Abstract
Because phosphoinositide 3-kinase (PI3K) plays a central role in cellular activation, proliferation, and survival, pharmacologic inhibitors targeting components of the PI3K pathway are actively being developed as therapeutics for the treatment of inflammatory disorders and cancer. These targeted drugs inhibit the activity of either PI3K itself or downstream protein kinases. However, a previously unexplored, alternate strategy is to activate the negative regulatory phosphatases in this pathway. The SH2-containing inositol-5′-phosphatase SHIP1 is a normal physiologic counter-regulator of PI3K in immune/hematopoietic cells that hydrolyzes the PI3K product phosphatidylinositiol-3,4,5-trisphosphate (PIP3). We now describe the identification and characterization of potent and specific small-molecule activators of SHIP1. These compounds represent the first small-molecule activators of a phosphatase, and are able to activate recombinant SHIP1 enzyme in vitro and stimulate SHIP1 activity in intact macrophage and mast cells. Mechanism of activation studies with these compounds suggest that they bind a previously undescribed, allosteric activation domain within SHIP1. Furthermore, in vivo administration of these compounds was protective in mouse models of endotoxemia and acute cutaneous anaphylaxis, suggesting that SHIP1 agonists could be used therapeutically to inhibit the PI3K pathway.
Introduction
In response to extracellular signals, phosphoinositide 3-kinase (PI3K) becomes activated and phosphorylates phosphatidylinositol-4,5-bisphosphate (PI-4,5-P2) within the plasma membrane to generate phosphatidylinositol-3,4,5-bisphosphate (PIP3). PIP3 then initiates a cascade of downstream signaling pathways by interacting with pleckstrin homology (PH) domain-containing proteins, such as protein kinase B (PKB, also known as Akt), that regulate cellular activation, proliferation and/or survival, depending on the cell type and stimulus.1 Cellular levels of PIP3 are normally tightly regulated by both PI3K and the 5′ inositol phosphatases SHIP1 (SH2 domain–containing inositol phosphatase) and SHIP2, as well as the 3′ inositol phosphatase PTEN, which dephosphorylates PIP3.2,3 Of these, SHIP1 is unique in that its expression is restricted primarily to immune and hematopoietic cells.2,4 SHIP's role in immune cell homeostasis is shown both by the myeloproliferative syndrome observed in SHIP1−/− mice, as well as the hypersensitivity of SHIP1−/− mice and cells to immune stimulation.5,6 SHIP1 mediates signaling from the inhibitory FcγRIIB receptor,7 and is important in terminating signal transduction from activating immune/hematopoietic cell receptor systems.8 Diminished SHIP1 activity or expression has been observed in human inflammatory diseases9 and hematopoietic malignancies.10–13
Because dysregulated activation of the PI3K pathway contributes to inflammatory/immune disorders and cancer, intense efforts have been invested into the development of inhibitors of PI3K itself, as well as downstream protein kinases.14–17 The precedent for discovery and biologic efficacy of kinase inhibitors is well established, and a number of promising new PI3K isoform–specific inhibitors have recently been developed and used in mouse models of inflammatory disease18,19 and glioma20 with minimal toxicities. However, because of the dynamic interplay between phosphatases and kinases in regulating biologic processes, inositol phosphatase activators represent a complementary, alternative approach to reduce cellular PIP3 levels (discussed in Knight and Shokat17 ). Of the phosphoinositol phosphatases that degrade PIP3, SHIP1 is a particularly ideal target for development of potential therapeutics for treating immune and hemopoetic disorders because its hematopoietic-restricted expression would limit the effects of a specific SHIP1 agonist to target cells, and hence would probably minimize off-target tissue effects.
In search of small molecule modifiers of SHIP1, we developed a chromogenic enzyme assay to monitor SHIP phosphatase activity, and isolated the meroterpenoid pelorol as a potent SHIP1 activator from a marine invertebrate extract library. Structural analogs of pelorol, AQX-016A and AQX-MN100, were synthesized that exhibited greater SHIP1-activating activity than the parent compound. We show that these small-molecule agonists were able to selectively activate SHIP1 through a novel allosteric activation mechanism, and are protective when administered to cell and mouse models of inflammation.
Materials and methods
The animal studies were approved by the University of British Columbia Animal Care Committee.
Formulation of compounds
For in vitro testing in the SHIP1 enzyme assay, AQX-016A and AQX-MN100 were dissolved in EtOH and diluted into aqueous buffer (20 mM Tris HCl [pH 7.5] and 10 mM MgCl2). The actual concentration of the drug in solution was determined by optical density measurement at 280 nm (λmax for both compounds) after high-speed centrifugation at 14 000g for 30 minutes to remove precipitated drug. For testing on cells, compounds were formulated in the carrier cyclodextrin (Cyclodex Technologies, High Springs, FL) at 6 mM (2 mg/mL). For oral administration to animals, compounds were dissolved in 100% cremophore EL (Sigma-Aldrich Canada, Oakville, ON, Canada) at 150 mM (50 mg/mL) prior to dilution to 6 mM in phosphate-buffered saline. However, while these compounds caged in cyclodextrin or formulated in cremophore EL micelles were very soluble in aqueous solution, they could not be used in the SHIP1 enzyme assays because of interference from both cyclodextrin and cremophore EL.
Production of recombinant SHIP1 enzyme and SHIP1 C2 domain
N-terminal His6-tagged SHIP1 enzyme was expressed in mammalian 293T cells by transient transfection with pME18S-His-SHIP1 plasmid and purified to more than 95% homogeneity by Ni-chelating bead chromatography (Qiagen, Mississauga, ON, Canada). Recombinant SHIP1 C2 domain (amino acid residues 725 to 863) was expressed in Escherichia coli transformed with a pET28C expression vector constructed as described in “Construction of the SHIP1 ΔC2 mutant and isolated C2 domain.” Recombinant protein purified from the cell lysates by Ni-chelating bead chromatography was more than 95% pure.
In vitro SHIP1 enzyme assay.
SHIP1 enzyme assays were performed in 96-well microtiter plates with 10 ng enzyme/well in a total volume of 25 μL of 20 mM Tris HCl (pH 7.5) and 10 mM MgCl2. SHIP1 enzyme was incubated with test extracts (provided in DMSO [dimethyl sulfoxide]) or vehicle for 15 minutes at 23°C before the addition of 100 μM inositol-1,3,4,5-tetrakisphosphate (IP4; Echelon Biosciences, Salt Lake City, UT). After 20 minutes at 37°C, the amount of inorganic phosphate released was assessed by the addition of Malachite Green reagent, and absorbance measurement was at 650 nm.21 For enzyme kinetics determinations, enzyme reaction progress was measured every minute for 15 minutes at concentrations of IP4 ranging from 10 to 100 μM. Initial reaction velocities were determined from the slope of the linear portion of the resulting time courses and plotted against IP4 concentration. SHIP2 enzyme was purchased from Echelon Biosciences, and an equivalent amount of inositol phosphatase activity was used in the in vitro enzyme assay. Enzyme data are expressed as the mean of triplicates (± SEM). Experiments were performed at least 3 times.
Production of SHIP1+/+ and SHIP1−/− bone marrow–derived macrophages and mast cells.
Bone marrow cells were aspirated from 4- to 8-week-old C57Bl6 × 129Sv mixed background mice and SHIP1+/+ and SHIP1−/− mast cells prepared as described previously.22 Bone marrow–derived macrophages from SHIP1+/+ and SHIP1−/− mice were obtained as described previously6 and maintained in Iscove modified Dulbecco medium (IMDM) supplemented with 10% fetal calf serum (FCS), 150 μM monothiolgylcerol (MTG), and 2% C127 cell-conditioned medium as a source of macrophage colony stimulating factor (M-CSF; macrophage medium).
LPS stimulation of macrophages.
For the analysis of lipopolysaccharide (LPS)–stimulated tumor necrosis factor α (TNFα) production, 2 × 105 cells were plated the night before in 24-well plates in macrophage medium. The next day, the medium was changed, and AQX-016A or carrier was added to cells at the indicated concentrations for 30 minutes prior to the addition of 10 ng/mL LPS. Supernatants were collected for TNFα determination by enzyme-linked immunosorbent assay (ELISA; BD Biosciences, Mississauga, ON, Canada). For analysis of intracellular signaling, 2 × 106 cells were plated the night before in 6-cm tissue-culture plates. The next day, the cells were cultured in macrophage medium without M-CSF for 1 hour at 37°C and then pretreated with AQX-016A or carrier for 30 minutes prior to the addition of 10 ng/mL LPS for 15 minutes. Cells were washed with 4°C PBS and resuspended in lysis buffer (50 mM HEPES, 2 mM EDTA, 1 mM NaVO4, 100 mM NaF, 50 mM NaPPi, and 1% NP40) supplemented with Complete Protease Inhibitor Cocktail (Roche, Montreal, QC, Canada). Lysates were rocked at 4°C for 30 minutes and clarified by centrifuging 20 minutes at 12 000g. Lysates were then made 1 × in Laemmli buffer, boiled 2 minutes, and loaded onto 7.5% SDS-polyacrylamide cells. Immunoblot analysis for phospho-PKB (Cell Signaling, Mississauga, ON, Canada), SHIP1, and actin (Santa Cruz Biotechnology, Santa Cruz, CA) were carried out as described previously.6
Stimulation of mast cells by FcϵRI crosslinking.
Mast cells were preloaded overnight in bone marrow mast cell (BMMC) medium lacking IL-3 with 0.1 μg/mL anti-DNP IgE (SPE-7; Sigma-Aldrich Canada). For calcium flux measurements, cells were incubated with 2 μM fura 2-acetoxymethyl ester (Molecular Probes, Eugene, OR) in Tyrode buffer at 23°C for 45 minutes. Cells were then washed and incubated in the presence of vehicle control, LY294002, or AQX-016A 30 minutes prior to stimulation with the indicated concentration of DNP–human serum albumin (DNP-HSA). Calcium influx was monitored by spectrofluorometry as described previously.23 For analysis of intracellular signaling, cells were preloaded with anti-DNP IgE as for calcium flux measurements, pretreated with AQX-016A or buffer control for 30 minutes at 37°C, and stimulated with 20 ng/mL DNP-HSA for 5 minutes. Total-cell lysates were then prepared and analyzed for phospho-PKB, phospho-p38MAPK, phospho-MAPK, Grb-2 (Cell Signaling), and SHIP16 by immunoblot analysis as described previously.24
Mouse endotoxemia model.
C57Bl6 mice aged 6 to 8 weeks (Vancouver Coastal Health Research Institute [VCHRI], Mammalian Model of Human Disease Core Facility, Vancouver, BC, Canada) were orally administered the indicated dose of AQX-016A, AQX-MN100, dexamethasone, or carrier 30 minutes prior to an intraperitoneal injection of 2 mg/kg LPS (E coli serotype 0111:B4; Sigma-Aldrich Canada). Blood was drawn 2 hours later for determination of plasma TNFα by ELISA. Results are representative of 3 independent experiments.
Mouse acute cutaneous anaphylaxis model.
CD1 mice aged 6 to 8 weeks (University of British Columbia Animal Facility, Vancouver, BC, Canada) were sensitized to the hapten DNP by cutaneous application of 25 μL 0.5% dinitroflourobenzene (DNFB; Sigma-Aldrich Canada) in acetone to the shaved abdomen of mice for 2 consecutive days. At 24 hours later, test substances (dissolved in 10 μL of 1:2 DMSO/MeOH) were painted on the right ear, while the left ear received vehicle control. At 30 minutes after drug application, DNFB (2,4-dinitrofluorobenzene) was applied to both ears to induce mast cell degranulation. A 6-mm punch was taken from the ear and immediately frozen on dry ice for subsequent determination of neutrophil myeloperoxidase (MPO) activity as described.25
Construction of the SHIP1 ΔC2 mutant and isolated C2 domain
A His6-tagged SHIP1 ΔC2 domain deletion mutant (deleting residues 725 to 863) in the mammalian expression vector pME18S was generated by a standard polymerase chain reaction (PCR)–based methodology. An N-terminal His6 C2 domain construct was also generated by PCR inserted into the pET28C bacterial expression vector using EcoRI and NdeI restriction sites.
PLO assays
Protein lipid overlay (PLO) assays were performed essentially as described26 with minor modifications. Lyophilized phosphatidylinositol-3,4-bisphosphate diC16 (PIP2; Echelon Biosciences) was reconstituted in a 2:1.8 solution of methanol and water. PVDF membranes (Millipore, Missisauga, ON, Canada) were initially wetted in methanol for 1 minute, washed 3 times for 5 minutes each with water, and gently agitated in TBST buffer (20 mM Tris [pH 7.5], 0.15 M NaCl [TBS] with 0.05% Tween 20) at 23°C overnight. Treated membranes were air-dried, and dilutions of reconstituted lipids were spotted in 1-μL aliquots to give the indicated amount of PIP2 per membrane spot. Membranes were dried completely and blocked with blocking buffer (3% BSA in TBS with 0.05% NaN3) for 1 hour at 23°C. Purified, recombinant C2 domain was diluted into blocking buffer (5 μM final) and treated with 4 μM AQX-MN100 or EtOH control for 30 minutes at 23°C prior to overnight incubation with the PIP2-spotted membranes. Membranes were washed 10 times over 50 minutes in TBST buffer at 23°C and incubated with anti-His6 mouse IgG (Qiagen, Missisauga, ON, Canada) for 1 hour at 23°C. Membranes were washed 10 times over 50 minutes and incubated with Alexa Fluor 660 goat antimouse IgG (Invitrogen, Burlington, ON, Canada) for 1 hour at 23°C. After washing, bound proteins were detected and quantified on a Li-Cor Odyssey scanner (Lincoln, NE).
Scintillation proximity assays
AQX-MN100 was radiolabeled with tritium by GE Healthcare (Piscataway, NJ) to a specific activity of 155.4 × 1010 Bq (42 Ci/mmol). Copper chelate (His-Tag) YSi SPA Scintillation Beads (GE Healthcare, Piscataway, NJ) were diluted in 0.25% BSA/TBS to 1.5 mg/mL, and recombinant, His6-tagged protein was added at the indicated concentrations: wild-type (1 pM), ΔC2 SHIP1 enzyme (1 pM), or C2 domain (10 nM). Protein was allowed to bind for 1 hour at 23°C, and 250 μg of beads were aliquoted per well of a 96-well plate. A total of 0.185 MBq (5 μCi) of [3H]-AQX-MN100 was added per well, the plate was gently agitated for 30 minutes, and the amount of bead-associated radioactivity was quantified by counting in a Wallac (Perkin-Elmer, Waltham, MA) BetaPlate plate scintillation counter.
Results
Identification of small-molecule SHIP1 agonists
Recombinant SHIP1 enzyme was used in a chromogenic assay to identify small-molecule modifiers of SHIP1 phosphatase activity from a marine invertebrate extract library. Of approximately 2000 extracts tested, several exhibiting SHIP1 agonistic activity were identified (Figure S1, available on the Blood website; see the Supplemental Figures link at the top of the online article). Using enzyme-guided fractionation, the active component was purified from extract B8, derived from the Papua New Guinea sponge, Dactylospongia elegans. We solved the structure of the active component and identified it as the meroterpenoid pelorol,27 which was also independently identified by Golcik et al28 from D. elegans and by Kwak et al29 from Petrosasapongia metachromia. While the interesting structure of pelorol was noted by both groups, and Golcik et al reported antiprotozoan activity of this compound,28 neither group appreciated the ability of pelorol to activate SHIP1 or modify the activity of mammalian cells as we now describe.
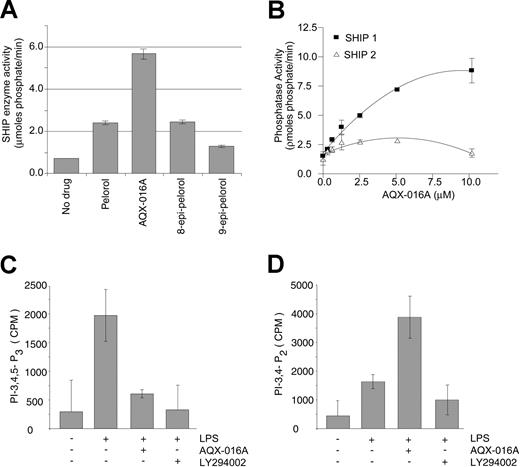
An efficient chemical synthetic protocol was then developed for the production of pelorol from the commercially available terpenoid sclareolide.27 This allowed production of sufficient quantities for analysis in biologic assays. In addition, certain synthetic intermediates, as well as structural analogs of pelorol, were tested for their SHIP1-activating ability. One of these, a synthetic intermediate designated AQX-016A (compound 22 generated in 7 chemical steps from sclareolide in Yang et al27 ) was 3 times more potent than the natural product pelorol at the same molar concentration (Figure 1A). Most of our subsequent studies were therefore performed with AQX-016A.
AQX-016A increases SHIP1 enzyme activity in vitro and in intact cells. (A) Purified pelorol, AQX-016A, 8-epi-pelorol, and 9-epi-pelorol were tested at 2 μM for their ability to enhance SHIP's enzyme activity. (B) The effect of AQX-016A on SHIP1 (♦) and SHIP2 (○) enzyme activity was compared in in vitro enzyme assays. In panels C and D, J16 cells were treated for 30 minutes with 15 μM AQX-016A, 25 μM LY294002, or carrier prior to stimulation with 50 ng/mL of LPS for 15 minutes at 37°C. Cellular lipids were extracted and analyzed for PIP3 (C) and PI-3,4-P2 (D) levels as described in “Materials and methods.” Data are expressed as means (± SEM) and are representative of 3 independent experiments.
AQX-016A increases SHIP1 enzyme activity in vitro and in intact cells. (A) Purified pelorol, AQX-016A, 8-epi-pelorol, and 9-epi-pelorol were tested at 2 μM for their ability to enhance SHIP's enzyme activity. (B) The effect of AQX-016A on SHIP1 (♦) and SHIP2 (○) enzyme activity was compared in in vitro enzyme assays. In panels C and D, J16 cells were treated for 30 minutes with 15 μM AQX-016A, 25 μM LY294002, or carrier prior to stimulation with 50 ng/mL of LPS for 15 minutes at 37°C. Cellular lipids were extracted and analyzed for PIP3 (C) and PI-3,4-P2 (D) levels as described in “Materials and methods.” Data are expressed as means (± SEM) and are representative of 3 independent experiments.
To evaluate the specificity of AQX-016A for SHIP1, we assessed AQX-016A's ability to activate its most closely related phosphoinositol phosphatase, SHIP2.2 As shown in Figure 1B, AQX-016A preferentially activated SHIP1 over SHIP2. We then determined whether AQX-016A was able to activate SHIP's enzyme activity in intact cells by analyzing the inositol phospholipid content of macrophages stimulated with LPS in the presence or absence of AQX-016A (Figure 1C). LPS stimulation of macrophages results in activation of PI3K and, as shown in Figure 1C, LPS induced a 3- to 5-fold increase in PIP3 levels. Treatment of cells with AQX-016A abolished this increase in PIP3 levels (Figure 1C) and resulted in a corresponding increase in the SHIP1 hydrolysis product PI-3,4-P2 (Figure 1D). For comparison, we also treated LPS-stimulated cells with the PI3K inhibitor LY294002 and, as expected, PIP3 levels were diminished to levels indistinguishable from cells not stimulated with LPS (Figure 1C) without a corresponding increase in PI-3,4-P2 levels (Figure 1D). Thus, both AQX-016A and LY294002 inhibited the PI3K-mediated increase in intracellular PIP3 levels, but through different mechanisms. The increased levels of the specific SHIP1 product, PI-3,4-P2, observed in AQX-016A–treated cells, was consistent with an activation of SHIP-mediated 5′-dephosphorylation of PIP3.
AQX-016A inhibits macrophage and mast-cell activation
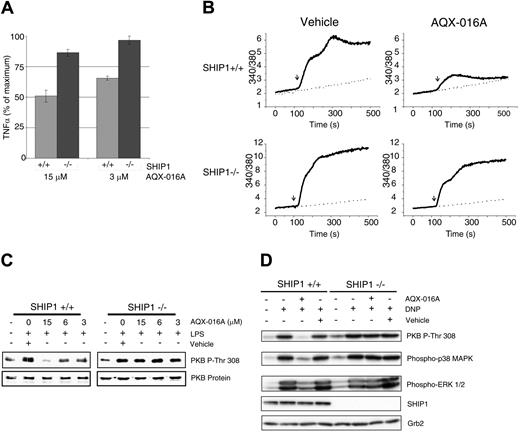
We next assessed the target specificity and biologic efficacy of AQX-016A by comparing its effects on PI3K-regulated processes in primary SHIP1+/+ versus SHIP1−/− macrophages and mast cells. Both LPS-induced macrophage6 and IgE-induced mast-cell activation22–24 involve activation of PI3K-dependent pathways which have previously been shown to be negatively regulated by SHIP1. LPS stimulation of macrophages is associated with a PIP3-dependent release of proinflammatory mediators such as TNFα.6 We examined the action of AQX-016A on SHIP1+/+ versus SHIP1−/− bone marrow–derived macrophages. Cells were pretreated for 30 minutes with AQX-016A prior to stimulation with 10 ng/mL of LPS for 2 hours. AQX-016A was able to suppress TNFα production in SHIP1+/+ cells by 30% at 3 μM and 50% at 15 μM (Figure 2A). In contrast, SHIP1−/− cells, TNFα production was indistinguishable from non–AQX-016A–treated cells at 3 μM and was suppressed 15% at 15 μM. For comparison, the PI3K inhibitor LY294002 inhibited both SHIP1+/+ and SHIP1−/− macrophages to the same extent (up to approximately 40% at 15 μM; Figure S2). Activation of mast cells via IgE plus antigen crosslinking of their IgE receptors results in elevation of intracellular calcium levels.22,30 As shown in Figure 2B, AQX-016A selectively inhibited IgE plus antigen-induced calcium entry to a substantially greater degree in SHIP1+/+ than in SHIP1−/− bone marrow–derived mast cells, whereas LY294002 inhibited both SHIP1+/+ and SHIP1−/− mast cells to the same extent (Figure S3). These data were consistent with AQX-016A inhibiting PI3K-dependent macrophage and mast-cell responses in a SHIP-dependent manner.
AQX-016A inhibits immune cell activation. (A) SHIP1+/+ (▩) and SHIP1−/− (■) macrophages were pretreated with AQX-016A or carrier 30 minutes prior to stimulation with 10 ng/mL of LPS at 37°C for 2 hours and TNFα production determination by ELISA. Absolute TNFα levels for SHIP1+/+ and SHIP1−/− cells were 623 ± 30 and 812 ± 20 pg/mL, respectively. Data are expressed as means (± SEM) and are representative of 3 independent experiments. (B) SHIP1+/+ and SHIP1−/− mast cells were preloaded with IgE and Fura-2 as described in “Materials and methods, Stimulation of mast cells by FcϵRI crosslinking” and treated for 30 minutes with 15 μM AQX-016A or carrier. Cells were then stimulated (as indicated by ↓) with 0 ng/mL (- - -) or 10 ng/mL (––) DNP-HSA, and intracellular calcium levels were monitored over time by spectrofluorometry.23 (C) SHIP1+/+ and SHIP1−/− macrophages were pretreated for 30 minutes with AQX-016A or carrier prior to stimulation with 10 ng/mL of LPS for 15 minutes at 37°C. Total-cell lysates were analyzed for the indicated phospho-proteins or proteins by immunoblot analysis. (D) Anti–DNP-IgE loaded SHIP1+/+ and SHIP1−/− mast cells were treated for 30 minutes with 30 μM AQX-016A or carrier prior to stimulation with 20 ng/mL DNP-HSA for 5 minutes at 37°C; cell lysates were analyzed as in panel C.
AQX-016A inhibits immune cell activation. (A) SHIP1+/+ (▩) and SHIP1−/− (■) macrophages were pretreated with AQX-016A or carrier 30 minutes prior to stimulation with 10 ng/mL of LPS at 37°C for 2 hours and TNFα production determination by ELISA. Absolute TNFα levels for SHIP1+/+ and SHIP1−/− cells were 623 ± 30 and 812 ± 20 pg/mL, respectively. Data are expressed as means (± SEM) and are representative of 3 independent experiments. (B) SHIP1+/+ and SHIP1−/− mast cells were preloaded with IgE and Fura-2 as described in “Materials and methods, Stimulation of mast cells by FcϵRI crosslinking” and treated for 30 minutes with 15 μM AQX-016A or carrier. Cells were then stimulated (as indicated by ↓) with 0 ng/mL (- - -) or 10 ng/mL (––) DNP-HSA, and intracellular calcium levels were monitored over time by spectrofluorometry.23 (C) SHIP1+/+ and SHIP1−/− macrophages were pretreated for 30 minutes with AQX-016A or carrier prior to stimulation with 10 ng/mL of LPS for 15 minutes at 37°C. Total-cell lysates were analyzed for the indicated phospho-proteins or proteins by immunoblot analysis. (D) Anti–DNP-IgE loaded SHIP1+/+ and SHIP1−/− mast cells were treated for 30 minutes with 30 μM AQX-016A or carrier prior to stimulation with 20 ng/mL DNP-HSA for 5 minutes at 37°C; cell lysates were analyzed as in panel C.
Next, we compared the ability of AQX-016A to inhibit activation of PIP3-dependent downstream signaling proteins in SHIP1+/+ versus SHIP1−/− cells. LPS stimulation of macrophages results in PKB phosphorylation. As shown in Figure 2C, AQX-016A preferentially inhibited, in a dose-dependent manner, LPS-stimulated PKB phosphorylation in SHIP1+/+ but not in SHIP1−/− macrophages. Similarly, AQX-016A inhibited the phosphorylation of PKB, p38MAPK, and extracellular signal–regulated kinase (ERK) in SHIP1+/+ but not in SHIP1−/− mast cells (Figure 2D). Similar protein loading was confirmed by reblotting with either antibodies to PKB or Grb2. We also examined the ability of AQX-016A to inhibit PKB activation in nonhematopoietic, prostate epithelial LNCaP cells, which do not express SHIP1. The human prostate cancer cell line LNCaP exhibits constitutive activation of PKB due to the loss of PTEN expression.31 As shown in Figure S4, LY294002 efficiently suppressed PKB phosphorylation, whereas AQX-016A had no effect at doses up to 60 μM. Thus, AQX-016A inhibits PIP3-regulated intracellular signal transduction events in SHIP-expressing hematopoietic cells, but not in SHIP-deficient hematopoietic or nonhematopoietic cells.
AQX-016A inhibits inflammation in vivo
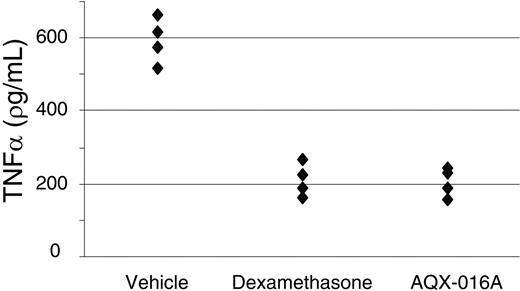
We went on to test whether AQX-016A would be effective in inhibiting inflammatory reactions in vivo by assessing its ability to provide protection in mouse models. The mouse model of endotoxic shock involves intraperitoneal injection of bacterial LPS and measurement of serum TNFα levels 2 hours later.32 We orally administered AQX-016A or the steroidal drug dexamethasone to mice 30 minutes prior to the LPS challenge. As predicted for an activator of SHIP1 and an inhibitor of macrophage activation, AQX-016A reduced the level of serum TNFα and did so to the same extent as dexamethasone (Figure 3).
AQX-016A inhibits inflammation in a mouse model endotoxemia. (A) Mice were administered 20 mg/kg AQX-016A or 0.4 mg/kg dexamethasone orally 30 minutes prior to an intraperitoneal injection of 2 mg/kg LPS. Blood was collected 2 hours later for TNFα determination by ELISA. Each symbol indicates 1 mouse, and data are representative of 3 independent experiments.
AQX-016A inhibits inflammation in a mouse model endotoxemia. (A) Mice were administered 20 mg/kg AQX-016A or 0.4 mg/kg dexamethasone orally 30 minutes prior to an intraperitoneal injection of 2 mg/kg LPS. Blood was collected 2 hours later for TNFα determination by ELISA. Each symbol indicates 1 mouse, and data are representative of 3 independent experiments.
AQX-MN100 is as biologically active as AQX-016A
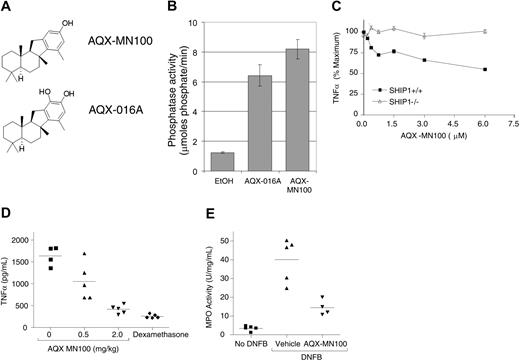
Our observation that AQX-016A was substantially more active on SHIP1+/+ than SHIP1−/− cells suggested it acted by specifically targeting SHIP1. However, the presence of a catechol moiety within AQX-016A (Figure 4A) was potentially problematic because catechols can exhibit activities independent of their specific protein pocket binding interaction.17 For example, catechols can bind metals or be oxidized to an orthoquinone, which can lead to covalent modification of proteins through Michael reactions.33 To rule out these possibilities, we synthesized a monophenolic version of AQX-016A, designated AQX-MN100 (M.N. and R.J.A., manuscript in preparation; Figure 4A). Analogous to AQX-016A, AQX-MN100 enhanced SHIP1 enzyme activity in vitro (Figure 4B). Like AQX-016A, AQX-MN100 also selectively inhibited TNFα production from SHIP1+/+ but not SHIP1−/− macrophages (Figure 4C). The maximum inhibition of TNFα production by AQX-MN100 in this in vitro assay is approximately 50%, a level comparable with the maximum inhibition exerted by LY292004 (Figure S2) and the macrophage inhibitory cytokine IL-10.34 Thus, the calculated EC50 for this inhibition was 0.3 to 0.6 μM. Oral administration of AQX-MN100 also efficiently inhibited the LPS-induced elevation of plasma TNFα levels in the mouse endotoxemia model (Figure 4D).
AQX-MN100 has the same biologic activities as AQX-016A. (A) Structures of AQX-MN100 and AQX-016A. (B) AQX-MN100 activates SHIP1 enzyme activity in vitro. Assays performed as in Figure 1A. (C) AQX-MN100 inhibits TNFα production from LPS stimulated SHIP1+/+ but not SHIP1−/− BMmφs. Cells and treatments were as described in Figure 2A. (D) AQX-MN100 inhibits LPS-induced plasma TNFα levels in mice. Mice were treated as in Figure 2E. (E) AQX-MN100 inhibits DNFB-induced neutrophil-specific myeloperoxidase (MPO) in sensitized mice. Mice were sensitized as in Figure 2F, and vehicle or AQX-MN100 applied to pairs of ears prior to DNFB challenge. Some mice were not challenged with DNFB (no DNFB). Ears were harvested, and MPO levels were determined. P < .02 for the AQX-MN100 versus the vehicle-treated groups. Data are expressed as the mean (±SEM) and are representative of 3 independent experiments.
AQX-MN100 has the same biologic activities as AQX-016A. (A) Structures of AQX-MN100 and AQX-016A. (B) AQX-MN100 activates SHIP1 enzyme activity in vitro. Assays performed as in Figure 1A. (C) AQX-MN100 inhibits TNFα production from LPS stimulated SHIP1+/+ but not SHIP1−/− BMmφs. Cells and treatments were as described in Figure 2A. (D) AQX-MN100 inhibits LPS-induced plasma TNFα levels in mice. Mice were treated as in Figure 2E. (E) AQX-MN100 inhibits DNFB-induced neutrophil-specific myeloperoxidase (MPO) in sensitized mice. Mice were sensitized as in Figure 2F, and vehicle or AQX-MN100 applied to pairs of ears prior to DNFB challenge. Some mice were not challenged with DNFB (no DNFB). Ears were harvested, and MPO levels were determined. P < .02 for the AQX-MN100 versus the vehicle-treated groups. Data are expressed as the mean (±SEM) and are representative of 3 independent experiments.
We also tested the ability of AQX-MN100 to inhibit cutaneous anaphylaxis. Anaphylactic or allergic responses are mediated by allergen-induced degranulation of presensitized mast cells.30 The mouse ear edema/cutaneous anaphylaxis model35 involves presensitization of mice with the haptenizing agent dinitrofluorobenzene (DNFB). At 1 week later, the allergic reaction is elicited by painting DNFB onto the ears of the mice. The efficacy of potential anti-inflammatory compounds is tested by topical application of the test substance to 1 ear and comparing the resulting ear edema or inflammation of the 2 ears. As shown in Figure 4E, topically applied AQX-MN100 dramatically inhibited allergen-induced inflammation compared with the vehicle control–treated ear. AQX-016A was also able to inhibit DNFB-induced inflammation in this model (Figure S5).
SHIP1 is an allosterically activated enzyme
The allosteric regulation of enzymes has remained underappreciated primarily because allosteric effectors are not easy to find. While most allosteric regulators have been discovered through serendipity, a few allosteric regulators have been deduced from discovery of activators or inhibitors of enzymes as a result of high-throughput chemical screens (HTSs).36 For example, the allostery of glucokinase was discovered from an HTS in search of activators for the treatment of diabetes.36 By analogy, our discovery of small-molecule activators of SHIP1 led us to postulate that SHIP1 might in fact be allosterically regulated. To this end, we investigated the molecular mechanism by which AQX-MN100 activated SHIP1, first by performing classical enzyme kinetic analysis of its phosphatase activity. Activity measurements were performed with substrate concentrations from 10 to 100 μM. Plots of the initial reaction velocity at each substrate concentration are predicted to exhibit a hyperbolic profile if SHIP1 obeys conventional Michaelis-Menten kinetics.37 However, we found SHIP1 in fact displayed sigmoidal reaction kinetics, suggesting allosteric activation by its end product (Figure 5A). Indeed, addition of the SHIP1 product PI-3,4-P2 to the enzyme reaction activated wild-type SHIP1 enzyme to the same extent as AQX-MN100 (Figure 5B). Interestingly, the 3′ inositol phosphatases PTEN38 and myotubularin (MTM)39 have also been recently shown to be allosterically activated by their phosphatidylinositol products (PI-4,5-P2 and PI-5-P, respectively).
The C2 domain is required for end-product allosteric activation of SHIP1 and binding of AQX-MN100. (A) SHIP1 enzyme initial velocities were determined at the indicated concentration of inositol-1,2,4,5-tetrakisphosphate (IP4) substrate. The equation Y = 49 + 150 / (1 + 10log(80 − ×) − 0.22) describes the line with R2 = 0.98. (B) The ability of product PI-3,4-P2 (20 μM) or AQX-MN100 (3 μM) to activate wild-type (WT) and C2 domain–deleted (ΔC2) SHIP1 enzyme was determined at 30 μM IP4. (C) Recombinant C2 domain was preincubated for 30 minutes at 23°C with 4.0 μM AQX-MN100 or EtOH control and allowed to bind to PI-3,4-P2 immobilized on membrane strips in a protein overlay assay as described in “Materials and methods, PLO assays.” (D) Recombinant C2 domain (10 nM) was coated onto copper chelate (His-Tag) YSi SPA Scintillation Beads in the presence of 0.25% BSA. Beads were then incubated with 0.185 MBq (5 μCi) of [3H]-AQX-MN100, and the bead-associated radioactivity was measured as described in “Materials and methods.” Data are expressed as means (±SEM) and are representative of at least 3 independent experiments.
The C2 domain is required for end-product allosteric activation of SHIP1 and binding of AQX-MN100. (A) SHIP1 enzyme initial velocities were determined at the indicated concentration of inositol-1,2,4,5-tetrakisphosphate (IP4) substrate. The equation Y = 49 + 150 / (1 + 10log(80 − ×) − 0.22) describes the line with R2 = 0.98. (B) The ability of product PI-3,4-P2 (20 μM) or AQX-MN100 (3 μM) to activate wild-type (WT) and C2 domain–deleted (ΔC2) SHIP1 enzyme was determined at 30 μM IP4. (C) Recombinant C2 domain was preincubated for 30 minutes at 23°C with 4.0 μM AQX-MN100 or EtOH control and allowed to bind to PI-3,4-P2 immobilized on membrane strips in a protein overlay assay as described in “Materials and methods, PLO assays.” (D) Recombinant C2 domain (10 nM) was coated onto copper chelate (His-Tag) YSi SPA Scintillation Beads in the presence of 0.25% BSA. Beads were then incubated with 0.185 MBq (5 μCi) of [3H]-AQX-MN100, and the bead-associated radioactivity was measured as described in “Materials and methods.” Data are expressed as means (±SEM) and are representative of at least 3 independent experiments.
In PTEN and MTM, the allosteric binding sites for their products were mapped to lipid-binding motifs in each protein. The SHIP1 protein contains a C2 domain located at the carboxy-terminal end of its phosphatase domain4 that we hypothesized could be its allosteric binding site. C2 domains were first described in the protein kinase C family where it serves to bind Ca2+,40 but C2 domains have since been identified in other proteins, where they have been shown to bind to a variety of ligands, including lipids.41,42 To test this possibility, we generated SHIP1 lacking its C2 domain (ΔC2 SHIP). As shown in Figure 5B, although ΔC2 SHIP1 was as active as the wild-type SHIP1, its activity could not be enhanced by the addition of either PI-3,4-P2 or AQX-MN100. This suggested that the C2 domain was required for the allosteric activation of SHIP1 activity and that it might be the binding site for its allosteric activators such as PI-3,4-P2 and AQX-MN100.
To examine whether the C2 domain could bind PI-3,4-P2, we expressed and isolated recombinant, His6-tagged C2 domain and determined its PI-3,4-P2–binding ability using PLO assays.26 Purified C2 domain was incubated with membrane strips spotted with PI-3,4-P2, and bound protein was detected using an anti-His6 antibody. As shown in Figure 5C, the C2 domain bound PI-3,4-P2; this binding was inhibited by AQX-MN100, consistent with both AQX-MN100 and PI-3,4-P2 interacting with the C2 domain at a common binding site. AQX-MN100 was verified to directly bind the C2 domain using scintillation proximity assays (SPAs) in which SPA beads were coated with either the C2 domain or control protein (BSA) prior to incubation with [3H]-AQX-MN100. As shown in Figure 5D, the C2 domain did indeed interact with [3H]-AQX-MN100. In complementary studies, we observed that [3H]-AQX-MN100 bound to wild-type SHIP1 but not to SHIP1 lacking its C2 domain (Figure S6). Together, these data are consistent with AQX-MN100 directly binding to SHIP's C2 domain, resulting in allosteric activation of the enzyme.
Discussion
The PI3K pathway has been the target of intense efforts to develop therapeutics.14–16,43 The PI3K family consists of multiple isoforms that vary in tissue distribution and in the receptor systems to which they are coupled.16 The classic PI3K inhibitors wortmannin and LY292004 have been excellent experimental tools for probing PI3K function, but they have not been successful in clinical development partly because they globally inhibit all members of the PI3K family.44 Recently however, isoform-specific inhibitors of PI3K have been developed. For example, an isoform-selective inhibitor that preferentially inhibits PI3Kγ has been described. PI3Kγ mediates signaling from G-protein–coupled receptors (GPCRs), and although its expression can be detected in endothelium, heart, and brain, it is primarily expressed in immune cells.44 Selective PI3Kγ inhibitors thus benefit from PI3Kγ's relatively restricted expression compared with the other PI3K isoforms, and the fact that many (though not all45 ) inflammatory processes involve GPCR-dependent steps.44 This inhibitor was found to be protective in mouse models of rheumatoid arthritis18 and glomerulonephritis.19 A dual PI3Kα/mTOR inhibitor17 was also recently found to have efficacy in a human glioma xenograft model20 without any undue toxicities, even though PI3Kα is expressed in all tissues and plays an important role in insulin signaling.15,46 To account for this observation, it has been postulated that a sufficient therapeutic window exists because cancer cells become very dependent on particular signaling pathways (called “oncogene addiction”14 ), and thus pharmacologic inhibitors show selectivity toward cancer versus normal cells.14 In addition to PI3K itself, downstream protein kinases are also being targeted with mixed results. Work is continuing on PKB inhibitors47–49 with limited success, perhaps because of dose limitations due to toxicities.50,51 The mTOR protein complex (mTORC1) inhibitor rapamycin, on the other hand, is currently approved as an immunosuppressive agent with manageable side effects.52 The toxicities of both PKB and mTORC1 inhibitors are partly related to the ubiquitous expression of both targets.
As a complementary approach to inhibiting PI3K and downstream protein kinases, we describe a novel paradigm for inhibiting PI3K signaling through activation of the phosphatases that negatively regulate this pathway. SHIP1 is a particularly good target for immune/hematopoietic disorders because of its restricted expression to hematopoietic cells. Because the relative activity of phosphatases present in a cell will influence the efficacy of kinase inhibitors, as discussed by Knight and Shokat,17 SHIP1 agonists could also be used to potentiate the activation of PI3K inhibitors and promote tissue targeting of PI3K inhibitors to the hematopoietic/immune cell compartment. Herein, we report the isolation of pelerol as a small-molecule SHIP1 agonist and the synthesis of more potent analogs (AQX-016A and AQX-MN100) that inhibit macrophage and mast-cell activation in vitro and inflammation in mouse models. Work is now under way to produce large quantities of compound to test AQX-MN100 in additional inflammatory/autoimmune disease models. Initial toxicology studies suggest both AQX-016A and AQX-MN100 are well tolerated and do not significantly affect peripheral blood cell counts, bone marrow progenitor numbers, and liver and kidney function (data not shown).
Our results suggest that the current model for SHIP1 activation, involving translocation of SHIP1 via its SH2 or its phosphorylated NPXY motifs from the cytoplasm to the plasma membrane without any change in its intrinsic phosphatase activity,7,8 needs to be modified. Specifically, we postulate that upon recruitment to the plasma membrane, SHIP1 hydrolyzes a small amount of PIP3 at a low, basal rate. This generates some PI-3,4-P2, which then binds to the C2 domain, leading to a conformation change which enhances its catalytic activity. Interestingly, as mentioned earlier, end product activation has also been reported for 2 3′ inositol lipid phosphatases. PTEN binds its product (PI-4,5-P2) using an N-terminal lipid binding motif, resulting in enhancement of phosphatase activity.38 Similarly, the MTM phosphatase binds its product (PI-5-P) via a divergent PH domain which allosterically activates its function.39 In the case of PTEN, the requirement for PI-4,5-P2 helps localize PTEN protein to specific regions of the membrane.53–55 In a similar fashion, PI-3,4-P2 may also similarly serve to localize SHIP1 to the plasma membrane in addition to allosterically activating its phosphatase activity, providing a positive feedback mechanism to quickly accelerate SHIP's activity to degrade PIP3. Regardless of whether PI-3,4-P2 binding regulates SHIP1 localization, this newly described allosteric activation domain within SHIP1 may be exploited therapeutically by pharmacologic agents such as AQX-MN100.
From its first description, allosteric sites have been considered to be better drug targets than active sites, and allosteric regulators have been predicted to possess more selectivity than active site modulators.36 While it remains challenging to prove that a specific drug-target interaction is responsible for mediating its biologic effects, our observation that AQX-016A and AQX-MN100 had minimal effects on SHIP1−/− mast cells and macrophages provides compelling support for the notion that the PI3K pathway inhibitory effects observed are indeed mediated through SHIP1. Furthermore, our observation that AQX-MN100 exhibits efficacy at a submicromolar EC50 (Figure 4C) suggests that it possesses a low likelihood of off-target effects based on calculations by Knight and Shokat.17 Indeed, we have found that AQX-MN100 had minimal off-target effects on a screen of 100 other kinases and phosphatases (Figure S7). Several of the targets included in this screen are C2 domain–containing proteins such as protein kinase C, Raf-1, and SHIP2. The lack of effect of AQX-MN100 on these enzymes suggest AQX-MN100 interacts with residues in the SHIP1 C2 domain that are not conserved in other C2 domains. In fact, the C2 domains of SHIP1 and SHIP2 only share 62% similarity (42% identity) at the amino acid level. Studies are under way to characterize the residues in the SHIP1 C2 domain important for interacting with AQX-MN100.
In summary, we show proof-of-principle that small-molecule activators of lipid phosphatases can be found, and that they provide a new paradigm for inhibition of PI3K-dependent processes. Small-molecule agonists of the hematopoietic cell–restricted SHIP1 enzyme, in particular, represent potential therapeutics for treatment of immune/hematopoietic disorders in which the PI3K pathway is dysregulated. Because of their unique target and mechanism of action, this novel class of SHIP1 agonistic compounds may also be powerful synergistic agents in combination with current therapies. Agonists of other allosterically regulated phosphatases, such as PTEN38 and MTM,39 may similarly be useful for diseases in which their impaired activity has been implicated.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by research grants from the Canadian Institutes of Health Research (CIHR) to A.L.-F.M., and the National Cancer Institute of Canada (NCIC) to G.K. and R.J.A. A.M.L. and P.Q. held CIHR/Michael Smith Foundation for Health Research (MSFHR) Training Program in Transplantation scholarships. J.K. held a CIHR TRID (Translational Research in Infectious Disease) scholarship. Aquinox Pharmaceuticals funded the radiolabeling of AQX-MN100 and the experiment shown in Figure S7.
National Institutes of Health
Authorship
Contribution: C.J.O., A.M.L., and A.L.-F.M. designed and performed research, analyzed data, and wrote the paper. M.N., L.Y., D.E.W., R.J.A., and G.K. provided new reagents. A.G., J.K., L.D., P.Q., J.R., L.P.C., and K.M. performed experiment(s). S.W.C., V.D., R.J.A., and G.K. analyzed data and contributed to manuscript writing. C.J.O. and A.M.L. contributed equally to this study.
Conflict-of-interest disclosure: The 4 principal scientists (A.L.-F.M., C.J.O., R.J.A., and G.K.) founded a start-up company (Aquinox Pharmaceuticals) to develop SHIP1 activators for the treatment of human disease. The 4 scientists do not receive compensation from Aquinox, nor do they play a role in the day-to-day operations of the company.
Correspondence: Alice L.-F. Mui, Jack Bell Research Centre, 2660 Oak Street, Vancouver, BC V6H 3Z6, Canada; e-mail: amui@interchange.ubc.ca.

![Figure 5. The C2 domain is required for end-product allosteric activation of SHIP1 and binding of AQX-MN100. (A) SHIP1 enzyme initial velocities were determined at the indicated concentration of inositol-1,2,4,5-tetrakisphosphate (IP4) substrate. The equation Y = 49 + 150 / (1 + 10log(80 − ×) − 0.22) describes the line with R2 = 0.98. (B) The ability of product PI-3,4-P2 (20 μM) or AQX-MN100 (3 μM) to activate wild-type (WT) and C2 domain–deleted (ΔC2) SHIP1 enzyme was determined at 30 μM IP4. (C) Recombinant C2 domain was preincubated for 30 minutes at 23°C with 4.0 μM AQX-MN100 or EtOH control and allowed to bind to PI-3,4-P2 immobilized on membrane strips in a protein overlay assay as described in “Materials and methods, PLO assays.” (D) Recombinant C2 domain (10 nM) was coated onto copper chelate (His-Tag) YSi SPA Scintillation Beads in the presence of 0.25% BSA. Beads were then incubated with 0.185 MBq (5 μCi) of [3H]-AQX-MN100, and the bead-associated radioactivity was measured as described in “Materials and methods.” Data are expressed as means (±SEM) and are representative of at least 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/6/10.1182_blood-2007-03-079699/7/m_zh80170706250005.jpeg?Expires=1769120756&Signature=lZdiKnIPv78t~1AHyCI3e2bfet7xvFvOlQKkgjEA8QR3DHcjDgDg~fkWp0Ea8oUiQe2CXL7q8E98w-1EAz7aywZ4RkWsAH68wAyO-ZNtXZwGtTI5r02cnzrBKjpDrnSaD1PTe1gVelzjCdCz5L7ylg0jQCVAAK5k4tY4hHnG7a-r7Kl5U2DRPs8vm2jV8wC3SNTPzXi48aBxvV0epd~MzXrSP8O1fwtiHxF5RQuKHQ34I1aWI-yhsSZGQNXD2wjT6xIT9YIfb6ZTukCHXlo4C9M1Qj7hLxQKEynB0MJHAzVVps3HOIsmko1GAEmT3rfl2JfSQ66mf4OpiqKo5TGAZw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal