Abstract
The Lutheran blood group glycoprotein, first discovered on erythrocytes, is widely expressed in human tissues. It is a ligand for the α5 subunit of Laminin 511/521, an extracellular matrix protein. This interaction may contribute to vaso-occlusive events that are an important cause of morbidity in sickle cell disease. Using x-ray crystallography, small-angle x-ray scattering, and site-directed mutagenesis, we show that the extracellular region of Lutheran forms an extended structure with a distinctive bend between the second and third immunoglobulin-like domains. The linker between domains 2 and 3 appears to be flexible and is a critical determinant in maintaining an overall conformation for Lutheran that is capable of binding to Laminin. Mutagenesis studies indicate that Asp312 of Lutheran and the surrounding cluster of negatively charged residues in this linker region form the Laminin-binding site. Unusually, receptor binding is therefore not a function of the domains expected to be furthermost from the plasma membrane. These studies imply that structural flexibility of Lutheran may be essential for its interaction with Laminin and present a novel opportunity for the development of therapeutics for sickle cell disease.
Introduction
The Lutheran glycoprotein (Lu gp) cellular adhesion molecule is widely expressed in human tissues1 and is known for carrying antigens of the Lutheran blood group system. On erythrocytes, Lu gp is expressed as 2 isoforms of 78 and 85 kDa.2 Both share a common extracellular portion that has previously been predicted to comprise 5 immunoglobulin superfamily (IgSF) domains.1 The 78-kDa isoform (also known as BCAM3 or Lu[v13]4 ) results from alternative splicing and lacks 40 C-terminal amino acids within the cytoplasmic domain that contain an SH3-binding motif, a dileucine motif responsible for basolateral targeting,5 and 5 potential phosphorylation sites.1
Lu gp binds specifically and with high affinity to the extracellular matrix (ECM) protein Laminin (Ln) containing the α5 subunit6-9 (Laminin 511 and Laminin 521 (Ln511/521) (numbering as in Aumailley et al10 ). This interaction plays a direct role in the pathophysiology of sickle cell disease by mediating adhesion of sickle cells, via Lu gp, to exposed Ln511/521 of inflamed or damaged vascular endothelium.9,11,12 Recent studies have shown that higher than normal intracellular levels of cAMP in sickle erythrocytes influence a protein kinase A–mediated or Rap1-mediated signaling pathway resulting in increased adhesion of sickle cells to the basement membrane glycoproteins Ln511/521.13,14 Furthermore, phosphorylation of the 85-kDa Lu gp at S621 in epinephrine-stimulated K562 cells alters adhesion to Ln511/521.15
The Ln-binding activity of Lu gp is a property of the amino terminal region of Lu gp, and has been localized to a region predicted to form the first 3 IgSF domains (D1D2D3).6,16 The complementary binding site on Ln is located within a large carboxyl-terminal globular domain of the α5 subunit, known to comprise 5 Laminin-type G (LG) modules, LG1 to LG5.17,18 These modules, present at the carboxy termini of all Ln α chains, have also been shown to regulate binding to at least 6 integrins18-20 in addition to α-dystroglycan and heparin.21-23 Binding to the latter 2 ligands results from positively charged residues on Ln and regions of negative charge (acidic sugar residues) on heparin and α-dystroglycan.24,25
Here we use a multidisciplinary approach to explore the molecular basis of the interaction between Lu gp and Ln511/521. We describe the elucidation of a structure for the Ln-binding portion of the Lu gp molecule. This structure comprises a high-resolution crystal structure for domains 1 and 2 (D1D2) to which has been added a predicted model for domain 3 (D3) by fitting to a molecular envelope calculated from small-angle x-ray scattering (SAXS) data. The location of these domains within the overall extracellular form of Lu is also demonstrated by SAXS. Secondly, site-directed mutagenesis has been used to examine residues suspected to be involved in the interaction with Ln. This led to the identification of amino acid residues comprising the Ln511/521-binding site on Lu gp. An unexpected finding is that, unlike most cell-cell interactions that characteristically result from the formation of cell adhesion pairs between the membrane-distal domains of cell adhesion molecules (eg, CD2:CD5826 and CD2:CD4827 ), Lu gp uses a more membrane-proximal interdomain junction region to form a critical part of its Ln-binding site.
Materials and methods
Synthesis, purification, and crystallization of Lu gp
The extracellular domains of human Lu gp were expressed as Fc fusion proteins in NS0 cells using the procedures as described.28 Residues 1 to 330 (D1D2D3), residues 1 to 515 (Lu gp), were included in the expressed proteins, immediately followed at the C-terminus by the tobacco etch virus protease (TEV) cleavage site VDENLYFQG and human IgG Fc immunoglobulin fragment. Secreted fusion proteins were purified from cell supernatant using Protein A Sepharose, and the Fc fragment was liberated by digestion with recombinant TEV (Invitrogen, Paisley, United Kingdom) and removed on Protein A Sepharose.
The D1D2 fragment was liberated by chymotrypsin digestion of the purified D1D2D3, using a 1:100 enzyme/Lu ratio at pH 7.4 for 18 hours at 18°C. Chymotrypsin was removed with a benzamidine column (Amersham Biosciences, Amersham, United Kingdom) prior to a final gel filtration purification step (HiLoad 16/60, Superdex 75 prep grade; Amersham Biosciences).
For crystallization trials, proteins were concentrated to 8 mg/mL in 20 mM HEPES (pH 7) buffer. Diffraction quality hexagonal crystals were grown from a mother liquor containing 18% (wt/vol) PEG4000, 0.1 M sodium citrate (pH 5.6), 0.2 M ammonium acetate. A second monoclinic crystal form was obtained from 12% (wt/vol) PEG20000, 0.1 M MES (pH 6.4), 6% (wt/vol) hexane-1,2-diol. Heavy atom derivatives of crystals were prepared by soaking in well solutions supplemented with either 10 mM potassium tetrachloro platinate (II) for 24 hours or 100 mM potassium dicyanoaurate (I) for 72 hours. Diffraction data were collected at the Daresbury synchrotron radiation source (SRS) on beamlines 14.1 and 10.1 (Table 1). Crystals of the monoclinic form (one molecule per asymmetric unit) generally produced higher quality diffraction data and were used for the experimental phase determination. Heavy atom sites were located with SHELXD29 and refined with MLPHARE.30 Density modification was performed using DM30 and ARP/warp.31 The model was refined against the native amplitudes and experimental phases using REFMAC5.30 The hexagonal crystal form was solved by molecular replacement using PHASER32 using separate domains from the monoclinic structure as search models. The structure was initially refined with CNS,33 using a simulated annealing protocol, then fully refined with REFMAC5. The validity of the structures was assessed with MOLPROBITY.34 Refinement statistics are summarized in Table 1.
Diffraction data processing and refinement statistics for D1D2 crystal forms
| . | Hexagonal native . | Monoclinic native . | Au derivative, high-energy remote . | Au derivative, low-energy remote . | Pt derivative, high-energy remote . | Pt derivative, low-energy remote . |
|---|---|---|---|---|---|---|
| Wavelength, Å | 1.488 | 1.285 | 1.01 | 1.042 | 1.071 | 1.08 |
| Spacegroup | P3221 | P21 | P21 | P21 | P21 | P21 |
| Resolution range, Å | 44.6-2.2 (2.28-2.2) | 35.3-1.7 (1.76-1.7) | 35.7-1.8 (1.86-1.8) | 35.7-1.8 (1.86-1.8) | 36.1-2.5 (2.59-2.5) | 33.5-2.5 (2.59-2.5) |
| Redundancy | 5.6 (1.8) | 6.4 (2.0) | 8.6 (2.7) | 8.9 (3.3) | 6.8 (3.0) | 5.9 (1.8) |
| Completeness | 90.1 (41.3) | 89.1 (48.3) | 89.5 (50.3) | 90.5 (49.6) | 93.6 (70.3) | 85.4 (32.6) |
| Rmerge | 10.2 (48.2) | 5.9 (26.4) | 9.8 (23.3) | 6.9 (20.0) | 11.8 (26.3) | 8.9 (32.6) |
| I/sigI | 14.1 (1.2) | 26.5 (2.5) | 19.7 (2.8) | 29.2 (3.7) | 16.5 (1.1) | 15.9 (2.6) |
| Isomorphous Rcullis | — | — | 0.51/0.65 | 0.52/0.67 | 0.85/0.91 | 0.87/0.92 |
| (acentrics/centrics) | ||||||
| Anomalous Rcullis | — | — | 0.83 | 0.88 | 0.95 | 0.97 |
| Initial phase figure-of- | — | 0.73 | — | — | — | — |
| merit | ||||||
| Unique reflections | 18741 | 23563 | — | — | — | — |
| Rwork | 0.205 | 0.168 | — | — | — | — |
| Rfree | 0.251 | 0.21 | — | — | — | — |
| Non-H atoms | 3643 | 2096 | — | — | — | — |
| Bond length RMSD, Å | 0.018 | 0.015 | — | — | — | — |
| Bond angle RMSD, ° | 1.498 | 1.678 | — | — | — | — |
| . | Hexagonal native . | Monoclinic native . | Au derivative, high-energy remote . | Au derivative, low-energy remote . | Pt derivative, high-energy remote . | Pt derivative, low-energy remote . |
|---|---|---|---|---|---|---|
| Wavelength, Å | 1.488 | 1.285 | 1.01 | 1.042 | 1.071 | 1.08 |
| Spacegroup | P3221 | P21 | P21 | P21 | P21 | P21 |
| Resolution range, Å | 44.6-2.2 (2.28-2.2) | 35.3-1.7 (1.76-1.7) | 35.7-1.8 (1.86-1.8) | 35.7-1.8 (1.86-1.8) | 36.1-2.5 (2.59-2.5) | 33.5-2.5 (2.59-2.5) |
| Redundancy | 5.6 (1.8) | 6.4 (2.0) | 8.6 (2.7) | 8.9 (3.3) | 6.8 (3.0) | 5.9 (1.8) |
| Completeness | 90.1 (41.3) | 89.1 (48.3) | 89.5 (50.3) | 90.5 (49.6) | 93.6 (70.3) | 85.4 (32.6) |
| Rmerge | 10.2 (48.2) | 5.9 (26.4) | 9.8 (23.3) | 6.9 (20.0) | 11.8 (26.3) | 8.9 (32.6) |
| I/sigI | 14.1 (1.2) | 26.5 (2.5) | 19.7 (2.8) | 29.2 (3.7) | 16.5 (1.1) | 15.9 (2.6) |
| Isomorphous Rcullis | — | — | 0.51/0.65 | 0.52/0.67 | 0.85/0.91 | 0.87/0.92 |
| (acentrics/centrics) | ||||||
| Anomalous Rcullis | — | — | 0.83 | 0.88 | 0.95 | 0.97 |
| Initial phase figure-of- | — | 0.73 | — | — | — | — |
| merit | ||||||
| Unique reflections | 18741 | 23563 | — | — | — | — |
| Rwork | 0.205 | 0.168 | — | — | — | — |
| Rfree | 0.251 | 0.21 | — | — | — | — |
| Non-H atoms | 3643 | 2096 | — | — | — | — |
| Bond length RMSD, Å | 0.018 | 0.015 | — | — | — | — |
| Bond angle RMSD, ° | 1.498 | 1.678 | — | — | — | — |
Values in parentheses refer to the highest resolution shell.
— indicates not applicable.
Lu gp D3 homology model
Sequence comparisons indicated that D3 was most similar to the Ig3 domain of perlecan (PDB ID 1gl4, chain B35 ), a known I-set IgSF domain, and this was used as the structural template for the model of D3. Sequences were aligned with ClustalW and adjusted manually; secondary structure was predicted with the Jpred36,37 and SAMT0238 servers. The sequence alignment and secondary structure predictions were used as input for Modeller39 to generate the 3-dimensional model.
Small-angle x-ray scattering analysis
Full SAXS methods can be found in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Briefly, SAXS data of D1D2D3 were collected on beamline X33 at the European Molecular Biology Laboratory (EMBL) of the Deutsches Elektronen Synchrotron (DESY) using a wavelength of λ = 0.15 nm, covering a scattering range of 0.16 nm−1 < q < 4.85 nm−1, where q = 4π sinθ/λ; 2θ is the scattering vector. Sample concentrations were 4.375 mg/mL, 8.75 mg/mL, and 17.5 mg/mL; samples were supplemented with 1 mM dithiothreitol and exposed for 60 seconds; scattering data from the buffer solution was also collected. The scattering images were integrated to one-dimensional linear profiles using in-house software at station X33. Scattering data were processed with the ATSAS2.1 software suite as previously described.40
SAXS data for the entire extracellular portion of Lu (D1-D5) were collected using the modified version of a NanoSTAR (Bruker AXS, Karlsruhe, Germany) at the University of Aarhus; see Pedersen41 for details. The sample concentration was 15 mg/mL and the scattering range was 0.085 < q < 3.5 nm−1. The 2-dimensional data were azimuthally averaged to one-dimensional linear profiles using Bruker SAXS software. Data were processed as described in the previous paragraph, except that buffer scattering subtraction and transformation to absolute units using the scattering of water as a standard were performed with in-house software at the University of Aarhus.
Preparation of mutant and native Lu gp Fc fusion proteins (Lu gpFc)
Point mutations were inserted into human Lu gp cDNA clones42 encoding the 5 extracellular domains in pIg vector by polymerase chain reaction (PCR) amplification as described.43 Mutant clones were confirmed by DNA sequence analysis. Native and mutant Lu gpFc and Muc-18 Fc (a gift from Dr Simmons [Glaxo SmithKline, Harlow, United Kingdom]) were expressed in COS-7 cells, purified using protein A Sepharose, and quantified as described.43
Lu gp binding Ln511/521 ELISA
Washes and protein dilutions were performed in phosphate-buffered saline with 0.2% bovine serum albumin (assay buffer). All protein dilutions were added in 50 μL and incubations were at 37°C with shaking for one hour unless stated otherwise. Immulon-4 96 well plates (Dynes Technologies, West Sussex, United Kingdom) were coated with 0.25 μg/well goat-anti–human-Fc (Jackson ImmunoResearch, Cambridgeshire, United Kingdom) in 0.1 M bicarbonate buffer (pH 9.6) for 24 hours at 4°C. After 3 washes 5, 0.5, 0.05, and 0.005 nM native or mutated Lu gpFc was added and the plate incubated. After one wash, the plate was blocked for 30 minutes at room temperature in assay buffer containing 5% human AB serum followed by one wash. Ln511/521 (Chemicon, Hampshire, United Kingdom) at 5 nM was added and the plate incubated. After 3 washes, a 1 in 100 dilution of rabbit antilaminin (Sigma, Dorset, United Kingdom) was added and the plate incubated, and after 3 washes a 1 in 1000 dilution of horseradish peroxidase–linked swine antirabbit (DAKO, Cambridgeshire, United Kingdom) was added, incubated, and washed a further 3 times. The plate was developed using 3,3′,5,5′-tetramethylbenzidine and 3% H2O2 in 0.1 M acetate/citrate buffer (pH 6), stopped with 2 M H2SO4, and read at 450 nm. The enzyme-linked immunosorbent assay (ELISA) was controlled by coating wells with 5 nM Ln511/521 and by addition of 5 nM Ln511/521 to captured Muc18 Fc.
Surface plasmon resonance assays
All assays were performed at 25°C using a Biacore X (Biacore, Stevenage, United Kingdom), a CM5 chip with Protein A immobilized on its surface and a flow rate of 30 μL per minute. Running buffer was phosphate-buffered saline (pH 7.4) containing 0.05% Tween 20, and regeneration buffer was 0.1 M glycine (pH 2). Native or mutant Lu gpFc was captured onto the chip until a change of 40 response units was observed (0.25 pm). Ln511/521 was added in a 100-μL injection of 10 nM (1 pm).
Results
Crystal structure of Lu gp D1D2
Extensive attempts to crystallize fragments of Lu gp comprising either all 3 N-terminal domains (residues 1-330) or the entire extracellular region were unsuccessful, consistent with innate molecular flexibility in Lu gp. Instead, a stable fragment of Lu gp consisting of domains 1 and 2 (D1D2) was produced by chymotryptic digestion of the 3-domain construct. This fragment was purified and crystallized in 2 crystal forms (Table 1). In the refined crystal structures, the asymmetric unit of the monoclinic form contains one molecule, whereas there are 2 molecules in the hexagonal asymmetric unit. Together these provide 3 separate views of Lu gp D1D2. Coordinates and structure factors for the D1D2 crystal structures are available from the Protein Data Base (accession codes 2PET [monoclinic form] and 2PF6 [hexagonal form]).
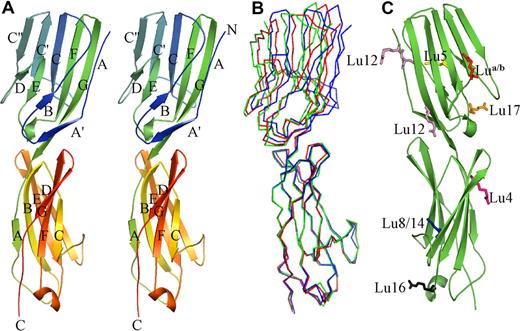
The crystal structures show the amino terminal portion of Lu gp consists of 2 IgSF domains (Figure 1), which together form a monomeric straight rod approximately 85-Å long and 25-Å wide. There is limited flexibility between the 2 domains, the relative positions of which are seen to vary by up to 12° when the 3 nonidentical molecules from the 2 crystal forms are compared (Figure 1B). Domain 1 (D1) is formed from residues 1 to 111 and is, as predicted, a member of the V-set of immunoglobulin domains (Figure S1). It contains the conserved disulphide bond between strands B and F, each of the B/C, C′/C″ and F/G loops—the equivalent of complementarity determining loops in the antibody V-domains—are relatively short. The C/C′ loop projects away from the body of the domain and appears to be held in this orientation primarily by a salt bridge formed between Arg44 and Glu105. The base of the domain is intimately associated with domain 2 (D2). The domains are linked by a short run of hydrophobic amino acids (112Val-Phe-Ala114) that are buried within the interface. Unexpectedly D2 (residues 115-229) lacks A′, C′, and C″ strands, ruling out its original sequence-based classification as a V-set domain (Figure S1). Despite the increased length of the polypeptide chain in this domain, it is most like a C1-set domain, unusual outside of immune system proteins. The additional residues are incorporated primarily within extended A/B, C/D, and E/F loops, all of which are at the base of the domain. One notable feature is the presence of a short helical segment within the E/F loop. All of these features lead to an extended surface at the C-terminal end of the domain, which lies adjacent to the interface with D3.
Structure of Lu gp D1D2. (A) Graphic representation in stereo showing the Lu gp structure. D1 is shown at the top and D2 at the bottom. The strands in each domain are labeled, as are the amino (N) and carboxy termini (C). (B) Overlay of Cα traces from the 3 independent copies of D1D2 in the 2 crystal structures. Orientation is as in panel A. (C) Location of residues that are altered in the known inherited forms of Lu gp that can be attributed to D1D2. Mutated residues are shown in stick form, and are colored as follows: Lu a/b is shown in red; Lu4, purple; Lu5, yellow; Lu8/14, blue; Lu12, pink; Lu16, gray; and Lu17, orange.
Structure of Lu gp D1D2. (A) Graphic representation in stereo showing the Lu gp structure. D1 is shown at the top and D2 at the bottom. The strands in each domain are labeled, as are the amino (N) and carboxy termini (C). (B) Overlay of Cα traces from the 3 independent copies of D1D2 in the 2 crystal structures. Orientation is as in panel A. (C) Location of residues that are altered in the known inherited forms of Lu gp that can be attributed to D1D2. Mutated residues are shown in stick form, and are colored as follows: Lu a/b is shown in red; Lu4, purple; Lu5, yellow; Lu8/14, blue; Lu12, pink; Lu16, gray; and Lu17, orange.
The 18 identified antigens of the Lutheran blood group system are predominantly associated with single-nucleotide polymorphisms resulting in single amino acid changes within Lu gp.44 A study of 11 of these antigens revealed that in 7 cases these changes could be located to residues in D1 or D2,44 the sites of which may now be visualized (Figure 1C). The prevalence of these changes in the 2 amino-terminal domains is consistent with their ready exposure at the cell surface, implying that they may lie furthermost from the membrane. These allelic changes appear to be distributed across all of the available faces of D1D2 and all occur at surface-exposed regions including both loop and sheet locations. In all cases, the observed amino acid changes are not expected to be deleterious for the overall Lu gp structure.
Construction of an homology model for D3
Chymotryptic digestion of D1D2D3 consistently resulted in proteolysis of D3, preventing an experimental determination of its structure. We therefore constructed a molecular model of D3 (residues 236-324), based on its sequence homology with known I1-set domains. Note that this differs from the original classification of D3 as a C2-set domain.1 The 2 single-point mutations (S244 and T271) associated with D3 inherited blood group antigens (LU6/LU9 and LU20) were both found to be located on the surface of the final model, an assumption not made throughout the modeling process.
SAXS analysis of Lu gp
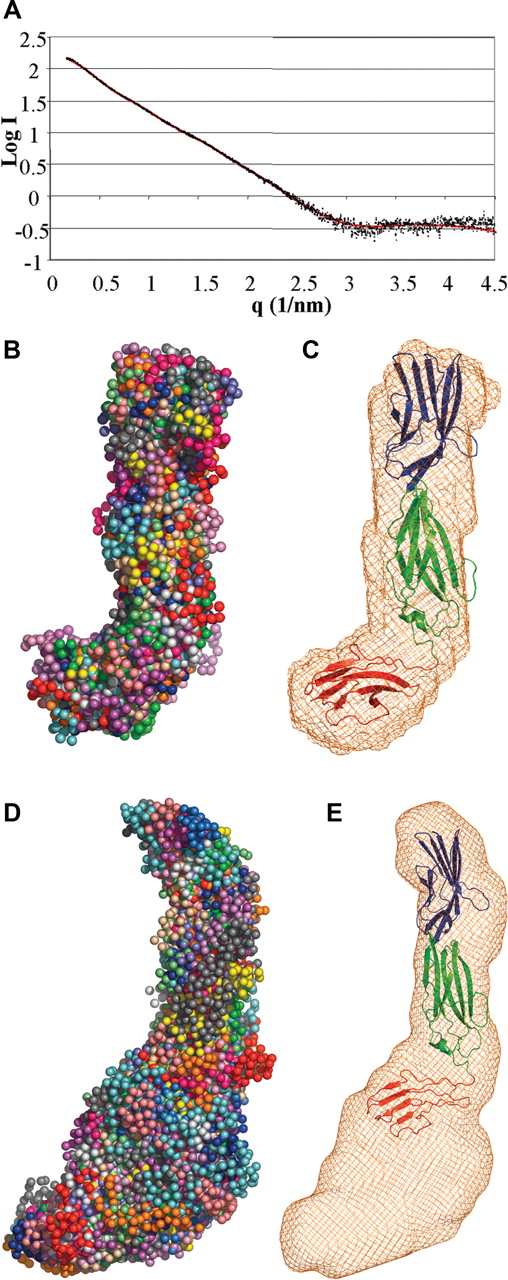
To delineate the placement of D3 with respect to D1D2, we performed a SAXS analysis of the D1D2D3 construct. Two methods were used to independently derive an overall conformation of D1D2D3. First, the scattering data (Figure 2A) were fitted with ab initio bead models using 2 separate programs, DAMMIN45 and GASBOR.47 In each case, chi values of approximately 1.9 were obtained, and both methods generated very similar hockey-stick shapes (Figure 2B). The chi value is a measure of the discrepancy between the experimental data and the theoretic scattering generated by the model; a good fit should return a value less than 3 (with absolute equivalence = 1). Second, the crystal structures of Lu gpD1D2 and the model of D3 were fitted as separate rigid bodies to the scattering data using the software SASREF.46 The resulting chi values ranged from 1.7 to 1.9. All 3 crystal structures of D1D2 gave similar results. When superimposed, the individual fits show very similar gross conformations with a significant bend angle between domains 2 and 3 (Figure 2C), although the exact orientation of D3 with respect to D2 is insufficiently defined to assign a single unique conformation. Similarly, the conformation of the D2-D3 linker peptide is not distinguished in this analysis.
SAXS analysis of Lu gp D1D2D3. (A) Experimental scattering data for Lu gp D1D2D3 plotted in black as a function of q and compared with a theoretic fit generated with SASREF.46 (B) Shapes obtained using a bead model using the program GASBOR47 from 20 independent simulations, each differently colored. The average shape is shown as orange wire-frame density in (C) representative rigid-body fit of the D1D2 crystal structure and D3 model to the scattering data, overlaid on average density obtained as in panel B. D1 is shown in blue; D2, green; and D3, red. (D) Shapes obtained from the scattering curve for Lu gp (D1-D5) as described in panel B. The average shape is shown as orange wire-frame density in (E) overlay of the D1D2D3 model on the envelope derived for Lu gp (D1-D5). D4 and D5 are expected to occupy the additional density at the base of the figure, proximal to the cell membrane.
SAXS analysis of Lu gp D1D2D3. (A) Experimental scattering data for Lu gp D1D2D3 plotted in black as a function of q and compared with a theoretic fit generated with SASREF.46 (B) Shapes obtained using a bead model using the program GASBOR47 from 20 independent simulations, each differently colored. The average shape is shown as orange wire-frame density in (C) representative rigid-body fit of the D1D2 crystal structure and D3 model to the scattering data, overlaid on average density obtained as in panel B. D1 is shown in blue; D2, green; and D3, red. (D) Shapes obtained from the scattering curve for Lu gp (D1-D5) as described in panel B. The average shape is shown as orange wire-frame density in (E) overlay of the D1D2D3 model on the envelope derived for Lu gp (D1-D5). D4 and D5 are expected to occupy the additional density at the base of the figure, proximal to the cell membrane.
A similar SAXS analysis was also performed on the overall Lu gp (D1-D5) construct. It is evident from the calculated envelopes and shapes that Lu gp forms an extended structure with a distinctive bend to one side of the midregion (Figure 2D). The derived D1D2 and D1D2D3 structures overlay well at one end of this shape (Figure 2E). Domains 4 and 5 are also expected to adopt IgSF fold, however, due to limited experimental evidence we feel it would be too speculative to include them in our model. Nonetheless, their approximate placement relative to D1D2D3 is evident.
Mutagenesis of acidic residues on D2 and D3 diminishes Ln511/521 binding
There are known areas of positively charged amino acids on LG domains, so we hypothesized that negatively charged residues on Lu gp would be responsible for Ln binding. This is consistent with the observations that interactions between positively charged residues on Ln and negative charges on both heparin and α-dystroglycan provide the basis for these molecular interactions.24,25 Analysis of a homology model of Lu gp initially allowed us to target surface-exposed clusters of aspartic and glutamic acid residues for mutation; the precise locations of acidic residues on D2 were subsequently demonstrated by the crystal structures. The majority of residues selected are conserved or identical in the mouse Lu gp homologue. Each mutant Lu gp was detected by Western blotting with monoclonal anti-Lu (BRIC 221 and BRIC 224) and an anti-Fc antibody (Figure S2). Because the epitopes for BRIC 221 and 224 are conformationally dependent and are destroyed in reducing conditions, these results show none of the mutations markedly altered the structure of Lu gpFc.
The Ln511/521-binding properties of the 13 mutant Lu proteins were examined by an ELISA using 4 Lu gp coating concentrations. Over the complete range of Lu gpFc concentrations, the D312A mutation caused a severe reduction in Ln511/521 binding, E309A and D310A showed a marked reduction, whereas E132A/D133A, D198A/D199A, E269A, E281A, and D316A caused only a slight decrease in binding (Figure 3A; Figure S3). At 0.05 nM, the native Lu gp was exposed to saturating amounts of Ln511/521, so this concentration was used to determine the different levels of adhesion to Ln511/521 of the various Lu gp mutants (Figure 3B; Figure S3). All other mutant proteins had the same or similar levels of binding to Ln511/521 as that obtained with native Lu gpFc (Figure 3B).
Mutation of acidic residues of Lu gp reduces Ln511/521 binding. (A) Representative ELISA titrations of 5, 0.5, 0.05, and 0.005 nM Lu gp binding to 5 nM Ln511/521 for the E309A, D310A, D312A, and D315A (solid line) mutations in comparison with native Lu gp (dashed line). Standard deviation for each point is less than 0.1. ELISA results of all other mutations can be seen in Figure S3. (B) The level of binding to Ln511/521 of mutant Lu gp compared with the native protein at 0.05 nM measured in an ELISA. Proteins were assayed in duplicate and the results shown are the mean of 2 separate ELISA plates and are expressed as the percentage of the absorption seen from wells containing native Lu gpFc (OD450 = 0.95). (C) Ln511/521 binding assessed by surface plasmon resonance using a Biacore X. Shown is the mean (± SEM) change in response units (RU) over the course of a 100-μL injection of 10 nM Ln511/521 for 2 assays per protein. In both panels B-C, Ln511/521 binding to Muc18 acted as a negative control. (D) Overlaid sensorgrams for a representative sample of the different proteins showing the association and dissociation curves of Ln511/521 binding to Lu gpFc. Vertical arrows indicate the beginning and end points of the Ln511/521 injection.
Mutation of acidic residues of Lu gp reduces Ln511/521 binding. (A) Representative ELISA titrations of 5, 0.5, 0.05, and 0.005 nM Lu gp binding to 5 nM Ln511/521 for the E309A, D310A, D312A, and D315A (solid line) mutations in comparison with native Lu gp (dashed line). Standard deviation for each point is less than 0.1. ELISA results of all other mutations can be seen in Figure S3. (B) The level of binding to Ln511/521 of mutant Lu gp compared with the native protein at 0.05 nM measured in an ELISA. Proteins were assayed in duplicate and the results shown are the mean of 2 separate ELISA plates and are expressed as the percentage of the absorption seen from wells containing native Lu gpFc (OD450 = 0.95). (C) Ln511/521 binding assessed by surface plasmon resonance using a Biacore X. Shown is the mean (± SEM) change in response units (RU) over the course of a 100-μL injection of 10 nM Ln511/521 for 2 assays per protein. In both panels B-C, Ln511/521 binding to Muc18 acted as a negative control. (D) Overlaid sensorgrams for a representative sample of the different proteins showing the association and dissociation curves of Ln511/521 binding to Lu gpFc. Vertical arrows indicate the beginning and end points of the Ln511/521 injection.
The mutant proteins were also assayed for Ln511/521 binding using a Biacore X (Figure 3C,D) and sensorgrams showed that the effect caused by each mutation is similar to that observed by ELISA (Figure 3A-B). The Biacore assays indicate that mutations affecting Ln511/521 binding slow the rate at which Ln511/521 binds to Lu gpFc, whereas the rate at which Ln511/521 dissociates is not affected (Figure 3C). Compared with our earlier study,6 the rate of dissociation of Lu gpFc and Ln511/521 is somewhat slower. In the previous published study,6 an Iasys optical biosensor and a different source of Ln511/521 were used, making direct comparisons between the 2 studies difficult. Association and dissociation of Lu gp and the Ln511/521 used in this study was measured on the Biacore X using Ln511/521 injections at 50 nM, 20 nM, 10 nM, 5 nM, and 2 nM and the calculated KD was 7.2 nM, which is comparable with previously reported values (7.9 nM6 and 10.8 nM16 ). It was not possible to obtain a set of binding curves to calculate KD for the mutant forms of Lu gp that showed a reduction in Ln binding because of the constraints of the limited availability and solubility of Ln511/521.
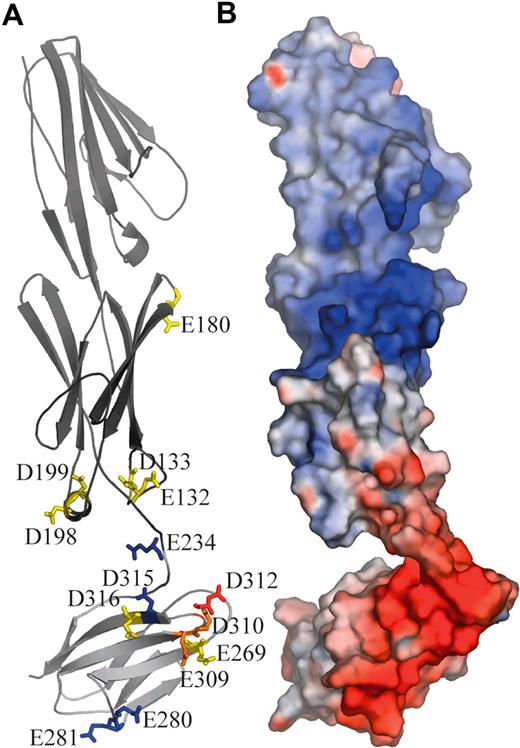
The results show E132A/D133A, D198A/D199A, E269A, D316A, and to a much greater extent E309A, D310A, and D312A, inhibit Ln511/521 binding to Lu gp. These residues cluster within the A/B and E/F loops at the base of D2 and B/C and F/G loops at the top of D3. The location and importance of each amino acid is depicted in Figure 4.
Location of charged amino acid residues involved in interactions with Ln511/521 on the surface of Lu gp. (A) Lu gp domains D1D2 (dark gray) and D3 (light gray) are shown in an orientation consistent with the SAXS envelope. Indicated are the positions of acidic residues that have been mutated to alanine. The residues are color coded according to their effect on Ln511/521 binding: blue indicates no effect; yellow, slight effect; orange, marked effect; and red, severe effect. (B) Lu D1D2D3 in the same orientation, shown as an electrostatic surface.
Location of charged amino acid residues involved in interactions with Ln511/521 on the surface of Lu gp. (A) Lu gp domains D1D2 (dark gray) and D3 (light gray) are shown in an orientation consistent with the SAXS envelope. Indicated are the positions of acidic residues that have been mutated to alanine. The residues are color coded according to their effect on Ln511/521 binding: blue indicates no effect; yellow, slight effect; orange, marked effect; and red, severe effect. (B) Lu D1D2D3 in the same orientation, shown as an electrostatic surface.
Mutagenesis of the D2-D3 linker affects Ln511/521 binding
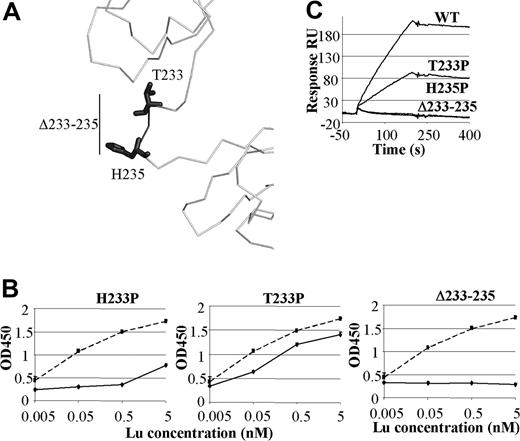
The results of site-directed mutagenesis described above are consistent with previous studies that used domain-deletion mutants of Lu gp to show that the binding site for Ln511/521 is within the first 3 domains.6,16 Despite containing key residues involved in binding Ln511/521 Lu gp proteins containing D2 without D3, and more significantly, D3 without D2, were unable to support binding of Ln511/521.6,16 The observation that both D2 and D3 are required for Ln511/521 binding raises the question of the importance of the linker or hinge sequence between D2 and D3 in defining the structure of Lu gp. We constructed 2 Lu gpFc mutants in which residues in the hinge were replaced with a proline (T233P and H235P) and a third mutant that had 3 residues removed from the hinge (Δ233-235) (Figure 5A). Western blotting with BRIC 221 and 224 confirmed these molecules were folded correctly (Figure S2). Ln511/521 binding by mutant T233P was markedly reduced and that of H235P and Δ233-235 was abolished (Figures 4B,C). Interestingly, even at high Lu gpFc concentrations in the ELISA, the Δ233-235 mutation did not bind Ln511/521 (Figure 5B). These data are consistent with an essential role for the linker region in allowing D2 and D3, and the rest of the Lu gp molecule, to adopt a conformation capable of binding Ln511/521.
Ln511/521 binding to D2-D3 linker mutants. (A) Three proteins, T233P, H235P, and Δ233-235 (darker shade), containing mutations within the domain 2 to 3 hinge were made. (B) Representative ELISA titrations of hinge region mutants to Ln511/521 in comparison with binding to the native protein. Standard deviations were less than 0.1 OD units for any point. (C) The sensorgrams of representative Biacore assays with these mutants are shown.
Ln511/521 binding to D2-D3 linker mutants. (A) Three proteins, T233P, H235P, and Δ233-235 (darker shade), containing mutations within the domain 2 to 3 hinge were made. (B) Representative ELISA titrations of hinge region mutants to Ln511/521 in comparison with binding to the native protein. Standard deviations were less than 0.1 OD units for any point. (C) The sensorgrams of representative Biacore assays with these mutants are shown.
Taken in their entirety, these mutagenesis data suggest the primary binding site for the Ln α5 chain is defined by negatively charged residues at the top of D3 and to a lesser extent at the base of D2. An appropriate presentation of both D2 and D3 for Ln511/521 binding is essential and appears to be dictated by the linker between these domains.
Discussion
Ln has previously been shown to bind to its other ligands (heparin, α-dystroglycan) via positively charged basic residues on the LG domains adhering to areas of negative charge on its ligands.21,22 The results presented here are consistent with a similar mechanism for the Ln α5 chain-Lu gp interaction. Inspection of the crystallographic structures of D1D2 and the model of D3 of Lu gp revealed dominant areas of negative electrostatic potential that we hypothesized form the Ln-binding site. Many of the acidic residues in this region are conserved in the murine homologue that also binds to human Ln511/521.6 When negatively charged residues in the patch were mutated to alanine and the mutant proteins assessed for Ln511/521 binding by both ELISA and Biacore assay, residue D312 and, to a lesser extent, E309 and D310 were identified as critical residues. Other negatively charged residues in the vicinity of these 3 also had small effects on binding. The multiple mutation E309A/D310A/D312A/ D315A/D316A in an ELISA assay did not bind to Ln511/521 at all, whereas a multiple mutation within another area of D3 (D277A/D278A/E280A/E281A) had little effect on Ln511/521 binding (Figure S3). This is further evidence of the importance of this area on Lu gp for Ln512/521 binding. When the positions of all the mutations made in this study are mapped to the crystallographic structure of D2 and the homology model of D3, those that affect the binding to Ln511/521 form a cluster at the domain interface (Figure 4), with the single exception of E180, the mutation of which caused an effect only in the surface plasmon resonance assay and not the ELISA. In the orientation of the D1D2D3 structure, along with the D1-D5 structure, shown in Figure 4 this cluster corresponds to a protrusion formed by the bend between D2 and D3, akin to a spur on the heel of a boot. We speculate that the negative charge on D312 interacts with a positively charged amino acid on the Ln α5 chain and that this charged interaction is the primary determinant of adhesion between the 2 molecules. The other acidic amino acids identified here could potentially produce a localized negative charge on Lu gp around D312 and facilitate docking by interaction with other positive charges on the Ln α5 chain. The close homology between the sequence of human and murine Lu gp6 suggests that murine Lu gp has a similar structure to its human counterpart and binds Ln511/521 in an identical manner.
Our results demonstrate not only that the binding site for the Ln α5 chain is defined by negatively charged residues on D2 and D3 but also that the linker region between these domains is critical for binding. The approximately 8 amino acids that comprise the link between D2 and D3 are likely to have an open conformation as evidenced by the susceptibility of this region to proteolysis and by SAXS analysis that shows that the Lu gp structure is nonlinear in this region. Unlike at the D1D2 junction, these residues are not predominantly hydrophobic (HYPTEHVQ), indicating the linker is unlikely to be buried within a tight interdomain interface. We have also shown that by placing a proline in this linker region (H235P) or by removing 3 amino acids (Δ233-235) and shortening the linker, Lu gp binding to Ln511/521 is totally abolished. These data could be interpreted as indicating residues T233, E234, and H235 are directly involved in binding to Ln511/521. However, this seems unlikely because mutation of E234A has only a minor effect on binding (Figure 3A-B). It is more plausible that to adhere to the Ln α5 chain Lu gp D1D2 needs to be in a suitable spatial orientation relative to D3 and possibly also the stem formed by D4 and D5.
One interpretation of an extended conformation at the D2/D3 interface is that this region is likely to exhibit interdomain flexibility. Although the crystal structures also show a degree of flexibility at the D1/D2 interface, the demonstration that domains 2 and 3 can readily fold when up to 3 amino acids are removed from the linker region implies a more open association between these domains. This is reminiscent of the 4-domain tandem structure of CD448 where domains 1 and 2 are tightly associated, as are domains 3 and 4, whereas an extended linker at the domain 2/domain 3 interface leads to molecular flexibility at this junction. Similarly, CD2, which has a 4 amino acid linker between IgSF domains 1 and 2, is also believed to undergo hinge bending shifts affecting domain orientation49 and exposing a buried antibody epitope during T-cell activation.50 In Lu gp, it is unclear whether the potential flexibility this extended linker may generate is essential for Ln binding. Nonetheless, the demonstration that the binding site straddles the adjacent ends of D2 and D3 implies that only a discrete arrangement of the domains would form a coherent binding site; variations from this position would be expected to be deleterious for Ln binding. The assumption that the D2/D3 boundary is a source of flexibility therefore suggests an association between these movements and effective Ln binding. A precedent for this proposed mechanism is provided by the killer cell inhibitory receptors in which a linker region between 2 IgSF domains forms the MHC-binding interface, with binding modulated by differences in the relative domain orientations.51 The potential for domain-domain movement within Lu gp provides a possible explanation for the increase in adhesion to Ln511/521 observed in cells expressing Lu gp in response to “inside-out” signaling events.13-15
A common feature of cell-cell adhesion pairs for which structures are available is the exploitation of the broad faces presented by amino-terminal IgSF domains (usually formed by the C″C′CFG sheet) to generate extensive contact surfaces.52 These are typified in the CD2:CD58 pair26 where some 1200 Å2 of surface area is buried in a relatively flat interface within the complex. A recent exception is the SIRPα receptor that instead uses the complementarity-determining loop regions as the adhesion site, although these are at the amino terminal end of the receptor and assumed to be furthermost from the cell membrane.52 Nonetheless, these and similar electrostatic interactions frequently generate only low-affinity binding. The Lu gp:Ln511/521 interaction differs substantially. The data presented here show this to be a high-affinity (KD ∼ 10 nM) interaction arising from a cluster of residues contributed from adjacent loop regions of 2 domains. It is feasible that enclosure of the ligand through domain movements may contribute to the slow off-rate of the complex. Indeed the Lu gp point mutations constructed in this study seemed only to have an effect on the on-rate of Ln binding and not the off-rate (Figure 3D). Unlike many cell adhesion molecules, though not all,53 the binding surfaces in Lu gp are not located on the amino-terminal domain, which would be most distal from the cell surface in the tandem arrangement implied by the overall shape of Lu gp (D1-D5). Access to the binding site would therefore require either interdigitation of extended Ln and Lu gp proteins when cells are located on the ECM or, alternatively, considerable flexing of Lu gp at the D2/D3 boundary to present the binding surface to the ligand. The extended structure observed for D1-D5 suggests that Lu gp does not adopt a compact structure akin to the horseshoe arrangement observed in both axonin54 and hemolin55 that could place the D2/D3 boundary furthermost from the cell surface. The predominance of sites for Lutheran blood group antigens on all faces of D1 and D2 (Figure 1C) also implies that these domains are fully exposed at the erythrocyte surface, again inconsistent with a compact structure. The angle generated by the D2-D3 junction as described in this study may promote interactions between the D1D2 fragment and other ECM components. This angle is similar to that observed between the IgSF domains 2 and 3 of the neural cell adhesion molecule NCAM, for which a heparin-binding site has also been located to domain 2.56 There is currently no evidence, however, that Lu gp can undergo the zipperlike formation of homophilic interactions described for NCAM to build multimolecular adhesion complexes.
Our findings provide clear evidence that Lu gp interaction with the Ln α5 chain is mediated by a negatively charged patch on the tip of Lu gp extracellular D3 and base of D2 in a manner analogous to that occurring for the other ligands of Ln, heparin, and α-dystroglycan. In this case, the negatively charged patch is composed of glutamic and aspartic acid residues rather than sulphated sugars. Our results further suggest that the Ln511/521-binding site on Lu gp is located at a potentially flexible interdomain linker region, and it is possible that the amino-terminal D1 and D2 may be required to fold back to expose the binding site for Ln511/521. Confirmation of this mechanism awaits further structural analysis of Lu gp and identification of the Lu gp–binding site in the LG1-3 region of the Ln α5 chain. Nevertheless, these findings pave the way for development of inhibitors of the Lu gp:Ln511/521 interaction and thereby potential novel therapies ameliorating vaso-occlusion in sickle cell disease.
The online version of this article contains a data supplement.
Presented in part in abstract form at the 48th annual meeting of the American Society of Hematology, Orlando, FL, December 9-12, 2006.57
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by National Institutes of Health Grants DK56267 and DK32094; by the Director, Office of Health and Environment Research Division, U.S. Department of Energy, under Contract DE-AC03–76SF00098; and by the United Kingdom National Health Service Research and Development Directorate. N.B. is supported by a studentship from the UK Medical Research Council.
We thank Dr Susan Ortlepp (CellTech, Slough, United Kingdom) for expert help with Lu gp overexpression; Dr Becky Conners, University of Bristol, for assistance with x-ray data collection and advice; and Hugh Beedie and James Osborne (Information Services, Cardiff University) for implementation of distributed software through Condor. We are grateful to the staff at the Daresbury SRS and Hamburg DESY synchrotrons for support and access to these facilities
National Institutes of Health
Authorship
Contribution: T.J.M. and N.B. designed research, performed research, analyzed and interpreted data, and drafted the paper; F.O.S., J.S.P., C.L.P.O., D.L., and T.W. performed research; F.A.S. drafted the paper; S.F.P. performed research and drafted the paper; N.M., J.A.C., R.L.B., and D.J.A. designed and directed research and drafted the paper. T.J.M. and N.B. contributed equally to this work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: R. Leo Brady, Department of Biochemistry, University of Bristol, Bristol BS8 1TD United Kingdom; e-mail: l.brady@bris.ac.uk or Tosti J. Mankelow, BITS, Southmead Road, Bristol, BS10 5ND, United Kingdom; e-mail: tosti.mankelow@nbs.nhs.uk.