Abstract
Global gene expression profiling of highly purified 5q-deleted CD34+CD38−Thy1+ cells in 5q− myelodysplastic syndromes (MDSs) supported that they might originate from and outcompete normal CD34+CD38−Thy1+ hematopoietic stem cells. Few but distinct differences in gene expression distinguished MDS and normal stem cells. Expression of BMI1, encoding a critical regulator of self-renewal, was up-regulated in 5q− stem cells. Whereas multiple previous MDS genetic screens failed to identify altered expression of the gene encoding the myeloid transcription factor CEBPA, stage-specific and extensive down-regulation of CEBPA was specifically observed in MDS progenitors. These studies establish the importance of molecular characterization of distinct stages of cancer stem and progenitor cells to enhance the resolution of stage-specific dysregulated gene expression.
Introduction
Although the existence of cancer stem cells (CSCs), in particular leukemic stem cells (LSCs), has been established for more than a decade,1,2 fundamentally important questions remain unanswered, relating to the exact identity and normal cellular origin of human CSCs, knowledge likely to have a major impact toward a better understanding of the evolution, prognosis, and therapeutic targeting of CSCs
Hematopoietic stem cells (HSCs) possess lifelong self-renewal capacity and are therefore at high risk of acquiring the multiple mutations thought to be required for ultimate leukemic transformation. However, conclusive evidence for a normal HSC origin of LSCs would require proof of clonal involvement of all blood cell lineages derived from the multipotent HSCs. However, as yet, it has only been possible to obtain direct evidence for multilineage involvement in myeloproliferative disorders.3,4 Although this could mean that leukemias rarely originate in normal HSCs or multipotent progenitors, it is as possible that in most cases it is not feasible to prove through this approach, as most leukemias are lineage restricted in nature. Specifically, it is likely that a transforming event in an HSC will not only promote the development of the dominating leukemic lineage but also simultaneously suppress the development of other lineages.5
Instead, an alternative and novel approach would be to apply genomics to better identify the origin of LSCs. As normal HSCs have been highly purified and found to have a distinct gene-expression profile when compared with normal progenitors,6,7 a similar approach should be possible to better establish whether LSCs have a stem or progenitor cell identity. It would however require the identity of the LSC population to be well established as well as conclusive evidence for a high clonal involvement of the purified LSCs to be investigated. So far, these criteria would be fulfilled only in rare cases, one being myelodysplastic syndromes (MDSs) or preleukemia, a group of clonal malignant hematopoietic disorders that frequently progress to acute myeloid leukemia (AML).8 In patients with 5q− syndrome, a distinct clinical subgroup of MDS characterized by an isolated deletion of the long arm of chromosome 5 (del(5q) or 5q−),9 we recently demonstrated that the 5q− stem cells reside in the minor CD34+CD38−Thy1+ compartment, with definitive evidence for clonal myeloid as well as B-lymphoid involvement in some patients, suggesting that 5q− syndrome at least in part originates at a multipotent stem/progenitor cell level.10,11 However, in most patients, no evidence was obtained for B-cell involvement and T-cell involvement could never be established.
In the present studies, we purified and performed global gene-expression profiling of normal and 5q− CD34+CD38−Thy1+ cells,12,13 typically representing 0.1% of the total MDS bone marrow (BM) cells, and CD34+CD38+Thy1− progenitors. This approach allowed us in all investigated cases to implicate the 5q− syndrome as a likely true HSC disease and to establish stage-specific dysregulated gene expression.
Patients, materials, and methods
Patient samples
BM samples from 11 MDS patients with typical 5q− syndrome9 (Table 1) and from 10 healthy subjects were collected at the Hematology Departments at Karolinska (Solna), Karolinska (Huddinge), South Hospital, Helsingborg Hospital, and Lund University Hospitals, Sweden. The investigation was approved by the Research Ethics Committees at the respective University Hospitals, and informed consent was obtained in accordance with the Declaration of Helsinki. The MDS patients were chosen based on the finding of an isolated del(5q) by conventional cytogenetic analysis in combination with clinical and morphologic characteristics typical for the 5q−syndrome (an indolent clinical course with macrocytic anemia and a variable need for red blood cell transfusions, normal to elevated platelet counts, normal to slightly reduced white blood cell [WBC] counts, hypolobulated megakaryocytes, and less than 5% blasts in the BM), resulting in MDS with refractory anemia (RA; French-American-British [FAB]) and a 5q− syndrome (World Health Organization [WHO]) diagnosis and a low-risk score according to the International Prognostic Scoring System (IPSS) for included patients. Patient nos. 2, 6, and 9 had been treated unsuccessfully with erythropoietin (EPO) patient nos. 10 and 11 had ongoing EPO treatment; and 6 of 11 patients needed erythrocyte transfusions regularly (Table 1).
Clinical, hematologic, and cytogenetic characteristics of the 5q− syndrome patients
| Patient no. . | Age, y . | Years since diagnosis . | Hb level, g/L . | WBC count, ×109/L . | Plt count, ×109/L . | Ongoing treatment . | CD34+CD38+ cells . | CD34+CD38−Thy-1+ cells . | ||
|---|---|---|---|---|---|---|---|---|---|---|
| % of MNCs . | % of FISH 5q− (no. of nuclei evaluated by FISH) . | % of MNCs . | % of FISH 5q− (no. of nuclei evaluated by FISH) . | |||||||
| 1 | 83 | 3 | 96 | 3.3 | 374 | E | 1.50 | 100 (216) | 0.16 | 99 (156) |
| 2 | 77 | 8 | 100 | 3.8 | 130 | E | 4.00 | 99 (262) | 0.10 | 100 (43) |
| 3 | 54 | 1 | 108 | 6.1 | 231 | E | 0.60 | 100 (224) | 0.01 | 96 (95) |
| 4 | 89 | 2 | 85 | 5.3 | 236 | E | 0.45 | 100 (221) | 0.08 | 100 (145) |
| 5 | 60 | 1 | 120 | 9.2 | 1234 | NONE | 1.60 | 100 (244) | 0.05 | 95 (264) |
| 6 | 58 | 4 | 122 | 3.3 | 448 | E | 3.20 | 99 (140) | 0.08 | 99 (165) |
| 7 | 64 | 3 | 100 | 4.9 | 267 | NONE | 3.20 | 98 (259) | 0.11 | 92 (76) |
| 8 | 85 | 1 | 102 | 3.6 | 309 | NONE | 0.50 | 93 (323) | 0.09 | 98 (251) |
| 9 | 85 | 7 | 76 | 1.3 | 95 | P | 0.43 | 100 (47) | 0.50 | 97 (73) |
| 10 | 82 | 3 | 103 | 6.4 | 443 | EPO, P | 0.34 | 96 (24) | 0.20 | 100 (51) |
| 11 | 86 | 8 | 129 | 4.7 | 204 | EPO | 1.50 | ND | 0.15 | 95 (40) |
| MDS mean [SEM] | 104 [4.7] | 4.7 [0.6] | 361 [93] | 1.57 [0.40] | 98.5 [0.73] | 0.14 [0.04] | 97.4 [0.79] | |||
| Normal mean* [SEM] | 135 | 7.0 | 270 | 1.80 [0.20] | 6.2† | 0.04 [0.01] | 6.2† | |||
| Patient no. . | Age, y . | Years since diagnosis . | Hb level, g/L . | WBC count, ×109/L . | Plt count, ×109/L . | Ongoing treatment . | CD34+CD38+ cells . | CD34+CD38−Thy-1+ cells . | ||
|---|---|---|---|---|---|---|---|---|---|---|
| % of MNCs . | % of FISH 5q− (no. of nuclei evaluated by FISH) . | % of MNCs . | % of FISH 5q− (no. of nuclei evaluated by FISH) . | |||||||
| 1 | 83 | 3 | 96 | 3.3 | 374 | E | 1.50 | 100 (216) | 0.16 | 99 (156) |
| 2 | 77 | 8 | 100 | 3.8 | 130 | E | 4.00 | 99 (262) | 0.10 | 100 (43) |
| 3 | 54 | 1 | 108 | 6.1 | 231 | E | 0.60 | 100 (224) | 0.01 | 96 (95) |
| 4 | 89 | 2 | 85 | 5.3 | 236 | E | 0.45 | 100 (221) | 0.08 | 100 (145) |
| 5 | 60 | 1 | 120 | 9.2 | 1234 | NONE | 1.60 | 100 (244) | 0.05 | 95 (264) |
| 6 | 58 | 4 | 122 | 3.3 | 448 | E | 3.20 | 99 (140) | 0.08 | 99 (165) |
| 7 | 64 | 3 | 100 | 4.9 | 267 | NONE | 3.20 | 98 (259) | 0.11 | 92 (76) |
| 8 | 85 | 1 | 102 | 3.6 | 309 | NONE | 0.50 | 93 (323) | 0.09 | 98 (251) |
| 9 | 85 | 7 | 76 | 1.3 | 95 | P | 0.43 | 100 (47) | 0.50 | 97 (73) |
| 10 | 82 | 3 | 103 | 6.4 | 443 | EPO, P | 0.34 | 96 (24) | 0.20 | 100 (51) |
| 11 | 86 | 8 | 129 | 4.7 | 204 | EPO | 1.50 | ND | 0.15 | 95 (40) |
| MDS mean [SEM] | 104 [4.7] | 4.7 [0.6] | 361 [93] | 1.57 [0.40] | 98.5 [0.73] | 0.14 [0.04] | 97.4 [0.79] | |||
| Normal mean* [SEM] | 135 | 7.0 | 270 | 1.80 [0.20] | 6.2† | 0.04 [0.01] | 6.2† | |||
BM morphology and blood values were investigated in connection with BM aspiration for the present studies. All patients were females and had a verified diagnosis of 5q− syndrome15 according to the WHO classification, with a typical BM morphology including hypolobular megakaryocytes and a low BM blast count (<5%; RA according to FAB classification), an isolated 5q deletion encompassing q13-q15 to q31-q35, normal to elevated platelet counts, and normal to slightly reduced WBC count. The relative frequencies of CD34+CD38+ and CD34+CD38−Thy-1+ cells were calculated from the number of BM MNCs and the fluorescence-activated cell sorter (FACS) profiles. If there were sufficient cells, at least 100 and 200 nuclei were always counted.
Hb indicates hemoglobin; WBC, white blood cell; Plt, platelet; E, erythrocyte transfusions; EPO, erythropoietin; P, prednisolone; and ND, not determined.
n=10.
Cut-off for FISH del(5q); median plus 2SD.
Purification of BM cell populations
BM mononuclear cells (MNCs) were isolated by Lymphoprep (Nycomed, Oslo, Norway) gradient centrifugation. Positive selection of CD34+ BM cells was performed using a magnetically activated cell sorting (MACS) CD34 isolation kit (Miltenyi Biotec, Bergish Gladbach, Germany) as previously described.11 The mean purity of enriched CD34+ cells was 81%. Normal and 5q− MNCs or CD34+ cells were cryopreserved in 10% dimethylsulfoxid (DMSO; Merck, Darmstadt, Germany) and 50% fetal calf serum (FCS; BioWhittaker, Walkersville, MD), thawed swiftly in a 37°C water bath, and washed twice in Dulbecco phosphate-buffered saline (PBS; PAA Laboratories, Pasching, Austria) and 5% FCS before staining. The CD34-enriched cells were incubated with CD38-allophycocyanin (APC), CD34–fluorescein isothiocyanate (FITC), and Thy-1 (CD90)–phycoerythrin (PE) monoclonal antibodies. The 30% CD34+ cells expressing the highest levels of CD38 and lacking expression of Thy-1 (CD34+CD38+Thy-1−) and the 5% CD34+ cells with the lowest expression of CD38 and coexpression of Thy-1 (CD34+CD38−Thy-1+) were sorted on a FACSDiVa (Becton Dickinson, San Jose, CA). All samples were stained with 7-amino-actinomyocin D (7-AAD; Sigma, St Louis, MO) to exclude nonviable cells. Both sorted populations reproducibly had a purity of more than 98% with regard to all 3 antigens. All antibodies were from BD Pharmingen (San Jose, CA) unless otherwise indicated.
Hematopoietic growth factors
Recombinant human (rh) granulocyte colony-stimulating factor (G-CSF), rh stem cell factor (SCF), rh interleukin-3 (IL-3), and rh granulocyte-macrophage colony-stimulating factor (GM-CSF) were generously provided by Amgen (Thousand Oaks, CA). Rh erythropoietin (EPO) was supplied by Boehringer Mannheim (Mannheim, Germany), thrombopoietin (THPO) by Genentech (San Francisco, CA), and rh FLT3 ligand (FL) by Immunex (Seattle, WA).
LTC-IC assay
Murine stromal feeders engineered to produce human growth factors (M2–10B4 and Sl/Sl mixed 1:1; kindly provided by Dr D. E. Hogge, Vancouver, BC, Canada) were used to support growth of long-term culture-initiating cells (LTC-ICs) as previously described.11 Cultures were established in 96-well collagen-coated microtiter plates with 5000 cells/well of each cell line, after irradiation with 8000 cGy, and cultured in long-term culture medium (MyeloCult H5100; Stem Cell Technologies, Vancouver, BC, Canada) with 10−6 M hydrocortisone 21-hemisuccinate. Seventy-five to 750 CD34+CD38+Thy-1− and 75-750 CD34+CD38−Thy-1+ cells from MDS patients and healthy subjects were added to the stroma layers, and cocultures were maintained at 37°C in high humidity and with 50% medium exchange every week. After 6 weeks, nonadherent and adherent cells were plated in methylcellulose cultures supplemented with SCF, GM-CSF, G-CSF, FL, IL-3 (all at 10 ng/mL), and EPO (5 U/mL). Colony-forming cells (CFCs; read-out of LTC-IC assay) were scored after an additional 12 days in culture. Individual colonies were then picked and transferred to slides for subsequent fluorescence in situ hybridization (FISH) analysis.
FISH probes and analyses
Interphase FISH analyses, using probes hybridizing to 5q31 (SpectrumOrange LSI EGRI) and 5p15.2 (SpectrumGreen LSI D5S721:D5S23), were performed essentially as described previously.10 All probes were obtained from Abbott (Stockholm, Sweden), and the signals were analyzed with the Chromofluor System (Applied Imaging, Newcastle, United Kingdom). The number of nuclei analyzed varied depending on the number of available cells, but whenever possible at least 200 nuclei were analyzed per cell population. In the nuclei of normal cells, the probes appear as 4 distinct signals, 2 orange and 2 green, whereas patients with del(5q) typically show 1 orange and 2 green signals. Based on FISH analyses on control cytospin preparations, the cut-off value (median + 2SD) for del(5q) was 6.2%. However, taking the purity of the sorted populations into account (> 98%), findings of less than 10% del(5q) were considered to be negative or inconclusive.
Purification of total RNA
Normal and MDS CD34+CD38+Thy-1− and CD34+CD38−Thy-1+ cells were sorted directly into RLT lysis buffer (Qiagen, Hilden, Germany) and snap-frozen at −80°C immediately after the sorting procedure. Samples were then thawed and, after homogenization by vortexing, total RNA was purified from 1 × 104 CD34+CD38+Thy-1− and CD34+CD38−Thy-1+ cells with RNeasy Micro Kit (Qiagen) following the manufacturer's protocol for isolation of RNA from animal cells including the optional on-column DNase treatment.
Double linear amplification of total RNA and probe preparation
Before amplification, 200 ng of Poly (dI-dC; Sigma) was added as nucleic acid carrier to each sample of total RNA. The total RNA isolated from 1 × 104 CD34+CD38+Thy-1− and CD34+CD38−Thy-1+ cells was double linear amplified with a combination of RiboAmp OA RNA Amplification Kit (Arcturus, Mountain View, CA) for the first round of amplification and the following generation of double-stranded cDNA, and Low RNA Input Fluorescent Linear Amplification Kit (Agilent, Palo Alto, CA) to generate the second-round amplified labeled aRNA, following the manufacturer's protocols. A pool of double linear amplified labeled Universal Human Reference RNA (Stratagene, La Jolla, CA) was used as reference to all samples at the hybridization. Samples were labeled with Cy3-CTP (PerkinElmer, Boston, MA) and reference reactions with Cy5-CTP (PerkinElmer). A nanodrop Spectrophotometer (NanoDrop Technologies, Wilmington, DE) was used to determine the dye concentration, and appropriate amounts of sample and reference aRNA were combined, lyophilized to dryness in an Eppendorf Concentrator (Eppendorf AG, Hamburg, Germany), and stored at −80°C.
Preparation and printing of oligonucleotide microarrays
Oligonucleotide microarrays were produced by the Swegene DNA Microarray Resource Center, Department of Oncology, Lund University, Sweden (http://swegene.onk.lu.se; Swegene Center Home page). Array-ready oligolibraries Human Genome Oligo Version 2.1 (containing 21 329 70mer probes, catalog no. 810518) and Human Genome Oligo Set Version 2.1 Upgrade (containing 5462 70mer probes, catalog no. 810518) were obtained from Operon Biotechnologies (Huntsville, AL). Lyophilized probes were resuspended in Pronto! Universal Spotting solution (Corning, Corning, NY) to a concentration of 24 mM. The entire probe set, in addition to a number of positive and negative controls, was printed in duplicate on aminosilane-coated UltraGAPS slides (Corning Incorporated) using a BioRobotics MicroGrid2 R600 robot (Genomic Solutions, Ann Arbor, MI) equipped with MicroSpot 10K quill pins (Genomic Solutions). Printing was performed in a temperature (18 to 20°C) and humidity (44%-49% relative humidity [RH]) controlled area, and the printed slides were left in a vacuum desiccator to dry for at least 48 hours before use.
Hybridization, image processing, and image acquisition
Microarray slides were rehydrated over steaming water for 1 to 2 seconds, snap-dried on a hot plate (98°C), and then UV cross-linked (800 mJ/cm2) using a Stratalinker (Stratagene). Prehybridization treatment, hybridization, and posthybridization washes of slides were performed according to the manufacturer's protocols provided with the Universal Microarray Hybridization Kit (Corning). In short, the array slides were presoaked with a sodium borohydride–reducing solution, prehybridized with a BSA-containing solution, washed, and dried by centrifugation. Prepared labeled aRNA was resuspended in hybridization solution and incubated at 65°C for 5 minutes and then cooled to ambient temperature before it was applied to the array and covered with a glass coverslip. The array slides were hybridized in Corning hybridization chambers (Corning) at 42°C for 17 to 20 hours, washed, and finally dried by centrifugation.
The Agilent G2565AA Microarray Scanner was used to measure the fluorescence intensities at photo multiplicator tube gain 100%, and, at 10 μM pixel resolution, data were collected at 2 different wavelengths (for Cy3 and Cy5), stored as multi-TIFF images, and analyzed by using the GenePix Pro 4.1.1 software (Axon Instruments, Foster City, CA) with standard flagging criteria. The quantified data matrix from GenePix was saved as a GenePix Results File (gpr) and loaded into the Bio Array Software Environment (BASE) for further data analysis. Background subtractions for Cy3 and Cy5 intensities were calculated using the median spot pixel intensity and median local background intensity provided in the GenePix result file.
Data extraction
In an initial spot quality filter, spots flagged by the criteria above, with a diameter of 40 pixels or less, an intensity of 0 units or less in either channel, or 10% or more saturated pixels in either channel, were removed. Intensity-dependent lowess14 fits were used to normalize intensity ratios on each assay. For each spot, the uncertainty of the expression value was estimated as u = (SNR1)−2 +(SNR2)−2, where SNRi is the signal to background noise ratio for channel i. Replicate measurements of the same reporter on an assay were merged and represented by a weighted mean. The weighted mean of a set of values xi was defined as m = Σiwixi/Σiwi, where the weight wi is exp(−3ui1/2/xi − m). This set of equations was solved numerically by simple iteration. The error of the merged value was defined as U = 1/Σi(1/ui) + Σiwi2(xi − m)2/(Σiwi)2. All reporters with a GenBank accession number or that were RefSeq specified were associated to genes using Array Clone Information Database (ACID).15 The 4 different types of assays (normal and 5q− CD34+CD38+Thy-1− progenitors and CD34+CD38−Thy-1+ stem cells) were compared pair-wise in different ways, and for each comparison the error model, presence filter (requiring 85% presence), and variation filter were applied to the assays involved, resulting in an optimal data extraction for each comparison. Association was based on UniGene Homo Sapiens build 176. Expression values for reporters representing the same gene were merged in the weighted fashion. Reporters without association to a known gene were excluded from the analysis. The merged data were transformed again to a modified expression value x′i = wi (xi − m). A presence filter and a variation filter across assays were also applied to the data, keeping only spots with expression in both channels in at least 21 of 23 assays and with a standard deviation of modified expression values greater than 0.3. The 23 assays represented 18 different samples, of which 3 were done in duplicate and 1 in triplicate. Hierarchical clustering revealed that replicate assays ended up in the same cluster (data not shown). This means that the experimental variability was small compared with differences between assay types, though not significantly smaller than variations among assays of the same type, which may indicate large homogeneity within the assay types. As the rest of the study concerns comparisons of different types, replicates were merged in the weighted fashion before continued analysis.
Statistical analysis
As a measure of difference between 2 types of assays, we used the false discovery rate in ranked gene lists. In each of the pair-wise comparisons, the genes that passed the filters were ranked according to the Fisher linear discriminant; F = (m1 − m0)/(σ12 + σ02)1/2, where m1 and m0 are the (unweighted) mean values for subgroups 1 and 0, respectively, whereas σ1 and σ0 are the standard deviations for the same subgroups. A permutation test with all possible permutations of sample labels was performed, and for each score, the average number of genes in a permutation list with a higher score was divided by the number of genes in the true list above to get the false discovery rate.
Q-PCR
Ten thousand BM CD34+CD38−Thy-1+ and CD34+CD38+Thy-1− cells were sorted directly into 330 μL RLT lysis buffer (Qiagen) and snap-frozen at −80°C. RNA extraction and DNase treatment were performed with the RNeasy Micro kit (Qiagen) according to the manufacturer's instructions for samples containing 105 or fewer cells. Eluted RNA samples were reverse transcribed using SuperScript II and random hexamers (Invitrogen, Carlsbad, CA) according to the protocol supplied by the manufacturer. Newly synthesized cDNA was diluted to approximately contain cDNA from 50 cells/μL and frozen at −20°C. Quantitative reverse transcriptase–polymerase chain reaction (Q-PCR) reactions were performed by mixing 2 × TaqMan universal PCR master mix, 20 × Assays-on-Demand (primer/MGB-probe mix), RNase-free H2O, and 5μL of cDNA to a final reaction volume of 20 μL. All experiments were performed in triplicates, and differences in cDNA input were compensated by normalizing against HPRT expression levels. The TaqMan Assays-on-Demand probes used were as follows: AREG, Hs00155832_m1; BMI1, Hs00180411_m1; CDC42SE2, Hs00184113_m1; CEBPΑ, Hs00269972_s1; CTNNA1, Hs00426996_m1; CTNNB1, Hs00170025_m1; DLK1, Hs00171584_m1; HLF, Hs00171406_m1; HPRT1, Hs99999909_m1; IFITM1, Hs00705137_s1; and TAF7, Hs00538821_s1 (all from Applied Biosystems, Foster City, CA).
Results
Extensive clonal involvement of the CD34+CD38−Thy-1+ HSC compartment and replacement of normal HSCs in 5q− syndrome
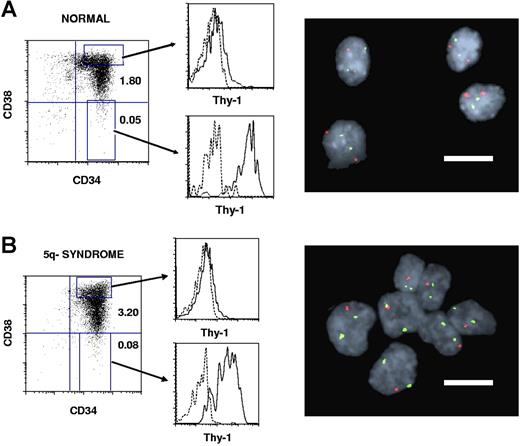
The size of the CD34+CD38−Thy1+ HSC compartment in 11 patients with 5q− syndrome9 represented on average 0.14% of total BM cells, only slightly increased over healthy individuals (mean 0.04%; Table 1; Figure 1). Notably, as much as 92% to 100% of the CD34+CD38−Thy1+ cells were part of the 5q− clone.
Thy-1 expression within CD34+CD38+ progenitor and CD34+CD38− stem cell compartments in normal and 5q− syndrome BM. CD34-enriched normal (A) and 5q− (B, patient no. 6) BM cells were stained with monoclonal antibodies (MAbs) against CD34, CD38, and Thy-1 or irrelevant isotype control Abs (“Purification of BM cell populations”). Left panels show expression profiles for already CD34-enriched cells, with the size of the CD34+CD38+ and CD34+CD38− populations given as percentages of total MNCs. Shown are also the gates for CD34+CD38+ and CD34+CD38− cells used for sorting and for further investigation of Thy-1 expression (middle panels). Dotted lines represent the negative isotype control and solid lines the specific Thy-1 expression. Note that Thy-1 expression is much higher in CD34+CD38− than in CD34+CD38+ cells for both healthy and 5q− subjects. Sorted CD34+CD38−Thy-1+ candidate HSCs and CD34+CD38+Thy-1− candidate progenitors were analyzed by FISH for the 5q deletion. Normal CD34+CD38−Thy-1+ cells (A) show 2 green and 2 red signals, whereas CD34+CD38−Thy-1+ cells with 5q deletion (B) show 2 green and 1 red signal (right panels). Bars in the FISH pictures represent 10 μm.
Thy-1 expression within CD34+CD38+ progenitor and CD34+CD38− stem cell compartments in normal and 5q− syndrome BM. CD34-enriched normal (A) and 5q− (B, patient no. 6) BM cells were stained with monoclonal antibodies (MAbs) against CD34, CD38, and Thy-1 or irrelevant isotype control Abs (“Purification of BM cell populations”). Left panels show expression profiles for already CD34-enriched cells, with the size of the CD34+CD38+ and CD34+CD38− populations given as percentages of total MNCs. Shown are also the gates for CD34+CD38+ and CD34+CD38− cells used for sorting and for further investigation of Thy-1 expression (middle panels). Dotted lines represent the negative isotype control and solid lines the specific Thy-1 expression. Note that Thy-1 expression is much higher in CD34+CD38− than in CD34+CD38+ cells for both healthy and 5q− subjects. Sorted CD34+CD38−Thy-1+ candidate HSCs and CD34+CD38+Thy-1− candidate progenitors were analyzed by FISH for the 5q deletion. Normal CD34+CD38−Thy-1+ cells (A) show 2 green and 2 red signals, whereas CD34+CD38−Thy-1+ cells with 5q deletion (B) show 2 green and 1 red signal (right panels). Bars in the FISH pictures represent 10 μm.
The content of functionally defined normal and MDS LTC-ICs, as an assay for stem cell activity,16,17 was next investigated in 4 patients. Although MDS BM cells, reflecting MDS as a disease of inefficient hematopoiesis, in almost all cases fail to long-term reconstitute MDS in immune-deficient mice in vivo,18 we have previously demonstrated that MDS-initiating LTC-IC activity, when detected, is exclusively contained within the minor CD34+CD38− HSC compartment.10,11,18 In agreement with this, in 2 of the 4 investigated patients with 5q− syndrome, we detected LTC-IC activity, and this was exclusively derived from CD34+CD38−Thy1+ cells (Table 2), despite investigating up to 10-fold more CD34+CD38+Thy-1− cells. Noteworthy, all LTC-CFCs generated from 5q− CD34+CD38−Thy1+ BM cells were shown to harbor the del(5q) (Table 2). Thus, neither CD34+CD38−Thy1+ nor CD34+CD38+Thy1− cells from 5q− patients displayed detectable normal HSC activity, supporting that the normal HSC compartment in 5q− syndrome patients has been largely replaced by a clonally expanded 5q− compartment with the same CD34+CD38−Thy1+ phenotype.
Normal and 5q− clonal LTC-IC activity of 5q− CD34+CD38+Thy-1− and CD34+CD38−Thy-1+ cells
| Patient no. . | LTC-CFC/1000 cells . | |||
|---|---|---|---|---|
| CD34+CD38+Thy-1− cells . | CD34+CD38−Thy-1+ cells . | |||
| Normal . | 5q deleted . | Normal . | 5q deleted . | |
| 1 | 0 | 0 | 0 | 29 |
| 2 | 0 | 0 | 0 | 0 |
| 4 | 0 | 0 | 0 | 0 |
| 6 | 0 | 0 | 0 | 51 |
| Normal mean, n=5 | 0 | 0 | 1838 | 0 |
| Patient no. . | LTC-CFC/1000 cells . | |||
|---|---|---|---|---|
| CD34+CD38+Thy-1− cells . | CD34+CD38−Thy-1+ cells . | |||
| Normal . | 5q deleted . | Normal . | 5q deleted . | |
| 1 | 0 | 0 | 0 | 29 |
| 2 | 0 | 0 | 0 | 0 |
| 4 | 0 | 0 | 0 | 0 |
| 6 | 0 | 0 | 0 | 51 |
| Normal mean, n=5 | 0 | 0 | 1838 | 0 |
Six-week LTC-CFC activity of CD34+CD38−Thy-1+ (75-750 cells/well) and CD34+CD38+Thy-1− (750 cells/well) cells from healthy controls and 5q− syndrome patients were evaluated as described in “LTC-IC assay.” After 6 weeks of coculture, the number of CFCs produced after an additional 2 weeks in methylcellulose were counted. Results are the means of 3 to 6 replicate wells from each patient and cell population. No LTC-CFCs were derived from normal or 5q− CD34+CD38+Thy-1− candidate progenitor cells.
Global gene-expression profiling implicates that the 5q− syndrome originates in normal CD34+CD38−Thy1+ HSCs
Although MDS stem cells share a CD34+CD38−Thy1+ phenotype with normal HSCs,1,10,13,19 this does not necessarily indicate the malignancy initiated in normal HSCs, as it might rather reflect acquisition of a stem cell cell-surface phenotype by a transformed MDS progenitor.19 Thus, here we used a novel global gene-profiling strategy in an attempt to better distinguish between a stem and progenitor cell identity of the 5q− stem cells, taking advantage of the distinct gene-expression patterns of normal HSCs and progenitors.6
While no previous studies have performed a global gene-expression profiling of CD34+CD38−Thy1+ BM cells, Georgantas et al reported genes significantly up- or down-regulated in normal BM CD34+CD38−Lin− HSCs compared with CD34+CD38+Lin+ progenitor cells.6 We extracted data from our normal CD34+CD38−Thy-1+ and CD34+CD38+Thy-1− data sets, ranked genes using Fisher linear discriminant scores,20 and found a high degree of agreement with the data set of Georgantas et al (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article),6 in which 1190 reporters were found to be significantly up-regulated in CD34+CD38−Lin− cells. We updated all gene identities for these reporters according to UniGene build 176 (http://www.ncbi.nlm.nih.gov/UniGene/) and found that 276 genes were also present in our list after filtration. Of those, 197 had positive Fisher scores in our data set. Eight of the 276 genes were found among our 20 most up-regulated genes, whereas the other 12 were not mentioned. To further evaluate the overall agreement, using our whole ranked list, we calculated the area under the receiver operating characteristic (ROC) curve,21 which is a linear transformation of the Wilcoxon rank sum.22 The ROC area for the 276 genes up-regulated in CD34+CD38−Lin− cells was 0.73 (Figure S1), with a P value of less than .001 (random gene permutations). Similarly, the 1159 reporters found down-regulated by Georgantas et al6 were associated to genes using UniGene build 176, and 374 of the genes were found in our ranked list as well. Turning our ranked list upside down, so that down-regulated genes received top ranks, we found that 11 of our top 20 down-regulated genes were down-regulated in the studies of Georgantas et al as well,6 whereas the other 9 were not mentioned. The ROC area was 0.82, with a P value of less than .001 (Figure S1). The complete list of genes differentially expressed between normal CD34+CD38−Thy-1+ and CD34+CD38+Thy-1− cells is available in Table S1.
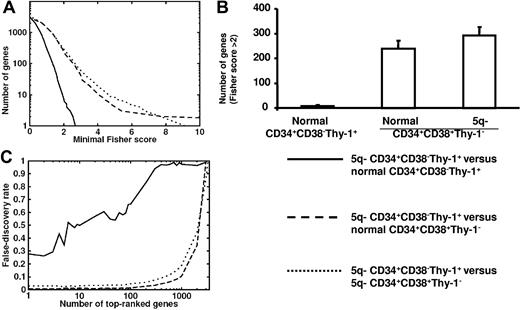
We next compared the global expression profiles of CD34+CD38−Thy-1+ MDS stem cells from 4 5q− syndrome patients (nos. 1, 2, 3, and 5) with those of CD34+CD38−Thy-1+ and CD34+CD38+Thy-1− cells from 5 healthy subjects. Notably, the number of differentially expressed genes was considerably less when comparing 5q− CD34+CD38−Thy-1+ cells with normal CD34+CD38−Thy-1+ cells than when comparing with normal or 5q− CD34+CD38+Thy-1− progenitors (Figure 2). Furthermore, the Fisher scores of 5q− CD34+CD38−Thy-1+ compared with normal CD34+CD38−Thy-1+ cells were comparable to the random expectation, represented by permutation test results, demonstrating a very close identity between normal and 5q− CD34+CD38−Thy-1+ cells (Figure 2A-B).
Comparison of the global gene-expression profiles of normal and 5q− CD34+CD38−Thy-1+ and CD34+CD38+Thy-1− cell populations. (A) Number of genes versus minimal Fisher score for 5q− CD34+CD38−Thy-1+ candidate HSCs when compared with normal CD34+CD38−Thy-1+ HSCs (solid line), normal CD34+CD38+Thy-1− progenitors (dashed line), and 5q− CD34+CD38+Thy-1− progenitors (dotted line). (B) Number of differentially expressed genes with Fisher score above 2 for 5q− CD34+CD38−Thy-1+ candidate HSCs when compared with normal CD34+CD38−Thy-1+ HSCs, normal CD34+CD38+Thy-1− progenitors, and 5q− CD34+CD38+Thy-1− progenitors, respectively. Error bars indicate 95% confidence interval (Poisson statistics). (C) False discovery rate as a function of number of top-ranked genes using the Fisher score. Results are shown for 5q− CD34+CD38−Thy-1+ candidate HSCs when compared with normal CD34+CD38−Thy-1+ HSCs (solid line), normal CD34+CD38+Thy-1− progenitors (dashed line), and 5q− CD34+CD38+Thy-1− progenitors (dotted line). The false discovery rate (ie, number of accepted genes in a permutation test divided by the same number in the correct list) was high when comparing 5q− HSCs with normal HSCs but essentially zero when compared with normal progenitors. A high false discovery rate implies that the compared populations do not differ in many more genes than expected by random fluctuations, whereas a low false discovery rate implies significant differences in gene expression between the compared cell types, reflecting more extensive differences than expected by random fluctuation.
Comparison of the global gene-expression profiles of normal and 5q− CD34+CD38−Thy-1+ and CD34+CD38+Thy-1− cell populations. (A) Number of genes versus minimal Fisher score for 5q− CD34+CD38−Thy-1+ candidate HSCs when compared with normal CD34+CD38−Thy-1+ HSCs (solid line), normal CD34+CD38+Thy-1− progenitors (dashed line), and 5q− CD34+CD38+Thy-1− progenitors (dotted line). (B) Number of differentially expressed genes with Fisher score above 2 for 5q− CD34+CD38−Thy-1+ candidate HSCs when compared with normal CD34+CD38−Thy-1+ HSCs, normal CD34+CD38+Thy-1− progenitors, and 5q− CD34+CD38+Thy-1− progenitors, respectively. Error bars indicate 95% confidence interval (Poisson statistics). (C) False discovery rate as a function of number of top-ranked genes using the Fisher score. Results are shown for 5q− CD34+CD38−Thy-1+ candidate HSCs when compared with normal CD34+CD38−Thy-1+ HSCs (solid line), normal CD34+CD38+Thy-1− progenitors (dashed line), and 5q− CD34+CD38+Thy-1− progenitors (dotted line). The false discovery rate (ie, number of accepted genes in a permutation test divided by the same number in the correct list) was high when comparing 5q− HSCs with normal HSCs but essentially zero when compared with normal progenitors. A high false discovery rate implies that the compared populations do not differ in many more genes than expected by random fluctuations, whereas a low false discovery rate implies significant differences in gene expression between the compared cell types, reflecting more extensive differences than expected by random fluctuation.
The false discovery rate was high when comparing 5q− CD34+CD38−Thy-1+ cells with normal CD34+CD38−Thy-1+ cells but essentially zero when compared with normal CD34+CD38+Thy-1− progenitors (Figure 1C), further reflecting the high similarity between 5q− and normal CD34+CD38−Thy-1+ cells.
Differentially expressed genes in 5q-deleted CD34+CD38−Thy1+ stem cells
To confirm the expected down-regulation of genes located at 5q31q32, we investigated 37 such genes found in our lists, of which 34 were down-regulated in CD34+CD38−Thy-1+ cells in at least 3 of the 4 investigated patients (Table S2).
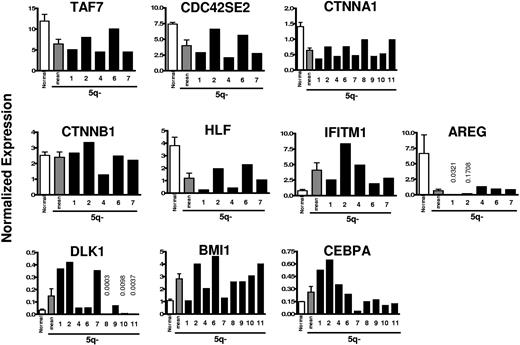
Based on the array data, a total of 10 potentially interesting genes were investigated in further detail using Q-PCR analysis, in part to confirm and better quantify some of the array findings on CD34+CD38−Thy-1+ cells (patient nos. 1 and 2) and in part to extend the analysis to additional patients. Table 3 shows array data on selected genes that were up- or down-regulated with high Fisher scores in 5q− CD34+CD38−Thy-1+ cells (complete list of genes in Table S3). Importantly, the Q-PCR analyses of patients 1 and 2 confirmed all array findings. In all patients investigated by Q-PCR, all 3 examined genes situated on the involved region of 5q (TAF7 at 5q31, CDC42SE2 at 5q23, and CTNNA1 at 5q31) were, as expected, down-regulated in CD34+CD38−Thy-1+ cells (Figure 3). Thus, TAF7,23 CDC42SE2,24 and CTNNA1 (alpha catenin; implicated in the Wnt signaling pathway as a tumor suppressor gene)25 were all down-regulated by approximately 50% (46%, 46%, and 55%, respectively; Figure 3). However, the functionally related CTNNB1 (beta catenin),26 implicated in the regulation of normal and leukemic HSC self-renewal27 (not located at 5q), was not significantly affected in investigated patients (Figure 3).
Differentially expressed genes in 5q− versus normal CD34+CD38−Thy-1+ cells
| Gene name . | Gene symbol . | Accession no. . | Known function . | Fisher score . |
|---|---|---|---|---|
| Up-regulated | ||||
| Interferon-induced transmembrane protein 1 | IFITM1* | NM_003641 | Inhibition of cell proliferation | 2.7 |
| Serine/threonine kinase 19 | STK19 | NM_032454 | Transcriptional regulation | 2.5 |
| Kelch-like ECT2 interacting protein | KLEIP | NM_014458 | Cytoskeleton organization | 2.0 |
| Kinesin family member 2C | KIF2C | NM_006845 | Chromosome movements during cell division | 2.0 |
| Growth arrest–specific 2 | GAS2 | NM_005256 | p53-dependent apoptosis | 2.0 |
| Cytochrome b-245, beta polypeptide | CYBB | NM_000397 | Microbicidal phagocytic oxidative system | 1.9 |
| Solute carrier family 9 | SLC9A3R1 | NM_004252 | Wnt signaling | 1.9 |
| Gp25L2 protein | HSGP25L2G | BC001123 | Located at 5q35.3; unknown function | 1.9 |
| TAF5 RNA polymerase II | TAF5* | NM_006951 | Transcriptional regulation | 1.8 |
| Thiosulfate sulfurtransferase, rhodanese | TST | NM_003312 | Mitochondrial enzyme | 1.8 |
| Topoisomerase (DNA) II binding protein | TOPBP1 | NM_007027 | DNA damage response protein | 1.7 |
| Golgin-67 | GOLGIN-67 | AF204231 | Golgi apparatus protein | 1.6 |
| CHK2 checkpoint homolog, S pombe | CHEK2 | NM_007194 | Putative tumor suppressor | 1.6 |
| Enah/Vasp-like | EVL | NM_016337 | Cytoskeleton rearrangement and signal transduction | 1.6 |
| Cell division cycle–associated 7 | CDCA7 | NM_031942 | C-myc target gene; lymphoblastoid cell transformation | 1.6 |
| CCAAT/enhancer binding protein (C/EBP), alpha | CEBPA* | NM_004364 | TF; neutrophil differentiation; mutated in AML | 1.5 |
| Cell division cycle 25 B | CDC25B | NM_021874 | Cell cycle regulator; oncogenic properties | 1.5 |
| Polymerase (D directed), epsilon 2, p59 subunit | POLE2 | NM_021874 | Cell cycle regulator; oncogenic properties | 1.5 |
| Polymerase (D directed), epsilon 2, p59 subunit | POLE2 | NM_002692 | DNA replication | 1.5 |
| CD99 antigen | CD99 | NM_002414 | T-cell apoptosis; involved in gastric adenocarcinomas | 1.4 |
| Bloom syndrome | BLM | NM_000057 | Suppressor of inappropriate recombination. | 1.4 |
| Defective in sister chromatid cohesion homolog 1 | DCC1 | NM_024094 | Faithful inheritance of chromosomes to daughter cells | 1.4 |
| D (cytosine-5-)-methyltransferase 3, alpha | DNMT3 | NM_022552 | De novo methylation | 1.4 |
| Inhibitor of DNA binding 1 | ID1 | NM_002165 | Malignant progression | 1.2 |
| Delta-like homolog | DLK1* | NM_003836 | Notch-related differentiation associated | 1.1 |
| Tumor protein p53 binding protein 1 | TP53BP1 | NM_005657 | Tumor suppressor gene | 1.0 |
| V-myc myelocytoma | MYC | NM_002467 | Oncogene | 1.0 |
| B lymphoma Mo-MLV insertion region | BMI1* | NM_005180 | Implicated in HSC self-renewal | 0.9 |
| Down-regulated | ||||
| Solute carrier family 25A, member 4 | SLC25A4 | NM_001151 | Apoptosis inhibitor | 2.4 |
| Ribosomal protein S21 | RPS21 | NM_001024 | Component of the small 40S ribosomal subunit | 2.2 |
| Arachidonate 12-lipooxygenase, 12R type | ALOX12B | NM_001139 | Iron ion binding | 1.9 |
| Amphiregulin schwannoma–derived growth factor | AREG* | NM_001657 | Interacts with EGF/TGF-α; inhibits carcinoma | 1.8 |
| Hepatic leukemia factor | HLF* | M95585 | TF involved in t(17;19) translocation in ALL | 1.7 |
| TAF7 RNA polymerase II | TAF7* | NM_005642 | Located at 5q31; transcriptional regulation | 1.7 |
| Pleckstrin homolog domain | PLEKHA6 | BC010522 | Signal transduction | 1.7 |
| WAS protein family | WASF3 | AB020707 | Member of the Wiskott-Aldrich syndrome protein family | 1.7 |
| Myotrophin | MTPN | AK055660 | Regulation of NFκB | 1.6 |
| Glycoprotein Ib (platelet), beta-polypeptide | GP1BB | L20860 | Associated with Bernard-Soulier or giant platelet disorder | 1.6 |
| Target of myb1-like 1 (chicken) | TOM1L1 | NM_005486 | Signal transduction | 1.6 |
| F-box and leucine-rich repeat protein 5 | FBXL5 | NM_033535 | Proteasome degradation | 1.6 |
| Activated leukocyte cell adhesion molecule | ALCAM* | Y10183 | Associated with tumor progression | 1.6 |
| Zinc finger protein 366 | ZNF366 | NM_152625 | Located at 5q13.2; transcriptional regulation | 1.6 |
| Forkhead box O1A | FOXO1A | NM_002015 | TF; involved in malignant transformation | 1.6 |
| MADS box transcription enhancer factor 2 | MEF2C | L08895 | Located at 5q14; transcriptional regulation | 1.5 |
| CREBB/EP300 inhibitor 1 | CRI1 | NM_014335 | CREBB/EP300 implicated as tumor suppressor gene | 1.5 |
| Endoplasmic reticulum chaperone (SIL1) | SIL1 | NM_022464 | Located at 5q31; enhances nucleotide exchange | 1.5 |
| CDC42 small effector 2 | CDC42SE2* | AK025620 | Located at 5q23.3; Rho GTPase; regulates actin and kinase signaling | 1.5 |
| Tyrosine kinase with Ig/EGF homology domains | TIE | NM_005424 | High expression correlates with shorter survival in CML | 1.4 |
| Transcription factor 8 | TCF8 | NM_030751 | Involved in TGF and BMP signaling | 1.4 |
| Transforming growth factor beta 1 | TGFB1 | NM_000660 | Multifunctional; proliferation/differentiation, transformation | 1.3 |
| Stem cell growth factor | SCGF | NM_002975 | Growth factor for primitive hematopoietic stem cells | 1.3 |
| Suppression of tumorigenicity 13 | ST13 | U17714 | Candidate tumor suppressor gene | 1.3 |
| R binding motif protein 27 | RBM27 | AB037732 | Located at 5q32; unknown function | 1.2 |
| Tumor necrosis factor, alpha-induced protein 8 | TNFAIP8 | NM_014350 | Located at 5q23.1; oncogenic factor in cancer cells | 1.1 |
| Bone morphogenetic protein 6 | BMP6 | NM_001718 | Secreted signaling protein involved in bone formation | 0.9 |
| Gene name . | Gene symbol . | Accession no. . | Known function . | Fisher score . |
|---|---|---|---|---|
| Up-regulated | ||||
| Interferon-induced transmembrane protein 1 | IFITM1* | NM_003641 | Inhibition of cell proliferation | 2.7 |
| Serine/threonine kinase 19 | STK19 | NM_032454 | Transcriptional regulation | 2.5 |
| Kelch-like ECT2 interacting protein | KLEIP | NM_014458 | Cytoskeleton organization | 2.0 |
| Kinesin family member 2C | KIF2C | NM_006845 | Chromosome movements during cell division | 2.0 |
| Growth arrest–specific 2 | GAS2 | NM_005256 | p53-dependent apoptosis | 2.0 |
| Cytochrome b-245, beta polypeptide | CYBB | NM_000397 | Microbicidal phagocytic oxidative system | 1.9 |
| Solute carrier family 9 | SLC9A3R1 | NM_004252 | Wnt signaling | 1.9 |
| Gp25L2 protein | HSGP25L2G | BC001123 | Located at 5q35.3; unknown function | 1.9 |
| TAF5 RNA polymerase II | TAF5* | NM_006951 | Transcriptional regulation | 1.8 |
| Thiosulfate sulfurtransferase, rhodanese | TST | NM_003312 | Mitochondrial enzyme | 1.8 |
| Topoisomerase (DNA) II binding protein | TOPBP1 | NM_007027 | DNA damage response protein | 1.7 |
| Golgin-67 | GOLGIN-67 | AF204231 | Golgi apparatus protein | 1.6 |
| CHK2 checkpoint homolog, S pombe | CHEK2 | NM_007194 | Putative tumor suppressor | 1.6 |
| Enah/Vasp-like | EVL | NM_016337 | Cytoskeleton rearrangement and signal transduction | 1.6 |
| Cell division cycle–associated 7 | CDCA7 | NM_031942 | C-myc target gene; lymphoblastoid cell transformation | 1.6 |
| CCAAT/enhancer binding protein (C/EBP), alpha | CEBPA* | NM_004364 | TF; neutrophil differentiation; mutated in AML | 1.5 |
| Cell division cycle 25 B | CDC25B | NM_021874 | Cell cycle regulator; oncogenic properties | 1.5 |
| Polymerase (D directed), epsilon 2, p59 subunit | POLE2 | NM_021874 | Cell cycle regulator; oncogenic properties | 1.5 |
| Polymerase (D directed), epsilon 2, p59 subunit | POLE2 | NM_002692 | DNA replication | 1.5 |
| CD99 antigen | CD99 | NM_002414 | T-cell apoptosis; involved in gastric adenocarcinomas | 1.4 |
| Bloom syndrome | BLM | NM_000057 | Suppressor of inappropriate recombination. | 1.4 |
| Defective in sister chromatid cohesion homolog 1 | DCC1 | NM_024094 | Faithful inheritance of chromosomes to daughter cells | 1.4 |
| D (cytosine-5-)-methyltransferase 3, alpha | DNMT3 | NM_022552 | De novo methylation | 1.4 |
| Inhibitor of DNA binding 1 | ID1 | NM_002165 | Malignant progression | 1.2 |
| Delta-like homolog | DLK1* | NM_003836 | Notch-related differentiation associated | 1.1 |
| Tumor protein p53 binding protein 1 | TP53BP1 | NM_005657 | Tumor suppressor gene | 1.0 |
| V-myc myelocytoma | MYC | NM_002467 | Oncogene | 1.0 |
| B lymphoma Mo-MLV insertion region | BMI1* | NM_005180 | Implicated in HSC self-renewal | 0.9 |
| Down-regulated | ||||
| Solute carrier family 25A, member 4 | SLC25A4 | NM_001151 | Apoptosis inhibitor | 2.4 |
| Ribosomal protein S21 | RPS21 | NM_001024 | Component of the small 40S ribosomal subunit | 2.2 |
| Arachidonate 12-lipooxygenase, 12R type | ALOX12B | NM_001139 | Iron ion binding | 1.9 |
| Amphiregulin schwannoma–derived growth factor | AREG* | NM_001657 | Interacts with EGF/TGF-α; inhibits carcinoma | 1.8 |
| Hepatic leukemia factor | HLF* | M95585 | TF involved in t(17;19) translocation in ALL | 1.7 |
| TAF7 RNA polymerase II | TAF7* | NM_005642 | Located at 5q31; transcriptional regulation | 1.7 |
| Pleckstrin homolog domain | PLEKHA6 | BC010522 | Signal transduction | 1.7 |
| WAS protein family | WASF3 | AB020707 | Member of the Wiskott-Aldrich syndrome protein family | 1.7 |
| Myotrophin | MTPN | AK055660 | Regulation of NFκB | 1.6 |
| Glycoprotein Ib (platelet), beta-polypeptide | GP1BB | L20860 | Associated with Bernard-Soulier or giant platelet disorder | 1.6 |
| Target of myb1-like 1 (chicken) | TOM1L1 | NM_005486 | Signal transduction | 1.6 |
| F-box and leucine-rich repeat protein 5 | FBXL5 | NM_033535 | Proteasome degradation | 1.6 |
| Activated leukocyte cell adhesion molecule | ALCAM* | Y10183 | Associated with tumor progression | 1.6 |
| Zinc finger protein 366 | ZNF366 | NM_152625 | Located at 5q13.2; transcriptional regulation | 1.6 |
| Forkhead box O1A | FOXO1A | NM_002015 | TF; involved in malignant transformation | 1.6 |
| MADS box transcription enhancer factor 2 | MEF2C | L08895 | Located at 5q14; transcriptional regulation | 1.5 |
| CREBB/EP300 inhibitor 1 | CRI1 | NM_014335 | CREBB/EP300 implicated as tumor suppressor gene | 1.5 |
| Endoplasmic reticulum chaperone (SIL1) | SIL1 | NM_022464 | Located at 5q31; enhances nucleotide exchange | 1.5 |
| CDC42 small effector 2 | CDC42SE2* | AK025620 | Located at 5q23.3; Rho GTPase; regulates actin and kinase signaling | 1.5 |
| Tyrosine kinase with Ig/EGF homology domains | TIE | NM_005424 | High expression correlates with shorter survival in CML | 1.4 |
| Transcription factor 8 | TCF8 | NM_030751 | Involved in TGF and BMP signaling | 1.4 |
| Transforming growth factor beta 1 | TGFB1 | NM_000660 | Multifunctional; proliferation/differentiation, transformation | 1.3 |
| Stem cell growth factor | SCGF | NM_002975 | Growth factor for primitive hematopoietic stem cells | 1.3 |
| Suppression of tumorigenicity 13 | ST13 | U17714 | Candidate tumor suppressor gene | 1.3 |
| R binding motif protein 27 | RBM27 | AB037732 | Located at 5q32; unknown function | 1.2 |
| Tumor necrosis factor, alpha-induced protein 8 | TNFAIP8 | NM_014350 | Located at 5q23.1; oncogenic factor in cancer cells | 1.1 |
| Bone morphogenetic protein 6 | BMP6 | NM_001718 | Secreted signaling protein involved in bone formation | 0.9 |
Selected list of genes up- or down-regulated in 5q− (n=4) CD34+CD38−Thy-1+ candidate HSCs when compared with normal (n=5) CD34+CD38−Thy-1+ cells (for complete list see Table S3).
TF indicates transcription factor; and CML, chronic myelogenous leukemia.
Microarray-based expression validated by Q-PCR.
Quantitative PCR expression analysis of selected genes in 5q− CD34+ CD38− Thy-1+ HSCs compared with normal CD34+CD38− Thy-1+ HSCs. RNA was isolated from highly purified normal and 5q− CD34+CD38−Thy-1+ cells and analyzed for quantitative expression of 10 genes, selected based on the array analysis. The mean expression of 5 healthy subjects (normal; □) compared with the mean (▩) and individual (patient nos. 1, 2, 4, 6, 7, 8, 9, 10, and 11; ■) expression of each gene for the 5q− syndrome patients normalized against HPRT expression levels. Error bars are SEM.
Quantitative PCR expression analysis of selected genes in 5q− CD34+ CD38− Thy-1+ HSCs compared with normal CD34+CD38− Thy-1+ HSCs. RNA was isolated from highly purified normal and 5q− CD34+CD38−Thy-1+ cells and analyzed for quantitative expression of 10 genes, selected based on the array analysis. The mean expression of 5 healthy subjects (normal; □) compared with the mean (▩) and individual (patient nos. 1, 2, 4, 6, 7, 8, 9, 10, and 11; ■) expression of each gene for the 5q− syndrome patients normalized against HPRT expression levels. Error bars are SEM.
HLF, a transcription factor involved in t(17;19)/TCF3-HLF–positive (previously E2A-HLF–positive) acute lymphoblastic leukemia (ALL),28 reported to be specifically expressed in normal human HSCs,6 was down-regulated by a mean of 3.2-fold in all 7 patients investigated (5 by Q-PCR and 2 additional by array). Interferon-induced transmembrane protein-1 (IFITM1), suggested to play a role in the antiproliferative activity of interferons,29 was up-regulated in all 7 investigated patients (in the 5 investigated by Q-PCR, by a mean of 5.3-fold), whereas Amphiregulin (AREG), an apoptosis inhibitor,30 was down-regulated in all 7 patients by as much as 10-fold or more (Figure 3).
Delta-like homolog (DLK1), involved in Notch signaling,31 was found to be up-regulated 1.5- to 12.6-fold in 6 of 9 patients investigated by Q-PCR analysis (Figure 3).
Of particular interest, BMI1, critically involved in regulation of HSC self-renewal,32,33 was up-regulated in all but 1 of 9 patients analyzed by Q-PCR by a mean of 2.5-fold (Figure 3).
CCAAT enhancer binding protein-alpha (CEBPA), essential for normal myeloid development and implicated in the transformation of AML,34,35 was up-regulated in all 4 patients (nos. 1, 2, 3, and 5) in the array analysis. This was confirmed by Q-PCR for 2 patients (nos. 1 and 2) and extended to an additional 2 patients (nos. 4 and 6). However, in 4 other patients (nos. 8, 9, 10, and 11), the expression was not significantly different from normal CD34+CD38−Thy-1+ cells, and in 1 patient (no. 7) the expression was down-regulated.
Enhanced resolution of dysregulated gene expression in purified CD34+CD38−Thy-1+ stem and CD34+CD38+Thy-1− progenitor cells with del(5q)
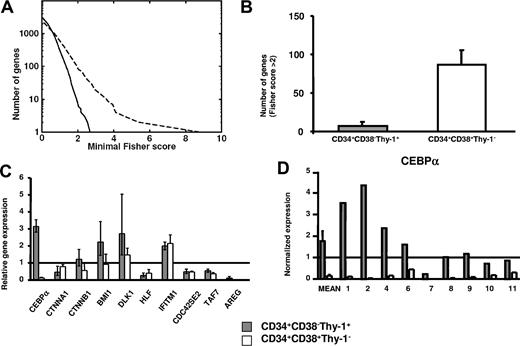
To evaluate (1) the potential benefit of global gene profiling of purified and distinct MDS stem and progenitor cell populations, rather than mixed populations,36-39 and (2) the identification of stage-specific changes in gene expression, we next compared differentially expressed genes between purified 5q− and normal CD34+CD38−Thy-1+ stem cells and between 5q− and normal CD34+CD38+Thy-1− progenitors (Figure 4). Whereas 5q− and normal CD34+CD38+Thy-1− progenitors showed a relatively high number of differentially expressed genes, much fewer and smaller differences were observed when comparing the profiles of normal and 5q− CD34+CD38−Thy-1+ stem cells (Figure 4A,B).
Comparison of differentially expressed genes in MDS stem cells and progenitors. (A) Number of genes versus minimal Fisher scores for 5q− versus normal CD34+CD38+Thy-1− progenitors (dashed line) and 5q− versus normal CD34+CD38−Thy-1+ HSCs (solid line), respectively. (B) Number of differentially expressed genes with Fisher score above 2 when comparing 5q− and normal CD34+CD38−Thy-1+ stem cells and when comparing 5q− and normal CD34+CD38+Thy-1− progenitors. Error bars indicate 95% confidence interval (Poisson statistics). (C) Microarray-based gene expression of 10 selected genes in 5q− CD34+CD38−Thy-1+ stem cells and 5q− CD34+CD38+Thy-1− progenitors relative to the mean expression in their normal counterparts (from 5 healthy subjects and 4 5q− patients). Error bars show SEM. (D) Q-PCR analysis of CEBPA expression in 5q− CD34+CD38−Thy-1+ stem cells and 5q− CD34+CD38+Thy-1− progenitors. Shown are the mean (SEM) differential expression for 5 healthy subjects and 9 5q− patients and individual expression levels for the investigated 5q− patients (nos. 1, 2, 4, 6, 7, 8, 9, 10, and 11) normalized against HPRT expression levels. Gray staples for HSCs and white staples for progenitors throughout the figure.
Comparison of differentially expressed genes in MDS stem cells and progenitors. (A) Number of genes versus minimal Fisher scores for 5q− versus normal CD34+CD38+Thy-1− progenitors (dashed line) and 5q− versus normal CD34+CD38−Thy-1+ HSCs (solid line), respectively. (B) Number of differentially expressed genes with Fisher score above 2 when comparing 5q− and normal CD34+CD38−Thy-1+ stem cells and when comparing 5q− and normal CD34+CD38+Thy-1− progenitors. Error bars indicate 95% confidence interval (Poisson statistics). (C) Microarray-based gene expression of 10 selected genes in 5q− CD34+CD38−Thy-1+ stem cells and 5q− CD34+CD38+Thy-1− progenitors relative to the mean expression in their normal counterparts (from 5 healthy subjects and 4 5q− patients). Error bars show SEM. (D) Q-PCR analysis of CEBPA expression in 5q− CD34+CD38−Thy-1+ stem cells and 5q− CD34+CD38+Thy-1− progenitors. Shown are the mean (SEM) differential expression for 5 healthy subjects and 9 5q− patients and individual expression levels for the investigated 5q− patients (nos. 1, 2, 4, 6, 7, 8, 9, 10, and 11) normalized against HPRT expression levels. Gray staples for HSCs and white staples for progenitors throughout the figure.
Strikingly, expression of CEBPA, which was either up-regulated or normal in CD34+CD38−Thy-1+ cells in most 5q− patients (Figures 3 and 4C,D), was rather consistently and dramatically down-regulated in 5q− CD34+CD38+Thy-1− progenitors (Figure 4C,D), as determined by microarray analysis. CTNNA1, BMI1, and DLK1 were also differentially affected in the 5q− CD34+CD38+Thy-1+ stem and CD34+CD38+Thy-1− progenitor populations (Figure 4C), whereas other investigated genes (TAF7, CDC42SE2, HLF, IFITM1, and AREG) showed a similar pattern of expression between CD34+CD38−Thy-1+ and CD34+CD38+Thy-1− cells (Figure 4C).
In contrast to recent studies,40 we did not find CTNNA1 to be down-regulated more than the expected 50%, perhaps reflecting that the reported hypermethylation of CTNNA1 in patients with 5q deletions might be a late event not typically observed in patients with 5q−syndrome.
Nine patients were investigated by Q-PCR to confirm and extend the contrasting expression of CEBPA in 5q− CD34+CD38−Thy-1+ stem and CD34+CD38+Thy-1− progenitor cells. Noteworthy, when compared with their normal counterparts, CEPBA was dramatically down-regulated in all 9 investigated patients (range, 2.2-1236 fold) in 5q− CD34+CD38+Thy-1− progenitors but not in CD34+CD38−Thy-1+ stem cells (Figure 4D). Although CEBPA was down-regulated (4.3-fold) in 1 case (no. 7) in 5q− CD34+CD38−Thy-1+ cells, its expression was reduced even more in 5q− CD34+CD38+Thy-1− progenitors (as much as 1236-fold) when compared with normal CD34+CD38+Thy-1− progenitors (Figure 4D).
Discussion
The present studies exemplify the importance of performing global gene-expression profiling of purified cancer stem and progenitor cell populations. Not only did the molecular fingerprinting of 5q− CD34+CD38−Thy-1+ cells provide stronger support for 5q− syndrome potentially originating from normal HSCs, but it also resulted in a much higher resolution of specific gene-expression changes when comparing MDS and normal stem and progenitor cells, rather than more heterogeneous populations.36-39
Although it has been postulated that LSCs and other CSCs may frequently originate in the corresponding rare normal multipotent stem cell populations,19,41 conclusive evidence for such a model has only been obtained for myeloproliferative disorders.3,4 In other leukemias the evidence has at best been circumstantial, typically limited to the leukemic and normal stem cells sharing a CD34+CD38− cell-surface phenotype.42 However, the expression of CD34 and CD38 antigens, although instrumental for identification of normal HSCs in steady-state hematopoiesis,13 has been demonstrated to fluctuate considerably in mice as well as in humans.43,44
In the current studies, we adopted a novel approach applying genomics to better determine the identity of the 5q− stem cell population. In all investigated patients, greater than 92% of CD34+CD38−Thy1+ cells were shown to harbor the del(5q), demonstrating that virtually all of the cells in the CD34+CD38−Thy1+ compartment of these patients are part of the MDS clone. The 5q− CD34+CD38−Thy1+ compartment represented 0.14% of all BM cells, only slightly expanded when compared with the CD34+CD38−Thy-1+ HSC compartment in healthy subjects, demonstrating that the normal CD34+CD38−Thy1+ HSC compartment had almost been completely replaced with 5q− stem cells, a conclusion further substantiated by the lack of detectable normal LTC-IC activity in all investigated patients. Thus, 5q− CD34+CD38−Thy1+ cells appear to largely outcompete and hence deplete their normal counterparts, a finding also supported by up to 99% of granulocytes in these patients with del(5q).10 However, the mechanisms or niches that are in place to limit the size of the normal HSC pool45,46 also appear to be able to restrict the size of the 5q− HSC compartment at this stage of the disease (up to 8 years after diagnosis).
In 2 of the 4 investigated 5q− patients, we observed 5q− LTC-CFC activity from CD34+CD38−Thy1+ but not CD34+CD38+Thy-1− cells, supporting that the infrequent 5q− CD34+CD38−Thy1+ cells are the MDS stem cells in these patients. However, in 2 other patients in which we also observed a virtually complete replacement of the normal CD34+CD38−Thy1+ compartment with 5q− CD34+CD38−Thy1+ cells, we failed to detect not only normal but also 5q− LTC-CFC activity. Although we could also show in these patients that normal HSCs have been outcompeted by 5q− MDS stem cells, the functional data failed in these cases to conclusively show that 5q− CD34+CD38−Thy1+ cells are in fact the MDS stem cells. The most likely interpretation of the lack of 5q− LTC-CFC activity in these 2 patients is that the readout of this assay requires efficient myeloid differentiation from investigated HSC populations, which is variably affected in MDS.
Strikingly, 5q− and normal CD34+CD38−Thy1+ stem cells showed an almost perfect match in global gene-expression patterns, as the Fisher scores were low and comparable to the random expectation. In contrast, 5q− CD34+CD38−Thy1+ cells displayed extensive differences to normal as well as 5q− CD34+CD38+Thy1− progenitors. Thus, the gene-expression pattern of 5q− CD34+CD38−Thy1+ cells further supports that the 5q− syndrome might originate in the normal CD34+CD38−Thy1+ HSC compartment, even though an origin in an even more primitive, not-yet identified cell population cannot be ruled out. Although previous studies suggested the possibility of an HSC origin of 5q− syndrome by demonstrating involvement of B cells in a few patients,10,47 the present results are compatible with most, if not all, 5q− syndromes potentially originating in the HSC pool. As LSCs and other CSCs will typically be biased in their differentiation toward one particular lineage and inefficient or incapable of generating other lineages, we predict that global gene-expression profiling will become an important complementary tool for determining the cellular origin of LSCs as well as other CSCs. Although our studies unequivocally establish a very close identity between normal and MDS stem cells, it cannot however be ruled out that this could result from reacquisition of a stem cell phenotype of a targeted normal progenitor cell population, rather than origin in a normal HSC.
The most salient finding in the present study was the many and pronounced differences observed in gene expression between 5q− and normal CD34+CD38+Thy1− progenitors compared with the few but distinct differences detected between 5q− and normal CD34+CD38−Thy1+ stem cells. Previous global expression profiling studies on MDS cases have, at most, been performed on CD34+ or CD133+ cells,36-39 containing both CD34+CD38−Thy1+ stem and CD34+CD38+Thy1− progenitor cells. The present approach allowed us to identify stage-specific changes in gene expression.
Multiple previous gene-profiling studies of MDS (including 5q−) patients had failed to find consistent changes in expression of CEBPA,36-39 despite mouse studies implicating the potential involvement of reduced CEBPA expression in development of MDS.48 In contrast, in the present studies we found in all 9 investigated 5q− patients that CEBPA was down-regulated in the MDS progenitors, at a minimum 2.2-fold. Our finding of consistently reduced CEBPA expression in 5q− progenitors supports an involvement of dysregulated CEBPA expression in the inefficient granulopoiesis characteristic of MDS.48
BMI1 was preferentially up-regulated in 5q− CD34+CD38−Thy1+ cells. Identified as an essential regulator of normal HSC self renewal and involved in leukemogenesis if deregulated,32 the up-regulation of BMI1 could potentially explain the competitive advantage of 5q− CD34+CD38−Thy1+ stem cells over normal HSCs.
In conclusion, the present studies highlight the importance of molecular characterization of distinct stages of cancer stem and progenitor cells to identify and characterize stage-specific dysregulated gene expression in cancer.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The expert advice and assistance of Kees-Jan Pronk, Anna Fossum, Zhi Ma, Klas Raaschau-Jensen, and Gunilla Gärdebring for enrichment procedures and cell sorting are highly appreciated. We thank Associate Professor Thoas Fioretos and members of the Lund Stem Cell Center, especially Professor Carsten Peterson, for helpful discussions and Ramin Tehranchi for help with figures. We thank all patients and BM volunteers and the staff at the Departments of Hematology for assistance with aspirations and Amgen and Genentech for generous contribution of cytokines.
This work was supported by grants from The Swedish Cancer Society, The Ingabritt and Arne Lundberg Foundation, The Knut and Alice Wallenberg Foundation via the SWEGENE program, The Georg Danielsson Foundation, The Gunnar Nilsson's Cancer Foundation, The Crafoordska Foundation, The Åke Wiberg Foundation, The Tobias Foundation, Government Public Health (ALF) Grants, Region Skåne, and the Medical Faculty, University of Lund. The Lund Stem Cell Center is supported by a Center of Excellence grant in Life Sciences from the Swedish Foundation for Strategic Research (SSF). P.E. has an Assistant Professor position supported by an SSF Center of Excellence grant.
Authorship
Contribution: L.N. designed research, performed research, collected data, analyzed and interpreted data, and wrote the manuscript. P.E. performed research, analyzed and interpreted data, performed statistical analysis, and drafted the manuscript. E.O., R.M., I.Å.-G., and B.S., performed research, collected data, and analyzed and interpreted data. K.T.-M. designed research, performed research, collected data, and analyzed and interpreted data. K.A. analyzed and interpreted data. R.H. and E.H.-L. provided clinical material, collected data, and analyzed and interpreted data. J.S. and G.B. provided clinical material and collected data. C.N., B.J., M.S., and Å.B. designed research, analyzed and interpreted data, and contributed to writing of the manuscript. S.E.W.J. designed research, analyzed and interpreted data, and wrote the manuscript.
P.E. and E.O. contributed equally to this work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sten Eirik W. Jacobsen, Lund Strategic Research Center for Stem Cell Biology and Cell Therapy, BMC B10, Sweden; e-mail: sten.jacobsen@med.lu.se.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal