Abstract
The Krüppel-like C2/H2 zinc finger transcription factors (KLFs) control development and differentiation. Erythroid Krüppel-like factor (EKLF or KLF1) regulates adult β-globin gene expression and is necessary for normal definitive erythropoiesis. KLF2 is required for normal embryonic Ey- and βh1-, but not adult βglobin, gene expression in mice. Both EKLF and KLF2 play roles in primitive erythroid cell development. To investigate potential interactions between these genes, EKLF/KLF2 double-mutant embryos were analyzed. EKLF−/−KLF2−/− mice appear anemic at embryonic day 10.5 (E10.5) and die before E11.5, whereas single-knockout EKLF−/− or KLF2−/− embryos are grossly normal at E10.5 and die later than EKLF−/−KLF2−/− embryos. At E10.5, Ey- and βh1-globin mRNA is greatly reduced in EKLF−/−KLF2−/−, compared with EKLF−/− or KLF2−/− embryos, consistent with the observed anemia. Light and electron microscopic analyses of E9.5 EKLF−/−KLF2−/− yolk sacs, and cytospins, indicate that erythroid and endothelial cells are morphologically more abnormal than in either single knockout. EKLF−/−KLF2−/− erythroid cells are markedly irregularly shaped, suggesting membrane abnormalities. EKLF and KLF2 may have coordinate roles in a common progenitor to erythroid and endothelial cells. The data indicate that EKLF and KLF2 have redundant functions in embryonic β-like globin gene expression, primitive erythropoiesis, and endothelial development.
Introduction
Krüppel-Like factors (KLFs) are a family of DNA-binding proteins with sequence homology to the Drosophila transcription factor, Krüppel. KLFs have 3 C2/H2 zinc finger domains and share conserved residues located primarily within these domains.1,2 Erythroid Krüppel-Like factor (EKLF or KLF1) was the first of 17 KLFs to be identified in mouse and man.3 It is expressed specifically in erythroid cells and positively regulates the adult β-globin gene.3,4 EKLF−/− mice develop fatal anemia during definitive (fetal liver) erythropoiesis, due to a defect in the maturation of red blood cells, and die by embryonic day 16 (E16).5-7 Several other members of the KLF family, including KLF2 (lung Krüppel-like factor, LKLF), also are expressed in erythroid cells.8-11
Based on phylogenetic analyses, the zinc finger domains of KLF2 and EKLF are very similar.1,2,12,13 KLF2−/− mice die between E12.5 and E14.5 due to heart failure and severe hemorrhaging, caused by defects in vascular endothelial cells and in stabilization of immature vessels by recruited smooth muscle cells.14,15 Prior to E12.5, KLF2−/− embryos have normal vasculogenesis and angiogenesis.14,15
KLF2 also plays an important role in hematopoietic cell biology. We reported that KLF2 is essential for primitive (embryonic yolk sac) erythropoiesis and positively regulates the embryonic β-like globin genes in vivo. E10.5 KLF2−/− primitive erythroid cells have abnormal morphology.9 KLF2 also regulates T-cell activation. Deficiency of KLF2 leads to a decrease in the peripheral T-cell pool16 due to defective thymocyte emigration.17 Overexpression of KLF2 in mice inhibits proinflammatory activation of peripheral blood monocytes.18
It was initially reported that EKLF does not affect embryonic/fetal globin gene expression. Interestingly, however, EKLF is expressed very early in mouse and chicken development, as early as the primitive streak stage, followed by expression in the yolk sac blood islands where primitive erythroid cells develop.19,20 EKLF also plays an essential role in hemoglobin metabolism and membrane stability in primitive erythroid cells. EKLF−/− primitive red blood cells have an accumulation of Heinz bodies reminiscent of α-globin chain aggregation, and defective membrane morphology.21,22 Also, in EKLF−/− embryos that carry a YAC transgene with the entire human β-globin locus, the human embryonic ϵ-globin gene is down-regulated.23 Recently, ChIP analyses have shown that EKLF binds to the CACCC boxes in the Ey- and βh1-globin gene promoters in primitive erythroid cells.24 These studies establish the important role of EKLF in primitive erythropoiesis.
The roles of transcription factors in the developmental regulation of the globin genes are under active investigation. The murine β-globin locus has 2 embryonic globin genes, Ey- and βh1-, and 2 adult globin genes, βmaj- and βmin-globin. We have shown that KLF2 positively regulates the human (ϵ-) and murine (Ey- and βh1-) embryonic β-like globin genes in the yolk sac, the site of primitive erythropoiesis. However, expression of these genes is diminished only by about 50% in KLF2−/− mice, and this suggests that an additional factor(s) is involved. We speculated that EKLF and KLF2 could have compensatory roles in primitive erythropoiesis and globin gene regulation. In the present study, we have shown that the simultaneous ablation of the EKLF and KLF2 genes has a severely negative impact on both primitive erythropoiesis and endothelial cell development in vivo.
Materials and methods
This study was approved by the Virginia Commonwealth University Institutional Animal Care and Use Committee (VCU IACUC).
Generating EKLF and KLF2 double-knockout mice
KLF2 knockout mice were generated by Wani et al as described previously.15 EKLF knockout mice were generated by Perkins et al,6 and heterozygotes were obtained from Jackson Labs, Bar Harbor, ME. Embryos and adult mice were screened for the KLF2 knockout allele using polymerase chain reaction (PCR) primers 5′-TGCTTACAACCTCCTAAATGTTCTGA-3′ and 5′-CCTACCCGCTTCCATTGCTC-3′, and for the normal allele using primers 5′-TTGTTTAGGTCCTCATCCGTGCCG-3′ and 5′-TTGCCGTCCTTTGCCACTTTCG-3′. For the EKLF knockout allele, primers from the neomycin gene were used: 5′-CCTGAGCTACTTAGTGAGATCC-3′ and 5′-TTCTTCTCCCATCTCTAACCC-3′. The normal EKLF allele was detected using primers 5′-AGAGGCTATTCGGCTATGACTG-3′ and 5′-TTCGTCCAGATCATCCTGATC-3′. The EKLF and KLF2 genes are close together on mouse chromosome 8. EKLF+/− KLF2+/− males were generated by breeding EKLF+/− and KLF2+/− mice and were bred with FVB/N female mice. The progeny were screened to obtain a mouse line with recombination between the EKLF and KLF2 mutant alleles, placing them both on the same chromosomal homolog (recombinant double heterozygotes).
Analysis of EKLF−/−KLF2−/− embryos
Recombinant double-heterozygous (EKLF+/−KLF2+/−) males and females were used in timed matings to generate wild-type, EKLF+/−KLF2+/−, and EKLF−/−KLF2−/− embryos. E10.5 embryos were photographed as whole mounts on an Olympus SZ2-ILST (Olympus America, Center Valley, PA) microscope at 16× magnification using an Olympus Q-Color 3 (Olympus America) camera and QCapture 2.81.0 software (Quantitative Imaging, Surrey, BC). The E10.5 embryos were collected for genotyping, and the yolk sacs were quick frozen. Total RNA was isolated from the yolk sacs for quantitative reverse transcriptase polymerase chain reaction (RT-PCR), using Trizol reagent (Sigma, St Louis, MO).
For light microscopy, E9.5 embryos and yolk sacs were fixed in 2% paraformaldehyde and 2.5% glutaraldehyde solution overnight, and embedded in eponate 12 plastic. Prior to fixation, a small portion of the tail was collected for genotyping. Sections were cut on an LKB 2128 Ultramicrotome at a thickness of 2 μm for yolk sacs and 6 μm for embryos. Sections were stained with toluidine blue and images made using a Nikon Optiphot bright field microscope (Nikon USA, Melville, NY) equipped with an Olympus Fluor 40 ×/0.85 brightfield objective (400 × total magnification). Images were taken with an Olympus DP70 digital camera (Olympus America) with Olympus DP Controller acquisition software (Olympus America). For identifying and counting abnormal erythroid cells in E9.5 yolk sac tissue sections, the following criteria were used: only complete cell sections, not fragments of cells, were counted; densely packed immature erythroid cells without space between them were not counted; and a cell was considered to be abnormally shaped if 25% of its volume was irregular, that is, formed a cytoplasmic projection(s). For electron microscopy, 2 EKLF−/−KLF2−/− and one wild-type yolk sac were fixed and embedded as described above. Sections were cut at 100 nm with an LKB 2128 Ultratome and stained with 5% Uranyl Acetate and Reynold's Lead Citrate. Images were made using the JEOL JEM-1230 TEM (JEOL USA, Peabody, MA) and a Gatan Ultrascan 4000 digital camera (Gatan, Pleasanton, CA).
Quantitative RT- PCR
Real-time PCR conditions and primer and probe sequences have previously been described.9 The number of embryos tested for each genotype ranged from 4 to 8. To quantify globin mRNA, the mean globin-to-GPA mRNA ratio in the wild-type embryos was designated as 100%, and the means for the other genotypes were expressed relative to wild-type littermates. To quantify GPA mRNA, the GPA-to-cyclophilin A mRNA ratio in wild-type embryos was designated as 100%. For statistical analyses, the Student t test of comparison of means was performed, and differences were considered significant at P values less than .025.
Flow cytometry and cytospin analyses
E9.5 embryos with intact yolk sacs were rinsed in FACS buffer (1× phosphate buffered saline + 5% fetal bovine serum). Blood from the embryo and yolk sac was allowed to drain into FACS buffer with heparin (0.2 U/mL). The blood cells were double-labeled with phycoerythrin (PE)–conjugated anti-TER119 and fluorescein isothiocyanate (FITC)–conjugated anti-CD71 (transferrin receptor) (eBiosciences, San Diego, CA). The samples were gently pushed through 75 μm nylon mesh and run on an EPICS Elite ESP Flow Cytometer (Coulter, Fullerton, CA). Forward and side scatter plots were used to identify and analyze embryonic blood cells and reduce contamination with maternal blood cells for the cytospin and flow cytometry assays. A density plot of relative logarithmic fluorescence units for PE versus FITC was generated. Five gates (R1 through R5) were drawn as previously described.25,26 The total of the average percentages of cells in R1 through R5 for KLF2−/−, EKLF−/−, and KLF2−/−EKLF−/− were normalized to the wild-type total. Blood samples containing 5000 to 20 000 embryonic blood cells were prepared using a SCA0081 cytospin apparatus (Shandon Southern, Thermo Scientific, Waltham, MA) and stained with Giemsa (Sigma Aldrich). The cytospins were viewed using a Nikon Eclipse 80i microscope (Nikon USA) with a Nikon Plan Apo 100×/1.40 objective (1000 × total magnification). Micrographs were taken with a Nikon DXM1200C camera (Nikon USA) with Nikon ACT-1C acquisition software (Nikon USA). Semiquantitative morphological analyses of the cytospins were performed blind to genotype. Ten cells per sample were scored for each genotype (wild-type, n = 4 samples; KLF2−/−, n = 2; EKLF−/−, n = 4; and EKLF−/−KLF2−/−, n = 4). The abnormal criteria scored were cytoplasmic blebbing and nuclear atypia, the latter included irregularity of nuclear contours, chromatin condensation, and binucleation. Cells that were normal for cytoplasmic or nuclear features scored 1, moderately abnormal cells scored 2, and the most abnormal cells scored 3.
Results
EKLF−/−KLF2−/− embryos display severe anemia
The EKLF and KLF2 genes are approximately 6 Mb apart on mouse chromosome 8. So, to obtain double-knockout embryos, EKLF+/− and KLF2+/− mice were first bred to generate double heterozygotes. These double heterozygotes were then bred with normal mice and screened for recombination between the EKLF and KLF2 loci, to obtain animals with both null alleles on the same homolog. EKLF+/−KLF2+/− mice with the recombined alleles (recombinant double heterozygotes) were observed at a frequency of 2 in 104 births from these matings. An EKLF+/−KLF2+/− mouse with the recombined alleles was bred with normal mice to generate a line of recombinant double-heterozygous mice.
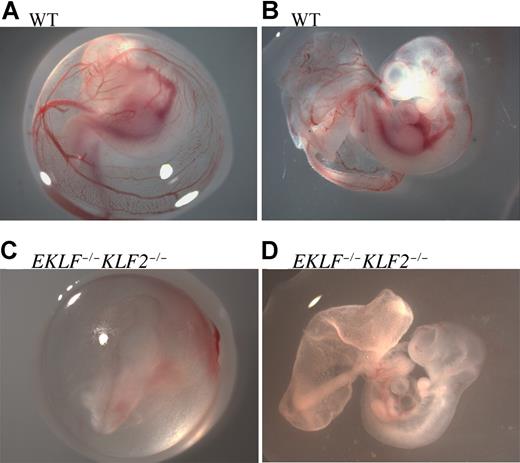
Recombinant double-heterozygous males and females from this line were bred in timed matings to obtain E10.5 wild-type, EKLF+/−KLF2+/−, and EKLF−/−KLF2−/− embryos. Embryos were photographed within the embryonic yolk sac (Figure 1A,C) and also after removing the embryonic yolk sac carefully, without disrupting the large vitelline and umbilical vessels, to limit bleeding (Figure 1B,D). At E10.5, EKLF−/− or KLF2−/− whole-mount embryos have apparently normal morphology.5,9 Compared with wild-type (Figure 1A,B), EKLF−/−KLF2−/− E10.5 yolk sacs and embryos are pale and anemic (Figure 1C,D). The deep red aorti evident in wild-type embryos were either less prominent or could not be detected in double mutants. Approximately 80% of E10.5 double-knockout embryos had reduced forebrain and/or hindbrain, probably due to anemia (n = 9). The results indicate an interaction between the EKLF and KLF2 genes. Interestingly, E9.5 EKLF−/−KLF2−/− yolk sacs were indistinguishable from wild-type in color, appearing pink as is normal (data not shown). The anemia that is obvious by E10.5 in EKLF−/−KLF2−/− embryos must be due to an inability of red blood cells to expand in number and/or hemoglobinize normally.
E10.5 wild-type and EKLF−/−KLF2−/− whole-mount embryos. Panels A and B are wild-type (WT), and panels C and D are EKLF−/−KLF2−/− E10.5 embryos. In panels A and C, embryos are surrounded by yolk sacs. Panels B and D are the same embryos, respectively, with yolk sac teased away, but vitelline and umbilical vessels still intact. Photographs were taken at 16× magnification. See “Analysis of EKLF−/−KLF2−/− embryos” for more image information.
E10.5 wild-type and EKLF−/−KLF2−/− whole-mount embryos. Panels A and B are wild-type (WT), and panels C and D are EKLF−/−KLF2−/− E10.5 embryos. In panels A and C, embryos are surrounded by yolk sacs. Panels B and D are the same embryos, respectively, with yolk sac teased away, but vitelline and umbilical vessels still intact. Photographs were taken at 16× magnification. See “Analysis of EKLF−/−KLF2−/− embryos” for more image information.
Embryos from matings between EKLF+/−KLF2+/− mice were obtained for χ-square analyses at E9.5, E10.5, and E11.5 (Table 1). Although EKLF−/−KLF2−/− embryos are severely compromised at E10.5, the expected ratio of genotypes was observed at E10.5 (χ-square value not significant at 0.27) and at E9.5 (χ-square of 0.40). However, there were no live EKLF−/−KLF2−/− embryos observed at E11.5, and this result was significantly different from expected (χ-square of 6.82, P < .05). It was possible to genotype several embryos that were already dead at E11.5 and to confirm that there were the expected number of EKLF−/−KLF2−/− conceptuses (χ-square of 0.88). This clearly establishes that EKLF−/−KLF2−/− embryos die earlier than EKLF−/−5,6 or KLF2−/−14,15 embryos, and before E11.5.
Number of live embryos observed from EKLF+/−KLF2+/− matings
| Stage . | Total . | WT . | EKLF+/−KLF2+/− . | EKLF−/−KLF2−/− . |
|---|---|---|---|---|
| E9.5 | 41 | 9 | 18 | 14 |
| E10.5 | 66 | 14 | 35 | 17 |
| E11.5* | 23 (12) | 6 (2) | 17 (0) | 0 (10) |
| Stage . | Total . | WT . | EKLF+/−KLF2+/− . | EKLF−/−KLF2−/− . |
|---|---|---|---|---|
| E9.5 | 41 | 9 | 18 | 14 |
| E10.5 | 66 | 14 | 35 | 17 |
| E11.5* | 23 (12) | 6 (2) | 17 (0) | 0 (10) |
Parentheses indicate number of dead embryos
EKLF−/−KLF2−/− mice have a dramatic reduction in embryonic globin mRNA
We studied the effect of simultaneous ablation of EKLF and KLF2 on endogenous embryonic α-like and β-like globin gene expression. We used quantitative real-time PCR to measure the amount of embryonic Ey-, βh1-, and ζ-globin mRNA in the E10.5 yolk sac. Glycophorin A (GPA) mRNA was used as an internal control to normalize the data. GPA is an erythroid-specific membrane marker and was selected to ensure that any changes in the amount of globin mRNA were not simply due to a reduction in the number of red blood cells.
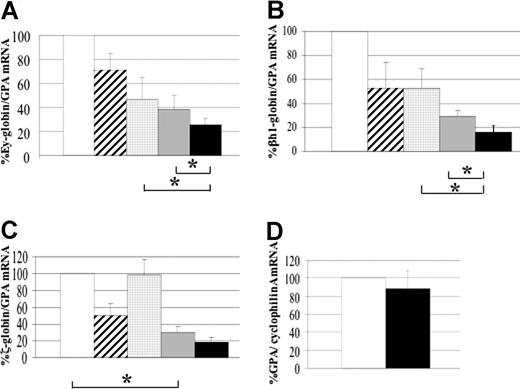
Double-heterozygous (EKLF+/−KLF2+/−), single-knockout (EKLF−/− or KLF2−/−), and double-knockout (EKLF−/−KLF2−/−) embryos were tested. All mutant embryos were compared with wild-type (EKLF+/+KLF2+/+) littermates, to greatly reduce differences in genetic background or stage of development. The data for the single-knockout KLF2 embryos is from a previous study and is shown here for comparison.9 Compared with wild-type, a significant reduction of Ey- and βh1-globin gene expression (P < .025) was observed in both the single-knockouts and double-knockouts (Figure 2A,B). This had previously been reported for KLF2−/− embryos.9 The mouse embryonic β-like globin genes are down-regulated in EKLF null mice. This had not previously been reported, but the human embryonic ϵ-globin gene, located on a transgene, is down-regulated in EKLF−/− mice.23 Interestingly, Ey- and βh1-globin gene expression was significantly reduced in the double-knockout embryos compared with either the EKLF or KLF2 single knockouts (P < .025, see asterisks in Figure 2A,B). In the double-knockout embryos, the amounts of β-like globin mRNA are 4- to 5-fold less than in the wild type, a level that is consistent with the severe anemia that is observed. This indicates that both EKLF and KLF2 are required for normal mouse embryonic β-like globin gene expression and that these 2 KLF genes can partially compensate for each other in regulating β-like globin gene expression in the embryo.
Mouse embryonic globin gene expression in yolk sacs from EKLF−/−KLF2−/− compared with wild-type and single knockouts. (A) Ey-globin mRNA, (B) βh1-globin mRNA, (C) ζ-globin mRNA. Glycophorin A (GPA) mRNA was used as an internal standard for quantitative RT-PCR. The globin-to-GPA mRNA ratio for wild-type (□) was taken as 100%, and for the other genotypes is expressed compared with 100%. To maintain similar genetic backgrounds between test and control samples, all mutants are compared with wild-type littermates. The other genotypes are EKLF+/−KLF2+/− (▨, n = 8), KLF2−/− (▩, n = 7), EKLF−/− ( , n = 4), and EKLF−/−KLF2−/− (■, n = 6). n represents the number of embryonic yolk sacs of each genotype used to determine the mean globin-to-GPA mRNA ratio. Error bars represent the standard deviation from the mean. The asterisks in panels A and B indicate a significant reduction in Ey- and βh1-globin mRNA (P < .025) in double- compared with single-knockout embryos. The asterisk in panel C indicates a significant reduction in ζ-globin mRNA in EKLF−/− compared with wild-type. (D) Mouse glycophorin A (GPA) gene expression in E10.5 EKLF−/−KLF2−/− yolk sacs. Cyclophilin A mRNA was used as an internal standard for quantitative RT-PCR. Wild-type (□, n = 5) and EKLF−/−KLF2−/− (■, n = 5) embryonic yolk sac RNAs were tested to determine the mean GPA-to-cyclophilin A mRNA ratio. Error bar is standard deviation, and wild-type was taken as 100%.
, n = 4), and EKLF−/−KLF2−/− (■, n = 6). n represents the number of embryonic yolk sacs of each genotype used to determine the mean globin-to-GPA mRNA ratio. Error bars represent the standard deviation from the mean. The asterisks in panels A and B indicate a significant reduction in Ey- and βh1-globin mRNA (P < .025) in double- compared with single-knockout embryos. The asterisk in panel C indicates a significant reduction in ζ-globin mRNA in EKLF−/− compared with wild-type. (D) Mouse glycophorin A (GPA) gene expression in E10.5 EKLF−/−KLF2−/− yolk sacs. Cyclophilin A mRNA was used as an internal standard for quantitative RT-PCR. Wild-type (□, n = 5) and EKLF−/−KLF2−/− (■, n = 5) embryonic yolk sac RNAs were tested to determine the mean GPA-to-cyclophilin A mRNA ratio. Error bar is standard deviation, and wild-type was taken as 100%.
Mouse embryonic globin gene expression in yolk sacs from EKLF−/−KLF2−/− compared with wild-type and single knockouts. (A) Ey-globin mRNA, (B) βh1-globin mRNA, (C) ζ-globin mRNA. Glycophorin A (GPA) mRNA was used as an internal standard for quantitative RT-PCR. The globin-to-GPA mRNA ratio for wild-type (□) was taken as 100%, and for the other genotypes is expressed compared with 100%. To maintain similar genetic backgrounds between test and control samples, all mutants are compared with wild-type littermates. The other genotypes are EKLF+/−KLF2+/− (▨, n = 8), KLF2−/− (▩, n = 7), EKLF−/− ( , n = 4), and EKLF−/−KLF2−/− (■, n = 6). n represents the number of embryonic yolk sacs of each genotype used to determine the mean globin-to-GPA mRNA ratio. Error bars represent the standard deviation from the mean. The asterisks in panels A and B indicate a significant reduction in Ey- and βh1-globin mRNA (P < .025) in double- compared with single-knockout embryos. The asterisk in panel C indicates a significant reduction in ζ-globin mRNA in EKLF−/− compared with wild-type. (D) Mouse glycophorin A (GPA) gene expression in E10.5 EKLF−/−KLF2−/− yolk sacs. Cyclophilin A mRNA was used as an internal standard for quantitative RT-PCR. Wild-type (□, n = 5) and EKLF−/−KLF2−/− (■, n = 5) embryonic yolk sac RNAs were tested to determine the mean GPA-to-cyclophilin A mRNA ratio. Error bar is standard deviation, and wild-type was taken as 100%.
, n = 4), and EKLF−/−KLF2−/− (■, n = 6). n represents the number of embryonic yolk sacs of each genotype used to determine the mean globin-to-GPA mRNA ratio. Error bars represent the standard deviation from the mean. The asterisks in panels A and B indicate a significant reduction in Ey- and βh1-globin mRNA (P < .025) in double- compared with single-knockout embryos. The asterisk in panel C indicates a significant reduction in ζ-globin mRNA in EKLF−/− compared with wild-type. (D) Mouse glycophorin A (GPA) gene expression in E10.5 EKLF−/−KLF2−/− yolk sacs. Cyclophilin A mRNA was used as an internal standard for quantitative RT-PCR. Wild-type (□, n = 5) and EKLF−/−KLF2−/− (■, n = 5) embryonic yolk sac RNAs were tested to determine the mean GPA-to-cyclophilin A mRNA ratio. Error bar is standard deviation, and wild-type was taken as 100%.
We had previously reported that ζ-globin gene expression is not affected in embryos with the KLF2 knockout.9 However, it is demonstrated here that EKLF−/− embryos have about 3-fold lower ζ-globin gene expression than wild type (P < .025), which is a new finding (Figure 2C, see asterisk). EKLF−/−KLF2−/− embryos may have slightly less ζ-globin mRNA than EKLF−/− embryos (P < .05), but this is not significant by our stated criterion of P being less than .025. Therefore, ζ-globin expression appears to be primarily dependent on EKLF and not KLF2.
Because glycophorin A (GPA) mRNA was the normalization control in the previously described quantitative RT-PCR assays, it was important to establish whether the amount of GPA mRNA in the EKLF−/−KLF2−/− E10.5 yolk sacs was the same as in wild type. Cyclophilin A mRNA amounts were used as an internal control to standardize the data. In Figure 2D, RNA from 5 wild-type and 5 EKLF−/−KLF2−/− E10.5 yolk sacs was tested to compare the GPA-to-cyclophilin A mRNA ratios. There was no significant difference in the amounts of GPA mRNA between the 2 genotypes. This indicates that in EKLF−/−KLF2−/− embryos, erythroid differentiation progresses at least to the point of producing GPA mRNA. These results suggest that primitive erythroid cells lacking KLF2 and EKLF reach at least the proerythroblast stage27 but are deficient in final maturation and hemoglobinization.
Circulating red blood cells from KLF2−/−, EKLF−/−, and EKLF−/−KLF2−/− embryos have abnormal morphology
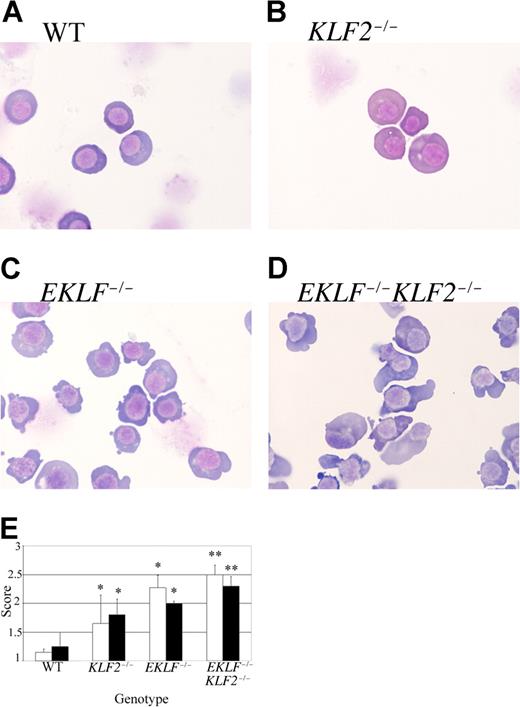
To further explore the effects of the EKLF and KLF2 mutations on mouse erythropoiesis, E9.5 blood was collected for cytospins and stained with Giemsa (Figure 3A-D). It is obvious that both the EKLF−/− (Figure 3C) and EKLF−/−KLF2−/− (Figure 3D) erythroid cells look substantially different from wild type (Figure 3A). Although it was previously shown that EKLF−/− erythroid cells have atypical features at E11.5 and E12.5,21,22 this is the first demonstration that they are abnormal as early as E9.5. KLF2−/− erythroid cells (Figure 3B) show less pronounced differences compared with wild-type cells. Semiquantitative morphological analyses were performed on between 2 and 4 blood samples per genotype, assessing 10 cells per sample for a total of 20 to 40 cells (Figure 3E). The scoring was done blind to genotype. KLF2−/−, EKLF−/−, and EKLF−/−KLF2−/− cells have progressively more severe cytoplasmic blebbing and nuclear atypia that include irregular nuclear contours, chromatin condensation, and binucleation (Figure 3E). KLF2−/− and EKLF−/− cells scored significantly higher than wild type for cytoplasmic and nuclear abnormalities (Figure 3E). EKLF−/−KLF2−/− cells are significantly more severely affected than KLF2−/− or EKLF−/− cells (Figure 3E) and additionally exhibit more elongated shapes than the cells of the other 3 genotypes (Figure 3D). The data further indicate that there are interactions between the EKLF and KLF2 genes and that the genes can compensate for each other in the single mutants.
Morphology of WT, KLF2−/−, EKLF−/−, and EKLF−/−KLF2−/− E9.5 circulating blood cells. The total number of embryo samples examined for each genotype is indicated in the Figure 4 legend. (A-D) Representative cytospins of Giemsa-stained primitive erythroid cells. Cytospins were prepared from blood samples collected from E9.5 yolk sacs and embryos. (A) Wild-type. (B) KLF2−/−. (C) EKLF−/−. (D) EKLF−/−KLF2−/−. Photographs were taken at 1000 × magnification. See “Analysis of EKLF−/−KLF2−/− embryos” for more image information. (E) Semiquantitative assessment of abnormal cytoplasmic and nuclear morphology in WT, KLF2−/−, EKLF−/−, and EKLF−/−KLF2−/− cytospins. Ten primitive blood cells from each sample were scored from 1 to 3, based on severity of cytoplasmic pleiomorphism and nuclear atypia (white and black bars, respectively). A score of 1 indicates the most normal, and of 3 the most abnormal morphology. Error bars indicate standard deviation from the mean. For WT, EKLF−/− and EKLF−/−KLF2−/−, n = 40 cells, and for KLF2−/− samples, n = 20. * indicates a significant difference from WT at P < .001, and ** is significantly different from WT, KLF2−/−, and EKLF−/− at P < .001 using Student t test of comparison of means.
Morphology of WT, KLF2−/−, EKLF−/−, and EKLF−/−KLF2−/− E9.5 circulating blood cells. The total number of embryo samples examined for each genotype is indicated in the Figure 4 legend. (A-D) Representative cytospins of Giemsa-stained primitive erythroid cells. Cytospins were prepared from blood samples collected from E9.5 yolk sacs and embryos. (A) Wild-type. (B) KLF2−/−. (C) EKLF−/−. (D) EKLF−/−KLF2−/−. Photographs were taken at 1000 × magnification. See “Analysis of EKLF−/−KLF2−/− embryos” for more image information. (E) Semiquantitative assessment of abnormal cytoplasmic and nuclear morphology in WT, KLF2−/−, EKLF−/−, and EKLF−/−KLF2−/− cytospins. Ten primitive blood cells from each sample were scored from 1 to 3, based on severity of cytoplasmic pleiomorphism and nuclear atypia (white and black bars, respectively). A score of 1 indicates the most normal, and of 3 the most abnormal morphology. Error bars indicate standard deviation from the mean. For WT, EKLF−/− and EKLF−/−KLF2−/−, n = 40 cells, and for KLF2−/− samples, n = 20. * indicates a significant difference from WT at P < .001, and ** is significantly different from WT, KLF2−/−, and EKLF−/− at P < .001 using Student t test of comparison of means.
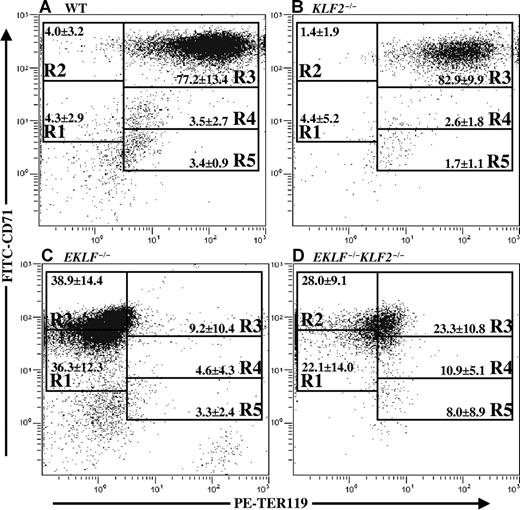
Because the cell membranes of KLF2−/−, EKLF−/−, and EKLF−/−KLF2−/− E9.5 erythroid cells were potentially abnormal based on morphology, a flow cytometric assay was used to quantify cell surface markers. Wild-type, EKLF−/−, KLF2−/−, and EKLF−/−KLF2−/− primitive erythroid cells were labeled with 2 monoclonal antibodies specific for red cell antigens, PE-conjugated anti-TER119, and FITC-conjugated anti-CD71 (anti-transferrin receptor). The cells were flow sorted, and a density plot of relative logarithmic fluorescence units for PE versus FITC was generated (Figure 4). According to Zhang et al,25 regions R1 through R5 are defined by a characteristic pattern of staining of the cells, such that these gates contain increasingly more mature erythroid cells. The assay has been validated for use in studying primitive erythropoiesis.26 At E9.5, we expected that more than 70% of normal primitive erythroid cells would be in R3 (early and late basophilic erythoblasts),26 and this was indeed the case (Figure 4A). KLF2−/− cells do not appear to distribute differently than wild type and are also predominantly in R3 (Figure 4B). Although EKLF−/− and EKLF−/−KLF2−/− cells are mostly in R1 and R2 (Figure 4C,D), and therefore appear to be less mature than wild-type or KLF2−/− cells, it may not be valid to draw conclusions from an assay performed with compromised mutant cells. The data does indicate that EKLF−/− and EKLF−/−KLF2−/− primitive erythroid cells have lower amounts of the epitopes recognized by the TER119 and CD71 antibodies on their cell surfaces than do wild-type or KLF2−/− cells (Figure 4). So, interestingly, the cell morphology abnormalities observed in EKLF−/− and EKLF−/−KLF2−/− cells are accompanied by changes in their complement of membrane proteins. Although it is difficult to be quantitative in collecting E9.5 blood, there was no correlation observed between genotype and the total number of primitive red blood cells collected per embryo, as determined by the flow cytometry analysis (data not shown). This further supports the hypothesis that the stability and/or maturation of red blood cells, and not their number, is defective in the EKLF−/− and EKLF−/−KLF2−/− embryos.
Representative double-labeled FITC-CD71 and PE-TER119 histoplots from E9.5 blood. Flow cytometry was used to determine CD71 and TER119 staining intensity of blood cells from E9.5 yolk sac and embryos. Mean percentages plus or minus standard deviations are indicated in each gate R1-R5 for each genotype. Regions R1 through R5 are defined respectively as CD71medTER119low, CD71highTER119low, CD71highTER119high, CD71medTER119high, and CD71lowTER119high. R1 contains predominantly progenitor cells, R2 contains proerythroblasts and early basophilic erythroblasts, R3 has early and late basophilic erythroblasts, R4 has chromatophilic and orthochromatophilic erythroblasts, and R5 has late orthochromatophilic erythroblasts and reticulocytes.25 (A) Wild type, mean percentages calculated from n = 32 embryos. (B) KLF2−/−, n = 8. (C) EKLF−/−, n = 10. (D) EKLF−/−KLF2−/−, n = 8.
Representative double-labeled FITC-CD71 and PE-TER119 histoplots from E9.5 blood. Flow cytometry was used to determine CD71 and TER119 staining intensity of blood cells from E9.5 yolk sac and embryos. Mean percentages plus or minus standard deviations are indicated in each gate R1-R5 for each genotype. Regions R1 through R5 are defined respectively as CD71medTER119low, CD71highTER119low, CD71highTER119high, CD71medTER119high, and CD71lowTER119high. R1 contains predominantly progenitor cells, R2 contains proerythroblasts and early basophilic erythroblasts, R3 has early and late basophilic erythroblasts, R4 has chromatophilic and orthochromatophilic erythroblasts, and R5 has late orthochromatophilic erythroblasts and reticulocytes.25 (A) Wild type, mean percentages calculated from n = 32 embryos. (B) KLF2−/−, n = 8. (C) EKLF−/−, n = 10. (D) EKLF−/−KLF2−/−, n = 8.
Abnormal morphology of the mouse E9.5 EKLF−/−KLF2−/− yolk sac
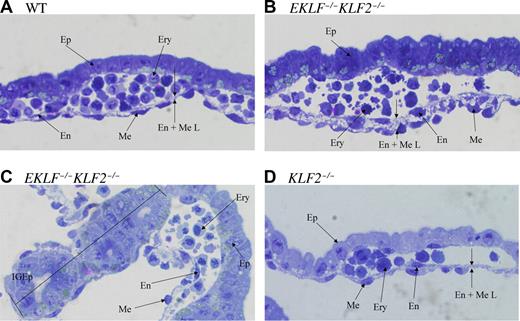
At E10.5, we had previously observed that while the nucleated primitive erythroid cells in wild-type embryonic yolk sac tissue sections are mainly round or elliptical shaped, KLF2−/− cells are primarily irregularly shaped.9 To determine whether mutant erythroid precursors in E9.5 visceral yolk sacs show abnormal morphology, representative tissue sections of wild-type, EKLF−/−KLF2−/−, and KLF2−/− are shown in Figure 5A-D. The visceral layer of the mouse yolk sac is derived from both endoderm and mesoderm. The endoderm forms a simple cuboidal to columnar epithelium (Ep) with microvilli that face the yolk sac cavity (Figure 5A, wild type). The mesodermally derived portion first forms angioblastic cords that further differentiate into developing blood cells (Ery), surrounded by endothelial cells (En) and a layer of simple squamous epithelium, the mesothelium (Me), that faces the extra-embryonic coelom (Figure 5A).
E9.5 EKLF−/−KLF2−/− yolk sac blood islands have abnormal morphology. Panel A is a representative section from a wild type, panels B and C are from 2 different EKLF/KLF2 double knockout, and panel D is from a KLF2−/− embryonic yolk sac. The abbreviations used are columnar epithelium (Ep), erythroid cells (Ery), endothelial cells (En), mesothelial cells (Me), endothelial/mesothelial layer (En + Me L), and ingrowth of epithelial cells (IGEp). Photographs were taken at 400 × magnification. See “Analysis of EKLF−/−KLF2−/− embryos” for more image information.
E9.5 EKLF−/−KLF2−/− yolk sac blood islands have abnormal morphology. Panel A is a representative section from a wild type, panels B and C are from 2 different EKLF/KLF2 double knockout, and panel D is from a KLF2−/− embryonic yolk sac. The abbreviations used are columnar epithelium (Ep), erythroid cells (Ery), endothelial cells (En), mesothelial cells (Me), endothelial/mesothelial layer (En + Me L), and ingrowth of epithelial cells (IGEp). Photographs were taken at 400 × magnification. See “Analysis of EKLF−/−KLF2−/− embryos” for more image information.
In EKLF−/−KLF2−/− E9.5 yolk sac sections from 2 different embryos (Figure 5B,C), the erythroid cells (Ery) have an abnormal morphology compared with wild type, with irregular shapes and multiple protruding projections. Also, the endothelial/mesothelial layer (En + Me L) is unusually thickened with more intercellular space, and the cells of each layer appear to have a more bulbous rather than the simple squamous shape typically observed in wild type (Figure 5B). In addition, the EKLF−/−KLF2−/− E9.5 yolk sacs have occasional abnormal ingrowths of epithelial cells (IGEp, Figure 5C). These unusual features are not observed in E9.5 KLF2−/− yolk sacs (Figure 5D), although erythroid but not other cells appear abnormal by E10.5 in KLF2−/− yolk sacs,9 and E9.5 circulating KLF2−/− blood cells have some abnormalities (Figure 3E). In E12.5 EKLF−/− yolk sacs, inclusion bodies in erythroid cells have been noted, but there is no indication of abnormalities in other cell types.21 EKLF−/−KLF2−/− yolk sacs have some aberrant features completely absent from single knockouts.
In quantitative studies of more than 280 erythroid cells from 3 E9.5 yolk sacs of each genotype, the percentage of abnormally shaped cells was significantly higher for EKLF−/−KLF2−/− than for KLF2−/− or wild type (54.2% compared with 1.2% or 2.9%, respectively, P < .001). In more than 148 endothelial cells examined, the percentage of bulbous-shaped cells was significantly higher for EKLF−/−KLF2−/− than for wild-type or KLF2−/− cells (32-fold more, P < .011; or 5-fold more, P < .024, respectively). Interestingly, the abnormal endothelial cells were almost exclusively located on the mesothelial, and not the epithelial, side of vessels. In more than 320 mesothelial cells examined, the percentage of bulbous cells was significantly higher for EKLF−/−KLF2−/− than for KLF2−/− or wild-type cells (4-fold more, P < .002). The average distance between the endothelial and mesothelial layers was significantly greater for EKLF−/−KLF2−/− yolk sacs, at 10.75 μm, than it was for wild type and KLF2−/−, at 1.75 μm and 2.25 μm, respectively (n = 4 measurements, P < .025). The average number of epithelial ingrowths per EKLF−/−KLF2−/− yolk sac was 2 (n = 3), so this is a relatively infrequent feature, though not observed in KLF2−/− or wild-type yolk sacs. In fact, this aberration could result from abnormalities in cell types other than epithelial cells. Our results indicate that EKLF and KLF2 have a pivotal role in erythroid cell development and are also required for the normal development of other cells in the yolk sac, particularly the other mesodermally derived cells, that is, endothelial and mesothelial cells.
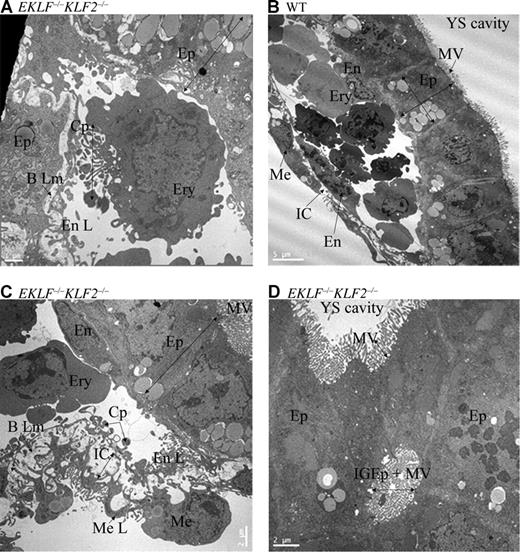
To examine yolk sac morphology in greater detail, wild-type and EKLF−/−KLF2−/− E9.5 yolk sacs were examined electron microscopically. Erythroid cells (Ery) in EKLF−/−KLF2−/− yolk sacs have abnormal morphology with the presence of large cytoplasmic projections (CPs) and filopodia-like structures (Figure 6A). The abnormal bulbous shape of the EKLF−/−KLF2−/− mesothelial cells (Me) is evident, as these cells protrude into the extra-embryonic coelom (compare wild type in Figure 6B and EKLF−/−KLF2−/− in Figure 6C). Due to the thickening of the intercellular space (IC) between the mesothelial (Me L) and endothelial layers (En L), the basal lamina (B Lm) frequently appears discontinuous rather than as a coherent delicate layer between the layers in the EKLF−/−KLF2−/− E9.5 yolk sac (Figure 6C). Additionally, the endothelial layer has a scalloped, lacey appearance due to the presence of multiple endothelial CPs and spaces (Figure 6C). In occasional areas of the EKLF−/−KLF2−/− yolk sac (Figure 6D), deep ingrowths of the epithelial cell layer (IGEp) have occurred with a resultant lumen or cavity within the epithelial layer. These results confirm that EKLF and KLF2 direct the development not only of erythroid cells, but also other mesodermally derived cells in the yolk sac.
Electron micrographs of E9.5 yolk sac cells. (A) Representative erythroid cell from EKLF/KLF2 double knockout. (B) Wild-type and (C) EKLF/KLF2 double-knockout sections indicating the cell types in the yolk sac. (D) Abnormal ingrowth of microvilli in epithelial cells in EKLF/KLF2 double knockout. The abbreviations used are erythroid cells (Ery), cytoplasmic projections (CP), mesothelial cells (Me), intercellular space (IC) between the mesothelial (Me L) and endothelial layers (En L), basal lamina (B Lm), microvilli (MV), and ingrowth of epithelial cells (IGEp). Scale is shown in each photograph. See “Analysis of EKLF−/−KLF2−/− embryos” for more image information.
Electron micrographs of E9.5 yolk sac cells. (A) Representative erythroid cell from EKLF/KLF2 double knockout. (B) Wild-type and (C) EKLF/KLF2 double-knockout sections indicating the cell types in the yolk sac. (D) Abnormal ingrowth of microvilli in epithelial cells in EKLF/KLF2 double knockout. The abbreviations used are erythroid cells (Ery), cytoplasmic projections (CP), mesothelial cells (Me), intercellular space (IC) between the mesothelial (Me L) and endothelial layers (En L), basal lamina (B Lm), microvilli (MV), and ingrowth of epithelial cells (IGEp). Scale is shown in each photograph. See “Analysis of EKLF−/−KLF2−/− embryos” for more image information.
Discussion
The KLF family of transcription factors has been implicated in various cellular processes, including development and differentiation.1,2 EKLF, the first KLF identified in mammals, has a central role in the regulation of the adult β-globin gene and is essential for the maturation and/or stability of definitive erythroid cells.3,5,6 We had previously shown that KLF2 positively regulates the human (ϵ-) and murine (Ey- and βh1-) embryonic globin genes during primitive erythropoiesis.9 Because expression of the embryonic β-like globin genes is not completely eliminated in KLF2−/− embryos, we predicted that an additional factor(s) is involved. Recently, the importance of EKLF in gene expression during primitive erythropoiesis has been strongly established.20-22 We have shown that KLF2 and EKLF can partially compensate for each other in regulating the embryonic globin genes. This is consistent with the work of Zhou et al,24 showing that EKLF binds the Ey- and βh1-globin genes in ChIP assays in embryonic erythroid cells. However, it is seemingly incongruous with the recent work of Shyu et al,28 suggesting that there is a limited amount of EKLF localized in the nuclei of primitive erythroid cells and that rather EKLF is primarily cytoplasmic.
This is the first report in knockout mice showing that KLF family members have compensatory roles in development. The observed reduction of both Ey- and βh1-globin mRNA is about 5-fold in the EKLF/KLF2 homozygous double-knockout embryos, leaving open the possibility that yet another KLF can partially compensate for the loss of EKLF and KLF2. Several other pairs of closely related transcription factors previously have been shown to either partially or fully functionally compensate for each other during development in double-knockout mouse models. This emphasizes the importance of testing the overlapping roles of transcription factors with closely related DNA binding regions, like EKLF and KLF2. For example, mice lacking both GATA-1 and GATA-2 have virtually no primitive erythroid cells, whereas the single-knockout models of these genes show more modest effects on primitive erythropoiesis.29 Multiple pairs of Hox genes have compensatory roles in developmental mouse models. For example, both Hoxa11 and Hoxd11, and Hoxa13 and Hoxd13, interact with each other in pattern formation of the limbs.30,31 In Pax1/Pax9 double-homozygous mutants, the sclerotomes in the developing vertebral column fail to undergo chondrogenesis, a phenotype more severe than the single-homozygous mutants.32 Double mutants of the myogenic bHLH genes MRF4 and MyoD display severe muscle deficiency, even though the single mutants of these genes show no defects in muscle development.33
Interestingly, this work shows that EKLF regulates mouse embryonic globin gene expression. The original reports describing EKLF knockout mice showed that Ey-, βh1-, and ζ-globin mRNA are present in EKLF−/− E10.5 and E11 yolk sacs.5,6 However, no quantitative data were included, and results from very few embryos were shown. The amounts of Ey-, βh1- and ζ-globin mRNA were described as normal. Surprisingly, quantitative data concerning mouse embryonic globin gene expression in the absence of EKLF appears to be lacking in the literature. Therefore, it appears that the 2-fold decrease in embryonic globin expression that we observe in EKLF−/− compared with wild-type embryos was previously overlooked. However, the expression of human ϵ-globin mRNA in transgenic mice lacking EKLF has been examined. Tanimoto et al23 showed that there was 2- to 3-fold less human ϵ-globin mRNA in EKLF−/− than in wild-type embryonic yolk sacs, which is consistent with our results for mouse embryonic globin mRNA. In similar experiments, Wijgerde et al34 did not consider a 2-fold decrease in human ϵ-globin mRNA, evident in quantitative graphs, to be significant.
Another unexpected finding from this work is that EKLF regulates embryonic ζ-globin gene expression. This is the first report of the regulation of an α-like globin gene by EKLF. Ablation of EKLF in the mouse apparently does not diminish adult α-globin gene expression, in fact, the α-globin gene is often used as a normalization control to assess reduced β-like globin expression.6 The ζ-globin gene has a functional CACCC promoter element,35 so it is possible that EKLF regulates the gene through this element. Both ζ- and α-globin are expressed simultaneously in mouse embryos. Therefore, the 5-fold decrease in ζ-globin mRNA that we observed in EKLF/KLF2 double-knockout embryos may not be deleterious.
It remains a possibility that EKLF and KLF2 directly regulate each other in cells. We observed a significant though modest decrease of EKLF mRNA expression in KLF2−/− E10.5 yolk sacs using qRT-PCR (data not shown). So, some of the decrease in globin expression observed in KLF2−/− embryos may be indirect, through a reduction of EKLF. In microarray analyses by Hodge et al,22 there was apparently no significant change in KLF2 gene expression in EKLF−/− compared with wild-type fetal livers, so EKLF may not affect KLF2 expression.
The morphology of E9.5 yolk sac endothelial cells is abnormal in EKLF−/−KLF2−/− mice. KLF2−/− embryos have apparently normal angiogenesis and vasculogenesis prior to E12.5, however by E12.5 hemorrhaging can begin, and some endothelial cells in the umbilical veins appeared abnormal and enlarged.14 Embryos with a conditional knockout of the KLF2 gene in hematopoietic and endothelial cells die at E12.5 to E14.5, like KLF2−/− embryos, further emphasizing the important role of KLF2 in endothelial cells.36 There are no reports of EKLF−/− yolk sac endothelial cell abnormalities, even at E12.5.21 Interestingly, endothelial abnormalities are observed much earlier in EKLF−/−KLF2−/− than in KLF2−/− embryos. The role of KLF2 in adult vascular endothelial cells is under intensive investigation. In cultured vascular endothelial cells, KLF2 mRNA is increased in response to fluid shear stress, and this response is induced through a defined KLF2 gene promoter region.37,38 Recently, evidence has been mounting for the role of KLF2 in endothelial physiology, including atherosclerosis,39 thrombosis,40 inflammatory response,41 and angiogenesis.42
Because EKLF is specifically expressed in erythroid and not endothelial cells, inductive signaling between the 2 cell types could explain the earlier onset of endothelial abnormalities in EKLF−/−KLF2−/− than in KLF2−/− embryos. It has previously been reported that endothelial cell lines derived from the mouse yolk sac induce the in vitro proliferation and differentiation of erythroid progenitors from the yolk sac.43 It also has been reported that adult bone marrow erythroblasts are a source of angiogenic factors.44 A second, nonmutually exclusive, possibility is that EKLF and KLF2 have an important interaction in a common mesodermal progenitor to both erythroid and endothelial cells, which must occur for normal erythroid and endothelial cell development. In support of this hypothesis, we have observed that in the chicken, EKLF is expressed in the posterior primitive streak, prior to blood island formation.20
The precise cause of death of EKLF−/−KLF2−/− embryos has not been determined. The double-knockout embryos could survive, even with only about 20% of the normal amount of hemoglobin. There is increasing evidence that KLF2−/− embryos die between E12.5 and E14.5 due to vascular endothelial cell defects.36 E9.5 (Figure 5D) and E11.514,15 KLF2−/− embryos apparently do not have abnormal endothelial cells. EKLF/KLF2 double-knockout embryos die before E11.5, have abnormal yolk sac erythroid and endothelial cells at E9.5, and are anemic at E10.5. It is plausible that the EKLF−/−KLF2−/− embryos die prior to E11.5, earlier than single knockouts, due to a combination of endothelial and erythroid defects.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to Megan Smith, Sean Fox, Sue Walker, Judy Williamson, Julie Farnsworth, Francis White, Andrea Bellamy, and Vinodhini Krishnan for excellent technical assistance. Flow cytometry was partially funded by NIH P30 CA16059. Dr Stuart Fraser provided invaluable advice on the flow cytometry assays. We thank Dr Gordon D. Ginder for critical evaluation of this work and valuable suggestions.
This work was supported by NIH grants HL60080 and DK62154 to J.A.L.
National Institutes of Health
Authorship
Contribution: P.B., T.K.L., T.G.S., J.L.H., and J.A.L. wrote the paper, designed research, performed research, and analyzed data. D.C.W. Jr, W.L., and M.B. designed and performed research and analyzed data. L.C.R. designed and performed research. J.BL. contributed vital reagents, provided advice, and reviewed data.
P.B. and T.K.L. contributed equally to this work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Joyce A. Lloyd, Department of Human Genetics, Virginia Commonwealth University, Massey Cancer Center, Goodwin Research Lab, 401 College St, PO Box 980035, Richmond, VA 23298-0035; e-mail: jlloyd@vcu.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal