 copies/mL]; P < .001; Figure 4A). There were sufficient values for virus-specific comparisons only for PIV. The results for 2 or more symptoms compared with no or one symptom were similar to the overall comparison (P = .004; Figure 4B). However, the comparison of one or more symptoms vs none was not significant (P = .10).
Quantitative viral load associated with number of symptoms in samples collected during respiratory virus infection episodes after HCT. (A) All samples that tested positive for respiratory viruses (P < .001). (B) Samples that tested positive for PIV (P = .004). The box plot shows the median and 75th and 25th percentiles; whiskers extend to the upper and lower adjacent values.
Quantitative viral load associated with number of symptoms in samples collected during respiratory virus infection episodes after HCT. (A) All samples that tested positive for respiratory viruses (P < .001). (B) Samples that tested positive for PIV (P = .004). The box plot shows the median and 75th and 25th percentiles; whiskers extend to the upper and lower adjacent values.
Risk factors for acquisition of respiratory virus infection
In univariate analysis, only recipient CMV seropositivity was associated with increased risk for respiratory virus acquisition. Other factors, including patient age, sex, donor type, stem cell source, donor CMV serostatus, conditioning regimen, underlying disease risk, seasonal allergies, time to lymphocyte engraftment, postengraftment lymphopenia, and acute GvHD, were not significantly associated with respiratory virus acquisition. Recipient CMV seropositivity remained significant after adjustment for age, conditioning regimen, and history of seasonal allergies in a multivariable model (hazard ratio = 4.1; 95% CI, 1.7-10.1; P = .002).
Discussion
Using sensitive and quantitative RT-PCR techniques in a weekly surveillance study, we have described an overall probability of 30% for infection episodes attributable to 4 common community-acquired respiratory viruses among a cohort of HCT recipients, regardless of the presence of respiratory symptoms, during the first 100 days after HCT. The probability of infection with RSV and influenza was similar in the present study to reports documented in the literature with a similar time frame.1,2,4 The cumulative incidence estimate for PIV infection of 17.9% at day 100 after HCT is higher than that shown in previous studies, although testing of respiratory specimens was performed only during the winter season and PIV circulates year-round.3,14 This higher incidence is probably a result of 3 factors: (1) RT-PCR testing identified half of the PIV-positive specimens that would have remained unidentified with conventional culture and DFA testing methods; (2) many of the episodes of PIV infection were asymptomatic; and (3) it seems that persistent shedding of asymptomatic parainfluenza may be relatively common in HCT populations, which provides a means of increasing transmission to others. Although prolonged shedding and persistence of PIV has been described in HCT recipients,3,14,36,37 ours is the first documentation of subclinical shedding in this population.
We found a 5.8% cumulative incidence of MPV, similar to previous studies that have described MPV infection in 5% to 9% of symptomatic adults with hematologic malignancy, including a prospective study of symptomatic HCT recipients after transplantation in which patients were followed for more than 1 year.9,22 The incidence of MPV infection in our study is lower, however, than that in a recent report describing MPV-positive nasopharyngeal aspirate samples in 86% of 21 adults in the first 180 days after HCT.9,38 The latter study also described detection of MPV in asymptomatic patients, a finding that we did not encounter. These authors suggested that the MPV infections in their patients after HCT may have originated in the hospital, on the basis of their finding of identical DNA sequences for every MPV-positive specimen.38 Therefore, the epidemiology of the MPV infections in their study would be different from that in our study, in which 4 of 6 MPV infections were acquired in the community. There was no documented transmission in our population, but 2 patients in the outpatient department tested positive within 4 days of one another. Differences in the severity of community outbreaks and strains of MPV may also be factors that affect the variation between studies. Experimental inoculation and longitudinal studies have documented the relatively frequent existence of asymptomatic viral shedding (RSV, PIV) among immunocompetent hosts,23,24,39 but controversy exists as to whether HCT recipients also exhibit asymptomatic viral shedding.6,25,27-29 Additional study using a prospective symptom-survey instrument and year-round serial testing for respiratory viruses is warranted to clarify the extent of subclinical shedding of respiratory viruses in HCT recipients.
PIV was the only virus detected in asymptomatic infection in the current study. Although we did not evaluate for transmission from asymptomatic carriers, it is interesting to note that 6 of the 17 PIV infections occurred within a 5-week period during April through May 2003; 4 of these infections were asymptomatic. Our description of subclinical infections raises several hypotheses regarding the potential for transmission. PIV is well described as a cause of severe and prolonged outbreaks of respiratory tract infections in hospital wards, neonatal nurseries, outpatient clinics, long-term care facilities, and other institutional settings.40-46 Outbreaks of PIV after HCT have been reported in both the inpatient and outpatient settings, and early infection-control measures are crucial.42,43 Even with good surveillance and isolation, prolonged outbreaks at our center and elsewhere suggest that symptom-based infection-control strategies that are successful for RSV or influenza may be less effective for the prevention of nosocomial PIV.42,43,45 To our knowledge, this is the first documentation of subclinical infection in which symptoms were never present. Because all of the asymptomatic infections in our current study occurred in the outpatient department, the issue of subclinical shedding has important implications for infection-control policies, which currently recommend surveillance and testing of symptomatic HCT patients while asymptomatic patients move freely about the outpatient clinics. Transmission to health care workers and family members in this setting may occur and perpetuate ongoing transmission.
Asymptomatic PIV shedding, subsequent to symptoms or subclinical infection alone, may also provide an explanation for airflow decline associated with PIV infection of the upper respiratory tract.3,15,16 Documentation of subclinical infection may be important for detecting and treating infection that might eventually progress to symptomatic respiratory disease and may have therapeutic implications if persistence of asymptomatic shedding is found to be associated with long-term decline in lung function. Even patients in our cohort who were initially symptomatic had prolonged persistence of symptoms with intermittent RT-PCR PIV3 positivity of respiratory specimens. This intermittent subclinical infection could be related to the sensitivity of the RT-PCR assay and suggests that PIV can persist below the level of detection and, therefore, might contribute to ongoing pathogenesis and transmission.
This study also evaluated quantitative respiratory virus load and measures of disease severity in immunocompromised patients. The results of previous studies in infants have differed as to whether there is an association between higher RSV viral loads measured with quantitative culture and increased disease severity.47,48 Considering RSV, PIV, MPV, and influenza infections collectively, our study supports the hypothesis that more severe disease, as described by the presence of an increased number of symptoms at the time of sample collection, correlates with increased quantitative RT-PCR viral load from upper respiratory samples. Although symptomatic and asymptomatic PIV infection episodes had similar median values for maximum viral load per infection episode (Figure 3), we found a significant difference in median viral load when we analyzed all PIV-positive samples using a cut-off of 0 or 1 symptom versus 2 or more symptoms instead of strict asymptomatic vs symptomatic categories (Figure 4). We chose this method of analysis to account for the use of our sensitive symptom survey, in which it was not unusual for a person to report a single symptom, such as “cough,” “runny nose,” or “watery eyes,” for an entire infection episode. Because virus was detected in the presence of even one symptom, clinical assessment of symptoms seemed to be a rather crude indicator of true viral infection in this study. Because of the overall small sample size and lack of specimens for RT-PCR testing and simultaneous symptom data for the 3 LRI episodes in this study, we are unable to comment on whether a difference in viral load is observed between upper and lower respiratory tract disease.
Because we describe relatively small numbers of each virus infection, collective analysis of all infection episodes may limit conclusions regarding specific viruses, such as with respect to quantitative viral load and risk factors for virus acquisition. Our study population represents approximately 15% of the total patients who received a transplant at our center during the study period. The protocol was quite involved without direct benefit to the patient; thus, many patients opted not to participate. The characteristics of the study population seem to be generally representative of the overall population of patients who received a transplant during the time period, with a slight predominance of allogeneic HCT recipients.49 This study did not include testing for other common respiratory viruses, including rhinoviruses, coronaviruses, and adenoviruses, which may also cause respiratory symptoms after HCT. Despite this, the yield of PIV detection doubled with RT-PCR compared with conventional methods. Prospective year-round testing for additional respiratory viruses in future studies will increase the overall incidence estimates to provide a more complete picture of the true burden of respiratory virus disease after HCT. Some of the samples that were classified as testing negative for a respiratory virus in the presence of symptoms may actually have been positive for yet-untested viruses. However, it is probable that misclassification of samples as negative would bias the findings toward the null and diminish any virus-specific symptom associations. The significant association between recipient CMV seropositivity and respiratory virus acquisition measured in our study has not been observed in other, larger cohort studies designed to examine risk factors for acquisition of respiratory viral infection after HCT. It is likely that our cohort was too small to provide an adequate assessment of risk factors.1,3,4
In conclusion, testing using RT-PCR increased the diagnosis of respiratory virus infections, especially of PIV, over that which would have been achieved by testing only symptomatic patients and using conventional testing methods. Prospective collection of respiratory samples from HCT recipients was well tolerated. We found that PIV causes asymptomatic infection in HCT recipients that is not preceded by clinical illness, which may contribute to pathogenesis in the infected patient and provide a means of transmission to others. Further study is needed to document rates of asymptomatic PIV infection in a larger cohort of HCT recipients, elucidate potential factors that may protect HCT recipients from developing symptomatic PIV infection, and identify whether subclinical infection in this patient population ultimately progresses to clinical infection or leads to transmission among other HCT recipients and immunocompetent persons such as health care workers.
Presented in abstract form at the annual meeting of the Infectious Diseases Society of America, San Francisco, CA, October 8, 2005 (Abstract 710), and a tandem meeting of the American Society of Bone Marrow Transplant/Center for International Blood and Marrow Transplant Research, Honolulu, HI, February 16, 2006 (Abstract 14).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Peter Choe and Nido Nguyen for database services; Cara Varley, Mary Garbowski, Patrick Sudour, Leo Tanaka, Debie T. Vu, Christina A. Stratis, and Greg Cardines for study coordination and assistance; and Nancy Wright, Terry Stevens-Ayers, Kristen White, Clara Bryan, and laboratory staff at the University of Washington Clinical Virology Laboratory for specimen processing, testing, and laboratory expertise.
This work was supported by National Institutes of Health grants CA 18029, CA 15704, and HL081595; A.J.P. received support from the Joel M. Meyers Endowment Fund, MedImmune Pediatric Fellowship Grant Award, and Pediatric Infectious Diseases Society Fellowship Award funded by MedImmune.
National Institutes of Health
Authorship
Contribution: A.J.P. performed the research, analyzed data, and wrote the manuscript; J.A.E. contributed to the analysis plan and critically reviewed the manuscript; J.K. performed PCR testing and critically reviewed the manuscript; K.A.G. performed statistical analyses and critically reviewed the manuscript; L.C. critically reviewed the manuscript and provided resources for the study; R.M. critically reviewed the manuscript and contributed laboratory resources; R.C.H. provided tissue samples, analyzed pathology specimens, and critically reviewed the manuscript; A.C. critically reviewed the manuscript and contributed laboratory resources; and M.B. designed and performed the research, collected data, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michael Boeckh, Fred Hutchinson Cancer Research Center, Program in Infectious Diseases, 1100 Fairview Ave N, D3-100, PO Box 19024, Seattle, WA 98109-1024; e-mail: mboeckh@fhcrc.org.