Abstract
The transcription factor STAT5 fulfills a distinct role in the hematopoietic system, but its precise role in primitive human hematopoietic cells remains to be elucidated. Therefore, we performed STAT5 RNAi in sorted cord blood (CB) and acute myeloid leukemia (AML) CD34+ cells by lentiviral transduction and investigated effects of STAT5 downmodulation on the normal stem/progenitor cell compartment and the leukemic counterpart. STAT5 RNAi cells displayed growth impairment, without affecting their differentiation in CB and AML cultures on MS5 stroma. In CB, limiting-dilution assays demonstrated a 3.9-fold reduction in progenitor numbers. Stem cells were enumerated in long-term culture-initiating cell (LTC-IC) assays, and the average LTC-IC frequency was 3.25-fold reduced from 0.13% to 0.04% by STAT5 down-regulation. Single-cell sorting experiments of CB CD34+/CD38− cells demonstrated a 2-fold reduced cytokine-driven expansion, with a subsequent 2.3-fold reduction of progenitors. In sorted CD34+ AML cells with constitutive STAT5 phosphorylation (5/8), STAT5 RNAi demonstrated a reduction in cell number (72% ± 17%) and a decreased expansion (17 ± 15 vs 80 ± 58 in control cultures) at week 6 on MS5 stroma. Together, our data indicate that STAT5 expression is required for the maintenance and expansion of primitive hematopoietic stem and progenitor cells, both in normal as well as leukemic hematopoiesis.
Introduction
The hematopoietic environment is a highly dynamic system, where multipotent hematopoietic stem cells (HSCs) perpetually make decisions regarding self-renewal, lineage commitment, and terminal differentiation to provide sufficient numbers of mature blood cells. An important transcription factor within the hematopoietic system, involved in self-renewal, proliferation, and apoptosis in response to a wide range of cytokines, is signal transducer and activator of transcription 5 (STAT5).1-3 Two isoforms of this STAT family member have been cloned, STAT5A and STAT5B, and like other STAT proteins STAT5 is a latent cytoplasmic transcription factor that dimerizes upon phosphorylation by a cytokine-activated receptor complex. The activated STAT complex then translocates to the nucleus where it binds to specific DNA sequences and increases transcription of target genes.4
In STAT5ABΔN/ΔN knockout mice, the presence of an almost normal peripheral blood count suggested that full-length STAT5 is either not necessary or redundant for adult hematopoiesis, even though colony assays demonstrated reduced numbers of cytokine-responsive myeloid progenitors.5 Instead, STAT5 was assigned a role in fetal liver and stress erythropoiesis due to Bcl-XL–mediated antiapoptosis.6,7 In competitive repopulation experiments, however, STAT5ABΔN/ΔN cells demonstrated a severe impairment in engrafting wild-type recipients, which did not appear to be due to a homing deficiency, but rather to a failure in postengraftment hematopoiesis.8,9 In addition, the cytokine responsiveness was severely impaired in STAT5ABΔN/ΔN knockout cells.10 Gain-of-function studies demonstrated that constitutively activated STAT51*6 imposed a self-renewal phenotype on human cord blood (CB)–derived CD34+ stem and progenitor cells.11 These observations were consistent with overexpression data in murine HSCs, where the self-renewal phenotype was coupled to the development of a myeloproliferative disease (MPD).12
In acute myeloid leukemia (AML), constitutive activation of STAT5 has been demonstrated in approximately 66% of the cases13,14 and may be attributed to activating mutations in upstream kinases, such as FLT3,15 KIT,16 or JAK217 or alternatively be due to autocrine growth factor production.14 This prompted us to ask whether AML cells are dependent upon STAT5 activation for their survival and proliferation. Recently, we have described an improved method for long-term culture of CD34+ AML cells. In this assay expansion, leukemic cobblestone area (L-CA) formation and self-renewal can be studied in vitro up to 22 weeks (D.v.G., H.S., Aleksandra Rizo, Dorina van der Kolk, E.V., and J.J.S., submitted manuscript). In the present study, this assay was combined with an optimized lentiviral transduction protocol to introduce STAT5 RNAi hairpins into these expanding CD34+ AML cells, and the results were compared with normal CD34+ CB cells.
Our data indicate that lentiviral-mediated RNA interference of STAT5 in CB CD34+ cells reduced the growth rate of primitive stem or progenitor cells, leading to decreased colony-forming cell (CFC) and long-term culture-initiating cell (LTC-IC) numbers and eventually resulted in lower expansion rates. Single-cell assays with the more primitive CB CD34+/CD38− cell population suggested that STAT5 is necessary for maintenance of hematopoiesis. Subsequent introduction of the STAT5 RNAi hairpin into CD34+ AML cells, a fraction enriched for L-CA–forming (stem) cells, demonstrated that self-renewal and expansion of leukemic stem cells was also dependent upon STAT5 expression. Thus, we conclude that STAT5 expression is required for the maintenance and expansion of human stem/progenitor cells in both normal and leukemic hematopoiesis.
Materials and methods
Long-term cultures on stroma
Cord blood (CB) CD34+ cells were derived from neonatal cord blood from healthy full-term pregnancies after informed consent from the obstetrics departments of the Martini Hospital and University Medical Center in Groningen, the Netherlands and isolated by MiniMACS (Miltenyi Biotec, Amsterdam, the Netherlands) selection. After transduction, 30 000 cells were expanded on MS5 stromal cells in long-term culture (LTC) medium (αMEM supplemented with heat-inactivated 12.5% FCS, heat-inactivated 12.5% horse serum [Sigma, Zwijndrecht, the Netherlands], penicillin and streptomycin, 200 mM glutamine, 57.2 μM β-mercaptoethanol [Sigma] and 1 μM hydrocortisone [Sigma]) as described previously.18 AML blasts from peripheral blood cells or bone marrow cells from untreated patients with AML were studied after informed consent was obtained in accordance with the Declaration of Helsinki, and the protocol was approved by the Medical Ethics Committee. AML mononuclear cells were isolated by density gradient centrifugation, and CD34+ cells were selected by MiniMACS. After transduction, at least 1.5 × 105 cells were plated onto T25 flasks preplated with MS5 stromal cells. AML cocultures were propagated as described in Rozenveld-Geugien et al,19 and details are described in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). AML cells were expanded in LTC medium supplemented with interleukin 3 (IL-3; Gist-Brocades, Delft, the Netherlands), granulocyte colony-stimulating factor (G-CSF; Rhone-Poulenc Rorer, Amstelveen, the Netherlands), and thrombopoietin (TPO; Kirin, Tokyo, Japan) (20 ng/mL each). Cultures were kept at 37°C and 5% CO2. Cultures were demipopulated weekly for analysis. TF-1 cells were cultured in RPMI 1640 (Biowhittaker, Verviers, Belgium) supplemented with 10% FCS and 10 ng/mL GM-CSF (Genetics Institute, Cambridge, MA).
Flow cytometry analysis
All fluorescence-activated cell sorter (FACS) analyses were performed on a FACScalibur LSR-II (Becton Dickinson [BD], Alpen a/d Rijn, the Netherlands) and data were analyzed using WinList 3D (Verity Software House, Topsham, ME). Cells were sorted on a MoFLo (DakoCytomation, Carpinteria, CA). Antibodies were obtained from BD. Annexin VAPC was obtained from IQ Products (Groningen, the Netherlands). Viability was assessed according to the manufacturer's recommendations. Briefly, at the designated times cells were harvested, resuspended in 100 μL calcium buffer containing 5 μL annexin V, and incubated for 20 minutes at 4°C in the dark. Cells were washed with 5 mL calcium buffer, and finally, binding of PE-conjugated annexin V was measured by fluorescence-activated cell sorting.
Lentiviral transductions
293T human embryonic kidney cells (2.5 × 106) were transfected with 3 μg pCMV Δ8.91, 0.7 μg VSV-G, and 3 μg pTRIP Renilla RNAi or pTRIP STAT5 RNAi (kind gifts of Prof Dr H. Spits, Department of Cell Biology and Histology, Amsterdam Medical Center, Division of Immunology, Netherlands Cancer Institute, Amsterdam, the Netherlands). Target sequences have been described by Scheeren et al20 (and Table S1). After 24 hours, medium was changed to HPGM (Cambrex, Verviers, Belgium), and after 12 hours supernatant containing lentiviral particles was harvested and stored at − 80°C. CD34+ AML blasts and CB CD34+ cells were isolated with MiniMACS columns and subsequently cultured in RPMI supplemented with 10% FCS, IL-3, G-CSF, and TPO (20 ng/mL each) for 4 hours at 37°C and 5% CO2 or in HPGM supplemented with c-Kit ligand, Flt-3 ligand (both from Amgen, Thousand Oaks, CA), and TPO (100 ng/mL each) for 16 hours at 37°C and 5% CO2, respectively. AML blasts were transduced as described previously21 and a detailed protocol is available in Document S1. Briefly, transductions were performed in 3 consecutive rounds of 8 to 12 hours with lentiviral supernatant supplemented with 10% FCS, IL-3, G-CSF, and TPO (20 ng/mL each), polybrene (4 μg/mL; Sigma), and CB CD34+ cells in 2 consecutive rounds of 8 to 12 hours with lentiviral supernatant supplemented with c-Kit ligand/Flt-3 ligand/TPO (100 ng/mL each) and polybrene (4 μg/mL). TF-1 cells were transduced in 1 round of 12 hours with lentiviral supernatant supplemented with 10% FCS, 10 ng/mL GM-CSF, and polybrene (8 μg/mL). Transduction efficiency was measured by FACS analysis and knockdown was investigated by Western blot using antibodies against phospho-STAT5A/B Tyr 694/699 (Millipore, Bedford, MA), STAT5 (C17), and STAT3 (C20) (Santa Cruz, CA) and actin (C4) (ICN Biomedicals, Zoetermeer, the Netherlands) in a 1:1000 dilution in PBS on an Odyssey infrared scanner (Li-Cor Biosciences, Lincoln, NE) or using enhanced chemiluminescence (ECL) according to the manufacturer's instructions (Roche Diagnostics, Almere, the Netherlands). Alternatively, knockdown was investigated by quantitative reverse-transcription–polymerase chain reaction (RT-PCR; primer sequences and quantitative PCR [Q-PCR] conditions are described in Document S1).
CFC, CFU-GM, BFU-E, and LTC-IC assays
CFC assays and LTC-IC assays on MS5 stromal cells were performed as described previously.22 Briefly, CFC assays were performed in 1.2% methylcellulose containing 30% FCS, 57.2 μM β-mercaptoethanol, and 2 mM glutamine, supplemented with 20 ng/mL IL-3, 20 ng/mL IL-6, 20 ng/mL G-CSF, 20 ng/mL c-Kit ligand, and 6 U/mL EPO (Cilag; Eprex, Brussels, Belgium). Cells were either plated in bulk by 1000 transduced cells per plate in duplicate or in 96-well plates in 1, 3, 9, 27, 81, or 243 cells per well. Colony-forming unit–granulocyte macrophage (CFU-GM) and burst-forming unit–erythrocyte (BFU-E) assays were performed in 1.2% methylcellulose containing 30% FCS, 57.2 μM β-mercaptoethanol, and 2 mM glutamine, supplemented with 10 ng/mL GM-CSF and 10 ng/mL IL-3 or 2 U/mL EPO, respectively. LTC-IC assays were performed by plating transduced CB CD34+ cells in limiting dilutions in the range of 10 to 2400 cells per well on MS5 stromal cells in 96-well plates in LTC medium.
Single-cell assays
CB CD34+ cells were isolated using MiniMACS columns and cultured in HPGM supplemented with c-Kit ligand/Flt-3 ligand/TPO (100 ng/mL each) for 8 hours at 37°C and 5% CO2 and subsequently transduced in 1 round of 8 hours with lentiviral supernatant supplemented with c-Kit ligand/Flt-3 ligand/TPO (100 ng/mL each) and polybrene (4 μg/mL). CD34+ CD38− YFP/GFP+ cells were single sorted into 96-well plates into 100 μL HPGM supplemented with c-Kit ligand/Flt-3 ligand/TPO (100 ng/mL each) and followed microscopically for 100 hours. After this period, methylcellulose for CFC assays was added and colonies were scored after 2 weeks.
(Quantitative) PCR
Target gene expression was investigated by (quantitative) RT-PCR (Q-PCR) (primer sequences and conditions are described in Document S1). For RT-PCR, total RNA was isolated from 1 × 105 to 1 × 106 cells using the RNeasy kit from Qiagen (Venlo, the Netherlands) according to the manufacturer's recommendations. RNA was reverse transcribed with Moloney murine leukemia virus (M-MuLV) reverse transcriptase (Roche Diagnostics). For PCR, 2 μL cDNA was amplified in a total volume of 25 μL using 2 units of Taq polymerase (Roche Diagnostics). As a negative control, RNA minus reverse transcriptase (−RT) prepared cDNA was used in PCR reactions. Aliquots (10 μL) were run on 1.5% agarose gels. For real-time RT-PCR, 2-μL aliquots of cDNA were real-time amplified using iQ SYBR Green supermix (Bio-Rad, Veenendaal, the Netherlands) on a MyIQ thermocycler (Bio-Rad) and quantified using MyIQ software. GAPDH expression was used to calculate and normalize expression of all genes investigated.
Results
STAT5 down-regulation impairs long-term expansion of CB CD34+ cells on MS5 bone marrow stroma
To address whether downmodulation of STAT5 expression affects the proliferation and survival of human stem and progenitor cells, CB CD34+ cells were sorted and transduced with lentiviral vectors containing short interfering hairpins against control (Renilla luciferase) or STAT5A/B mRNA (Figure 1A). Flow cytometric analysis demonstrated transduction efficiencies ranging between 50% and 95% (Figure 1B and data not shown). Western blot analysis confirmed efficient down-regulation of STAT5 in CB CD34+, up to 70% to 75% downmodulation relative to STAT5 expression in control cells (Figure 1C, normalized against STAT3 protein, with control cultures arbitrarily set at 100%). STAT3 protein, as a close homologue of STAT5, is shown to indicate specificity of the STAT5 hairpin. Knockdown of STAT5 protein levels correlated well with down-regulation of STAT5 mRNA levels in transduced cells (Figure 1D, STAT5 mRNA values were normalized against GAPDH mRNA values). In addition, in TF-1 cells transduced with STAT5 hairpins it was demonstrated that phospho-STAT5 and total STAT5 were comparably down-regulated, whereas phospho-STAT3 was not affected, furthermore indicating specificity of the down-regulation (Figure 1C).
Downmodulation of STAT5 by RNAi decreases growth of CD34+ stem/progenitor cells. (A) Schematic overview of the lentiviral RNAi vectors used in these studies. (B) Transduction efficiency of CD34+ CB stem/progenitor cells, performed as indicated in “Lentiviral transductions.” (C) Western blot analysis of STAT5 levels in control (Renilla) or STAT5 RNAi–transduced CD34+ cells. Percentages indicate expression of STAT5 relative to control cells, normalized against STAT3 levels. Phospho-STAT3 and -STAT5 levels are shown for the hematopoietic TF-1 cell line. Quantitative Western blot analysis was performed on an Odyssey infrared scanner or using Quantity One imaging software from Bio-Rad. (D) Quantitative PCR for STAT5 mRNA in transduced CB CD34+ stem/progenitor cells. STAT5 mRNA was normalized against GAPDH mRNA expression.
Downmodulation of STAT5 by RNAi decreases growth of CD34+ stem/progenitor cells. (A) Schematic overview of the lentiviral RNAi vectors used in these studies. (B) Transduction efficiency of CD34+ CB stem/progenitor cells, performed as indicated in “Lentiviral transductions.” (C) Western blot analysis of STAT5 levels in control (Renilla) or STAT5 RNAi–transduced CD34+ cells. Percentages indicate expression of STAT5 relative to control cells, normalized against STAT3 levels. Phospho-STAT3 and -STAT5 levels are shown for the hematopoietic TF-1 cell line. Quantitative Western blot analysis was performed on an Odyssey infrared scanner or using Quantity One imaging software from Bio-Rad. (D) Quantitative PCR for STAT5 mRNA in transduced CB CD34+ stem/progenitor cells. STAT5 mRNA was normalized against GAPDH mRNA expression.
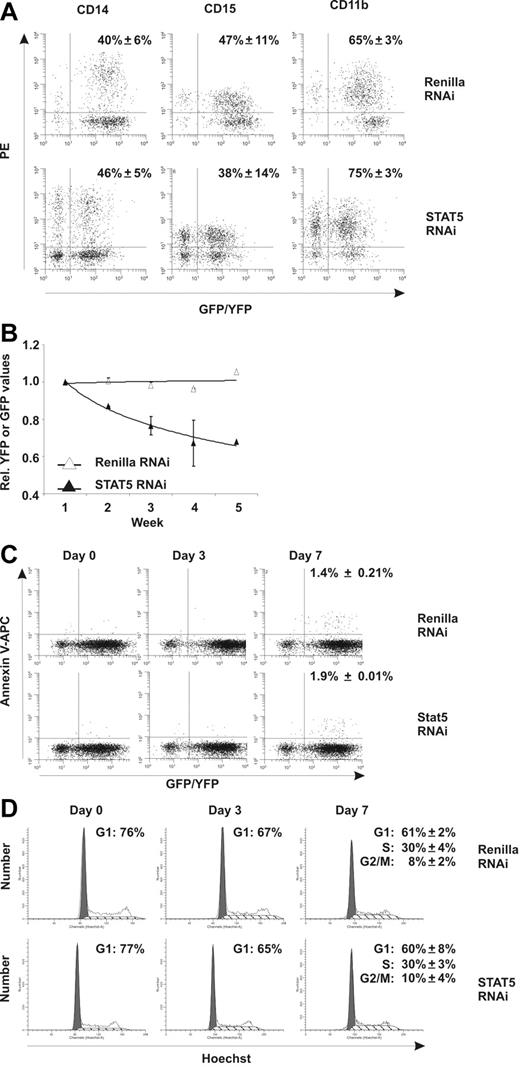
To evaluate the effects of STAT5 knockdown on CD34+ cell growth and differentiation, transduced cells were plated onto MS5 stromal cells and cultures were weekly demidepopulated and analyzed for 5 weeks by flow cytometry. No effects of STAT5 downmodulation were observed on the myeloid differentiation program, as no change in the percentage of GFP+ cells positive for CD14 (40% ± 6% vs 46% ± 5%), CD15 (47% ± 11% vs 38% ± 14%), and CD11b (65% ± 3% vs 75% ± 3%) was observed (Figure 2A, N = 3). STAT5 downmodulation also did not affect erythroid differentiation, because the percentage of cells positive for CD36, CD71, and glycophorin A was not different from control Renilla–transduced cells (data not shown). The percentage of YFP/GFP+ cells in suspension was also monitored for 5 weeks. Stable YFP percentages were observed over time in control Renilla cultures relative to the YFP percentage at week 1, indicating no negative effects of the transduction procedures (Figure 2B). The STAT5 hairpin (GFP), however, induced a pronounced reduction of GFP+ cells in suspension over time (29% ± 12.7%), indicating that STAT knockdown impairs cell survival or proliferation. To test whether increased apoptosis was responsible for the reduction in cell numbers, annexin V–APC [Allophycocyanin] labeling was performed. At days 0 and 3 after transduction, no difference in apoptosis within the YFP/GFP+ population between control and STAT5 RNAi MS5 cocultures was detected above background (Figure 2C), as well as in CD34+ cells cultured in 100 ng/mL c-Kit ligand/Flt-3 ligand/TPO (data not shown). Seven days after transduction, annexin V staining was marginally increased, but did not demonstrate any differences between Renilla and STAT5 RNAi (Figure 2C).
Downmodulation of STAT5 by RNAi decreases growth of CD34+ stem/progenitor cells. (A) FACS analysis of transduced CB cells on MS5 coculture. MS5 cocultures were analyzed at week 3 for cells positive for CD14, CD15, and CD11b. The average percentage (from 3 independent experiments) of YFP/GFP+ cells positive for the respective markers is shown with standard deviations. (B) Transduced CD34+ CB cells in long-term coculture on MS5 stromal cells. Shown are average YFP (Renilla control) or GFP STAT5 RNAi percentages relative to week 1. A trendline is indicated and error bars denote standard deviations. N = 4. (C) Annexin V–APC stain of transduced CB cells on the indicated days in MS5 coculture. For day 7, the average percentage of annexin V–positive cells within the YFP or GFP gate from 3 independent experiments is shown with standard deviations. (D) Hoechst (5 μg/mL) cell-cycle labeling of YFP+ or GFP+ CB cells on the indicated days in MS5 coculture. For day 7, the average percentages of cells in G1, S, or G2/M phase of the cell cycle are shown from 3 independent experiments with standard deviations.
Downmodulation of STAT5 by RNAi decreases growth of CD34+ stem/progenitor cells. (A) FACS analysis of transduced CB cells on MS5 coculture. MS5 cocultures were analyzed at week 3 for cells positive for CD14, CD15, and CD11b. The average percentage (from 3 independent experiments) of YFP/GFP+ cells positive for the respective markers is shown with standard deviations. (B) Transduced CD34+ CB cells in long-term coculture on MS5 stromal cells. Shown are average YFP (Renilla control) or GFP STAT5 RNAi percentages relative to week 1. A trendline is indicated and error bars denote standard deviations. N = 4. (C) Annexin V–APC stain of transduced CB cells on the indicated days in MS5 coculture. For day 7, the average percentage of annexin V–positive cells within the YFP or GFP gate from 3 independent experiments is shown with standard deviations. (D) Hoechst (5 μg/mL) cell-cycle labeling of YFP+ or GFP+ CB cells on the indicated days in MS5 coculture. For day 7, the average percentages of cells in G1, S, or G2/M phase of the cell cycle are shown from 3 independent experiments with standard deviations.
Next, we tested whether changes in cell-cycle status could account for the differences observed between Renilla YFP+ and STAT5 GFP+ cells. Figure 2D shows Hoechst labeling of YFP+ and GFP+ cells (from MS5 cocultures) on days 0, 3, and 7 after transduction, demonstrating a slight decrease in the fraction of cells in G1 phase, but again with no apparent differences between control and STAT5 RNAi cultures. Similar results were obtained with c-Kit ligand/Flt-3 ligand/TPO–treated cultures (data not shown). Together, the reduced growth of STAT5 knockdown cultures and the absence of differences in both apoptosis and cell-cycle assays suggest that knockdown of STAT5 impairs the outgrowth of a small fraction of primitive stem or progenitor cells.
STAT5 down-regulation reduces progenitor and stem-cell frequencies
To test the effects of STAT5 downmodulation on the progenitor pool, colony assays were performed with transduced CB CD34+ cells in methylcellulose. Downmodulation of STAT5 levels resulted in a 56.5% plus or minus 25% reduction of colony-forming unit–granulocyte macrophage (CFU-GM) colonies (P = .01; n = 3) and a 43.3% plus or minus 31.9% reduction of burst-forming unit–erythrocyte (BFU-E) colonies (P < .05; n = 3). In Figure 3A, a representative example is shown. The reduction after STAT5 RNAi of total colony numbers in colony-forming cell (CFC) assays relative to Renilla RNAi was 43.3% plus or minus 16.2% (P < .01; n = 3, data not shown).
Knockdown of STAT5 reduces CFC and LTC-IC frequencies of CB CD34+ cells. (A) CFU-GM and BFU-E assays with control and STAT5-transduced CB cells. Assays were performed 3 times in duplicate under conditions indicated in “CFC, CFU-GM, BFU-E, and LTC-IC assays.” Error bars denote standard deviations. (B) Limiting-dilution CFC assay performed as described in “CFC, CFU-FM, BFU-E, and LTC-IC assays.” Poisson statistics were used to calculate CFC progenitor frequencies. N = 3; a representative experiment is shown. (C) Percentage of CFC progenitors as calculated from the limiting-dilution assays. The average of 3 experiments is shown, with error bars representing standard deviations. A Student t test was performed to calculate statistical differences between the 2 groups. (D) Limiting-dilution LTC-IC assay performed as described in “CFC, CFU-GM, BFU-E, and LTC-IC assays.” Poisson statistics were used to calculate LTC-IC frequencies. N = 3; a representative experiment is shown. (E) The average percentage of LTC-IC cells as calculated from the limiting-dilution experiment shown in D. N = 3; error bars denote standard errors.
Knockdown of STAT5 reduces CFC and LTC-IC frequencies of CB CD34+ cells. (A) CFU-GM and BFU-E assays with control and STAT5-transduced CB cells. Assays were performed 3 times in duplicate under conditions indicated in “CFC, CFU-GM, BFU-E, and LTC-IC assays.” Error bars denote standard deviations. (B) Limiting-dilution CFC assay performed as described in “CFC, CFU-FM, BFU-E, and LTC-IC assays.” Poisson statistics were used to calculate CFC progenitor frequencies. N = 3; a representative experiment is shown. (C) Percentage of CFC progenitors as calculated from the limiting-dilution assays. The average of 3 experiments is shown, with error bars representing standard deviations. A Student t test was performed to calculate statistical differences between the 2 groups. (D) Limiting-dilution LTC-IC assay performed as described in “CFC, CFU-GM, BFU-E, and LTC-IC assays.” Poisson statistics were used to calculate LTC-IC frequencies. N = 3; a representative experiment is shown. (E) The average percentage of LTC-IC cells as calculated from the limiting-dilution experiment shown in D. N = 3; error bars denote standard errors.
The precise progenitor frequency was determined in limiting-dilution CFC assays using YFP- and GFP-sorted cells. Knockdown of STAT5 in CD34+ cells demonstrated an almost 4-fold reduction in the frequency of progenitors from 5.42% plus or minus 0.47% in Renilla control cells to 1.37% plus or minus 0.13% in STAT5 knockdown (Figure 3B,C; P < .02).
To investigate whether long-term culture-initiating cell (LTC-IC) frequencies might also be affected, limiting-dilution LTC-IC assays were performed with YFP- and GFP-sorted cells in 3 independent experiments. Data in Figure 3D,E demonstrate a reduction in the percentage of LTC-IC colonies formed from 0.23% to 0.037% upon knockdown of STAT5. In 2 additional independent experiments, a reduction from 0.044% to 0.023% and from 0.1% to 0.048% was observed (Renilla versus STAT5 RNAi, respectively). On average, this indicates a 3.25-fold reduction in LTC-IC frequency from 0.13% to 0.04% upon STAT5 RNAi. This decrease in the number of LTC-IC colonies after STAT5 RNAi was also observed when enumerating the number of cobblestone area-forming cells (CAFCs; Renilla versus STAT5, respectively, 0.097% and 0.020%; data not shown). Together these data suggest that a reduction of STAT5 leads to an impaired maintenance of stem and progenitor cells.
Knockdown of STAT5 decreases the production of offspring of CD34+/CD38− cells
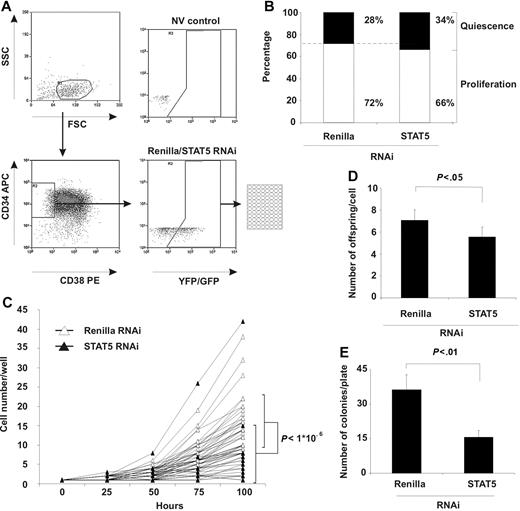
Apoptosis and the cell-cycle distribution analysis did not show a change in the majority of the CD34+ cells (Figure 2C,D). However, because both stem cell (LTC-IC) and progenitor frequencies decreased in STAT5 knockdown cells (Figure 3), we wondered what the effect of STAT5 RNAi would be in a subpopulation of CD34+ cells (eg, primitive CD34+/CD38− cells), a fraction that is highly enriched for cells with stem-cell activity.23 After transduction of CD34+ isolated cells, CD34+/CD38− YFP+ (Renilla) and GFP+ (STAT5) cells were single-cell sorted into 96-well plates (Figure 4A) and subsequently monitored for 100 hours by microscopic evaluation. The number of cells present per well was enumerated each 25 hours and classified “quiescent” when still containing only 1 cell and “proliferating” when multiple cells were observed. Figure 4B demonstrates that the number of quiescent cells was marginally increased in STAT5 knockdown cells while the number of wells that contained proliferating cells was slightly decreased, but both changes were not significant (P > .1, n = 3).
STAT5 knockdown impairs outgrowth of primitive CD34+ CD38− cells. (A) Sorting scheme for obtaining single CD34+ CD38− YFP+ (Renilla) or GFP+ (STAT5 RNAi) cells in 96-well format. NV indicates a nontransduced (no virus) control to set gates. (B) Percentage of wells quiescent or proliferating after STAT5 knockdown. Indicated percentages are averaged from 3 independent experiments. (C) Cell number per proliferating well at the indicated time points after sorting. Thirty individual wells per group are shown. Bars indicate average cell number with standard deviations at t = 100 hours. A Student t test was performed to calculate statistical differences between the 2 groups. N = 3. (D) The average number of daughter cells (offspring) produced per proliferating cell/well. A Student t test was performed to calculate statistical differences between the 2 groups. Error bars denote standard deviations; N = 3. (E) The average number of CFC colonies produced by proliferating cells per plate. A Student t test was performed to calculate statistical differences between the 2 groups. Error bars denote standard deviations; N = 3.
STAT5 knockdown impairs outgrowth of primitive CD34+ CD38− cells. (A) Sorting scheme for obtaining single CD34+ CD38− YFP+ (Renilla) or GFP+ (STAT5 RNAi) cells in 96-well format. NV indicates a nontransduced (no virus) control to set gates. (B) Percentage of wells quiescent or proliferating after STAT5 knockdown. Indicated percentages are averaged from 3 independent experiments. (C) Cell number per proliferating well at the indicated time points after sorting. Thirty individual wells per group are shown. Bars indicate average cell number with standard deviations at t = 100 hours. A Student t test was performed to calculate statistical differences between the 2 groups. N = 3. (D) The average number of daughter cells (offspring) produced per proliferating cell/well. A Student t test was performed to calculate statistical differences between the 2 groups. Error bars denote standard deviations; N = 3. (E) The average number of CFC colonies produced by proliferating cells per plate. A Student t test was performed to calculate statistical differences between the 2 groups. Error bars denote standard deviations; N = 3.
However, of the CD34+/CD38− cells that were capable of division, wells containing STAT5 knockdown cells demonstrated significantly fewer cells after 100 hours of culture than control wells (Figure 4C, 8 ± 8 vs 16 ± 7 cells, respectively, P < .001, 30 individual cells for each group are shown). Furthermore, the average number of daughter cells produced in each well by STAT5 knockdown cells was significantly decreased compared with control Renilla knockdown cells (Figure 4D, 5.6 ± 0.9 cells vs 7.1 ± 0.9 cells, respectively, P < .01), resulting in a lower total amount of cells per plate (289 ± 64 vs 198 ± 72 cells, respectively, for Renilla and STAT5 RNAi, P < .01, data not shown). In addition, methyl cellulose was added to these wells after 100 hours of culture, and after 2 weeks the number of CFC colonies was enumerated. The number of wells positive for colonies was 2.3-fold lower in STAT5 hairpin–transduced cells compared with control cells (36 ± 6.5 vs 16 ± 2.9 colonies, respectively, for Renilla and STAT5 RNAi, P < .05, n = 3, Figure 4E), which verified the outcome of the limiting-dilution CFC assays (Figure 3B,C). Together these data demonstrate that at the single-cell level in primitive CD34+/CD38− cells, STAT5 RNAi impairs the proliferation rate.
Activated STAT5 is frequently observed in AML cells, which is efficiently down-regulated after lentiviral-mediated RNAi
STAT5 knockdown affected the proliferation rate of CD34+/CD38− cells. For malignant AML cells, this primitive cell compartment was exclusively shown to contain severe combined immunodeficient (SCID) leukemia-initiating cells.24 Because constitutive phosphorylation and activation of STAT5 has been described for AML patients,25 we were interested in the effect of STAT5 knockdown in self-renewing leukemic AML stem/progenitor cells. The AMLs were classified, according to FAB classification, as M1 (N = 3), M2 (N = 3), M4 (N = 4), and M5 (N = 5). The percentage of CD34+ cells in the AML mononuclear cell fraction demonstrated distinct variability as depicted in Table 1. PCR studies demonstrated that 8 of 16 AMLs had FLT3-ITD mutations (Table 1). Figure 5A depicts a Western blot of 8 primary AML samples and demonstrated the presence of phospho-STAT5 in 5 patients (62%), thus confirming previously published results.13,14 The phosphorylation of STAT5, however, did not correlate with Flt3 mutation status, potentially reflecting activation of other activators of STAT5 or the presence of autocrine growth factor production.14-17 Subsequently, the subpopulation of CD34+ AML cells (N = 10) was sorted and transduced with our lentiviral vectors. Flow cytometric analysis demonstrated comparable transduction efficiencies of AML CD34+ cells compared with CB CD34+ cells, ranging between 50% and 95% (Figures 1B,5B). Western blot analysis confirmed efficient down-regulation of STAT5 in these AML CD34+ cells (Figure 5C), which varied from 20% to 85% as normalized against STAT3 or actin protein. The functionality of STAT5 RNAi is shown in Figure 5D, demonstrating a downmodulation of the STAT5 target genes CIS, SOCS 1, and SOCS 2 in both the STAT5 RNAi-transduced TF1 cell line as well as in transduced AML CD34+ cells.
Summary of AML patient characteristics
| AML . | PB/BM . | % CD34 . | Flt3-ITD . | p-Stat5 . | FAB . | Risk group . | Karyotype . | Transduced . | CD34+ cells plated per T25 . | Growth until wk . | Expansion after Renilla RNAi . | Expansion after Stat5 RNAi . | Fold down-regulation . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | PB | 85 | − | + | M1 | Poor | +3q,−7, −10 | No | N/A | N/A | N/A | N/A | N/A |
| 2 | PB | 3 | − | − | M1 | Int | Normal | No | N/A | N/A | N/A | N/A | N/A |
| 3 | PB | 14 | − | + | M2 | Good | t(8;21) | No | N/A | N/A | N/A | N/A | N/A |
| 4 | PB | 15 | − | + | M4 | Poor | t(6;11)(q27;23) | No | N/A | N/A | N/A | N/A | N/A |
| 5 | PB | 69 | + | ± | M5 | Int | Normal | Yes | 1.5 × 105 | 6 | 25.0 | 15.6 | 1.6 |
| 6 | BM | 5 | + | − | M5b | Int | Normal | Yes | 2.2 × 105 | 4 | 4.2 | 5.7 | 0.7 |
| 7 | PB | 12 | + | + | M5 | Int | Normal | Yes | 1.5 × 105 | 6 | 9.7 | 4.0 | 2.4 |
| 8 | PB | 1 | + | + | M5 | Int | Normal | Yes | 1.5 × 105 | 6 | 8.1 | 2.4 | 3.4 |
| 9 | BM | nd | − | − | M4 | Int | Normal | Yes | 5.0 × 105 | 6 | 214.2 | 12.7 | 16.8 |
| 10 | BM | 80 | + | + | M1 | Good | inv(16) | Yes | 5.0 × 105 | 6 | 17.7 | 0.7 | 24.4 |
| 11 | BM | 5 | − | + | M4 | Poor | inv(16) | Yes | 5.0 × 105 | 6 | 207.1 | 67.1 | 3.1 |
| 12 | BM | 37 | nd | nd | M4 | Int | Normal | Yes | 2.2 × 105 | 4 | 7.6 | 1.6 | 4.8 |
| 13 | PB | 17 | + | nd | M2 | Poor | del 9, (q12;22) | Yes | 1.5 × 105 | 2, then † | N/A | N/A | N/A |
| 14 | BM | nd | − | − | M2 | Int | Normal | Yes | 5.0 × 105 | 2, then † | N/A | N/A | N/A |
| 15 | PB | 7 | + | − | M5b | Int | Normal | No | N/A | N/A | N/A | N/A | N/A |
| 16 | PB | 30 | + | nd | M1 | Int | Normal | Yes | N/A | N/A | N/A | N/A | N/A |
| AML . | PB/BM . | % CD34 . | Flt3-ITD . | p-Stat5 . | FAB . | Risk group . | Karyotype . | Transduced . | CD34+ cells plated per T25 . | Growth until wk . | Expansion after Renilla RNAi . | Expansion after Stat5 RNAi . | Fold down-regulation . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | PB | 85 | − | + | M1 | Poor | +3q,−7, −10 | No | N/A | N/A | N/A | N/A | N/A |
| 2 | PB | 3 | − | − | M1 | Int | Normal | No | N/A | N/A | N/A | N/A | N/A |
| 3 | PB | 14 | − | + | M2 | Good | t(8;21) | No | N/A | N/A | N/A | N/A | N/A |
| 4 | PB | 15 | − | + | M4 | Poor | t(6;11)(q27;23) | No | N/A | N/A | N/A | N/A | N/A |
| 5 | PB | 69 | + | ± | M5 | Int | Normal | Yes | 1.5 × 105 | 6 | 25.0 | 15.6 | 1.6 |
| 6 | BM | 5 | + | − | M5b | Int | Normal | Yes | 2.2 × 105 | 4 | 4.2 | 5.7 | 0.7 |
| 7 | PB | 12 | + | + | M5 | Int | Normal | Yes | 1.5 × 105 | 6 | 9.7 | 4.0 | 2.4 |
| 8 | PB | 1 | + | + | M5 | Int | Normal | Yes | 1.5 × 105 | 6 | 8.1 | 2.4 | 3.4 |
| 9 | BM | nd | − | − | M4 | Int | Normal | Yes | 5.0 × 105 | 6 | 214.2 | 12.7 | 16.8 |
| 10 | BM | 80 | + | + | M1 | Good | inv(16) | Yes | 5.0 × 105 | 6 | 17.7 | 0.7 | 24.4 |
| 11 | BM | 5 | − | + | M4 | Poor | inv(16) | Yes | 5.0 × 105 | 6 | 207.1 | 67.1 | 3.1 |
| 12 | BM | 37 | nd | nd | M4 | Int | Normal | Yes | 2.2 × 105 | 4 | 7.6 | 1.6 | 4.8 |
| 13 | PB | 17 | + | nd | M2 | Poor | del 9, (q12;22) | Yes | 1.5 × 105 | 2, then † | N/A | N/A | N/A |
| 14 | BM | nd | − | − | M2 | Int | Normal | Yes | 5.0 × 105 | 2, then † | N/A | N/A | N/A |
| 15 | PB | 7 | + | − | M5b | Int | Normal | No | N/A | N/A | N/A | N/A | N/A |
| 16 | PB | 30 | + | nd | M1 | Int | Normal | Yes | N/A | N/A | N/A | N/A | N/A |
PB indicates peripheral blood; BM, bone marrow; Flt3-ITD, Flt3 internal tandem duplication; FAB, French-American-British; nd, not determined; +, present; −, not present; and N/A, not applicable.
Efficient STAT5 knockdown in CD34+ AML cells impairs long-term growth on MS5 coculture. (A) Western blot analysis for phospho-STAT5 in 8 AML patients. Actin is shown as loading control. Flt3-ITD mutations are indicated, as well as the fold down-regulation of expansion after STAT5 RNAi. (B) Transduction efficiency of CD34+ AML cells, performed as indicated in “Lentiviral transductions.” (C) Western blot analysis of STAT5 levels in control (Renilla) or STAT5 RNAi–transduced CD34+ cells. Percentages indicate expression of STAT5 relative to control cells, normalized against STAT3 or actin levels. Quantitative Western blot analysis was performed on an Odyssey infrared scanner or using Quantity One imaging software from Bio-Rad. (D) Q-PCR analysis of the expression of the STAT5 target genes CIS, SOCS 1, and SOCS 2 after STAT5 RNAi in transduced TF1 and AML CD34+ cells. (E) Cumulative cell counts of control (Renilla) or STAT5 RNAi–transduced CD34+ AML cells on MS5 coculture. All cell counts were normalized against week 0, the time of plating. Four representative cultures are shown. N = 11. (F) Transduced CD34+ AML cells in long-term coculture on MS5 stromal cells. Shown are average YFP (Renilla control) or GFP STAT5 RNAi percentages relative to week 1. A trendline is indicated and error bars denote standard deviations. N = 7. (G) Averaged calculated expansion of 7 AML MS5 cocultures, with error bars indicating standard deviations. The average expansion at week 6 is indicated next to the experimental group.
Efficient STAT5 knockdown in CD34+ AML cells impairs long-term growth on MS5 coculture. (A) Western blot analysis for phospho-STAT5 in 8 AML patients. Actin is shown as loading control. Flt3-ITD mutations are indicated, as well as the fold down-regulation of expansion after STAT5 RNAi. (B) Transduction efficiency of CD34+ AML cells, performed as indicated in “Lentiviral transductions.” (C) Western blot analysis of STAT5 levels in control (Renilla) or STAT5 RNAi–transduced CD34+ cells. Percentages indicate expression of STAT5 relative to control cells, normalized against STAT3 or actin levels. Quantitative Western blot analysis was performed on an Odyssey infrared scanner or using Quantity One imaging software from Bio-Rad. (D) Q-PCR analysis of the expression of the STAT5 target genes CIS, SOCS 1, and SOCS 2 after STAT5 RNAi in transduced TF1 and AML CD34+ cells. (E) Cumulative cell counts of control (Renilla) or STAT5 RNAi–transduced CD34+ AML cells on MS5 coculture. All cell counts were normalized against week 0, the time of plating. Four representative cultures are shown. N = 11. (F) Transduced CD34+ AML cells in long-term coculture on MS5 stromal cells. Shown are average YFP (Renilla control) or GFP STAT5 RNAi percentages relative to week 1. A trendline is indicated and error bars denote standard deviations. N = 7. (G) Averaged calculated expansion of 7 AML MS5 cocultures, with error bars indicating standard deviations. The average expansion at week 6 is indicated next to the experimental group.
Knockdown of STAT5 impairs long-term growth of leukemic cells on MS5 cocultures
To evaluate the effects of STAT5 knockdown on the proliferation of AML CD34+ cells, transduced cells were plated onto MS5 stromal cells and subsequently followed for growth and expansion (N = 10). Cultures of AMLs with FAB classification M2 failed to expand and were terminated. Figure 5E demonstrates 4 representative cumulative growth cultures of the remaining cultures. All expanding CD34+ AML cultures transduced with the STAT5 hairpin (N = 8) demonstrated an impaired growth compared with control-transduced cultures, although the level of reduction in growth was variable. The average reduction in cell number after STAT5 RNAi compared with control cells was 72% plus or minus 17% (P < .001). This reduction was also observed when relative percentages of YFP+ (Renilla control) and GFP+ (STAT5 hairpin) cells were calculated in expanding AML cultures. GFP percentages indicated a decline similar to the decline in CB CD34+ MS5 cocultures (Figure 5F, data shown are an average of 7 AML cultures, compare with Figure 2B).
Furthermore, the expansion of these AML cultures was calculated. Figure 5G depicts that STAT5 knockdown resulted in an almost 5-fold decreased expansion from an average 80-fold expansion (± 58) of control-transduced cultures at week 6, to an average 17-fold expansion (± 15) for STAT5 hairpin–transduced cultures (P < .05, Table 1). Figure 5A and Table 1 indicate that the fold down-regulation of this expansion is slightly more pronounced for AML samples with STAT5 phosphorylation than for the ones without. This suggests that these AMLs are more dependent upon STAT5 signaling, although the number of AMLs analyzed is limited and additional future experiments in which more AML samples are analyzed are needed to explore this relationship in further detail. Taken together, these data suggest that CD34+ AML cells are also dependent upon STAT5 signaling for their long-term growth.
Discussion
The present study demonstrates that limited reduction of STAT5 expression especially affects the hematopoietic stem/progenitor pool. In our MS5 coculture experiments, we observed a growth reduction, without affecting the differentiation program of the myeloid and erythroid lineages. This finding is remarkable because a wide variety of hematopoietic growth factors and cytokines, including FLT3-L, SCF, G-CSF, GM-CSF, IL-3, EPO, and TPO, has been shown to induce STAT5 transactivation and affect the differentiation of human stem/progenitor cells.26-32 These differences in cell biologic effects downstream of STAT5 most likely find their origin in differences between cell types, (combinations of) cytokine signals, and concentrations of cytokines (reviewed in Smithgall et al2 and Coffer et al3 ). In contrast, distinct phenotypes were observed by downmodulation of STAT5 expression in the HSC and progenitor subpopulations, because a significant reduction in progenitor and LTC-IC frequencies was observed. Recently, Scherr et al have reported that normal human CD34+ progenitors were less sensitive to STAT5 RNAi then CML CD34+ progenitors,33 but their reductions of approximately 45% for normal CD34+ progenitors are comparable with our findings. Furthermore, their and our observed reduction in colony numbers after STAT5 RNAi corresponded with the reductions seen in CFU-mix colonies (∼ 43%) and in CFU-GM colonies (∼ 53%) in STAT5ABΔN/ΔN knockout mice.5 These findings suggest that the reduction in STAT5 levels, as obtained by RNA interference, is sufficient to induce similar hematopoietic phenotypes as the 100% STAT5 reduction in the STAT5AB knockout mice, suggesting that our observations are not due to differences in transduction rates or RNAi efficiency.
Both progenitors and more primitive cells, determined with LTC-IC assays, displayed reduced frequencies in STAT5A RNAi–transduced cells. In addition, single-cell experiments with CD34+CD38− cells demonstrated that the number of daughter cells produced by STAT5 RNAi cells is lower than in control cells and gives rise to fewer colonies in methylcellulose. Therefore, we speculate that a more primitive cell (eg, a common myeloid progenitor [CMP]) or an HSC is being affected by STAT5 reduction. Interestingly, HSCs from STAT5ABΔN/ΔN knockout mice with impaired repopulation potential displayed a reduction in spleen colony-forming units (CFU-S).8,9 Those CFU-Ss were smaller than those from wild-type cells, indicating fewer progeny cells produced per colony, which is in line with our findings. Whether STAT5 affects both primitive progenitors (CMPs) and HSCs or only HSCs, with subsequent reductions in downstream progenitors, needs further elucidation by prospective purification steps of STAT5 RNAi–transduced HSCs versus CMPs. How STAT5 affects these stem/progenitor cells remains unclear. In STAT5ABΔN/ΔN mice, increased apoptosis in erythroid progenitors has been assigned to decreased transcription of the Bcl-XL gene,6,7 and restoration of Bcl-XL could fully compensate for this effect.34 On the other hand, STAT5A-deficient mice demonstrate a defect in proliferation of bone marrow–derived macrophages.35 Further studies in purified CD34+CD38− cells transduced with STAT5 RNAi should hence provide more insight in the processes of apoptosis and proliferation.
Overexpression of a constitutively active STAT5 enhanced the self-renewal potential of CD34+ cells.11,12 In mice, this was coupled to the development of myeloproliferative disease (MPD) or multilineage leukemias.12,36 Detailed analysis of derivatives of this constitutive active STAT5 showed that the enhanced self-renewal or MPD/multilineage leukemia was potentially due to enhanced tetramer formation and hence more stable DNA binding of constitutive active STAT5 mutants to promoters of target genes.36 Interestingly, similar STAT5 tetramers were observed in various AML, chronic myeloid leukemia (CML), and acute lymphoid leukemia (ALL) patient samples.36 In addition, STAT5 activation has been shown to be critical for BCR-ABL–dependent transformation of hematopoietic cells.33,37-40
For AML, constitutive activation of STAT5 has been demonstrated,13,14 but the role of STAT5 signaling in the pathogenesis of AML remained so far elusive. This is predominantly hampered by experimental limitations. Gene transfer into primary AML has shown stable targeting efficiencies,41,42 but in liquid culture these cells lost their viability after approximately 15 days. Although AML SCID-leukemia initiating cells (AML SL-ICs) could sustain longer growth in (nonobese diabetic [NOD])–SCID mice24,43 this assay was so far not combined with molecular interference in signal transduction pathways. Furthermore, it was demonstrated that the in vivo NOD-SCID model also has limitations, as almost half of the investigated AML cases did not engraft in irradiated mice.44 Therefore, we used an AML culture assay in which expansion of transduced leukemic cells could be followed for prolonged periods of time. Analysis of persistence of mutations in secondary cultures and the presence of self-renewal capacity after serial replating confirmed that we were studying leukemic stem and progenitor cells (van Gosliga et al, submitted manuscript). The results indicate that by lentiviral transduction RNA interference allows us to study the importance of STAT5 activation in sorted CD34+AML cells. Knockdown of STAT5 in AML CD34+ cells shows a reduction in cumulative cell numbers within 5 to 6 weeks of coculture on MS5. This reduction in expansion of AML cells upon STAT5 RNAi was similar to the decline we observed in CD34+ CB cells, suggesting that a comparable cell type, either a primitive progenitor or a stem cell, is being affected. In addition, no changes for myeloid and erythroid markers between STAT5 RNAi and control AML cultures were observed (data not shown). This is in line with the results of CB CD34+ cells demonstrating that STAT5 RNAi cells are not affected in their differentiation program.
Our data indicate that the concomitant reduction in STAT5 phosphorylation in the STAT5 RNAi–transduced cells is sufficient to limit the expansion of the leukemic stem/progenitor pool. From a clinical point of view, this is an important observation, because a partial reduction of STAT5 already gives a distinct phenotype. This would imply that alternative strategies (eg, blocking kinase activity, which results in a partial reduction in STAT5 activity) will have distinct effects on the leukemic stem-cell compartment. However, this inhibitory effect was not exclusive for CD34+ AML cells, but was also noticed in normal CD34+ cells. Future studies will have to elucidate whether a therapeutic window exists for STAT5 downmodulation to specifically target AML CD34+ cells. Nevertheless, in vivo studies with the multitarget kinase inhibitor SU11248 have demonstrated marked reductions (> 50%) in STAT5 phosphorylation in peripheral blood of AML patients that coincided with a decreased blast cell count in some of those patients, in line with the results presented here.45 In conclusion, our data indicate that STAT5 expression is required for the maintenance and expansion of both normal as well as leukemic stem/progenitor cells.
The online version of this manuscript contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by Dutch Cancer Society (KWF) grant RUG 2000–2004 (E.V.) and an NWO-VENI [Nederlandse organisatie voor Wetenschappelijk Onderzoek-VENI] (2004–2008) grant (J.J.S.).
The authors would like to acknowledge Geert Mesander and Henk Moes for help with flow cytometry; Prof Dr H. Spits from the Department of Cell Biology and Histology, Amsterdam Medical Center, Division of Immunology, Netherlands Cancer Institute, Amsterdam, the Netherlands, for the kind gifts of pTRIP Renilla RNAi and pTRIP STAT5 RNAi; and Amgen and Kirin for providing cytokines. The authors greatly appreciate the help of Dr A. van Loon, Dr J. J. Erwich, and colleagues (Obstetrics departments from the Martini Hospital and UMCG) for collecting cord blood.
Authorship
Contribution: H.S. performed research, analyzed data, and wrote the paper; D.G. and A.T.J.W. contributed analytic tools; B.J.L.E. analyzed data; J.J.S. and E.V. designed research and analyzed data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Edo Vellenga, Division of Hematology, Department of Medicine, University Medical Center Groningen, Hanzeplein 1, 9713 GZ Groningen, the Netherlands; e-mail: e.vellenga@int.umcg.nl.