Abstract
The nuclear factor-κB (NF-κB) transcription factors play important roles in cancer development by preventing apoptosis and facilitating the tumor cell growth. However, the precise mechanisms by which NF-κB is constitutively activated in specific cancer cells remain largely unknown. In our current study, we now report that NF-κB–inducing kinase (NIK) is overexpressed at the pretranslational level in adult T-cell leukemia (ATL) and Hodgkin Reed-Sternberg cells (H-RS) that do not express viral regulatory proteins. The overexpression of NIK causes cell transformation in rat fibroblasts, which is abolished by a super-repressor form of IκBα. Notably, depletion of NIK in ATL cells by RNA interference reduces the DNA-binding activity of NF-κB and NF-κB–dependent transcriptional activity, and efficiently suppresses tumor growth in NOD/SCID/γcnull mice. These results indicate that the deregulated expression of NIK plays a critical role in constitutive NF-κB activation in ATL and H-RS cells, and suggest also that NIK is an attractive molecular target for cancer therapy.
Introduction
The nuclear factor-κB (NF-κB) transcription factors are known to regulate the expression of a wide range of genes involved in development, immune responses, apoptosis, and carcinogenesis as dimers of the REL family members, RelA, RelB, c-Rel, p50, and p52.1 The p50 and p52 proteins are generated by proteasome-mediated processing of their precursors, p105 and p100, respectively. In resting cells, Rel proteins are sequestered in the cytoplasm through their interactions with the ankyrin repeats of the inhibitory proteins IκBα, -β, and -ϵ, as well as the precursor proteins p105 and p100. On stimulation, signals converge at the multiprotein IκB kinase (IKK) complex, which is composed of 2 catalytic subunits, IKK1/α and IKK2/β, and the scaffolding proteins, NF-κB essential modulator (NEMO, also known as IKKγ) and ELKS.2 Phosphorylation by the IKK complex of specific serine residues on the IκB or precursor proteins results in their poly-ubiquitination and proteasome-dependent degradation or processing.2 Released NF-κB then translocates to the nucleus and regulates expression of target genes
NF-κB signaling pathways are largely classified as either canonical or noncanonical based on the stimuli and targets of the IKK complex.2 Canonical activation is induced by stimuli, such as tumor necrosis factor-α (TNFα) and interleukin-1β, and involves NEMO- and IKK2/β-dependent phosphorylation and the subsequent degradation of IκB proteins. Noncanonical NF-κB pathways are activated after the stimulation of a range of TNF receptor family members, such as B-cell activating factor belonging to the TNF family (BAFF) receptor, lymphotoxin-β receptor, Fn14 and CD40, and direct NF-κB–inducing kinase (NIK)- and IKK1/α-dependent phosphorylation and subsequent processing of p100, leading to activation of NF-κB complexes containing RelB.2,3 Of note in this context, the noncanonical pathways operate in a delayed fashion and are sensitive to protein synthesis inhibition.4,5
Compared with the mechanisms underlying the transduction of ligand-induced signaling to NF-κB activation, much less is known about how NF-κB is constitutively activated in a variety of cancer cells.6 Constitutively high NF-κB activity has typically been demonstrated in human hematopoietic cancer cells, including adult T-cell leukemia (ATL), Hodgkin lymphoma, and multiple myeloma cells.7,8 We have previously reported the aberrant expression of p52 in ATL and Hodgkin Reed-Sternberg (H-RS) cells that do not express viral regulatory proteins, such as Tax of the human T-cell leukemia virus or latent membrane protein 1 of the Epstein-Barr virus.9,10 In addition, IKK activation in ATL and H-RS cells was found to be sensitive to protein synthesis inhibition.10,11 These results indicate that the noncanonical pathways of NF-κB activation operate in these cancer cells. Aberrant p52 expression has also been reported in other types of cancer cells, including breast,12 prostate,13 pancreas,14 and colon.15 However, the actual triggers of noncanonical NF-κB activation in these cancer cells remain largely unknown except for certain multiple myeloma cells that have mutations in the NIK, TRAF3, and related genes.16,17
N1K is a serine-threonine kinase that is an essential participant in the induction of the IKK1-dependent processing of p100 as well as IκB degradation in response to stimuli, such as CD70, CD40 ligand, and BAFF.18 It has also been reported previously that the IKK complex is recruited to CD27 in a manner dependent on NIK function. However, the mechanism by which NIK activity is regulated thereafter was unknown until it was recently demonstrated that these stimuli protect basally translated endogenous NIK protein from proteasome-mediated degradation.19,20 Liao et al reported that the interaction of NIK with TNF receptor–associated factor 3 (TRAF3) is responsible for the rapid degradation of NIK and that noncanonical NF-κB stimuli induce the degradation of TRAF3 and the elevation of NIK expression.19 In a separate study, Qing et al have demonstrated that noncanonical NF-κB stimuli stabilize the NIK protein but do not modify its RNA expression or protein translation.20 The findings of these studies explain the delay in triggering the noncanonical pathway and its high sensitivity to protein synthesis inhibition.
Because NIK is a central regulator of the noncanonical pathway of NF-κB activation, we have investigated in our current study how this kinase is regulated in hematopoietic cancer cells, in which IKK is constitutively activated in the absence of viral regulators.
Methods
Cell culture
ED40515(−),21 ATL-43Tb(−),22 and TL-Om123 are human T-cell leukemia virus type-I (HTLV-I)–infected T-cell lines established from the leukemic cells of ATL patients. The H-RS cell lines, HDLM-2, L428, and L540, were purchased from the German Collection of Micro-organisms and Cell Cultures (Braunschweig, Germany). CEM24 and Jurkat25 are HTLV-I–free human T-lymphoblastic leukemia cell lines. A human B-cell line, Romas RG69,20 was a kind gift from Dr Gutian Xiao (State University of New Jersey, Piscataway, NJ). Primary leukemia cells derived from ATL patients were obtained under informed consent at Imamura Bun-in Hospital and supplied through the Joint Study on Predisposing Factors of ATL Development. The patients were diagnosed with ATL on the basis of clinical and hematologic features and the presence of antibodies to ATL-associated antigens in serum and of the HTLV-I proviral genome in the leukemia cells. Use of peripheral blood lymphocytes from ATL patients for research purposes was approved by the institutional review board of each institute. Peripheral blood mononuclear cells (PBMCs) derived from healthy donors were also obtained under informed consent. PBMCs were isolated from both ATL patients and healthy donors by density gradient separation with Ficoll-Plaque PLUS (Amersham Biosciences, Uppsala, Sweden). Cells were maintained in RPMI 1640 supplemented with 10% fetal bovine serum, 100 U/mL penicillin G, and 100 μg/mL streptomycin sulfate; 5R is a NEMO-deficient subline of the Rat-1 cell line and has been described previously.26 B5 and h12 are sublines of Rat-1 and 5R, respectively, express the blasticidin deaminase gene under the control of an NF-κB–dependent promoter, and have also been described previously.26,27 Plat-E packaging cells were described previously.28 B5, h12, Plat-E, 293T cells, and mouse embryonic fibroblasts were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, 100 U/mL penicillin G, and 100 μg/mL streptomycin sulfate. Anchorage-independent cell growth was examined essentially as described previously.29 Images were captured using an inverted microscope (IX70, Olympus, Tokyo, Japan) and processed with Openlab 3.0.2 software (Improvision, Coventry, United Kingdom). Cells used in this study were all maintained at 37°C in air containing 5% CO2.
Virus infection and transfection
Plat-E cells were transfected with pMRX-HA-NIK-ires-puro, pMRX-HA-kd-NIK-ires-puro, or pMRX-HA-ires-puro (EV1) (Document S1, available on the Blood website; see the Supplemental Materials link at the top of the online article) using the calcium phosphate precipitation method. Culture supernatants were collected 48 hours after transfection and filtered. B5 and h12 cells were infected for 2 hours in the presence of 10 μg/mL polybrene. Infected cells were then cultured in medium containing 2 μg/mL puromycin, and cell clones were isolated. Rat fibroblasts expressing SR-IκBα or its empty control vector (EV2) were established essentially as described previously.10 For production of lentiviruses, 293T cells were cotransfected with pCS-puro-Ctli, pCS-puro-NIKi-1, or pCS-puro-NIKi-2 (Document S1) together with the pCMVΔR8.2 packaging construct and pHCMV-VSV-G (kind gifts from Dr I.S.Y. Chen) using FuGENE 6 (Roche Applied Science, Indianapolis, IN). Culture supernatants were collected 48 hours after transfection and filtered. ED40515(−) and ATL-43Tb(−) cells were infected once or twice with 24 hours interval with these lentiviruses for 6 hours in the presence of 10 μg/mL polybrene. At 48 hours after the infection, cells were cultured in medium containing 2 μg/mL puromycin for an additional 48 hours. These infectants were subjected to immunoblotting, electrophoretic mobility shift assay (EMSA), and transient transfection with 2 μg of IgκCona-luc30 and pEF1-LacZ26 using DMRIE-C (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Assays for luciferase and β-galactosidase were performed 48 hours after transfection by standard methods. Luciferase activity was normalized on the basis of β-galactosidase activity. The growth of lentivirus-infected cells was determined by the trypan blue staining method.
Immunoprecipitation
For the immunoprecipitation of endogenous NIK, approximately 2 × 107 cells were lysed in buffer A (20 mM Tris-HCl, pH7.5, 0.5% Nonidet P-40, 150 mM NaCl supplemented with 1 μg/mL aprotinin, 1 μg/mL leupeptin, 0.57 mM phenylmethanesulphonylfluoride, 10 μM MG132, 10 μM MG115) followed by preclearing with purified rabbit IgG (Cedarlane Laboratories, Hornby, ON) and protein G-Sepharose beads (Pierce Biotechnology, Rockford, IL). After centrifugation at 14000 rpm for 3 minutes, supernatants were subjected to immunoprecipitation with purified nonimmune rabbit IgG or anti-NIK antibody (#4994) (Cell Signaling Technology, Danvers, MA). Immunoprecipitates were washed 3 times with TNT buffer (20 mM Tris-HCl, pH 7.5, 200 mM NaCl, and 1% Triton X-100). Endogenous NIK proteins were detected by immunoblotting with anti-NIK antibody (#4994). For the immunoprecipitation of HA-tagged NIK, 750 μg cell lysates prepared with buffer A was subjected to immunoprecipitation with anti-HA antibody (12CA5, a kind gift from Dr A. Israël, Institut Pasteur Paris, Paris, France). Immunoprecipitates were washed 3 times with TNT buffer. HA-tagged NIK proteins were detected by immunoblotting with anti-NIK antibody. For immunoprecipitation of endogenous IKK1/2, 1500 μg cell lysates prepared with buffer A were subjected to immunoprecipitation with anti-IKK1 monoclonal antibody (B78-1; BD PharMingen, San Diego, CA) or purified mouse IgG2b (MI10-104; Bethyl Laboratories, Montgomery, TX). Immunoprecipitates were washed 3 times with TNT buffer. Expression of endogenous proteins was detected by immunoblotting with antiphospho-IKK1/IKK2 (Ser180/Ser181) (#2681; Cell Signaling Technology), anti-IKK1 (H-744), or anti-IKK2 (H-470; Santa Cruz Biotechnology, Santa Cruz, CA) antibodies.
Quantitative RT-PCR
Total RNA was extracted using Isogen reagents (Nippon Gene, Tokyo, Japan) according to the manufacturer's instructions. Quantitative RT-PCR amplifications were performed with 100 ng total RNA, 0.3 μM of each primer, and 0.25 μM TaqMan probe using an ABI-7700 Sequence Detector (Applied Biosystems, Foster City, CA): reverse transcription was performed at 48°C for 30 minutes, Taq DNA polymerase was activated at 95°C for 10 minutes, followed by 45 amplification cycles of 95°C for 15 seconds, and annealing and extension at 60°C for 1 minute. The NIK, VEGF, ICAM-1, and MMP-9 mRNA levels were normalized based on the amount of 18S ribosomal RNA determined simultaneously by the real-time RT-PCR.
Mice and inoculation of cells
NOD/SCID/γcnull (NOG)31 mice were purchased from the Central Institute for Experimental Animals (Kawasaki, Japan). All mice were maintained under specific pathogen-free conditions in the Animal Center of Tokyo Medical and Dental University (Tokyo, Japan). The Ethical Review Committee of the institute approved the experimental protocol. ED40515(−) cells expressing Ctli or NIKi-1 and -2 were washed twice with serum-free RPMI 1640 and resuspended in the same medium. Mice were anesthetized with ether and inoculated subcutaneously in the postauricular region with 5 × 106 cells per mouse, as described previously.31 We measured tumor size and weight 2 weeks after cell inoculation.
Statistics
Statistical significance was evaluated using a 2-tailed, unpaired Student's t test. P values less than .05 were considered to be significant.
Results
NIK is aberrantly expressed in both adult T-cell leukemia and Hodgkin Reed-Sternberg cells
The constitutive processing of p100 to p52 in ATL and H-RS cells9,10 prompted us to examine whether NIK is aberrantly expressed in both established and primary ATL cells. Immunoblotting of whole-cell lysates prepared from ATL or H-RS cells did not show any detectable NIK signal (data not shown); however, when endogenous NIK was immunoprecipitated from approximately 20 million of these cells and subjected to immunoblotting, NIK was specifically detectable in anti-NIK immunoprecipitates from ATL and H-RS cells, but not from control cells, such as CEM and Jurkat (Figure 1A). Previous studies revealed that inhibition of the proteasome function allowed for detection of endogenous NIK in simple whole-cell lysates of B-cell lines.19,20 Treatment of ED-40515(−) cells with the MG132 proteasome inhibitor for 3 hours before harvesting enabled us to observe robust endogenous NIK expression at the expected position (Figure 1B). Lysates of 293T cells with or without exogenous NIK expression were used as the positive and negative controls, respectively. We next examined the NIK expression levels as well as those of p100 phosphorylated at serine residues 866 and 870 in a panel of ATL, H-RS, and control cells (Figure 1C). No appreciable NIK expression could be observed in control CEM and Jurkat T-cell lines treated with MG132, in which NF-κB is not constitutively activated. Proteasome inhibition induced strong NIK expression in other Tax-negative ATL-derived cell lines, ATL-43Tb(−) and TL-Om1. Proteasome inhibition also strongly augmented NIK expression in H-RS cells, but only weakly so in the control B-cell lines, RG69. These results indicate that the steady-state levels of NIK of the authentic size are elevated in ATL and H-RS cells, and suggest that NIK may be abundantly produced in ATL and H-RS cells, but is rapidly degraded by the proteasome. The levels of NIK expression correlated well with those of phosphorylated p100 (Figure 1C). Moreover, p52 and the phoshorylated form of IκBα were also abundant in ATL and H-RS cell lines, but not in the control T-cell lines (Figure 1C). These results indicate that the overexpression of NIK is closely linked to the downstream events leading to constitutive activation of the canonical and noncanonical NF-κB pathways in ATL and H-RS cells. A previous study suggested that L428 cells express a C-terminally truncated form of IκBα and that the phosphorylated form of this protein was accumulated after treatment of the cells with proteasome inhibitor or dexamethasone.32,33 In agreement with this, we did not detect IκBα expression with the antibody used in this study, which recognizes the C-terminus of the protein, but detected the phoshorylated form of this IκBα only after treatment with MG132 (data not shown).
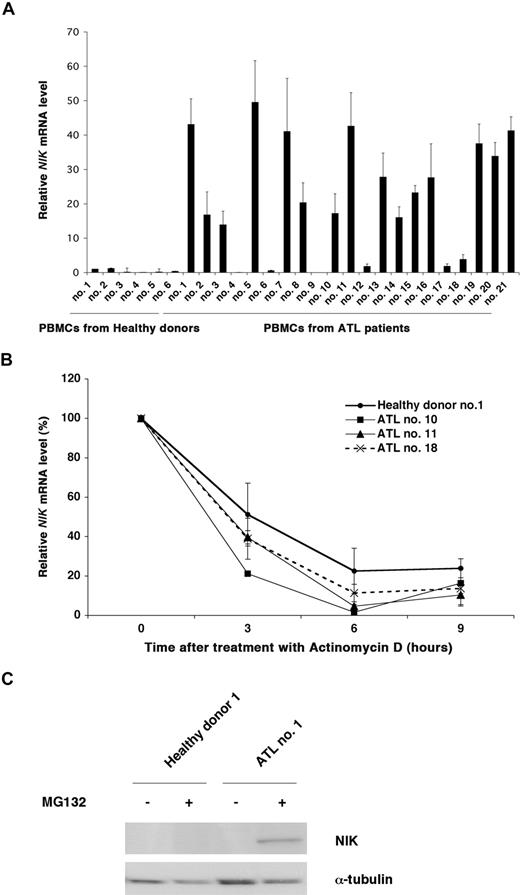
NIK protein is overexpressed in established ATL and Hodgkin Reed-Sternberg cells. (A) Steady-state levels of NIK expression in the ATL and H-RS cell lines were revealed by immunoprecipitation-coupled immunoblotting. Approximately 2 × 107 cells were lysed with buffer A. After preclearing, immunoprecipitation was performed at 4°C, using anti-NIK antibody (NIK) or its isotype IgG (IgG). After 3 washes with TNT buffer, immune complexes were analyzed by immunoblotting with anti-NIK antibody. (B) 293T cells were transfected with pMRX-HA-iresPuro or pMRX-HA-NIKiresPuro for 24 hours. Whole-cell lysates were used as negative and positive controls. ED40515(−) cells were pretreated with (+) or without (−) MG132 (20 μM) for 3 hours, lysed with RIPA buffer, and subjected to immunoblotting with anti-NIK or anti-α-tubulin antibodies. Immunoprecipitation-coupled immunoblotting was performed as in panel A. (C) Top panels: control T-cell lines (CEM and Jurkat), leukemic cell lines derived from ATL patients that do not express Tax (ED40515(−), ATL43-Tb(−), and TL-Om1, a control B-cell line (RG69), and H-RS cell lines (HDLM-2 and L540) were pretreated with (+) or without (−) MG132 (20 μM) for 3 hours, and 30 μg of the whole-cell extracts were subjected to Western blot analysis with the antibodies to the indicated proteins. Bottom panels: Whole-cell extracts from the indicated cell lines were analyzed by Western blotting with the antibodies to the indicated proteins. (D) Total RNA was extracted from the indicated cell lines and subjected to real-time RT-PCR to quantify the NIK mRNA levels. The NIK mRNA levels were normalized to 18S RNA. The relative NIK mRNA levels shown represent the fold increases in mRNA abundance, relative to that of the CEM cells (arbitrarily set at 1). (E) Cells were cultured in the presence of actinomycin D (5 μg/mL) for the times indicated, and then total RNA was isolated and subjected to quantitative RT-PCR as in panel D. Data are expressed as mean plus or minus SD of 3 independent experiments. The relative amounts of NIK mRNA shown represent the percentages in mRNA abundance, relative to that of each cell line before the addition of actinomycin D (arbitrarily set at 100%). IB indicates immunoblotting; IP, immunoprecipitation.
NIK protein is overexpressed in established ATL and Hodgkin Reed-Sternberg cells. (A) Steady-state levels of NIK expression in the ATL and H-RS cell lines were revealed by immunoprecipitation-coupled immunoblotting. Approximately 2 × 107 cells were lysed with buffer A. After preclearing, immunoprecipitation was performed at 4°C, using anti-NIK antibody (NIK) or its isotype IgG (IgG). After 3 washes with TNT buffer, immune complexes were analyzed by immunoblotting with anti-NIK antibody. (B) 293T cells were transfected with pMRX-HA-iresPuro or pMRX-HA-NIKiresPuro for 24 hours. Whole-cell lysates were used as negative and positive controls. ED40515(−) cells were pretreated with (+) or without (−) MG132 (20 μM) for 3 hours, lysed with RIPA buffer, and subjected to immunoblotting with anti-NIK or anti-α-tubulin antibodies. Immunoprecipitation-coupled immunoblotting was performed as in panel A. (C) Top panels: control T-cell lines (CEM and Jurkat), leukemic cell lines derived from ATL patients that do not express Tax (ED40515(−), ATL43-Tb(−), and TL-Om1, a control B-cell line (RG69), and H-RS cell lines (HDLM-2 and L540) were pretreated with (+) or without (−) MG132 (20 μM) for 3 hours, and 30 μg of the whole-cell extracts were subjected to Western blot analysis with the antibodies to the indicated proteins. Bottom panels: Whole-cell extracts from the indicated cell lines were analyzed by Western blotting with the antibodies to the indicated proteins. (D) Total RNA was extracted from the indicated cell lines and subjected to real-time RT-PCR to quantify the NIK mRNA levels. The NIK mRNA levels were normalized to 18S RNA. The relative NIK mRNA levels shown represent the fold increases in mRNA abundance, relative to that of the CEM cells (arbitrarily set at 1). (E) Cells were cultured in the presence of actinomycin D (5 μg/mL) for the times indicated, and then total RNA was isolated and subjected to quantitative RT-PCR as in panel D. Data are expressed as mean plus or minus SD of 3 independent experiments. The relative amounts of NIK mRNA shown represent the percentages in mRNA abundance, relative to that of each cell line before the addition of actinomycin D (arbitrarily set at 100%). IB indicates immunoblotting; IP, immunoprecipitation.
We next investigated NIK expression at the mRNA level by quantitative PCR (Figure 1D) and found that that NIK transcripts were at between 20- and 100-fold higher levels in ATL and H-RS cells, compared with CEM cells. Next, actinomycin D was used to block new mRNA synthesis, so that decay of existing transcripts could be detected. Quantitative PCR analyses revealed that the half-life of NIK mRNA was approximately 3 hours both in the ATL and control T cells (Figure 1E). Essentially similar results were obtained with the other cell lines shown in Figure 1D, including H-RS cell lines (data not shown). A previous report has demonstrated that NF-κB is constitutively activated in primary ATL cells in the peripheral blood.34 We therefore quantified the NIK mRNA levels in PBMCs from both healthy donors and ATL patients (Figure 2A), and found that NIK mRNA is overexpressed in PBMCs of 15 of 21 ATL patients. Actinomycin D treatment of PBMCs further revealed that NIK mRNA was not apparently stabilized in primary ATL cells (Figure 2B). Moreover, fluorescence in situ hybridization studies on primary ATL cells failed to detect amplification or translocation of the NIK gene (Figure S1; Table S2). Finally, when PBMCs were cultured for 3 hours in the presence of MG132, NIK protein was detectable in cells from an ATL patient showing abundant NIK mRNA expression, but not in those from a healthy donor (Figure 2C).
Overexpression of the NIK mRNA and protein in PBMCs from ATL patients. (A) Total RNA was extracted from PBMCs from healthy donors and ATL patients and then subjected to quantitative RT-PCR. The NIK mRNA levels were normalized to 18S RNA. The relative nik mRNA levels shown represent the fold increases in mRNA abundance relative to that of healthy donor 1 (arbitrarily set at 1). These data are expressed as the mean plus or minus SD of 3 independent experiments. (B) PBMCs were cultured in the presence of actinomycin D (5 μg/mL) for the times indicated, and then total RNA was isolated and subjected to quantitative RT-PCR. The relative amounts of NIK mRNA shown represent the percentages in mRNA abundance, relative to that of PBMCs before the addition of actinomycin D (arbitrarily set at 100%). (C) PBMCs from a healthy donor and an ATL patient were treated with (+) or without (−) MG132 (20 μM) for 3 hours, lysed with RIPA buffer, and subjected to immunoblotting with anti-NIK or anti-α-tubulin antibodies.
Overexpression of the NIK mRNA and protein in PBMCs from ATL patients. (A) Total RNA was extracted from PBMCs from healthy donors and ATL patients and then subjected to quantitative RT-PCR. The NIK mRNA levels were normalized to 18S RNA. The relative nik mRNA levels shown represent the fold increases in mRNA abundance relative to that of healthy donor 1 (arbitrarily set at 1). These data are expressed as the mean plus or minus SD of 3 independent experiments. (B) PBMCs were cultured in the presence of actinomycin D (5 μg/mL) for the times indicated, and then total RNA was isolated and subjected to quantitative RT-PCR. The relative amounts of NIK mRNA shown represent the percentages in mRNA abundance, relative to that of PBMCs before the addition of actinomycin D (arbitrarily set at 100%). (C) PBMCs from a healthy donor and an ATL patient were treated with (+) or without (−) MG132 (20 μM) for 3 hours, lysed with RIPA buffer, and subjected to immunoblotting with anti-NIK or anti-α-tubulin antibodies.
NIK transforms rat fibroblasts in an NF-κB–dependent manner
To further explore the roles for NIK during cell transformation, we infected the 3T3-like rat fibroblast cell line Rat-1 with a retroviral vector expressing human NIK and examined its oncogenic activity. As expected, cells transduced with this NIK vector exhibited strong NF-κB DNA binding activity within 36 hours (data not shown). Rat-1 cells transduced with a control retrovirus became resistant to the selection marker puromycin approximately 24 hours after infection and continued to proliferate rapidly. In contrast, Rat-1 cells transduced with the NIK expression vector expressed a readily detectable level of NIK, had a transformed morphology, but ceased proliferating and died within 3 to 4 days after becoming resistant to puromycin. Cells that survived 2 weeks of puromycin selection after NIK transduction eventually appeared indistinguishable from those transduced with the control vector and showed no detectable NIK expression or NF-κB DNA binding activity (data not shown).
Based on these observations, we speculate that the retroviral overexpression of NIK is toxic to the cells so that only cells that had lost its expression could emerge from the puromycin-resistant pools. To address this problem, we used B5 and h12 cells carrying an integrated Igκ2bsrH plasmid that confers resistance to the antibiotic blasticidin S when cells are constitutively expressing active NF-κB.26 B5 cells are derived from Rat-1 cells, and h12 cells are from 5R cells that lack NEMO expression. When the B5 and h12 cells were transduced with the wild-type NIK retroviral expression vector and subjected to selection with both puromycin and blasticidin S, the majority of the resultant cell clones maintained detectable NIK expression (Figure 3A), elevated catalytic activity of IKK (Figure 4), and the initial transformed morphology (Figure 5B). On the other hand, when B5 and h12 cells were transduced with a retrovirus vector expressing a catalytically inactive mutant form of NIK and selected with puromycin alone, the cells successfully expressed this protein (Figure 3A) without significant morphologic change (Figure 5B) or constitutive NF-κB activation (Figure 3C). As expected, these cells failed to survive selection with blasticidin S (data not shown).
NIK induces constitutive NF-κB activity in rat fibroblasts. (A) B5 and h12 cells were infected with retroviruses capable of expressing HA-tagged NIK (NIK) or catalytically inactive NIK (kd-NIK). Pools of B5 and h12 cells transduced with the control pMRX-HAiresPuro vector (EV1) were used as a control. Cytoplasmic extracts from EV1 and 2 independent cell clones (no. 1 and no. 2) were subjected to immunoprecipitation using antibody against the HA epitope. Immunoprecipitates were then resolved by 8% SDS-PAGE and subjected to immunoblotting with anti-NIK antibody. 293T cells were transiently transfected with the pMRX-HAiresPuro vector (EV1) or pMRX-HA-NIKiresPuro (NIK). Cytoplasmic extracts (30 μg) were then used for immunoblotting as negative and positive controls, respectively. (B) Elevated p52 production in rat fibroblasts. Whole-cell lysates from B5 and h12 cells expressing wild-type NIK or kd-NIK were subjected to SDS-PAGE and immunoblotting with anti-p52 for detection of p100 and p52 or antiactin antibodies. (C) Elevated NF-κB–DNA binding activity in rat fibroblasts; 5 μg of nuclear extracts prepared from B5 and h12 cells expressing wild-type NIK or kd-NIK were analyzed by EMSA, using oligonucleotides encoding an NF-κB–binding sequence or Oct-1–binding sequence as probes. (D) DNA-binding NF-κB components in B5 and h12 cells expressing wild-type NIK were analyzed by super-shift EMSA. Nuclear extracts (5 μg) from B5 NIK#1 and h12 NIK#2 cells were preincubated for 30 minutes with preimmune (PI), anti-p50, anti-RelA or anti-RelB sera, and then subjected to EMSA with the NF-κB–specific probe. IB indicates immunoblotting; IP, immunoprecipitation.
NIK induces constitutive NF-κB activity in rat fibroblasts. (A) B5 and h12 cells were infected with retroviruses capable of expressing HA-tagged NIK (NIK) or catalytically inactive NIK (kd-NIK). Pools of B5 and h12 cells transduced with the control pMRX-HAiresPuro vector (EV1) were used as a control. Cytoplasmic extracts from EV1 and 2 independent cell clones (no. 1 and no. 2) were subjected to immunoprecipitation using antibody against the HA epitope. Immunoprecipitates were then resolved by 8% SDS-PAGE and subjected to immunoblotting with anti-NIK antibody. 293T cells were transiently transfected with the pMRX-HAiresPuro vector (EV1) or pMRX-HA-NIKiresPuro (NIK). Cytoplasmic extracts (30 μg) were then used for immunoblotting as negative and positive controls, respectively. (B) Elevated p52 production in rat fibroblasts. Whole-cell lysates from B5 and h12 cells expressing wild-type NIK or kd-NIK were subjected to SDS-PAGE and immunoblotting with anti-p52 for detection of p100 and p52 or antiactin antibodies. (C) Elevated NF-κB–DNA binding activity in rat fibroblasts; 5 μg of nuclear extracts prepared from B5 and h12 cells expressing wild-type NIK or kd-NIK were analyzed by EMSA, using oligonucleotides encoding an NF-κB–binding sequence or Oct-1–binding sequence as probes. (D) DNA-binding NF-κB components in B5 and h12 cells expressing wild-type NIK were analyzed by super-shift EMSA. Nuclear extracts (5 μg) from B5 NIK#1 and h12 NIK#2 cells were preincubated for 30 minutes with preimmune (PI), anti-p50, anti-RelA or anti-RelB sera, and then subjected to EMSA with the NF-κB–specific probe. IB indicates immunoblotting; IP, immunoprecipitation.
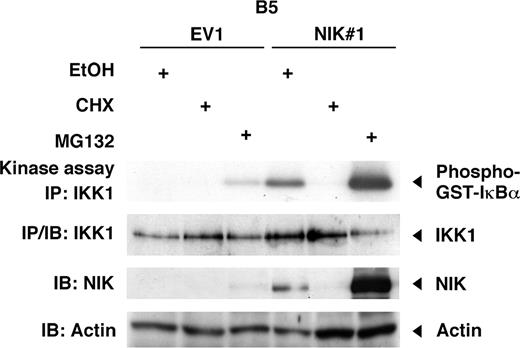
NIK expression parallels IKK activity after CHX or MG132 treatment. B5 cells transduced with the control vector (EV1) or B5 cells expressing wild-type NIK (NIK#1) were treated for 4 hours with either vehicle (ethanol, EtOH), cycloheximide (CHX; 50 μg/mL), or MG132 (20 μM). Cytoplasmic extracts were subjected to immunoprecipitation with IKK1-specific antibody, and then immunoprecipitates were used for an in vitro kinase assay. IKK1 expression in the immunoprecipitates was revealed by immunoblotting with IKK1-specific antibody. NIK and actin levels in the cytoplasmic extracts used for immunoprecipitation were determined by immunoblotting with anti-NIK or antiactin antibodies, respectively. IB indicates immunoblotting; IP, immunoprecipitation; GST, glutathione-S-transferase tag.
NIK expression parallels IKK activity after CHX or MG132 treatment. B5 cells transduced with the control vector (EV1) or B5 cells expressing wild-type NIK (NIK#1) were treated for 4 hours with either vehicle (ethanol, EtOH), cycloheximide (CHX; 50 μg/mL), or MG132 (20 μM). Cytoplasmic extracts were subjected to immunoprecipitation with IKK1-specific antibody, and then immunoprecipitates were used for an in vitro kinase assay. IKK1 expression in the immunoprecipitates was revealed by immunoblotting with IKK1-specific antibody. NIK and actin levels in the cytoplasmic extracts used for immunoprecipitation were determined by immunoblotting with anti-NIK or antiactin antibodies, respectively. IB indicates immunoblotting; IP, immunoprecipitation; GST, glutathione-S-transferase tag.
The overexpression of NIK transforms rat fibroblasts in an NF-κB–dependent manner. (A) Top 2 panels: 5 μg of nuclear extracts prepared from B5 and h12 cells transduced with empty vector (EV2) or SR-IκBα (SR) were analyzed by EMSA, using NF-κB and Oct-1 probes. Middle 5 panels: whole-cell extracts (30 μg) of B5 or h12 infectants were subjected to SDS-PAGE and immunoblotting with anti-p52, antiphospho-IκBα, anti-IκBα, or antiactin antibodies. Bottom panel: HA-tagged NIK was immunoprecipitated from B5 and h12 infectants with anti-HA antibody and detected by immunoblotting with anti-NIK antibody (H-248). (B) Phase-contrast micrographs of cells cultured on monolayers (top images) or in soft agar (bottom images). B5 or h12 cell clones expressing wild-type NIK (NIK#1 and NIK#2) or not (EV1) were cultured in soft agar for 3 weeks. These cells were further transduced with SR-IκBα, and then pooled cells were assayed for anchorage-independent growth in soft agar. B5 and h12 cell clones expressing kd-NIK were also examined. Original magnification ×100. Scale bar represents 100 μm. SR indicates super-repressor; kd-NIK, catalytically inactive NIK; IB, immunoblotting; IP, immunoprecipitation.
The overexpression of NIK transforms rat fibroblasts in an NF-κB–dependent manner. (A) Top 2 panels: 5 μg of nuclear extracts prepared from B5 and h12 cells transduced with empty vector (EV2) or SR-IκBα (SR) were analyzed by EMSA, using NF-κB and Oct-1 probes. Middle 5 panels: whole-cell extracts (30 μg) of B5 or h12 infectants were subjected to SDS-PAGE and immunoblotting with anti-p52, antiphospho-IκBα, anti-IκBα, or antiactin antibodies. Bottom panel: HA-tagged NIK was immunoprecipitated from B5 and h12 infectants with anti-HA antibody and detected by immunoblotting with anti-NIK antibody (H-248). (B) Phase-contrast micrographs of cells cultured on monolayers (top images) or in soft agar (bottom images). B5 or h12 cell clones expressing wild-type NIK (NIK#1 and NIK#2) or not (EV1) were cultured in soft agar for 3 weeks. These cells were further transduced with SR-IκBα, and then pooled cells were assayed for anchorage-independent growth in soft agar. B5 and h12 cell clones expressing kd-NIK were also examined. Original magnification ×100. Scale bar represents 100 μm. SR indicates super-repressor; kd-NIK, catalytically inactive NIK; IB, immunoblotting; IP, immunoprecipitation.
The expression of wild-type NIK in B5 and h12 cells potently induces p52 expression and NF-κB DNA binding activity, whereas the catalytically inactive NIK mutant does not (Figure 3B,C). We also found a specifically phosphorylated form of IκBα in cells expressing wild-type NIK (Figure 3A). Super-shift experiments revealed that the NF-κB–DNA binding complexes in B5 and h12 cells expressing NIK involve p50, RelB, and RelA (Figure 4D). The presence of p52 in the DNA binding complexes could not be examined, however, because an antibody recognizing rat p52 in super-shift assay is not currently available. Instead, we analyzed DNA-binding complexes induced by NIK expression in wild-type mouse embryonic fibroblasts (Figure S2). Retroviral overexpression of NIK indeed induced DNA-binding NF-κB complexes containing p52, and enhanced expression of p52 and phosphorylated form of IκBα. We have previously demonstrated that the treatment of ATL cells with MG132 greatly enhances IKK activity, whereas protein synthesis inhibition quickly abolished this activity.11 Figure 4 shows that the IKK activity in B5 cells stably expressing NIK (NIK#1) is modulated by MG132 and cycloheximide (CHX) in a manner that is very similar to that seen in ATL cells. In addition, treatment of NIK#1 cells with MG132 remarkably elevates the level of exogenous NIK expression. The constitutive NF-κB activation caused by the presence of exogenous NIK was found to be abolished by the retroviral expression of a super-repressor form of IκBα (SR-IκBα), without affecting exogenous NIK expression (Figure 5A). Interestingly, the forced expression of SR-IκBα also diminishes the p52 and p100 expression levels.
We next tested the ability of NIK to induce anchorage-independent growth of rat fibroblasts. B5 and h12 cells transduced with the control vector did not form colonies of significant size in soft agar, whereas those transduced with wild-type NIK expression vector formed a number of large colonies, as shown in Figure 5B and Table 1. Cells expressing catalytically inactive NIK failed to form colonies in soft agar. The expression of SR-IκBα completely abolished NIK-induced colony formation and also the morphologic alterations of B5 and h12 cells. Given that SR-IκBα specifically suppresses NF-κB activation, we conclude from these results that NIK transforms rat fibroblasts in an NF-κB–dependent manner.
Efficiency of colony formation in soft agar
| Cells* . | Colony-forming efficiency, % . | Average size of colonies, μm† . |
|---|---|---|
| B5-EV1 | 0.7 ± 0.5 | 62.6 ± 1.5 |
| B5-NIK#1 | 23.2 ± 2.0‡ | 236.2 ± 12.6‡ |
| B5-NIK#2 | 18.9 ± 2.4‡ | 184.1 ± 19.8‡ |
| B5-kd-NIK#1 | 1.5 ± 0.3 | 63.1 ± 1.4 |
| B5-kd-NIK#2 | 1.3 ± 0.1 | 62.8 ± 1.8 |
| h12-EV1 | 1.2 ± 0.3 | 60.5 ± 0.0 |
| h12-NIK#1 | 12.8 ± 1.7‡ | 146.9 ± 4.6‡ |
| h12-NIK#2 | 17.7 ± 1.7‡ | 154.9 ± 5.6‡ |
| h12-kd-NIK#1 | 1.4 ± 1.0 | 61.5 ± 2.1 |
| h12-kd-NIK#2 | 1.5 ± 0.4 | 62.5 ± 4.7 |
| B5-EV1-EV2 | 1.2 ± 0.3 | 61.8 ± 1.1 |
| B5-NIK#1-EV2 | 21.1 ± 1.0‡ | 193.8 ± 3.7‡ |
| B5-NIK#2-EV2 | 14.3 ± 1.0‡ | 150.4 ± 8.7‡ |
| h12-EV1-EV2 | 1.5 ± 0.7 | 60.8 ± 0.4 |
| h12-NIK#1-EV2 | 12.3 ± 1.7‡ | 119.4 ± 5.6‡ |
| h12-NIK#2-EV2 | 14.0 ± 1.8‡ | 160.3 ± 7.2‡ |
| B5-EV1-SR-IκBα | 1.5 ± 0.0 | 61.7 ± 0.5 |
| B5-NIK#1-SR-IκBα | 3.4 ± 0.0 | 64.8 ± 1.1 |
| B5-NIK#2-SR-IκBα | 3.9 ± 0.1 | 63.3 ± 0.4 |
| h12-EV1-SR-IκBα | 1.7 ± 1.0 | 61.3 ± 0.4 |
| h12-NIK#1-SR-IκBα | 2.7 ± 0.3 | 62.3 ± 0.1 |
| h12-NIK#2-SR-IκBα | 3.4 ± 1.4 | 61.4 ± 0.2 |
| Cells* . | Colony-forming efficiency, % . | Average size of colonies, μm† . |
|---|---|---|
| B5-EV1 | 0.7 ± 0.5 | 62.6 ± 1.5 |
| B5-NIK#1 | 23.2 ± 2.0‡ | 236.2 ± 12.6‡ |
| B5-NIK#2 | 18.9 ± 2.4‡ | 184.1 ± 19.8‡ |
| B5-kd-NIK#1 | 1.5 ± 0.3 | 63.1 ± 1.4 |
| B5-kd-NIK#2 | 1.3 ± 0.1 | 62.8 ± 1.8 |
| h12-EV1 | 1.2 ± 0.3 | 60.5 ± 0.0 |
| h12-NIK#1 | 12.8 ± 1.7‡ | 146.9 ± 4.6‡ |
| h12-NIK#2 | 17.7 ± 1.7‡ | 154.9 ± 5.6‡ |
| h12-kd-NIK#1 | 1.4 ± 1.0 | 61.5 ± 2.1 |
| h12-kd-NIK#2 | 1.5 ± 0.4 | 62.5 ± 4.7 |
| B5-EV1-EV2 | 1.2 ± 0.3 | 61.8 ± 1.1 |
| B5-NIK#1-EV2 | 21.1 ± 1.0‡ | 193.8 ± 3.7‡ |
| B5-NIK#2-EV2 | 14.3 ± 1.0‡ | 150.4 ± 8.7‡ |
| h12-EV1-EV2 | 1.5 ± 0.7 | 60.8 ± 0.4 |
| h12-NIK#1-EV2 | 12.3 ± 1.7‡ | 119.4 ± 5.6‡ |
| h12-NIK#2-EV2 | 14.0 ± 1.8‡ | 160.3 ± 7.2‡ |
| B5-EV1-SR-IκBα | 1.5 ± 0.0 | 61.7 ± 0.5 |
| B5-NIK#1-SR-IκBα | 3.4 ± 0.0 | 64.8 ± 1.1 |
| B5-NIK#2-SR-IκBα | 3.9 ± 0.1 | 63.3 ± 0.4 |
| h12-EV1-SR-IκBα | 1.7 ± 1.0 | 61.3 ± 0.4 |
| h12-NIK#1-SR-IκBα | 2.7 ± 0.3 | 62.3 ± 0.1 |
| h12-NIK#2-SR-IκBα | 3.4 ± 1.4 | 61.4 ± 0.2 |
kd-NIK indicates catalytically inactive NIK; SR, super-repressor; EV1, empty vector for NIK or kd-NIK; and EV2, empty vector for SR-IκBα.
Cells were inoculated in 0.33% soft agar and cultured for 3 weeks.
Colonies larger than 60 μm were counted as positive. The sizes of more than 100 positive colonies were averaged.
P < .05 vs B5-EV1.
NIK mediates constitutive NF-κB activation in ATL cells
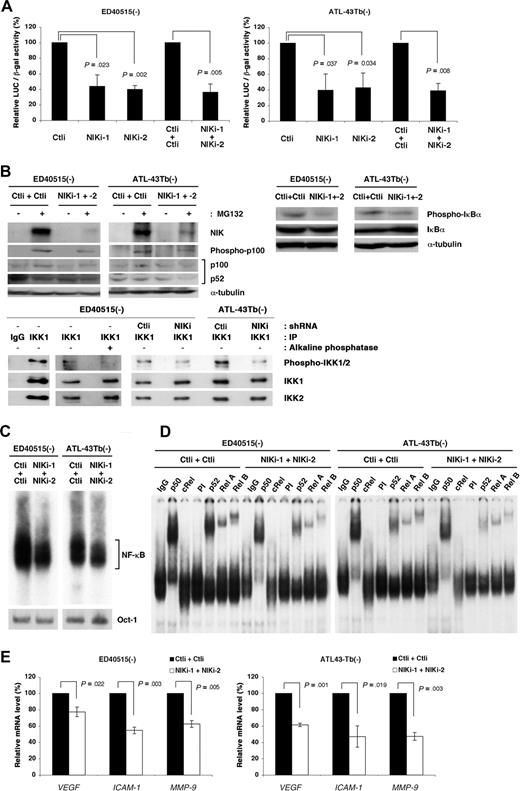
The similar modulation of IKK activity by CHX or MG132 in both ATL and B5 cells expressing NIK (Figure 4) suggests that NIK plays an important role in constitutive NF-κB activation in ATL cells. We therefore examined whether the RNA interference-mediated silencing of endogenous NIK gene expression would lower NF-κB–dependent transcription in these cells. ED-40515(−) and ATL-43Tb(−) cells were infected with lentiviral constructs that express short hairpin RNA (shRNA) molecules that target mRNA for either Renilla luciferase (Ctli) or NIK (NIKi), and then subjected to puromycin selection for 2 days. To suppress NIK expression maximally, we used independently or in combination 2 shRNAs (NIKi-1 and -2) that target different NIK sequences and reduce NIK expression. The infected cells were then assayed for transcriptional activity by transient transfection with an NF-κB–dependent reporter gene (Figure 6A). Lentiviral expression of NIKi constructs resulted in suppression of NF-κB–dependent reporter gene expression in ATL cells when independently used, and the combined use of the 2 NIKi constructs (NIKi-1 and -2) was found to be more effective. We then examined ATL cells transduced with NIKi-1 and -2 constructs for the expression of endogenous NIK and specifically phosphorylated forms of p100, IκBα, and IKKs by immunoblotting (Figure 6B) and for NF-κB DNA binding activity by EMSA (Figure 6C). NIK expression in ATL cells was found to be down-regulated by the shRNA-mediated silencing (Figure 6B). As expected, p52 and phosphorylated p100 were also reduced by NIK depletion, and interestingly, phoshorylation of IκBα was also suppressed. This is consistent with the results observed in NIK-transduced rat fibroblasts that express the phoshorylated form of IκBα (Figure 5A), indicating that NIK, when aberrantly and stably expressed, induces phosphorylation of IκBα. In addition, NIK depletion suppressed phosphorylation of the serine residues in the activation loop of IKKs, suggesting a key role for NIK in constitutive activation of IKKs in ATL cells (Figure 6B). Moreover, depletion of NIK resulted in suppression of NF-κB DNA binding activity (Figure 6C). Super-shift assays revealed that DNA-binding of NF-κB components, p50, p52, RelA, and RelB was reduced by NIK depletion (Figure 6D). As shown previously, c-Rel was not detected in ATL cells.34 We further investigated alterations in the expression of NF-κB target genes by NIK depletion. Vascular endothelial growth factor (VEGF), matrix metalloproteinase-9 (MMP-9), and intracellular adhesion molecule-1 (ICAM-1), the expression of which has been reported to be under the control of NF-κB,35-37 are highly expressed in ATL cells and suggested to contribute to their invasive properties.38-41 Quantitative RT-PCR studies reveal that depletion of NIK results in down-regulation of the expression of these NF-κB target genes (Figure 6E).
Depletion of NIK suppresses NF-κB-dependent transcription in ATL cells. (A) ED40515(−) and ATL-43Tb(−) cells were infected with lentiviral vectors expressing Renilla luciferase (Ctli) or NIK-specific shRNAs (NIKi-1 or NIKi-2). In parallel, ED40515(−) and ATL-43Tb(−) cells were infected with lentiviral vectors expressing Ctli or NIKi-1 shRNAs, and 24 hours later, these cells were super-infected with lentiviral vectors expressing Ctli or NIKi-2 shRNAs. Twenty-four hours after infection, cells were selected with puromycin for 2 days. Puromycin-resistant cells were then transfected with 2 μg of IgκCona-Luc and 2 μg EF1-LacZ. Luciferase (LUC) activity was determined 48 hours after transfection and normalized to β-gal activity. Relative luciferase activities, in comparison with control cells, 100 are shown. Data are expressed as mean plus or minus SD of 3 independent experiments. P values are versus control (Ctli). (B) Super-infected cells were treated with or without MG132 (20 μM) for 3 hours and subjected to SDS-PAGE and immunoblotting with anti-NIK (#4994), antiphosphorylated p100, or anti-α-tubulin antibodies. Whole-cell extracts (30 μg) from these cells were analyzed by SDS-PAGE and immunoblotting with antiphospho-IκBα, anti-IκBα, or anti-α-tubulin antibodies. Cytoplasmic extracts prepared from ED40515(−) cells infected or not with lentivirus were precleared and immunoprecipitation was performed, using anti-IKK1 monolconal antibody or its isotype IgG (IgG). After 3 washes with TNT buffer, immune complexes were treated or not with Shrimp Akaline Phosphatase (Takara Bio) and then subjected to SDS-PAGE and immunoblotting with antiphospho-IKK1/2, anti-IKK1, or anti-IKK2 antibodies. (C) A total of 5 μg of nuclear extracts prepared from lentivirus-infected cells shown in panel B were analyzed by EMSA, using oligonucleotides encoding the NF-κB–binding sequence or Oct-1–binding sequence as probes. (D) Nuclear extracts (5 μg) from lentivirus-infected cells shown in panel B were preincubated for 30 minutes with purified mouse IgG, anti-p50, anti-cRel antibody, preimmune (PI), anti-p50, anti-RelA or anti-RelB sera, and then subjected to EMSA with the NF-κB–specific probe. (E) Total RNAs from lentivirus-infected cells shown in panel B were examined by quantitative RT-PCR for VEGF, ICAM-1, and MMP-9 mRNA levels. Each mRNA level was normalized to 18S RNA. Relative mRNA levels, in comparison with control cells, 100 are shown. Data are expressed as mean plus or minus SD of 3 independent experiments. P values are versus control (Ctli + Ctli).
Depletion of NIK suppresses NF-κB-dependent transcription in ATL cells. (A) ED40515(−) and ATL-43Tb(−) cells were infected with lentiviral vectors expressing Renilla luciferase (Ctli) or NIK-specific shRNAs (NIKi-1 or NIKi-2). In parallel, ED40515(−) and ATL-43Tb(−) cells were infected with lentiviral vectors expressing Ctli or NIKi-1 shRNAs, and 24 hours later, these cells were super-infected with lentiviral vectors expressing Ctli or NIKi-2 shRNAs. Twenty-four hours after infection, cells were selected with puromycin for 2 days. Puromycin-resistant cells were then transfected with 2 μg of IgκCona-Luc and 2 μg EF1-LacZ. Luciferase (LUC) activity was determined 48 hours after transfection and normalized to β-gal activity. Relative luciferase activities, in comparison with control cells, 100 are shown. Data are expressed as mean plus or minus SD of 3 independent experiments. P values are versus control (Ctli). (B) Super-infected cells were treated with or without MG132 (20 μM) for 3 hours and subjected to SDS-PAGE and immunoblotting with anti-NIK (#4994), antiphosphorylated p100, or anti-α-tubulin antibodies. Whole-cell extracts (30 μg) from these cells were analyzed by SDS-PAGE and immunoblotting with antiphospho-IκBα, anti-IκBα, or anti-α-tubulin antibodies. Cytoplasmic extracts prepared from ED40515(−) cells infected or not with lentivirus were precleared and immunoprecipitation was performed, using anti-IKK1 monolconal antibody or its isotype IgG (IgG). After 3 washes with TNT buffer, immune complexes were treated or not with Shrimp Akaline Phosphatase (Takara Bio) and then subjected to SDS-PAGE and immunoblotting with antiphospho-IKK1/2, anti-IKK1, or anti-IKK2 antibodies. (C) A total of 5 μg of nuclear extracts prepared from lentivirus-infected cells shown in panel B were analyzed by EMSA, using oligonucleotides encoding the NF-κB–binding sequence or Oct-1–binding sequence as probes. (D) Nuclear extracts (5 μg) from lentivirus-infected cells shown in panel B were preincubated for 30 minutes with purified mouse IgG, anti-p50, anti-cRel antibody, preimmune (PI), anti-p50, anti-RelA or anti-RelB sera, and then subjected to EMSA with the NF-κB–specific probe. (E) Total RNAs from lentivirus-infected cells shown in panel B were examined by quantitative RT-PCR for VEGF, ICAM-1, and MMP-9 mRNA levels. Each mRNA level was normalized to 18S RNA. Relative mRNA levels, in comparison with control cells, 100 are shown. Data are expressed as mean plus or minus SD of 3 independent experiments. P values are versus control (Ctli + Ctli).
NIK regulates tumorigenicity of ATL cells in vivo
We finally investigated biologic effects of NIK depletion in ATL cells. NIK depletion did not significantly influence the growth of cells in culture (Figure 7A). We then examined whether depletion of NIK affects the tumorigenicity of ATL cells in a mouse model. NOD/SCID/γcnull mice were subcutaneously inoculated with ED-40515(−) cells that express Ctli or NIKi and are characterized in Figure 6B,C, and tumor formation was evaluated 2 weeks later. As expected, ED-40515(−) cells expressing Ctli efficiently formed large tumors, whereas tumors formed in mice inoculated with ED-40515(−) cells expressing NIKi were significantly smaller (Figure 7B-D), suggesting that NIK supports efficient tumor cell growth in vivo.
Depletion of NIK in ATL cells suppresses tumor formation in NOD-SCID/γcnull (NOG) mice. (A) Pools of ED40515(−) cells expressing Ctli or NIKi-1 and -2, shown in Figure 6B, C, D, and E, were analyzed for cell growth in vitro by the trypan blue staining method. Relative cell numbers, in comparison with control cells (arbitrarily set at 1), are shown. Data are expressed as mean plus or minus SD of 3 independent experiments. P values are vs control (Ctli + Ctli). n.s. indicates no significant difference. (B-D) NOG mice were inoculated subcutaneously in the postauricular region with the puromycin-resistant ED-40515(−) cells (5 × 106). Tumor formation in mice was evaluated 2 weeks after inoculation. Tumor weight (B) and size (C) relative to those of tumors formed in mice inoculated with ED40515(−) cells expressing Ctli are shown. (D) Photographs of tumors formed 2 weeks after cell inoculation. Each result was obtained from 5 different mice (means are shown [error bars]). P values are versus control (Ctli + Ctli).
Depletion of NIK in ATL cells suppresses tumor formation in NOD-SCID/γcnull (NOG) mice. (A) Pools of ED40515(−) cells expressing Ctli or NIKi-1 and -2, shown in Figure 6B, C, D, and E, were analyzed for cell growth in vitro by the trypan blue staining method. Relative cell numbers, in comparison with control cells (arbitrarily set at 1), are shown. Data are expressed as mean plus or minus SD of 3 independent experiments. P values are vs control (Ctli + Ctli). n.s. indicates no significant difference. (B-D) NOG mice were inoculated subcutaneously in the postauricular region with the puromycin-resistant ED-40515(−) cells (5 × 106). Tumor formation in mice was evaluated 2 weeks after inoculation. Tumor weight (B) and size (C) relative to those of tumors formed in mice inoculated with ED40515(−) cells expressing Ctli are shown. (D) Photographs of tumors formed 2 weeks after cell inoculation. Each result was obtained from 5 different mice (means are shown [error bars]). P values are versus control (Ctli + Ctli).
Discussion
Persistent activation of NF-κB has previously been reported to play an essential role in the growth and survival of specific cancer cell types, including ATL, H-RS, melanoma, and prostate cancer cells.9,42-45 Inappropriate NF-κB activation can also contribute to the resistance to the apoptotic responses induced by certain anticancer drugs.46 On the other hand, cancer cell apoptosis can be induced when persistent NF-κB activity is blocked by inhibitors, such as SR-IκBα, by drugs targeting IKK or the proteasome, via peptides targeting p50 or NEMO, and by double-stranded oligonucleotides containing NF-κB binding sites.47,48 One problem with such inhibitors, however, is their lack of specificity to cancer cells because they also necessarily block normal NF-κB activation. Hence, it would be desirable to specifically inhibit NF-κB activation in cancer cells by identifying molecular targets in each cancer type. Virally transformed cancer cells express a virus-derived regulatory protein(s) that targets critical molecules in a variety of key signaling pathways. Cytokine autocrine loops or genetic alterations to genes regulating the NF-κB signaling mechanisms that lead to persistent NF-κB activation have also been identified in some cancer cells.16,17,32,47,49 However, the mechanisms underlying persistent NF-κB activation in many types of cancer remain unknown.
Most primary ATL cells, although infected with HTLV-I, are characterized by the loss of viral protein expression, including Tax, probably because of the host immune surveillance during the long period of latency.50 Nevertheless, NF-κB is strongly and persistently activated in ATL cells through IKK,9 although the mechanism of IKK activation has remained unknown. The findings in our present study demonstrate the aberrant expression of NIK at the pretranslational level in ATL cells derived from 15 of 21 patients. This overexpression does not seem to correlate with the patients' age, sex, disease type, or percentage of abnormal lymphocytes (Table S1). Further studies will be required to clarify potentially NIK-independent NF-κB activation in the other 6 cases. The stable expression of functional NIK in fibroblasts, but not that of its catalytically inactive mutant, causes cellular transformation and persistent NF-κB activation with molecular features quite similar to those reported previously in ATL cells. These include the rapid loss of IKK activity after protein synthesis inhibition and the superinduction of IKK activity in the presence of MG132.11 Moreover, RNA interference studies have also indicated that the deregulated NIK expression is the principal cause of constitutive NF-κB activation in ATL cells. In line with a previous report by Ramakrishnan et al, which showed that the induction of IκBα degradation by CD70, CD40 ligand, and BLyS/BAFF is dependent on the function of NIK,18 we find in our present experiments that the stable expression of NIK induces IκBα phosphorylation and the formation of DNA binding complexes containing not only p50 and RelB, but also RelA both in wild-type and in NEMO-deficient rat fibroblasts. This indicates that NIK can stimulate the canonical pathway characterized by IκBα phosphorylation and RelA activation and that NIK does not require NEMO for it. Interestingly, the forced expression of SR-IκBα in these fibroblasts abolishes the transformed phenotype and suppresses constitutive NF-κB activity, with the p100 and p52 expression levels being diminished simultaneously, probably because p100 expression is largely dependent on NF-κB activity.51 RelB expression is also known to be controlled by NF-κB,52 suggesting that the noncanonical pathway of NF-κB activation does not work independently but rather coincides with NF-κB activation through the canonical pathway under stable conditions.
H-RS cells were also found to overexpress NIK, including its transcripts, in this study. Earlier reports have described 2 potential mechanisms of constitutive NF-κB activation in H-RS cells: persistent signaling from receptors that cause NF-κB activation, such as CD30, CD40, and RANK as well as a CD40-like molecule latent membrane protein 1 of the Epstein-Barr virus; and disruption of IκBα-dependent suppression resulting from the mutation of this gene.32,48 The H-RS cell lines used in this study are Epstein-Barr virus-negative, and neither HDLM-2 nor L540 cells harbor mutations in their IκB genes. Indeed, CD30, CD40, and RANK were all found to be expressed in the H-RS cell lines used in this study, but we envisage that the aberrant expression of NIK is a distinct mechanism underlying the persistent NF-κB activation in these cells. It is partly because these TNF family receptor molecules, when stimulated or overexpressed transiently in cultured cells, elevate the NIK protein expression levels with a concomitant reduction in TRAF3 but do not increase NIK mRNA.19,20
Whereas the transient stimulation of a B-cell line with BAFF or anti-CD40 antibody stabilizes the NIK protein at the posttranslational level and does not up-regulate its mRNA expression,20 NIK was observed to be constitutively overexpressed in ATL and H-RS cells at the pretranslational level. These differing mechanisms of NIK regulation may not be all that surprising, however, in light of the transient vs persistent nature of the activation of NF-κB. The barely detectable levels of steady-state NIK protein expression and its robust accumulation after proteasome inhibition in ATL and H-RS cells further suggest that the proteasome-dependent degradation of NIK occurs rapidly in tumor cells as in normal cells, although we cannot rule out the possibility that TNF family receptors known to be overexpressed in H-RS cells influence the stability of NIK to some extent. This point is currently very difficult to address because the protein amount of NIK in the absence of the proteasome inhibitor is quite limited. At least 3 mechanisms of pretranslational induction of NIK are plausible: the stabilization of NIK transcripts, transcriptional activation and/or amplification of the NIK gene. It should be noted that the stability of NIK mRNA in ATL cells was similar to that in control cells, suggesting that NIK expression is deregulated in ATL cells at the level of mRNA production. In this regard, we are currently analyzing the regulatory region of the NIK gene in normal and cancer cells.
We detected NIK in whole-cell lysates only when the cells themselves were treated with the proteasome inhibitor, MG132. It is possible that the expression of the NIK protein is tightly regulated under detectable levels in resting normal cells. However, in ATL and H-RS cells, enhanced NIK production, although still not detectable by simple immunoblotting, may be sufficient to cause its deregulated activity toward IKK. During the manuscript preparation, 2 reports demonstrated deregulated expression of NIK because of mutations in TRAF3, CYLD, or NIK itself in multiple myeloma cells.16,17 In case of ATL cells, formation of a fusion protein after genomic rearrangement seems to be unlikely based on the apparently normal size of the protein. At present, the mechanism of overproduction of NIK mRNA in ATL cells remains to be determined, but the fluorescence in situ hybridization results suggest that aberrant NIK expression in ATL cells is not the result of genomic abnormalities, such as amplification or translocation.
Successful anticancer drug or gene therapies can be conducted in a number of ways, including the general administration of particular reagents that mechanistically work exclusively on cancer cells, or delivering conventional anticancer reagents specifically to cancer cells. The former strategy is likely to be more promising in the case of hematopoietic cancers. In this regard, NIK could be an attractive molecular target for ATL and Hodgkin lymphoma therapy, although the physiologic functions of NIK in human adults remain unknown. Suppressing high NF-κB activity levels by targeting NIK may also sensitize these cancer cells to commonly used anticancer agents.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all of the ATL patients who donated blood samples for use in this study, Dr K. Yamaguchi and the Joint Study on Predisposing Factors of ATL Development for providing and analyzing sample blood, and the following researchers for donating invaluable reagents: Dr M. Maeda (Kyoto University, Kyoto, Japan) for the ED40515(−) and ATL-43Tb(−) cells, Dr D. Goeddel (Amgen, Thousand Oaks, CA) for NIK cDNAs, Dr N.R. Rice and Dr A. Israël (Institut Pasteur Paris, Paris, France) for p50, RelA, and RelB antisera, Dr T. Kitamura (University of Tokyo, Tokyo, Japan) for Plat-E cells, Dr I.S.Y. Chen (UCLA, Los Angeles, CA) for pHCMV-VSVG and pCMVΔR8.2 packaging plasmids, and Dr H. Miyoshi (RIKEN Tsukuba Institute, Tsukuba, Japan) for CS-CDF-CG-PRE plasmid. The authors also thank Dr G. Courtois (INSERM, Paris, France) and the members of the Department of Molecular Virology for helpful discussions.
This work was supported by research grants from the Ministry of Health and Labor Sciences (HIV/AIDS, H18-005) (Naoki Yamamoto) and from the Ministry of Education, Culture, Sports, Science and Technology of Japan (18390145; Naoki Yamamoto) and (17013029; S.Y.).
Authorship
Contribution: Y.S., T.S., and S.Y. designed the study; Y.S., Norio Yamamoto, H.S., V.J.M.B., Y.I., K.M., X.Q., I.I., J.I., and S.Y. carried out the research; M.Z.D. carried out the animal experiments; A.U. and T.W. collected and analyzed sample blood from ATL patients; T.M. contributed to lentiviral vector constructions; Y.S. and S.Y. analyzed the data; T.S., Naoki Yamamoto and S.Y. controlled the data; Y.S. and S.Y. wrote the paper; all authors checked the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Shoji Yamaoka, Department of Molecular Virology, Graduate School of Medicine, Tokyo Medical and Dental University, 1-5-45, Yushima, Bunkyo-ku, Tokyo, 113-8510, Japan; e-mail: shojmmb@tmd.ac.jp.




![Figure 7. Depletion of NIK in ATL cells suppresses tumor formation in NOD-SCID/γcnull (NOG) mice. (A) Pools of ED40515(−) cells expressing Ctli or NIKi-1 and -2, shown in Figure 6B, C, D, and E, were analyzed for cell growth in vitro by the trypan blue staining method. Relative cell numbers, in comparison with control cells (arbitrarily set at 1), are shown. Data are expressed as mean plus or minus SD of 3 independent experiments. P values are vs control (Ctli + Ctli). n.s. indicates no significant difference. (B-D) NOG mice were inoculated subcutaneously in the postauricular region with the puromycin-resistant ED-40515(−) cells (5 × 106). Tumor formation in mice was evaluated 2 weeks after inoculation. Tumor weight (B) and size (C) relative to those of tumors formed in mice inoculated with ED40515(−) cells expressing Ctli are shown. (D) Photographs of tumors formed 2 weeks after cell inoculation. Each result was obtained from 5 different mice (means are shown [error bars]). P values are versus control (Ctli + Ctli).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/111/10/10.1182_blood-2007-09-110635/6/m_zh80090818940007.jpeg?Expires=1763463703&Signature=ST2icChFq00yl1JT3HJWeP6oP7Ar133ufmJOeP5pEoulV76gnwi2Y9T53XPicitEjZJkghXmkLQuCmszPS9vYJesEZhU~9bfDmjVga515KUo30Nbauht6okF~ngOXbdk0pkehRmEt3EsRiC4MQRACOKybH5yUmGtJkN87H1~3MCAHZ~CQ5hQxnV5OWAhhbuTeglMFeuLaPmrbIt8P-yK7RMthW4k2jvp4NoN2Awt-My8hWMvPJprxBCELjcb25jYiwCAdwF1tOAxRWAZuL7r4LAYTKzlt~4rnaYGbj1Z9gKTenTakl3SJLoaPheNYSygwnrJWVy206neLqc9emZcSw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal