Abstract
Anaplastic large cell lymphomas (ALCLs) are highly proliferating tumors that commonly express the AP-1 transcription factor JunB. ALK fusions occur in approximately 50% of ALCLs, and among these, 80% have the t(2;5) translocation with NPM-ALK expression. We report greater activity of JunB in NPM-ALK–positive than in NPM-ALK–negative ALCLs. Specific knockdown of JUNB mRNA using small interfering RNA and small hairpin RNA in NPM-ALK–expressing cells decreases cellular proliferation as evidenced by a reduced cell count in the G2/M phase of the cell cycle. Expression of NPM-ALK results in ERK1/2 activation and transcriptional up-regulation of JUNB. Both NPM-ALK–positive and –negative ALCL tumors demonstrate active ERK1/2 signaling. In contrast to NPM-ALK–negative ALCL, the mTOR pathway is active in NPM-ALK–positive lymphomas. Pharmacological inhibition of mTOR in NPM-ALK–positive cells down-regulates JunB protein levels by shifting JUNB mRNA translation from large polysomes to monosomes and ribonucleic particles (RNPs), and decreases cellular proliferation. Thus, JunB is a critical target of mTOR and is translationally regulated in NPM-ALK–positive lymphomas. This is the first study demonstrating translational control of AP-1 transcription factors in human neoplasia. In conjunction with NPM-ALK, JunB enhances cell cycle progression and may therefore represent a therapeutic target.
Introduction
Anaplastic large cell lymphoma (ALCL) represents a group of highly malignant peripheral T-cell lymphomas characterized by sustained expression of CD30.1,2 More than 70% of ALCLs carry specific chromosomal translocations in which the 3′ tyrosine kinase coding portion of the ALK (anaplastic lymphoma kinase) gene on chromosome 2 is fused to the 5′ oligomerization domain coding a portion of one of multiple partners, most often the NPM1 (nucleophosmin) gene on chromosome 5.3 Parts of other genes such as TFG, TPM3, ATIC, CLTCL, RanBP2, and MSN occur less frequently as fusion partners.4 All ALK fusion proteins contain the c-terminal catalytic ALK domain required for cellular transformation.4,5 Aberrant expression of constitutively active ALK is directly implicated in the pathogenesis of ALCL.5-7 Furthermore, inhibition or knockdown of ALK kinase attenuates proliferation and survival of ALCL cells.8,9 However, despite the progress in the field, understanding of the detailed molecular mechanisms of ALK-mediated lymphomagenesis is still limited
Recent studies have established that JunB is overexpressed in CD30+ lymphomas including ALCL, Hodgkin lymphoma, and lymphomatoid papulosis.10,11 JunB belongs to the activator protein (AP-1) family. AP-1 is a sequence-specific DNA-binding transcription factor, which consists of homodimers or heterodimers formed by Jun (c-Jun, JunB, JunD), Fos (c-Fos, Fra-1, Fra-2), and ATF family proteins.12 AP-1 promotes mitogen-induced cell cycle progression and plays a role in the regulation of apoptosis. However, the function of JunB in neoplastic transformation is complex, and depending on the cellular context either proapoptotic or antiapoptotic functions have been reported.12-16 Although JunB was recently shown to mediate induction of the CD30 promoter in ALCL and Hodgkin lymphoma,17,18 its role in promoting neoplastic growth is not fully understood.
Recently, activation of phosphatidylinositol 3-kinase (PI3K) and consecutive phosphorylation of the serine/threonine kinase AKT have been linked to NPM-ALK signaling.19-21 An important target of AKT is the protein kinase designated as the mammalian target of rapamycin (mTOR). The immunosuppressant rapamycin binds to FKBP which in turn inhibits mTOR by binding its FK-rapamycin binding (FRB) domain, and thereby functions as a critical regulator of protein translation.22,23 mTOR can phosphorylate the downstream target p70S6-kinase(K) at Thr389, a critical site for catalytic activation of this serine/threonine protein kinase. Activation of P70S6K and of ribosomal protein S6 (rpS6) is critical for enhanced translation of certain mRNA molecules that contain an unusual oligopyrimidine tract at their transcriptional start site (5′TOP motif).24,25
Under certain conditions such as amino acid starvation or growth arrest, cells rapidly repress the biosynthesis of the translational machinery, which is in great part coded by capped (5′TOP containing) mRNAs, thus conserving energy. Likewise, when amino acids are replenished or mitogenic stimulation is applied, cells can rapidly respond by resuming biosynthesis of the translational apparatus. This control mechanism is thought to function through specific disaggregation of high-molecular-weight polysomes containing 5′TOP mRNAs, under starvation conditions. The 5′TOP mRNAs can thereafter be found in mRNPs or monosomes. This lowers the abundance of 5′TOP protein.25,26
We found that the transcription factor JunB is a critical target of mTOR and is translationally deregulated in NPM-ALK–positive lymphomas. In this context, JunB enhances cell cycle progression and may therefore represent a therapeutic target. Our data indicate a novel mechanism for regulation of AP-1 activity in general and suggest a potential new therapeutic approach by specifically modulating translation of an oncogene.
Materials and methods
Cell culture
Ba/F3 cells stably expressing NPM-ALK (Ba/F3-NPM-ALK) and doxycycline inducible NPM-ALK–expressing cell lines (TonBaF.1-NPM-ALK) were generated by retroviral transduction as described elsewhere.27 TonBaF.1-NPM-ALK cell lines stably expressing anti-JUNB sh-RNA or nonspecific scrambled control sh-RNA were produced by transduction of TonBaF.1-NPM-ALK cells with pSilencer-H1 vectors (Ambion, Austin, TX) using the phoenix eco (product no. SD 3444; American Type Culture Collection [ATCC]) packaging-cell line, which was deposited at the ATCC (Manassas, VA) Safe Deposit by Dr G. Nolan. For transfection with retroviral vectors, polyfect reagent from Quiagen (Hilden, Germany) was used. Ba/F3-NPM-ALK cells were routinely kept in RPMI medium containing 10% FBS, G418, and IL-3. For TonBaF.1-NPM-ALK cells, G418 and hygromycin B were added and when expressing sh-RNAs, G418, puromycin, and hygromycin were added. For all TonBaF.1-NPM-ALK–derived cells, expression of NPM-ALK was induced by weaning the cells from IL-3 for 24 hours and subsequent addition of 2 μg/mL doxycycline in the complete absence of IL-3 for 6 to 72 hours. The human ALCL cell lines DEL, FePD, JB-6, Karpas-299 (K299), SR-786, SU-DHL1, and SUP-M2 were acquired from the “Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ).” They were maintained in RPMI medium containing 10% FBS. For the expression of NPM-ALK and kinase-dead NPM-ALK HEK293 cells were transfected with pSBC-1-NPM-ALK and pSBC-2-PAC, or kinase-dead pSBC-1-NPM-ALK and pSBC-2-PAC, respectively, using Superfect (Quiagen). They were selected for stable expression using 0.7 μg/mL puromycin. Cell densities and viability were measured with a Casy TTC system (Schärfe System, Reutlingen, Germany). Cell viability was also assessed using trypan blue solution (Sigma-Aldrich, St Louis, MO). Relative proliferation of suspension cells was assayed using the Vybrant MTT Cell Proliferation Assay Kit from Molecular Probes (Invitrogen, Eugene, OR). If not otherwise stated, all pharmacologic inhibitors and antibiotics were used in the following concentrations and bought from the indicated manufacturers: LY294002 (48.8 μM) from Cell Signaling Technologies (Beverly, MA); rapamycin (20 nM) and UO126 (12.5 μM), both from Calbiochem (San Diego, CA); and cycloheximide (40 μg/mL), hygromycin B (250 μg/mL), G418 (1 mg/mL), and puromycin (2 μg/mL), all from Sigma-Aldrich. Doxycycline (2 μg/μL) was from Clontech Laboratories (Mountain View, CA).
Knockdown of JUNB, c-JUN, and GAPDH by siRNA
siRNAs were purchased form Sigma-Aldrich (JUNB) and Dharmacon (Chicago, IL) (c-JUN, GAPDH, scrambled control). The sequences or product names of siRNAs were as follows: JUNB, 5′-GCAUCAAAGUGGAGCGCAATT-3′ (sense) and 5′-UUGCGCUCCACUUUGAUGCTT-3′ (antisense); c-JUN, 5′-GAGCGGACCUUAUGGCUACUU-3′ (sense) and 5′-GUAGCCAUAAGGUCCGCUCUU-3′ (antisense); GAPDH, siCONTROL Human GAPD Duplex D-001140–01-20; Scrambled control, siCONTROL Non-Targeting siRNA no. 1 D-001210–01. Transient transfection of Karpas-299 cells was performed using the Amaxa Nucleofector II and the Amaxa cell line kit V (Amaxa, Gaithersburg, MD) according to the manufacturer's protocol. For each transfection, 3 × 106 cells were transfected with 0.1 nmol of the adequate siRNA. At 24 hours after transfection, JunB, c-Jun, and GAPDH protein levels were analyzed by Western blotting to confirm adequate gene silencing.
Plasmid constructs
For sh-RNA vectors, an sh construct (sh-JUNB) was inserted into the pSilencer 5.1-H1-Retro vector from Ambion. Top-strand sequence was as follows: 5′-GATCCGGTGAAGACACTCAAGGCTTTCAAGAGAAG-CCTTGAGTGTCTTCACCTTTTTTGGAAA-3′. As control, pSilencer 5.1-H1 Retro Scrambled vector (Ambion) was used. For construction of NPM-ALK and kinase-dead NPM-ALK expression vectors, an EcoRI HindIII fragment of human NPM-ALK cDNA (RNA source: KARPAS-299[DSMZ ACC 031]; cDNA primers: NPM-F, 5′-CCCGAATTCATGGAAGATTCGATGGACATGGACA and ALK-R, 5′-CCCAAGCTTCTA-GGGCGGAGCTCAGGCTCGTGCTGCC) was inserted into pSBC-1.28 pSBC-1-NPM-ALK and pSBC-2-PAC (PAC, puromycin acetylase) were fused to create bicistronic transcription units. The kinase-negative mutation was generated with the QuikChange site-directed mutagenesis kit (Stratagene Europe, Amsterdam, The Netherlands) using the sense oligonucleotide primer CTGCAAGTGGCTGTGAGGACGCTGCCTGAAGTG (the underlined G replaces an A in the nonmutated sequence), together with the corresponding antisense oligonucleotide primer and the pSBC-1-NPM-ALK construct as template. Sequence authenticity and single base mutation were verified by sequencing (MWG, Munich, Germany).
mRNA quantification
Total RNA was isolated from cell pellets or sucrose gradient fractions using Trizol Reagent (Invitrogen, Carlsbad, CA). For Northern blot analysis, RNAs were electrophoretically separated and transferred by capillary blot onto Hybond N+ membrane (Hybond, Escondido, CA). cDNA made from K299 was used to amplify a 493-bp ACTB and a 609-bp JUNB fragment (primers: JUNB N-FOR: 5′-CGGCAGCTACTTTTCTGGTC-3′; JUNB N-REV: 5′-CGCTCTTGGTCTTCCATGTT; ACTB N-FOR: 5′-AGCACAGAGCCTCGCCTTT-3′; ACTB N-REV: 5′-AGAGGCGTACAGGGATAGCA-3′). dCTP32-fragment labeling was performed using the Rediprime II DNA Labeling System (Amersham, GE Healthcare, Buckinghamshire, United Kingdom). For quantitative real-time PCR (qRT-PCR), cDNA was prepared by reverse transcription (superscript transcriptase was from Invitrogen, Carlsbad, CA) of 1 μg total RNA (purified by DNase digestion using TURBO DNase from Ambion). For detection of specific transcripts, Universal PCR Master Mix (Applied Biosystems, Foster City, CA) or Platinum sybr green qpcr SUPERMIX-UDG with ROX (Molecular Probes Invitrogen, Eugene, OR) and TaqMan-assays or conventional probes and/or primers were used (Document S1 for primer and probe sequences and TaqMan-assay designations, available on the Blood website; see the Supplemental Materials link at the top of the online article). All PCR-based assays showed PCR efficiency higher than 90% and were quantified with the ΔΔCT method.
cDNA microarray analysis
We generated a cDNA library representing NPM-ALK–regulated clones by subtracting a pool of NPM-ALK–negative (MAC-2A; FE-PD) from a pool of NPM-ALK–positive (JB-6, SUP-M2, SU-DHL-1, SR-786, DEL, Karpas299) ALCL lines using the PCR-Select cDNA Subtraction Kit (Clontech, Palo Alto, CA) according to the manufacturer's protocol. We selected 768 clones that were up-regulated and 768 that were down-regulated in NPM-ALK–positive ALCL. These cDNA clones and genes assumed to be relevant for cancer pathogenesis were spotted, generating specific cDNA microarrays comprising approximately 4992 clones.29 Microarray hybridization experiments and data analyses were performed as described previously.29 Gene trees were generated by hierarchic clustering with the standard correlation (Pearson correlation around zero) and a separation ratio of 0.5 and a minimum distance of 0.001 using Genesis 1.1.3 (Technical University, Graz, Austria).30
Immunohistochemistry
Immunohistochemistry (IHC) was performed on paraffin-embedded, formalin-fixed biopsy specimens. All primary antibodies were diluted in antibody diluent (Dako, Glostrup, Denmark). The following antibodies were used (concentration given in parentheses): ALK1 (1:100); p-Thr389 p70s6K (1:200); pSer240/244 rpS6 (1:100); p44/42 MAPK p-Thr202/p-Tyr204 (1:50); JunB (1:100); CD30 (1:50); and MIB1 (1:30). Detection systems used were as follows: Dako REAL Detectin System; Peroxidase/DAB+ or AEC Substrate Chromogen ready-to-use (both from Dako); or Benchmark XT Ultraview universal DAB detection kit from Ventana (Strasbourg, France). Specimens were counterstained with hematoxylin and were assessed by a pathologist (G.H.). For exact antibody information see Document S1. Acquisition of photomicrographs was performed using a Nikon Eclipse E600 microscope equipped with a 60×/0.95 plan apo objective lens and a Nikon DS 5M digital camera (Nikon, Vienna, Austria), and Photoshop CS (Adobe Systems, Munich, Germany) for white balance, contrast, and brightness correction.
Immunoblotting
Cells were grown to densities of 0.5 to 1.5 × 106 cells/mL and lysed in denaturing lysis buffer (50 mM Tris-HCl [tris(hydroxymethyl)aminomethane] pH 7.4, 150 mM NaCl, 0.1% triton X-100, 5 mM EDTA [ethylenediaminetetraacetic acid], 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride [PMSF], and protease inhibitors). Equal amounts of protein (20 μg) were separated by 12% SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred electrophoretically to nitrocellulose Trans-Blot Transfer Membranes (Bio-Rad, Hercules, CA). Antibody binding was detected using the enhanced chemiluminescence (ECL) kit (Amersham, GE Healthcare)
Electro mobility (super)shift analysis
Cell extracts were prepared using whole-cell extract buffer (20 mM HEPES, 20% glycerol, 50 mM KCl, 1 mM EDTA, 1 mM DTT, 400 mM NaCl, 5 μg/mL leupeptin, 0.2 units/mL aprotinin, 1 mM PMSF, 5 mM Na3VO4, 10 mM NaF, 5 mM glycerophosphate, pH 7.9). The double-stranded (ds) oligonucleotides used included the following: 5′-CGC TTG ATG ACT CAG CCG GAA-3′ (consensus AP-1–binding site underlined) or 5′-CGC TTG ATG ACT TGG CCG GAA-3′ (mutated AP-1–binding site underlined). The ds oligonucleotides (5 pmol) were end-labeled with [32P] γATP (25 μCi [0.925 MBq]; Harmann Analytic, Braunschweig, Germany). The binding reaction reaction mix (20 μL) contained 15 μg protein (whole-cell extract), 20 μg BSA, 2 μg poly dI-dC (Roche Diagnostics, Mannheim, Germany), 10 mM Tris, 1 mM PMSF, 0.1 mM EDTA, 5% glycerol, 50 mM NaCl, and 0.1% NP-40. End-labeled ds oligonucleotide (0.1 pmol) was added, and the mixture was incubated for 45 minutes at room temperature. When desired, 2 μL of the adequate trans cruz supershift reagent (Santa Cruz Biotechnology, Santa Cruz, CA) was added prior to incubation. DNA-protein complexes were resolved on a nondenaturating 5% polyacrylamide gel (prerun for 30 minutes) at 200 V for 3 hours.
Cell-cycle analysis
Cell-cycle analysis was performed using propidium iodide (PI) staining and flow cytometry. For each analysis, 0.5 × 106 cells were harvested 48 hours after siRNA treatment and fixed using 100% ice-cold ethanol. Analysis was performed with the Beckman Coulter (Fullerton, CA) DNA-Prep reagent kit and the FACSCalibur (Becton Dickinson, San Jose, CA) fluorescence-activated cell sorter (FACS) device.
Polysome gradient preparation
Cytoplasmic extracts were prepared essentially as described by Jefferies et al24 , except that 5 mM MgCl2 was present during cell lysis and in sucrose gradients. Typically, 15 OD260 units (1 U corresponds to the amount of RNA in 1 mL of OD260 = 1) of cytoplasmic extracts were applied to a 12-mL linear gradient from 17.1% to 51% sucrose (wt/vol). Centrifugation was carried out in an SW41 Beckman Coulter rotor at 270 000g for 150 minutes at 2°C. Gradient tubes were punctured from the bottom with a hollow needle and 12 fractions were harvested from top to bottom (80% sucrose was used to push the gradients up and through the OD254 flow cell, and finally to the fraction collector), while OD254 was monitored via an online flow cell of an ISCO density gradient fractionator (Teledyne ISCO, Lincoln, NE).
Results
Increased mRNA expression of AP-1 target genes but not of AP-1 itself in NPM-ALK–positive ALCL cell lines
To identify whether distinct gene expression signatures are associated with the presence of NPM-ALK, we screened 8 ALCL cell lines (NPM-ALK negative: MAC-2A and FE-PD; NPM-ALK positive: JB-6, SUP-M2, SU-DHL-1, SR-786, DEL, and Karpas-299) using subtractive cDNA libraries followed by cDNA microarray analysis. Unsupervised clustering using a set of 84 cDNAs with the greatest differential expression between NPM-ALK–positive and –negative ALCL cell lines classified the ALCL lines in 2 clusters with regard to their NPM-ALK status (Figure 1A; for detailed gene list see Table S1). Most interestingly, among this group of differentially expressed genes, 7.1% (6/84) were target genes of AP-1 transcription factors, which is significantly higher than the expected occurrence of 1.1% (49/4362 genes present on our microarray); P = .01, χ2 test. All of these known AP-1 target genes, such as GM-CSF alpha, GM-CSF beta, ARF-5, FAS, FASL, and BCL-3, were more highly expressed in NPM-ALK–positive than in –negative ALCL cell lines (Figure 1B). Conversely, AP-1 transcription factors themselves were not differentially expressed (Figure S1). This indicates that expression of NPM-ALK might play a role in the regulation of AP-1 activity without influencing its transcription.
AP-1 transcription factor activity is increased in NPM-ALK–containing cells. (A) Hierarchic clustering using gene expression data of 84 genes differentially expressed in NPM-ALK–negative (blue) and NPM-ALK–positive (red) ALCL cell lines. The dendrogram at the right demonstrates clustering of cell lines using complete linkage. (B) Hierarchic clustering using mRNA expression of 6 AP-1 target genes separating NPM-ALK–negative (blue) and NPM-ALK–positive (red) ALCL cell lines. (C) EMSA of NPM-ALK–negative (MAC-2A, FE-PD) and–positive (SR-786, Karpas 299) ALCL cell lines. Cell lysates were incubated with a [32P]-labeled AP-1 consensus DNA probe (A) or a [32P]-labeled mutated probe (M) (representative results of 3 independent experiments). (D top panel) Western blots using ALK and β-actin antibodies on lysates from vector control (VC) and NPM-ALK–expressing Ba/F3 cells, as well as TonBaF.1-NPM-ALK cells inducibly expressing NPM-ALK without (unind) and 48 hours after addition of 2 μg/mL doxycycline (+ dox). (Bottom panel) EMSA of cell lysates incubated with AP-1 consensus DNA probe (A) or mutated probe (M) (representative results of 3 independent experiments). (E top panel) Western blots of wild-type NPM-ALK and kinase-negative (KN) NPM-ALK–expressing HEK293 cells. Lysates of control (VC) and stably expressing NPM-ALK and KN transfectants were analyzed for presence of fusion protein (bottom) and for NPM-ALK phosphorylated at the pY342/pY343 position. (Bottom panel) EMSA of HEK293 KN and HEK293 NPM-ALK incubated with AP-1 consensus DNA probe (A) and mutated probe (M). (F) Supershift analysis. Cell lysates were incubated with an AP-1 consensus DNA probe and various anti–AP-1 antibodies (c-Jun, JunB, JunD, Fos, Fra-1, Fra-2, and ATF2) (representative results of 3 independent experiments). (G) JUNB mRNA and protein are increased in TonBaF.1-NPM-ALK cells induced for 48 hours with doxycycline. (Left) mRNA expression levels relative to a calibrator (c-JUN mRNA level of Mac2A cells compared with β-actin) are given to facilitate comparison between AP-1 factors (bars indicate standard deviation [SD] of triplicate experiments). (Right) Immunoblotting using antibodies indicated on lysates of uninduced and induced TonBaF.1-NPM-ALK cells.
AP-1 transcription factor activity is increased in NPM-ALK–containing cells. (A) Hierarchic clustering using gene expression data of 84 genes differentially expressed in NPM-ALK–negative (blue) and NPM-ALK–positive (red) ALCL cell lines. The dendrogram at the right demonstrates clustering of cell lines using complete linkage. (B) Hierarchic clustering using mRNA expression of 6 AP-1 target genes separating NPM-ALK–negative (blue) and NPM-ALK–positive (red) ALCL cell lines. (C) EMSA of NPM-ALK–negative (MAC-2A, FE-PD) and–positive (SR-786, Karpas 299) ALCL cell lines. Cell lysates were incubated with a [32P]-labeled AP-1 consensus DNA probe (A) or a [32P]-labeled mutated probe (M) (representative results of 3 independent experiments). (D top panel) Western blots using ALK and β-actin antibodies on lysates from vector control (VC) and NPM-ALK–expressing Ba/F3 cells, as well as TonBaF.1-NPM-ALK cells inducibly expressing NPM-ALK without (unind) and 48 hours after addition of 2 μg/mL doxycycline (+ dox). (Bottom panel) EMSA of cell lysates incubated with AP-1 consensus DNA probe (A) or mutated probe (M) (representative results of 3 independent experiments). (E top panel) Western blots of wild-type NPM-ALK and kinase-negative (KN) NPM-ALK–expressing HEK293 cells. Lysates of control (VC) and stably expressing NPM-ALK and KN transfectants were analyzed for presence of fusion protein (bottom) and for NPM-ALK phosphorylated at the pY342/pY343 position. (Bottom panel) EMSA of HEK293 KN and HEK293 NPM-ALK incubated with AP-1 consensus DNA probe (A) and mutated probe (M). (F) Supershift analysis. Cell lysates were incubated with an AP-1 consensus DNA probe and various anti–AP-1 antibodies (c-Jun, JunB, JunD, Fos, Fra-1, Fra-2, and ATF2) (representative results of 3 independent experiments). (G) JUNB mRNA and protein are increased in TonBaF.1-NPM-ALK cells induced for 48 hours with doxycycline. (Left) mRNA expression levels relative to a calibrator (c-JUN mRNA level of Mac2A cells compared with β-actin) are given to facilitate comparison between AP-1 factors (bars indicate standard deviation [SD] of triplicate experiments). (Right) Immunoblotting using antibodies indicated on lysates of uninduced and induced TonBaF.1-NPM-ALK cells.
Higher AP-1 activity in NPM-ALK–positive compared with –negative ALCL cell lines
Based on these findings, we investigated NPM-ALK–dependent AP-1 activity by electrophoretic mobility shift assay (EMSA). Protein extracts were incubated using a probe that contained the AP-1 consensus sequence TRE (12-O-tetradecanoylphorbol-13-acetate [TPA] response element).32 Lysates of both NPM-ALK–negative and –positive ALCL cell lines showed AP-1–binding activity. However, the NPM-ALK–negative cell lines MAC-2A and FE-PD showed relatively weak binding activity compared with the NPM-ALK–positive lines SR-786 and Karpas-299 (Figure 1C). The specificity of the assay was supported by a probe that contained a mutated TRE.
NPM-ALK expression enhances AP-1 DNA-binding activity
To investigate the potential of NPM-ALK to induce AP-1 activity, we used a modified Ba/F3 murine hematopoietic cell line that expresses NPM-ALK either in a constitutive or an inducible manner27 (Figure 1D top panel; ALK mRNA values in Figure S3). TonBaF.1-NPM-ALK cells express NPM-ALK from a reverse-Tet-(tetracycline)-response element. In these cells, NPM-ALK expression is induced upon addition of doxycycline.27 In agreement with previous reports,5 stable expression of NPM-ALK achieved by retroviral transduction with pMSCV-NPM-ALK-neo led to IL-3 independent outgrowth and survival (data not shown). The AP-1 complex is actively bound to the AP-1 consensus sequence in NPM-ALK–expressing Ba/F3 but not in control cells (Figure 1D bottom left panel). Moreover, TonBaF.1-NPM-ALK cells show pronounced AP-1–binding activity after 48 hours of induction with doxycycline. (Figure 1D bottom right panel). Thus, NPM-ALK is capable of inducing AP-1 activity.
The catalytic domain of ALK is essential for induction of AP-1 activity
The kinase domain of NPM-ALK was shown to be essential for NPM-ALK–induced mitogenesis.4,5 Using site-directed mutagenesis, a kinase-negative variant was generated by substitution of the invariant lysine residue 210, which is located in the ATP-binding pocket of the catalytic domain of the fusion gene by arginine. Upon expression of the constitutive active and kinase-defective chimera constructs in human embryonic HEK293 cells, autophosphorylation at the position Y342/Y343 site was abolished in the kinase-negative protein, as shown using a pY-specific antibody (Figure 1E top panel). In addition, EMSAs of lysates from HEK293 cells expressing wild-type NPM-ALK or kinase-negative NPM-ALK demonstrate that the kinase-negative NPM-ALK cells lack AP-1 DNA-binding activity. This indicates that the active catalytic domain of ALK is essential for inducing AP-1 activity (Figure 1E bottom panel).
JunB is the AP-1 transcription factor induced by NPM-ALK
To clarify which component(s) constitute the NPM-ALK–induced active AP-1 complex, we performed EMSA supershift analyses using anti–c-Jun, -JunB, -JunD, -Fos, –Fra-1, –Fra-2, and -ATF2 antibodies (Figure 1F). NPM-ALK–expressing cell lines such as the ALCL cell lines SR-786 and Karpas299, as well as the doxycycline-induced TonBaF.1-NPM-ALK cell line demonstrated a complete shift when anti-JunB antibody was used, while no other antibodies showed comparably shifted bands. Interestingly, the moderate AP-1 DNA-binding activity of NPM-ALK–negative ALCL lines, such as MAC-2A, could also be attributed mainly to JunB activation (Figure 1F bottom right panel).
To further examine the consequences of NPM-ALK expression on mRNA and protein induction of JunB and other major AP-1 factors, we analyzed TonBaF.1-NPM-ALK cells after doxycycline induction. Induced expression of NPM-ALK led to pronounced expression of JUNB mRNA and protein, whereas mRNA of other AP-1 factors was only moderately expressed (Figure 1G). Accordingly, the relative mRNA expression values of AP-1 factors in 4 human ALCL lines (Fe-PD, Mac-2A, Karpas-299, SR-786) showed prominent JUNB expression compared with other AP-1 factors (Figure S1). In contrast, HEK-293 cells of nonhematologic origin demonstrated a marked induction of FOS mRNA and protein and to a lesser extent JunB upon NPM-ALK induction, indicating that the composition of the AP-1 complex induced by NPM-ALK is dependent on the cellular origin (Figure S2). Hence, JunB is the major AP-1 transcription factor induced by NPM-ALK in all hematologic cell lines tested, whereas in a cell line of nonhematologic origin NPM-ALK primarily induces other AP-1 factors.
Knockdown of JUNB impairs cell-cycle progression of NPM-ALK–expressing cells
Although in some contexts JunB antagonizes c-Jun–containing AP-1 complexes and has antiproliferative activity,33 in other contexts JunB can substitute for c-Jun and rescue defects resulting from c-Jun deficiency.34 To investigate the functional role of JunB in the context of NPM-ALK, we chose a loss-of-function approach using RNA interference by sh-RNAs.35 We modified TonBaF.1-NPM-ALK cells by retroviral gene transfer of JUNB sh-RNA (sh-JUNB) and scrambled sh-RNA (sh-control). Twenty-four hours after addition of doxycycline to TonBaF.1-NPM-ALK-sh-JUNB and TonBaF.1-NPM-ALK-sh-control cells, NPM-ALK expression was highly stimulated (data not shown). sh-JUNB expression decreased JUNB mRNA levels on average by 77%, compared with sh-control cells, as measured by quantitative real-time PCR (qRT-PCR; Figure 2A left panel). Compared with control, proliferation of cells with suppressed JUNB was significantly reduced (P = .013; parametric t test over all time points, Figure 2A right panel). Caspase-3 activity was not different between sh-JUNB– and sh-control–expressing cells (Figure S4), consistent with the absence of apoptosis.
Knockdown of JUNB impairs cell-cycle progression in NPM-ALK–expressing cells. (A) Small hairpin (sh) RNA knockdown of JUNB in doxycycline-induced TonBaF.1-NPM-ALK cells. (Right) JUNB mRNA is decreased in 24-hour doxycycline-induced TonBaF.1-NPM-ALK cells stably transduced with JUNB sh-RNA–producing vector (sh-JUNB) compared with cells producing scrambled sh-RNA (sh-control). JUNB expression levels of sh-control–treated cells were set as 1 to facilitate comparison with sh-JUNB cells (bars indicate standard deviation [SD] of triplicate experiments). (Left) Cell counts (normalized to control at t = 0) demonstrate a decreased cellular proliferation of sh-JUNB–treated TonBaF.1-NPM-ALK cells at time points indicated (representative results of 3 independent experiments). (B) Knockdown experiments using small interfering (si) RNA in the human NPM-ALK–positive ALCL cell line Karpas-299. (Top panel left) Immunoblot demonstrating changes in protein levels upon transfection with scrambled siRNA (siControl), GAPDH siRNA (siGAPDH), JUNB siRNA (siJUNB), and c-JUN siRNA (si c-JUN), respectively. (Top panel right) Cell counts (normalized to control at t = 0) demonstrate a decreased cellular proliferation of Karpas-299 transfected with siJUNB at time points indicated (representative results of 6 independent experiments. SDs for 2 biologic replicates are given). (Bottom panel) Cell-cycle analysis assessed by propidium iodide staining and flow cytometry shows a decrease in the G2/M fraction associated with an increase of the G0/G1 fraction after siJUNB transfection (representative results of 3 independent experiments). (C) EMSA of Karpas-299 cytoplasmic lysates, 24 hours after siRNA transfection with siControl, siGAPDH, siJUNB, and si c-JUN. Cell lysates were incubated with a [32P]-labeled AP-1 consensus DNA (representative results of 3 independent experiments).
Knockdown of JUNB impairs cell-cycle progression in NPM-ALK–expressing cells. (A) Small hairpin (sh) RNA knockdown of JUNB in doxycycline-induced TonBaF.1-NPM-ALK cells. (Right) JUNB mRNA is decreased in 24-hour doxycycline-induced TonBaF.1-NPM-ALK cells stably transduced with JUNB sh-RNA–producing vector (sh-JUNB) compared with cells producing scrambled sh-RNA (sh-control). JUNB expression levels of sh-control–treated cells were set as 1 to facilitate comparison with sh-JUNB cells (bars indicate standard deviation [SD] of triplicate experiments). (Left) Cell counts (normalized to control at t = 0) demonstrate a decreased cellular proliferation of sh-JUNB–treated TonBaF.1-NPM-ALK cells at time points indicated (representative results of 3 independent experiments). (B) Knockdown experiments using small interfering (si) RNA in the human NPM-ALK–positive ALCL cell line Karpas-299. (Top panel left) Immunoblot demonstrating changes in protein levels upon transfection with scrambled siRNA (siControl), GAPDH siRNA (siGAPDH), JUNB siRNA (siJUNB), and c-JUN siRNA (si c-JUN), respectively. (Top panel right) Cell counts (normalized to control at t = 0) demonstrate a decreased cellular proliferation of Karpas-299 transfected with siJUNB at time points indicated (representative results of 6 independent experiments. SDs for 2 biologic replicates are given). (Bottom panel) Cell-cycle analysis assessed by propidium iodide staining and flow cytometry shows a decrease in the G2/M fraction associated with an increase of the G0/G1 fraction after siJUNB transfection (representative results of 3 independent experiments). (C) EMSA of Karpas-299 cytoplasmic lysates, 24 hours after siRNA transfection with siControl, siGAPDH, siJUNB, and si c-JUN. Cell lysates were incubated with a [32P]-labeled AP-1 consensus DNA (representative results of 3 independent experiments).
Since JunB and to a lesser extent c-Jun are induced by NPM-ALK (Figure 1G) and since expression of both AP-1 factors can be found in NPM-ALK–positive ALCL cell lines (Figure S1 and Mathas et al10 and Leventaki et al36 ), we examined the functional contribution of both AP factors in the human NPM-ALK–positive ALCL cell line Karpas-299. Transient knockdown of JUNB, c-JUN, and GAPDH using siRNA substantially decreased the respective protein levels (Figure 2B top left panel). Specific silencing of JUNB significantly decreased cellular proliferation compared with control, GAPDH, and c-JUN siRNA treatment (P = .007; parametric t test over all time points; Figure 2B top right panel). Cell-cycle analysis revealed that the decrease in proliferation after siJUNB transfection was associated with a decrease of the G2/M fraction and an increase in the G0/G1 fraction (Figure 2B bottom panel). Only downmodulation of JunB was followed by a marked decrease of AP-1 DNA-binding activity, whereas c-JUN knockdown showed no effect on AP-1 activity (Figure 2C). In NPM-ALK–positive ALCL, JunB can therefore be seen as the prominent factor of the active AP-1 complex leading to cell cycle progression in G2/M phase.
NPM-ALK regulates JunB through mTOR and MAPK signaling
Activated ERKs substantially contribute to the increased expression of AP-1 members in many tumor types.12 Recently, it has been shown that NPM-ALK induces ERK phosphorylation.37,38 Moreover, NPM-ALK activates the serine/threonine kinase AKT/PKB through activation of PI3 kinase.19-21 Accordingly, we demonstrate that activation of ERK and AKT is dependent on the expression of NPM-ALK in modified BA/F3 cells as determined by Western blot analysis using phosphorylation-specific antibodies (Figure 3A left panel). The mammalian target of rapamycin (mTOR) is a critical downstream target of AKT that phosphorylates p70S6K at Thr389 and at Thr421/Ser424.23,39 In NPM-ALK–expressing BA/F3 cells, we found abundant p70S6K phosphorylation at sites Thr389 and Thr421/Ser424, indicating NPM-ALK induced mTOR activity (Figure 3A right panel). Similarly, the NPM-ALK–positive ALCL cell lines Karpas-299 and SR-786 showed enhanced p70S6K phosphorylation at Thr389 compared with the NPM-ALK–negative cell line MAC2A (Figure S4).
NPM-ALK regulates JunB via mTOR and MAPK signaling. (A) NPM-ALK induces activation of AKT, ERK, and mTOR. Whole-cell lysates from vector control or NPM-ALK–expressing BA/F3 cells were immunoblotted with antibodies against proteins indicated on the right. (B) JunB protein expression is attenuated in NPM-ALK–expressing cells by mTOR or MEK inhibition. SR-786: NPM-ALK–positive ALCL cell line (top panel); TB-NA: TonBaF.1-NPM-ALK cells 24 hours after induction with 2 μg/mL doxycycline (+ dox), or without doxycycline (− dox) (bottom panel). Western blot analysis was performed with antibodies indicated on the right after treatment for 24 hours with 20 nM rapamycin, 12.5 μM U0126, both agents, or control (DMSO). (C) JUNB mRNA is decreased by MEK but not mTOR inhibition. qRT-PCR was performed on SR-786 cells and doxycycline-induced TB-NA cells upon treatment as indicated (20 nM rapamycin, 12.5 μM U0126, both agents, control [DMSO]). Logarithmic scale of mean JUNB expression levels related to expression of a housekeeping gene (β-actin) as 2−ΔCT (bars indicate SEM of triplicate experiments). (D) Following 24-hour treatment with 20 nM rapamycin, 12.5 μM U0126, both agents, 48.8 μM LY294002, or control (DMSO), whole-cell lysates from SR-786 cells were blotted with antibodies against proteins indicated on the right. (E) Cell counts (normalized to control at t = 0) at indicated time points: Treatment conditions as in panel D resulted in reduced cell growth.
NPM-ALK regulates JunB via mTOR and MAPK signaling. (A) NPM-ALK induces activation of AKT, ERK, and mTOR. Whole-cell lysates from vector control or NPM-ALK–expressing BA/F3 cells were immunoblotted with antibodies against proteins indicated on the right. (B) JunB protein expression is attenuated in NPM-ALK–expressing cells by mTOR or MEK inhibition. SR-786: NPM-ALK–positive ALCL cell line (top panel); TB-NA: TonBaF.1-NPM-ALK cells 24 hours after induction with 2 μg/mL doxycycline (+ dox), or without doxycycline (− dox) (bottom panel). Western blot analysis was performed with antibodies indicated on the right after treatment for 24 hours with 20 nM rapamycin, 12.5 μM U0126, both agents, or control (DMSO). (C) JUNB mRNA is decreased by MEK but not mTOR inhibition. qRT-PCR was performed on SR-786 cells and doxycycline-induced TB-NA cells upon treatment as indicated (20 nM rapamycin, 12.5 μM U0126, both agents, control [DMSO]). Logarithmic scale of mean JUNB expression levels related to expression of a housekeeping gene (β-actin) as 2−ΔCT (bars indicate SEM of triplicate experiments). (D) Following 24-hour treatment with 20 nM rapamycin, 12.5 μM U0126, both agents, 48.8 μM LY294002, or control (DMSO), whole-cell lysates from SR-786 cells were blotted with antibodies against proteins indicated on the right. (E) Cell counts (normalized to control at t = 0) at indicated time points: Treatment conditions as in panel D resulted in reduced cell growth.
Treatment with rapamycin impairs phosphorylation and activation of p70S6K and, subsequently, rpS6,23 thereby largely blocking the translational activation of 5′TOP mRNAs.25,26 Indeed, we noticed that JUNB harbors a 5′TOP-like motif in its 5′UTR near the transcriptional start site (data not shown). To determine the relative contributions of the mTOR- and MEK/ERK-dependent pathways on JunB regulation by NPM-ALK, we treated the ALCL cell line SR-786 with rapamycin, the MEK-inhibitor U0126, or both (Figure 3B top panel). We observed a significant decrease in JunB protein using rapamycin or UO126. A similar effect was seen when cells were treated with both rapamycin and UO126 (Figure 3B top panel). In TonBaF.1-NPM-ALK cells, addition of doxycycline induced NPM-ALK expression, which in turn induced JunB. Similarly to SR-786, treatment with rapamycin and UO126 caused a decrease of JunB protein in TonBaF.1-NPM-ALK cells as seen by Western blot analysis (Figure 3B bottom panel). The decrease of JunB protein upon rapamycin treatment was not related to increased protein degradation as evidenced by similar rates of degradation upon cycloheximide treatment in the presence or absence of rapamycin (Figure S6).
qRT-PCR expression analysis of SR-786 and doxycycline-induced TonBaF.1-NPM-ALK cells revealed decreased JUNB mRNA upon treatment with the MEK-inhibitor UO126, whereas rapamycin appeared to have no influence on JUNB mRNA content (Figure 3C). Thus, an activated MEK/ERK pathway induces JUNB transcription, whereas activated mTOR increases JunB translation. Both signaling cascades were found to be active in NPM-ALK–expressing cells. Treatment of SR-786 cells with rapamycin decreased phosphorylation of the mTOR target p70S6K and, subsequently, rpS6. Similarly, inhibition of the upstream PI3K/AKT pathway using the PI3K inhibitor, LY294002, decreased phosphorylation levels of p70S6K and rpS6. Treatment neither with rapamycin nor with LY294002 abolished ERK phosphorylation (Figure 3D). Treatment with UO126 inhibited ERK phosphorylation, but did not inhibit phosphorylation of p70S6K and only slightly inhibited phosphorylation of rpS6. In addition, when cells were treated with both UO126 and rapamycin, phosphorylation of ERK, as well as of p70S6K and rpS6, was inhibited (Figure 3D). The effect of LY294002 on ERK phosphorylation and UO126 on rpS6 phosphorylation can be explained by cross-links of the MEK/ERK and the mTOR pathways.40,41 Inhibition of either mTOR, MEK or both impaired proliferation compared with control (DMSO) (Figure 3F).
mTOR pathway activity in NPM-ALK–positive ALCL tumors
To investigate whether NPM-ALK–dependent activation of the mTOR-triggered p70S6K-rpS6 pathway also occurs in human lymphoma patient samples, we analyzed 10 NPM-ALK–positive and 15 NPM-ALK–negative ALCL patient samples using immunohistochemistry. Ninety percent (9/10) of NPM-ALK–positive ALCL samples compared with only 7% (1/15) of NPM-ALK–negative ALCL samples demonstrated phosphorylation of p70S6K at Thr389 (P < .01, χ2 test) (Figure 4). Phosphorylation of rpS6 was detected in 67% (4/6) of NPM-ALK–positive and in 40% (4/10) of NPM-ALK–negative samples (P = .02, χ2 test). These results are consistent with a recent report of a frequently activated mTOR pathway in ALK-positive ALCL tumors and cell lines.42 However, ERK phosphorylation was observed in all ALCL patient samples tested irrespective of NPM-ALK status (Figure 4). All ALCL tumors expressed JunB protein, although anti-JunB immunohistochemical staining of NPM-ALK–positive ALCLs on average appeared to be slightly stronger compared with NPM-ALK–negative ALCL samples (Figure 4).
Activation of mTOR and ERK in NPM-ALK–positive ALCL patient samples. Immunohistochemistry of ALCL samples using antibodies against (phospho-) proteins indicated on the left. Representative images from 9 NPM-ALK–positive ALCL (left) and 14 NPM-ALK–negative ALCL (right) patient samples. ALK antibody staining demonstrates expression of NPM-ALK in ALCL cells. JunB antibody staining is shown as minimum and maximum signal intensities of NPM-ALK–positive (left) and NPM-ALK–negative samples (right). See “Immunohistochemistry” for image acquisition information.
Activation of mTOR and ERK in NPM-ALK–positive ALCL patient samples. Immunohistochemistry of ALCL samples using antibodies against (phospho-) proteins indicated on the left. Representative images from 9 NPM-ALK–positive ALCL (left) and 14 NPM-ALK–negative ALCL (right) patient samples. ALK antibody staining demonstrates expression of NPM-ALK in ALCL cells. JunB antibody staining is shown as minimum and maximum signal intensities of NPM-ALK–positive (left) and NPM-ALK–negative samples (right). See “Immunohistochemistry” for image acquisition information.
NPM-ALK–induced mTOR signaling controls ribosome binding to JUNB mRNA
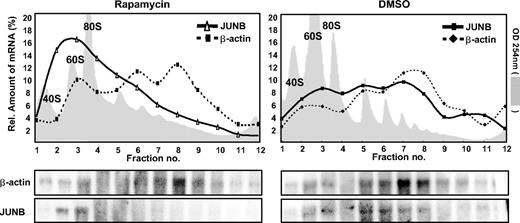
Rapamycin suppresses translation of 5′TOP motif–bearing mRNAs that are translated in heavy polysomes consisting of a relatively high number of ribosomes in proliferating cells.26 Based on the observation that JunB protein but not JUNB mRNA expression is reduced by rapamycin in NPM-ALK–expressing cells, we hypothesized that JUNB mRNA might be shifted from large polysomes into monosomes and ribonucleic particles (RNPs) upon rapamycin-triggered inhibition of the NPM-ALK–stimulated mTOR pathway. We therefore investigated the segregation of JUNB mRNAs between polysomes, monosomes, and RNPs upon inhibition of mTOR by rapamycin in NPM-ALK–positive ALCL cells. Cytoplasmic lysates from SR-786 cells, treated with rapamycin or DMSO (vehicle) for 24 hours, were applied on 17.1% to 51% linear sucrose gradients and ultracentrifuged. RNA content of the gradients was assessed by photometric online monitoring for absorbance at 254 nM, while the gradients were fractionated from top to bottom. The amount of JUNB mRNA in each of the 12 fractions was determined by Northern blot analysis. In preparations from cells treated with rapamycin, JUNB transcripts preferentially sedimented with monosomes or RNPs (Figure 5). In contrast, untreated cells (DMSO) demonstrated a significantly higher amount of JUNB mRNA in fractions representing disomes, trisomes, and heavier polysomal complexes. A probe against the non–5′TOP-regulated mRNA β-actin was used as an internal control. Translation of β-actin is known to occur via heavy polysomes in proliferating and quiescent cells.26 Accordingly, we did not detect a shift to monosomes and RNPs after rapamycin treatment. Thus, translation of JunB in NPM-ALK–positive ALCL cells is mediated by an mTOR-dependent shift of JUNB mRNA into larger ribosomal complexes that can be reversed by rapamycin.
Rapamycin inhibits polysomal partitioning of JUNB mRNA. Ribosomal 17.1% to 51% sucrose gradients were prepared from NPM-ALK–positive ALCL cell line SR-786 treated with 20 nM rapamycin or control (DMSO). Twelve fractions of each gradient were collected from bottom to top after ultracentrifugation, with online monitoring of RNA content via an OD254 UV-flow cell (gray shaded areas). RNA was isolated from each fraction and analyzed by gel electrophoresis (bottom panels), and quantities of JUNB as well as β-actin mRNA were determined using Northern blot analysis on the same blot membranes. As indicated by the photometric OD254 measurement, fractions 3 and 4 represent monosomes; fraction 5, disomes; and fractions 6 and higher, higher polysomes. Mono-polysomal β-actin and JUNB mRNA distribution correspond to the number of ribosomes that translate each molecule.
Rapamycin inhibits polysomal partitioning of JUNB mRNA. Ribosomal 17.1% to 51% sucrose gradients were prepared from NPM-ALK–positive ALCL cell line SR-786 treated with 20 nM rapamycin or control (DMSO). Twelve fractions of each gradient were collected from bottom to top after ultracentrifugation, with online monitoring of RNA content via an OD254 UV-flow cell (gray shaded areas). RNA was isolated from each fraction and analyzed by gel electrophoresis (bottom panels), and quantities of JUNB as well as β-actin mRNA were determined using Northern blot analysis on the same blot membranes. As indicated by the photometric OD254 measurement, fractions 3 and 4 represent monosomes; fraction 5, disomes; and fractions 6 and higher, higher polysomes. Mono-polysomal β-actin and JUNB mRNA distribution correspond to the number of ribosomes that translate each molecule.
Discussion
We elucidated a crucial function of the fusion tyrosine kinase NPM-ALK, which is expressed in most ALCL tumors. NPM-ALK activates JunB via both ERK1/2-MAPK and -mTOR pathways. Activated MAPK signaling is present in ALK-positive as well as -negative ALCL tumors and leads to transcription of JUNB. mTOR activity, however, is enhanced in NPM-ALK–expressing cells and induces JunB translation. Active mTOR signaling results in more effective translation of JUNB mRNA by a higher quantity of ribosomes.
A major role of c-Jun in NPM-ALK–positive ALCL was reported recently.36 We, however, propose that JunB is the main AP1 transcription factor involved in the pathogenesis of ALCL. This is evidenced by near-complete supershift by anti-JunB antibody only and decrease of AP1-binding activity after JUNB knockdown. Knockdown of c-JUN did not alter AP1-binding activity. Furthermore, relative mRNA expression levels of the major AP1 factors reveal JunB to be most abundant. RNA interference mediated knockdown of JUNB-impaired cellular proliferation and reduced the G2/M fraction, whereas c-JUN knockdown did not influence cell proliferation.
As NPM-ALK–positive and –negative ALCL tumors both express JunB,10,11 it was not obvious to link regulation of JunB to NPM-ALK expression. However, cDNA microarray analysis of NPM-ALK–positive and –negative ALCL cell lines led us to a difference in AP-1 activity as evidenced by differential AP-1 target gene expression (Figure 1B), whereas mRNA expression of AP-1 transcription factors was not affected. Correspondingly, we found that JunB protein expression appears to be more abundant in NPM-ALK–positive cells, as indicated by the difference in AP-1 DNA-binding activity in lysates from NPM-ALK–positive compared with NPM-ALK-negative ALCL cells (Figure 1C). Hence, we hypothesized that NPM-ALK–positive ALCL might have an additional mechanism to increase JunB at the posttranscriptional level. We demonstrate that the mTOR pathway is activated in NPM-ALK–positive ALCL tumors as well as cell lines and can be induced by introducing NPM-ALK into BA/F3 cells (Figures 3A,D and 4). Pharmacological inhibition of mTOR decreases JunB protein levels with no marked impairment of JUNB mRNA levels (Figure 3B,C). Since mTOR activity is lost and JunB is decreased with inhibition of PI3K, our results agree with the generally accepted model of the PI3K-AKT-mTOR signaling cascade25 and establish its crucial role in regulating JunB translation in NPM-ALK–expressing neoplasms.
In concordance with others,37,38 we show that NPM-ALK induces ERK1/2 phosphorylation (Figure 3A). Nevertheless, both NPM-ALK–positive and –negative ALCL tumors show phosphorylated ERK1/2 at the same expression pattern (Figure 4). Pathways that involve mitogen-activated protein kinases of the ERK, p38, and JUN amino-terminal kinase (JNK) families are known to regulate AP-1 activity at transcriptional and posttranslational stages.12 Decrease of JUNB mRNA upon inhibition of ERK1/2 MAPK signaling is in line with this generally accepted model (Figure 3C). However, to our knowledge, this is the first report to describe translational regulation of a human AP-1 transcription factor via mTOR. A similar translational control mechanism has been demonstrated in yeast for the AP1 family protein GCN4.43
mRNA of JUNB harbors various 5′TOP motifs in its 5′-UTR near the transcriptional start site. However, for 5′TOP transcripts, the absolute requirement for a C at the 5′-cap followed by an uninterrupted stretch of 4 to 14 pyrimidines was postulated by Meyuhas in 2000.25 This requirement is not fully met by the JUNB mRNA sequence. Nevertheless, we clearly demonstrate that upon inhibition of the NPM-ALK–stimulated mTOR pathway (Figure 5), JunB protein level is translationally regulated since JUNB mRNA is shifted from larger polysomes into monosomes as well as RNPs. Since we demonstrate that JUNB mRNA functionally behaves like 5′TOP mRNAs,26 we would like to propose a novel mechanism for the regulation of AP-1 activity in general.
Our data show that NPM-ALK promotes neoplasia at least in part by shifting the segregation of JUNB mRNAs from monosomes to polysomes; this can be reversed by treatment with rapamycin. Rapamycin, also known as sirolimus, and its analogues are clinically used as immunosuppressants, but have also been shown to provide antitumoral activity in some cancers. Current studies are evaluating the clinical effect of rapamycin derivates on advanced-stage cancers with various results.23,44-48 ALCL is a highly aggressive hematologic malignancy, and effective therapeutic strategies that minimize toxicity to the patient are needed. This study provides evidence that pharmacologic inhibition of mTOR might be an effective alternative or addition to the treatment of NPM-ALK–positive tumors.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Theodor Koerner Fonds (P.B.S.), Oesterreichische Nationalbank Jubilaeumsfonds 12147 (P.B.S.), the Austrian Science Fund (FWF) grants P-15300 (G.H.), P-18478-B12 (L.K.), P-15865 (V.S.), and the SFB 28.1000 (V.S.).
We thank Stephan W. Morris for providing MSCV-NPM-ALK vectors; Juliane Strauss and Montserrat Pinent for technical assistance; Tarek Moustafa for critical comments on our work; and the whole staff at Prof Michael Trauner's laboratory for technical advice. We further thank Gertrude Zisser and Lisa Kappel for their expertise on polysomic preparation and Eugenia Lamont for editing the paper.
Authorship
Contribution: P.B.S. and P.V. designed the project, performed most of the experiments, analyzed data, and wrote the paper; N.H. and K.F. performed most of the pharmacological treatment experiments on BaF3 cells; R.G.O. performed some of the EMSAs; I.B. and C.F. performed some immunoblots; W.L. provided some of the laboratory facilities. S.S. performed immunohistochemistry and most of the qRT-PCR analyses; A.H. performed cell cycle analysis; W.G.D. provided HEK293 cells with constructs including NPM-ALK kinase-dead mutants; V.S. designed experiments on AP-1 activity; H.B. supervised the gradient preparations and analyses; M.E.K. provided the MAC-2A cell line and contributed helpful ideas to the development of the paper; D.W.S. designed the pharmacological treatments; G.H. supervised the project and the final editing of the paper, and selected and evaluated all tumor specimens together with L.K. P.B.S. and P.V. contributed equally to this work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Philipp Bernhard Staber, Klinische Abteilung für Hämatologie, Medizinische Universitätsklinik Graz, Auenbruggerplatz 38, A-8036 Graz, Austria; e-mail: philipp.staber@meduni-graz.at.

![Figure 1. AP-1 transcription factor activity is increased in NPM-ALK–containing cells. (A) Hierarchic clustering using gene expression data of 84 genes differentially expressed in NPM-ALK–negative (blue) and NPM-ALK–positive (red) ALCL cell lines. The dendrogram at the right demonstrates clustering of cell lines using complete linkage. (B) Hierarchic clustering using mRNA expression of 6 AP-1 target genes separating NPM-ALK–negative (blue) and NPM-ALK–positive (red) ALCL cell lines. (C) EMSA of NPM-ALK–negative (MAC-2A, FE-PD) and–positive (SR-786, Karpas 299) ALCL cell lines. Cell lysates were incubated with a [32P]-labeled AP-1 consensus DNA probe (A) or a [32P]-labeled mutated probe (M) (representative results of 3 independent experiments). (D top panel) Western blots using ALK and β-actin antibodies on lysates from vector control (VC) and NPM-ALK–expressing Ba/F3 cells, as well as TonBaF.1-NPM-ALK cells inducibly expressing NPM-ALK without (unind) and 48 hours after addition of 2 μg/mL doxycycline (+ dox). (Bottom panel) EMSA of cell lysates incubated with AP-1 consensus DNA probe (A) or mutated probe (M) (representative results of 3 independent experiments). (E top panel) Western blots of wild-type NPM-ALK and kinase-negative (KN) NPM-ALK–expressing HEK293 cells. Lysates of control (VC) and stably expressing NPM-ALK and KN transfectants were analyzed for presence of fusion protein (bottom) and for NPM-ALK phosphorylated at the pY342/pY343 position. (Bottom panel) EMSA of HEK293 KN and HEK293 NPM-ALK incubated with AP-1 consensus DNA probe (A) and mutated probe (M). (F) Supershift analysis. Cell lysates were incubated with an AP-1 consensus DNA probe and various anti–AP-1 antibodies (c-Jun, JunB, JunD, Fos, Fra-1, Fra-2, and ATF2) (representative results of 3 independent experiments). (G) JUNB mRNA and protein are increased in TonBaF.1-NPM-ALK cells induced for 48 hours with doxycycline. (Left) mRNA expression levels relative to a calibrator (c-JUN mRNA level of Mac2A cells compared with β-actin) are given to facilitate comparison between AP-1 factors (bars indicate standard deviation [SD] of triplicate experiments). (Right) Immunoblotting using antibodies indicated on lysates of uninduced and induced TonBaF.1-NPM-ALK cells.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/9/10.1182_blood-2007-02-071258/7/m_zh80230709630001.jpeg?Expires=1767697641&Signature=YsHwWPuR4od~M1XL1YhaUciWW1YBlBlrYA3JD6McH-zSbKz-ZeTW08WmO4~tqempuAv7y8Co3RQtV68SJekvFuCNloIQ-sswqVu~wl8kPsYlDHScB4O4-VqYKlC4FqMmW3tdMR~mVorSVfPYII2O0AUjguG9mDvXEEiFYO~u~NG0QHSkrCNP7I2MNqluULW~7-IOuTXmHaLgyKOLWFPIbQGUrw8hFsVZ~NLXsj0N3kA9wyiKFBJ3Sl4~VkdMvMz8sCYJokXpGSjVnBd1kqFyqJlCMtl2U~fK57za2zpuJC1uSu7dicOJGkTWuE6cmKzkM9uP1W6-WqoAhhFUWinjbQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 2. Knockdown of JUNB impairs cell-cycle progression in NPM-ALK–expressing cells. (A) Small hairpin (sh) RNA knockdown of JUNB in doxycycline-induced TonBaF.1-NPM-ALK cells. (Right) JUNB mRNA is decreased in 24-hour doxycycline-induced TonBaF.1-NPM-ALK cells stably transduced with JUNB sh-RNA–producing vector (sh-JUNB) compared with cells producing scrambled sh-RNA (sh-control). JUNB expression levels of sh-control–treated cells were set as 1 to facilitate comparison with sh-JUNB cells (bars indicate standard deviation [SD] of triplicate experiments). (Left) Cell counts (normalized to control at t = 0) demonstrate a decreased cellular proliferation of sh-JUNB–treated TonBaF.1-NPM-ALK cells at time points indicated (representative results of 3 independent experiments). (B) Knockdown experiments using small interfering (si) RNA in the human NPM-ALK–positive ALCL cell line Karpas-299. (Top panel left) Immunoblot demonstrating changes in protein levels upon transfection with scrambled siRNA (siControl), GAPDH siRNA (siGAPDH), JUNB siRNA (siJUNB), and c-JUN siRNA (si c-JUN), respectively. (Top panel right) Cell counts (normalized to control at t = 0) demonstrate a decreased cellular proliferation of Karpas-299 transfected with siJUNB at time points indicated (representative results of 6 independent experiments. SDs for 2 biologic replicates are given). (Bottom panel) Cell-cycle analysis assessed by propidium iodide staining and flow cytometry shows a decrease in the G2/M fraction associated with an increase of the G0/G1 fraction after siJUNB transfection (representative results of 3 independent experiments). (C) EMSA of Karpas-299 cytoplasmic lysates, 24 hours after siRNA transfection with siControl, siGAPDH, siJUNB, and si c-JUN. Cell lysates were incubated with a [32P]-labeled AP-1 consensus DNA (representative results of 3 independent experiments).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/9/10.1182_blood-2007-02-071258/7/m_zh80230709630002.jpeg?Expires=1767697641&Signature=qcAY3WzeDJN2Ig73aRNCe9HuRq~070NNtLkoJ9Magkoh5Ii-MEbm26B8x5oyBIAwUrL8ZKKRGyHCb8u12bJlsuTp1CPZFzN-nfS5-FtQzX270TWlYG2joNtG628d8r~skUr1VHGyG1e6WDmOxQdKEuvol0-NVkEsZWMra255MO1KWqiwj0Ksw-0IQvIL84wHAmTAnYGo~RU3LV7hBjrfA~zEE4RtgUeXfdecy~WvXocU5~U4XT3IyN~LXVro4P1RQtdDdKxcGqIMAQipoM9Fx5BgpRBRkVnZRjC3KjrLU7SX6F9GNQlVkY3dOd5GJfhX1UBA4p~FkJTOhtwMhFIF9A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. NPM-ALK regulates JunB via mTOR and MAPK signaling. (A) NPM-ALK induces activation of AKT, ERK, and mTOR. Whole-cell lysates from vector control or NPM-ALK–expressing BA/F3 cells were immunoblotted with antibodies against proteins indicated on the right. (B) JunB protein expression is attenuated in NPM-ALK–expressing cells by mTOR or MEK inhibition. SR-786: NPM-ALK–positive ALCL cell line (top panel); TB-NA: TonBaF.1-NPM-ALK cells 24 hours after induction with 2 μg/mL doxycycline (+ dox), or without doxycycline (− dox) (bottom panel). Western blot analysis was performed with antibodies indicated on the right after treatment for 24 hours with 20 nM rapamycin, 12.5 μM U0126, both agents, or control (DMSO). (C) JUNB mRNA is decreased by MEK but not mTOR inhibition. qRT-PCR was performed on SR-786 cells and doxycycline-induced TB-NA cells upon treatment as indicated (20 nM rapamycin, 12.5 μM U0126, both agents, control [DMSO]). Logarithmic scale of mean JUNB expression levels related to expression of a housekeeping gene (β-actin) as 2−ΔCT (bars indicate SEM of triplicate experiments). (D) Following 24-hour treatment with 20 nM rapamycin, 12.5 μM U0126, both agents, 48.8 μM LY294002, or control (DMSO), whole-cell lysates from SR-786 cells were blotted with antibodies against proteins indicated on the right. (E) Cell counts (normalized to control at t = 0) at indicated time points: Treatment conditions as in panel D resulted in reduced cell growth.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/9/10.1182_blood-2007-02-071258/7/m_zh80230709630003.jpeg?Expires=1767697641&Signature=fDW7w1ZJYIBCTyM2NZnqqyGgmaYI-F-az8MDoEsvgqI6j0-dqretCiFnKK5q2nUJgbwe~Yeo7yqiozxu02~FTeWZkJEl2V~KfG5puRlV43PcPjzBcDiAqce1BoNFvu8TwWeNfJTEZt3JTSojcRUVQ24kgXmOSzGSYYyLWld~MvBuu5PIcUHFq5LtNk0iCcj1mmPnPv6J5dVVMAnzr-uxQvNTtQY-K1O9HgZKrh2VqnLB1ERHg3xHUL7J2wz~IgyTFP-fBxGoHyX9a06PZaF0WV7Ep-CCslmV62hmbPKUblyp3jg5vjxH8kTDmNGLEoUHsKZqDBarkS24XcO2QBrmlg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal