Abstract
Factor VIII (fVIII) is a serum protein in the coagulation cascade that nucleates the assembly of a membrane-bound protease complex on the surface of activated platelets at the site of a vascular injury. Hemophilia A is caused by a variety of mutations in the factor VIII gene and typically requires replacement therapy with purified protein. We have determined the structure of a fully active, recombinant form of factor VIII (r-fVIII), which consists of a heterodimer of peptides, respectively containing the A1-A2 and A3-C1-C2 domains. The structure permits unambiguous modeling of the relative orientations of the 5 domains of r-fVIII. Comparison of the structures of fVIII, fV, and ceruloplasmin indicates that the location of bound metal ions and of glycosylation, both of which are critical for domain stabilization and association, overlap at some positions but have diverged at others.
Introduction
The principal mechanism used to stop the loss of blood in mammals following vascular injury consists of a pair of overlapping proteolytic cascades called the extrinsic and intrinsic pathways.1-4 The process of blood coagulation requires extraordinary spatial and temporal regulation, which is accomplished by assembling and tethering the central proteolytic activities of these cascades at the location of transiently exposed biomolecules and cellular surfaces (Figure 1A). This includes an integral membrane protein called “tissue factor” that initiates the rapid up-regulation of the short-lived extrinsic pathway,5 and the surfaces of activated platelets, which modulate the activation of the longer-lived intrinsic pathway.6 A total of 2 homologous procoagulants, factors V and VIII (fV and fVIII), are each localized on the surface of these platelets, where they nucleate the assembly of multiprotein proteolytic complexes.
Factor VIII and the coagulation cascade. (A) The blood coagulation cascade consists of 2 pathways (extrinsic and intrinsic) that are initiated by the exposure of tissue factor (TF) or phosphatidylserine groups (PS) of activated platelet membranes to circulating protein factors, respectively. fVIII is a plasma glycoprotein that acts as an initiator and regulator of the intrinsic pathway. Upon proteolytic activation by either fXa or thrombin, fVIIIa dissociates from VWF, associates with the fIXa serine protease, and directs the localization of the resulting complex to the membrane surface of activated platelets via an interaction with its C-terminal C2 domain (structure in inset). The membrane-bound complex between fVIIIa and fIXa complex functions to proteolytically activate fX, which then activates thrombin (fII). (B) Domain structure of fVIII. fVIII is synthesized as a single polypeptide chain of 2332 residues. Based on sequence homology, fVIII has the domain structure A1-A2-B-A3-C1-C2. Linker regions between domains are denoted with lowercase letters (“a1,” etc). The location of domain boundaries and primary sites of proteolytic processing during secretion and activation are denoted by residue numbers. The circulating fVIII heterodimer is associated with VWF primarily through interactions with the “a3” acidic region at the light chain N-terminus and with the C2 domain. Various proteases interact with the activated heterotrimer at positions denoted at the bottom panel. Membrane association is primarily accomplished through the C2 domain; its deletion completely abrogates binding of fVIII to platelet surfaces.
Factor VIII and the coagulation cascade. (A) The blood coagulation cascade consists of 2 pathways (extrinsic and intrinsic) that are initiated by the exposure of tissue factor (TF) or phosphatidylserine groups (PS) of activated platelet membranes to circulating protein factors, respectively. fVIII is a plasma glycoprotein that acts as an initiator and regulator of the intrinsic pathway. Upon proteolytic activation by either fXa or thrombin, fVIIIa dissociates from VWF, associates with the fIXa serine protease, and directs the localization of the resulting complex to the membrane surface of activated platelets via an interaction with its C-terminal C2 domain (structure in inset). The membrane-bound complex between fVIIIa and fIXa complex functions to proteolytically activate fX, which then activates thrombin (fII). (B) Domain structure of fVIII. fVIII is synthesized as a single polypeptide chain of 2332 residues. Based on sequence homology, fVIII has the domain structure A1-A2-B-A3-C1-C2. Linker regions between domains are denoted with lowercase letters (“a1,” etc). The location of domain boundaries and primary sites of proteolytic processing during secretion and activation are denoted by residue numbers. The circulating fVIII heterodimer is associated with VWF primarily through interactions with the “a3” acidic region at the light chain N-terminus and with the C2 domain. Various proteases interact with the activated heterotrimer at positions denoted at the bottom panel. Membrane association is primarily accomplished through the C2 domain; its deletion completely abrogates binding of fVIII to platelet surfaces.
When fVIII is bound to activated platelets at the site of vascular injury, it recruits the serine protease fIXa into a complex that then catalyzes the proteolytic activation of fX.1,4,7 The proteolytic activity of fIXa is enhanced by approximately 200 000-fold through its interaction with fVIII, calcium, and the phospholipid bilayer,8 corresponding to an increase of approximately 109 in kcat/KM.
The full-length, unprocessed fVIII protein consists of 2332 amino acid residues and has the domain structure A1-A2-B-A3-C1-C29-12 (Figure 1B). The 3 A domains are each approximately 330 residues, and approximately 40% identical to each other and to the copper-binding protein ceruloplasmin.13 The C domains are smaller (approximately 160 residues) and are more distantly related to various members of the discoidin protein fold family, such as galactose oxidase.14-17 The B domain has no known structural homologs, is heavily glycosylated, and is relatively dispensible for procoagulant activity. fVIII is initially processed by proteolytic cleavage events that remove a large portion of the B domain, generating a heterodimer that circulates in a tight complex with von Willebrand factor (VWF).18 This interaction is essential for maintaining stable levels of fVIII in circulation.19 Upon vascular injury, further proteolytic processing generates activated factor VIIIa (fVIIIa), a heterotrimer (A1/A2/A3-C1-C2) that is released from VWF and binds to activated platelets.18
The carboxy-terminal 159 amino acids of fVIII comprise its C2 domain, which is involved in binding to VWF and primarily responsible for binding to platelet membrane surfaces. This latter binding interaction is dependent on the transient, specific exposure of phosphatidylserine (PS) head groups on the outer leaflet of activated platelet membranes.20-23 The VWF and membrane-binding activities of the C2 domain appear to be competitive and mutually exclusive.23-26
Several structural models are available to the coagulation community for analysis of fVIII. A hypothetical model of the fVIII “A” domains has been generated from the crystal structure of ceruloplasmin.13 The structures of the C2 domain of fV and fVIII have been solved to high resolution,16,27,28 and a homology model of the fVIII C1 domain has been described.29 Complementing those studies is a model of the full-length fVIII heterodimer, generated from 2D electron diffraction studies, which provides a low-resolution (approximately 15 Å) view of the overall disposition and orientations of the individual fVIII domains.30 Finally, a 2.8-Å resolution structure of a portion of fV (missing its A2 domain) has also been reported.31
Methods
A recombinant form of fVIII (r-fVIII) was expressed, secreted, and purified from Chinese hamster ovary (CHO) cells as a heterodimer in the presence of VWF as previously described.32 The heterodimeric species of r-fVIII consists of a heavy chain (A1 and A2 domains) and a light chain (residues 1563-1648 of the B domain and the entire A3, C1, and C2 domains). Purified r-fVIII was pooled, concentrated to 0.5 mg/mL, and stored at −70°C in storage buffer (50 mM imidazole [pH 6.7]; 410 mM NaCl, 4 mM CaCl2, 0.1% wt/vol PEG1000, and 0.001% wt/vol Tween 80). This truncated variant of fVIII maintains procoagulant activity and can be subsequently activated by thrombin.
For crystallization purposes, r-fVIII was concentrated to 2 mg/mL by vacuum dialysis at 4°C against the storage buffer listed. r-fVIII was subsequently crystallized by hanging drop reverse-vapor diffusion at 4°C against reservoirs containing 8% to 12% (wt/vol) PEG8000, 100 to 300 mM NaCl, and 50 mM Tris-HCl (pH 6.5-7.5). Reverse-vapor diffusion occurs due to the higher salt concentration residing in the r-fVIII storage buffer, leading to a reduction in salt concentration in the protein drop that causes crystallization. Prior to data collection, crystals were cryoprotected with either 30% (vol/vol) DMSO or 25% to 30% glycerol and flash-frozen in liquid nitrogen. Data collection was performed at Beamline 5.0.2 at the Advanced Light Source (Berkeley, CA). Data were indexed, refined, and integrated with D*TREK (Molecular Structure Corporation, The Woodlands, TX)33 and scaled using the program SCALA (CCP4, Daresbury Laboratory, United Kingdom).34 The space group was determined to be P41212 by analyses of symmetry and systematic absences, and by examination of unbiased anomalous difference, isomorphous difference and omit maps. Particularly obvious validating features of these maps, indicating the position of bound metals and oligosaccharide structures, are shown in Figure 2.
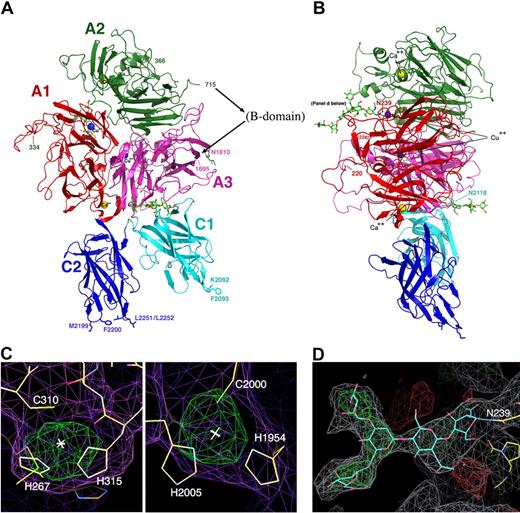
The structure of the B domain deleted fVIII heterodimer. (A,B) The fVIII domains are individually labeled and colored. The C-terminal end of the heavy chain and the N-terminal end of the light chain are indicated with residue numbers (715 and 1695). Residues 1563 to 1694 (the N-terminal 80 residues of the light chain) are present in the construct but are poorly ordered. The N- and C-termini of 2 additional disordered regions are also indicated with residue numbers: residues 220 and 228 that flank a surface loop in the A1 domain (right panel) and residues 334 and 366 that flank the linker region between the A1 and A2 domains (left panel). Shown are 2 bound calcium ions, 2 bound copper ions, and 3 well-ordered and visible N-linked oligosaccharide structures. Shown and labeled for reference are 4 residues on the C2 domain that are thought to be involved in membrane binding and a similarly positioned pair of residues on the C1 domain. (C) Anomalous difference peaks at the sites of bound copper ions buried in the A1 and A3 domains (contoured at 5σ; both are approximately 9σ peak height overall). The residues involved in metal binding at this site are conserved in the analogous copper-binding site in ceruloplasmin, but are diverged from fV. (D) Difference density for 1 of 3 N-linked oligosaccharide structures, which modifies Asn239 in the A1 domain. The density is readily apparent for the entire pentameric polysacharide mannose core of an N-linked sugar, and represents unbiased density prior to any modeling of the covalent modification.
The structure of the B domain deleted fVIII heterodimer. (A,B) The fVIII domains are individually labeled and colored. The C-terminal end of the heavy chain and the N-terminal end of the light chain are indicated with residue numbers (715 and 1695). Residues 1563 to 1694 (the N-terminal 80 residues of the light chain) are present in the construct but are poorly ordered. The N- and C-termini of 2 additional disordered regions are also indicated with residue numbers: residues 220 and 228 that flank a surface loop in the A1 domain (right panel) and residues 334 and 366 that flank the linker region between the A1 and A2 domains (left panel). Shown are 2 bound calcium ions, 2 bound copper ions, and 3 well-ordered and visible N-linked oligosaccharide structures. Shown and labeled for reference are 4 residues on the C2 domain that are thought to be involved in membrane binding and a similarly positioned pair of residues on the C1 domain. (C) Anomalous difference peaks at the sites of bound copper ions buried in the A1 and A3 domains (contoured at 5σ; both are approximately 9σ peak height overall). The residues involved in metal binding at this site are conserved in the analogous copper-binding site in ceruloplasmin, but are diverged from fV. (D) Difference density for 1 of 3 N-linked oligosaccharide structures, which modifies Asn239 in the A1 domain. The density is readily apparent for the entire pentameric polysacharide mannose core of an N-linked sugar, and represents unbiased density prior to any modeling of the covalent modification.
The initial crystals used for molecular replacement were cross-linked (by transient 5-minute exposure to 0.05% vol/vol glutaraldehyde, followed by cryocooling) and diffracted to approximately 4.5-Å resolution. Subsequent crystals were larger, more robust, and diffracted up to approximately 3.7 Å without requiring cross-linking. One of these specimens was used for final rebuilding and refinement (Tables 1,2). The unit cell parameters for the crystal used for the final refinement are a = b = 134.8 Å and c = 358.4 Å.
Data collection and processing statistics
| Crystal . | F8_w (cross-linked) . | Nat-7 . | Nat-4b . | Nat-4c . |
|---|---|---|---|---|
| Wavelength, Å | 1.0246 | 1.00 | 1.00 | 1.00 |
| Unit cell, Å | a = b = 134.7, c = 353.6 | a = b = 134.5, c = 359.7 | a = b = 134.6, c = 359.5 | a = b = 134.8, c = 358.4 |
| Resolution, Å | 30-4.5 (4.6-4.5) | 100-4.3 (4.4-4.3) | 100-3.9 (4.0-3.9) | 100-3.7 (3.76-3.70) |
| No. reflections (no. unique reflections) | 308 594 (20 241) | 133 617 (42 517) | 199 559 (28 509) | 189 184 (34 208) |
| Redundancy | 15.2 | 3.1 | 7.0 (2.9) | 5.5 (3.0) |
| I/σ (I)* | 19.5 (5.4) | 17.7 (3.5) | 12.9 (1.0) | 13.7 (0.91) |
| Completeness, %* | 100 (100) | 97.7 (98.6) | 92.5 (61.3) | 94.5 (67.3) |
| Rsym* | 0.161 (0.682) | 0.084 (0.553) | 0.092 (0.735) | 0.096 (0.798) |
| Crystal . | F8_w (cross-linked) . | Nat-7 . | Nat-4b . | Nat-4c . |
|---|---|---|---|---|
| Wavelength, Å | 1.0246 | 1.00 | 1.00 | 1.00 |
| Unit cell, Å | a = b = 134.7, c = 353.6 | a = b = 134.5, c = 359.7 | a = b = 134.6, c = 359.5 | a = b = 134.8, c = 358.4 |
| Resolution, Å | 30-4.5 (4.6-4.5) | 100-4.3 (4.4-4.3) | 100-3.9 (4.0-3.9) | 100-3.7 (3.76-3.70) |
| No. reflections (no. unique reflections) | 308 594 (20 241) | 133 617 (42 517) | 199 559 (28 509) | 189 184 (34 208) |
| Redundancy | 15.2 | 3.1 | 7.0 (2.9) | 5.5 (3.0) |
| I/σ (I)* | 19.5 (5.4) | 17.7 (3.5) | 12.9 (1.0) | 13.7 (0.91) |
| Completeness, %* | 100 (100) | 97.7 (98.6) | 92.5 (61.3) | 94.5 (67.3) |
| Rsym* | 0.161 (0.682) | 0.084 (0.553) | 0.092 (0.735) | 0.096 (0.798) |
Space group is P41212.
Numbers in parentheses are statistics from the highest-resolution shells.
Model refinement statistics
| Resolution, Å | 100-3.7 (3.76-3.7) |
| Rwork | 0.289 (0.470) |
| Rfree | 0.341 (0.485) |
| RMSD bond, Å | 0.0092 |
| RMSD angle, ° | 1.69 |
| Protein residues/average B-factor, no. | 1315/160.1 |
| Carbohydrate moieties/average B-factor, no. | 13/192.0 |
| Metal ions/average B-factor, no. | 4/158.5 |
| Ramachandran distribution, no. residues (%) | 992 (86.9), 107 (9.4), 43 (3.7) |
| Resolution, Å | 100-3.7 (3.76-3.7) |
| Rwork | 0.289 (0.470) |
| Rfree | 0.341 (0.485) |
| RMSD bond, Å | 0.0092 |
| RMSD angle, ° | 1.69 |
| Protein residues/average B-factor, no. | 1315/160.1 |
| Carbohydrate moieties/average B-factor, no. | 13/192.0 |
| Metal ions/average B-factor, no. | 4/158.5 |
| Ramachandran distribution, no. residues (%) | 992 (86.9), 107 (9.4), 43 (3.7) |
The phasing of the r-fVIII crystal structure was performed iteratively by molecular replacement, using the programs Phaser (CCP4) and EPMR.35 First, the 3 A domains of fVIII were simultaneously located and placed with the program Phaser using a search model consisting of a polyalanine peptide chain containing those domains, which were derived from the crystal structure of ceruloplasmin.13 The correct solution to this search was well above background, with the top 2 Z-scores for the rotation search corresponding to 7.6 and 3.1, respectively; the top 2 Z-scores for the translation search were 23.7 and 13.5. A similar search using the coordinates of the A1 and A3 domains from the structure of inactive “A2-deleted” fV31 was unsuccessful. The position of each of the A domains was further validated by removing each domain from the solution and performing a subsequent search to find each domain independently. The position of the C1 domain was then determined, using a model of C129 derived from the structure of the fVIII C2 domain16 as a search template. Last, the position of the C2 domain was determined using the program EPMR with the previous solutions combined and used as a static structure while the fVIII C2 domain crystal structure was used as a search model. The resulting solution for the fVIII C2 domain generated a correlation coefficient of 0.601 and an R-factor of 50.8.
The relative orientations of the fVIII domains resembles the orientations of the same domains that were independently determined in the crystal structure of a fragment of fV.31 That model was not used at any stage of phase determination in this study. Additional protein models that did not result in an obvious molecular replacement solution were all-atom ceruloplasmin36 and a model derived from electron diffraction data for fVIII.30 In the latter structure, the orientation of the 2 C domains differ significantly from those found in the crystal structure reported here.
Homology models of the 3 A domains and the C1 domain were constructed using the ROBETTA protein structure prediction server.37 The resulting models were superimposed on the molecular replacement solution and used as starting models for rigid body refinement using the CNS program,38 initially for the entire structure as one rigid group, and subsequently parsed into individual domains as individual rigid groups. After initial simulated annealing protocols, the values of Rwork and Rfree were 0.398 and 0.459, respectively, at which point model rebuilding was commenced against unbiased composite omit maps. During the iterative process of rebuilding and refinement, sequentially higher quality data sets were generated as shown in Tables 1,2, and were used accordingly.
Model building was done with the program COOT39 against SIGMAA-weighted Fourier and difference maps, and the structure was refined using CNS after randomly removing 5% of the measurements to monitor the free R-factor (Rfree). Due to the low resolution of the X-ray data in this study, all modifications of the structure during model building and refinement were evaluated and accepted based on their effect on the value of the Rfree and on the difference between Rfree and Rwork, rather than solely on the value Rwork (which is more subject to model bias). During the refinement, features of unbiased electron density that were correlated with previously unmodeled structural features (such as the obvious presence of an N-linked, bifurcated mannose core glycosyl modification of Asn1810, and strong features of anomalous density for bound metal ions, as shown in Figure 2) provided clear validation of the molecular replacement solution and subsequent refinement. The final model corresponds to Rwork/Rfree values of 0.289/0.341 and geometric root mean square deviation (RMSD) values of 0.0092 Å (bond distance) and 1.69° (bond angles). Data and refinement statistics are provided in Tables 1 and 2. A comparison of the refinement statistic with structures at similar resolution in the Protein Databank (PDB) database and the Ramachandran plot of the current structure are provided in Figures S1,S2 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Results
Structure and domain organization
We crystallized a fully active form of engineered r-fVIII, which consists of a heterodimer of the A1-A2 domains (the “heavy chain”) and the A3-C1-C2 domains (the “light chain”). The structure permits unambiguous modeling of the relative orientations of all 5 domains of r-fVIII as well as identification of sites of glycosylation and metal binding (Figure 2). While the C2 domain is loosely tethered to the structure and appears capable of significant motion, the C1 domain forms an extensive hydrophobic interface with the A3 domain and is likely locked into a single docked position and conformation.
The structures of the individual domains within fVIII are quite similar to available crystal structures of homologous proteins. The 3 A domains can be superimposed on those from ceruloplasmin13 with an overall RMSD of 1.7 Å, and can be similarly superimposed on the A1 and A3 domains from fV with an RMSD of 2.2 Å. The C2 domain is closely related to the previously determined crystal structure of the isolated domain16 (RMSD, 1.5 Å). Comparison of the 2 individual C domains with those from the crystal structure of fV gives RMSD values of approximiately1.9 Å.
Across the structure of the fVIII heterodimer, 4 regions are disordered (Figure 2): 2 short surface loops within the A1 domain (residues 34-38 and 213-227), the linker region between the A1 and A2 domains (residues 335-366), and a long 155-residue region spanning the end of the A2 domain, through the truncated portion of the B domain, and into the N-terminal region of the A3 domain. The remaining portion of the light chain (A3-C1-C2) is well ordered (except for its final 3 residues). The N-terminal residues of the light chain are critical for binding to VWF and are proteolytically truncated by thrombin during r-fVIII activation; it is therefore likely that they are only structurally ordered within the circulating r-fVIII/vWF complex.
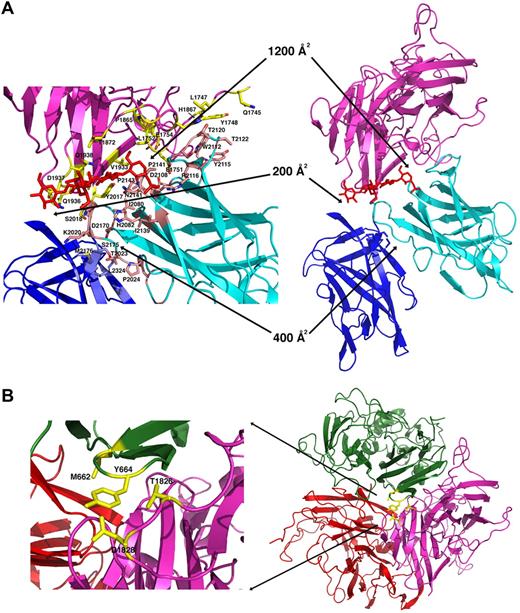
The orientation of the C1 domain is similar to that observed in the crystal structure of fV31 (which is missing its A2 domain), but is rotated by approximately 90° relative to the model of r-fVIII generated from previous 2D electron diffraction studies30 (Figure 3). The C1 domain is tightly associated with the A3 domain of the light chain, creating a 1200-Å2 aromatic/hydrophobic interface that buries approximately 20 residues and a well-ordered N-linked high-mannose glycosyl modification of N2118 (Figure 4A). This interface includes several aromatic residues (Y1748, Y2017, Y2105, and W2112), 3 prolines (P1865, P2142 and P2143), several aliphatic residues (L1747, L1752, V1933, L2015 and I2145), and a large number of hydrophilic residues, effectively locking the C1 domain into place within the fVIII light chain. Of the residues in the A3-C1 interface, at least 6 (L1752, N2015, and Y2017 and 2105, R2116, and T2122) are sites of missense mutations associated with hemophilia A.
Comparisons of fV and fVIII structures. The relative domain orientations of r-fVIII with a previously reported model generated from electron diffraction studies, and with 2.8-Å resoloution crystal structures of ceruloplasmin and of an inactive (A2-deleted) fV construct are shown. The structures are all oriented similarly with respect to the A domains. Metals ions modeled in the various structures as calcium and copper are indicated as yellow and blue spheres, respectively. The docked orientations of the C domains of r-fVIII, and their interactions with each other and with the A3 domain of the light chain, are both rotated by approximately 90° with respect to the electron diffraction model, but in excellent agreement with their homologous domains in fV. The box shows superposition of full-length fVIII (colored by domains) versus inactive (A2-deleted) fV (shown in orange). The RMSD for aligned α-carbons is approximately 2 Å.
Comparisons of fV and fVIII structures. The relative domain orientations of r-fVIII with a previously reported model generated from electron diffraction studies, and with 2.8-Å resoloution crystal structures of ceruloplasmin and of an inactive (A2-deleted) fV construct are shown. The structures are all oriented similarly with respect to the A domains. Metals ions modeled in the various structures as calcium and copper are indicated as yellow and blue spheres, respectively. The docked orientations of the C domains of r-fVIII, and their interactions with each other and with the A3 domain of the light chain, are both rotated by approximately 90° with respect to the electron diffraction model, but in excellent agreement with their homologous domains in fV. The box shows superposition of full-length fVIII (colored by domains) versus inactive (A2-deleted) fV (shown in orange). The RMSD for aligned α-carbons is approximately 2 Å.
Interface dimensions and packing in the r-fVIII molecule. (A) The light chain and packing of the C domains. The C1 domain (cyan ribbon and pink side chains) is engaged in an extensive docked interaction against the A3 domain (light magenta ribbon and yellow side chains) that involves multiple aromatic, aliphatic, and hydrophilic side chains, several of which are sites of missense mutations associated with protein dysfunction and hemophilia A. An N-linked glycosyl modification (red) is also involved in the C1-A3 interface. In contrast, the C2 domain (blue ribbon and side chains) is loosely tethered to the r-fVIII molecule, displaying small, entirely hydrophilic interfaces with the A3 and C1 domains. Interestingly, a number of residues in these interfaces are also associated with hemophilia. (B) Packing of the trimer of A domains and close up of the packing of the A2 domain (green) against the A3 domain, with residues that have been previously mutated to create engineered disulfide cross-links indicated. These residues are located on 2 adjacent loops in the A domains, which appear to display sufficient flexibility to permit covalent disulfide formation while maintaining r-fVIII activity in vivo.
Interface dimensions and packing in the r-fVIII molecule. (A) The light chain and packing of the C domains. The C1 domain (cyan ribbon and pink side chains) is engaged in an extensive docked interaction against the A3 domain (light magenta ribbon and yellow side chains) that involves multiple aromatic, aliphatic, and hydrophilic side chains, several of which are sites of missense mutations associated with protein dysfunction and hemophilia A. An N-linked glycosyl modification (red) is also involved in the C1-A3 interface. In contrast, the C2 domain (blue ribbon and side chains) is loosely tethered to the r-fVIII molecule, displaying small, entirely hydrophilic interfaces with the A3 and C1 domains. Interestingly, a number of residues in these interfaces are also associated with hemophilia. (B) Packing of the trimer of A domains and close up of the packing of the A2 domain (green) against the A3 domain, with residues that have been previously mutated to create engineered disulfide cross-links indicated. These residues are located on 2 adjacent loops in the A domains, which appear to display sufficient flexibility to permit covalent disulfide formation while maintaining r-fVIII activity in vivo.
In contrast, the C2 domain is relatively loosely tethered to the fVIII molecule (Figure 4A), displaying a small 400-Å2 interface to the C1 domain and a 200-Å2 interface with the A1 domain, both of which are comprised primarily of hydrophilic residues. However, the observed orientation of the C2 domain is similar to that observed in the fV crystal structure, indicating that this domain orientation is a reproducible feature of the full-length heterodimer (Figure 3). The respective interactions of the C1 and C2 domains within the fVIII molecule agree with their observed behavior in solution when expressed as isolated domains: the C1 domain is insoluble and cannot readily be purified, while the isolated C2 domain is well behaved in solution at high concentrations and displays specific binding to both plasma membranes and to recombinant VWF constructs (behaviors that mimic its role in the full-length coagulation factor).40,41
Bound metal ions and sites of glycosylation
In addition to the protein domains, 4 bound metal ions and 3 sites of glycosylation are observed in the structure (Figure 2). Acidic residues from individual A domains ligate two of the metals, which are modeled as calcium ions. The first calcium, found in the A1 domain, is tightly coordinated by a single glutamate (E110) and 3 aspartate residues (D116, D125, and D126), and is located at a position within the protein fold that is also occupied by calcium in both ceruloplasmin36 and in fV.31 The second modeled calcium is coordinated by 2 aspartate residues (D538 and D542) in the A2 domain.
The 2 additional metal ions, one each in the A1 and A3 domains, are observed in anomalous difference Fourier maps (Figure 2C) and are modeled as copper ions. Previous biochemical studies have indicated the presence of one or more bound copper ions in these domains in both human fVIII42 and in fV,43 and a functional role for bound copper in association of the A1 and A3 domains.44 This property reflects the relationship of both of these coagulation factors to the copper-binding protein ceruloplasmin, which contains multiple copper ions bound near the A1-A3 domain interface. Comparison of these domains among all 3 proteins indicates that their copper-binding functions are maintained, although with significant divergence in the identity and position of the side chains that coordinate the metal ion (Figure S3).
Density for the first bound copper in fVIII is observed in the A3 domain (Figures 2C,S3), coordinated by 2 histidine residues (H1954 and H2005) and a single cysteine (C2000). This metal binding site is conserved in ceruloplasmin, which is occupied by a similarly located copper ion.36 In fV, a single bound copper ion is also located near the A1-A3 boundary, again coordinated entirely by residues from the A3 domain (H1802, H1804, and D1844). These amino acid ligands differ from fVIII, but are also partially conserved with ceruloplasmin.
A second bound copper ion is buried in the A1 domain of fVIII, and is also coordinated by 2 histidine residues (H267 and H315) and a single cysteine (C310; Figure 2C). The metal-binding residues in this site are again conserved in ceruloplasmin, but not in fV (where the corresponding residues are F239, H207, and S282).
Overall, the metal-binding function of fVIII appears to be more closely related to ceruloplasmin than to fV. fV and fVIII each share homology in copper-binding residues with ceruloplasmin, but not with each other; this may indicate that the 2 coagulation factors have independently diverged from a common copper-binding ancestor. It is possible that additional or alternate copper binding sites may be occupied in various fVIII and/or fV constructs, depending on the precise nature of the expression system and cell line. The encorporation of copper ions in proteins, unlike transition metals such as calcium and magnesium, is known to often require specific chaperones that are highly specific to individual protein targets and expression pathways.
Of 4 potential sites of N-linked glycosylation in the r-fVIII molecule (N42 and N239 in the A1 domain, N1810 in the A3 domain, and N2118 in the C1 domain), density is clearly visible for an oligosaccharide structure consistent with a canonical N-linked modification at N239, N1810, and N2118. One of these 3 positions (N1810) is also observed to be glycosylated in the structure of fV.31 Of the glycosylation modifications in fVIII, unbiased difference density is particularly striking for extensive well-ordered oligosaccharide structures at both N239 (Figure 2D) and N2118. In both cases, the sugar moieties are located in a domain interface (between A1 and A2 for N239, and between A3 and C1 for N2118) and appear to participate in packing and stabilization. In contrast, the oligosaccharide group at N1810 is located near a surface of the A3 domain that is believed to be involved in binding both VWF (through interactions with its N-terminal acidic region) and with LDL receptors involved in fVIII clearance.45 Thus, this latter modification, if physiologically relevant, may be important for stabilization and/or clearance of fVIII in vivo.
Discussion
Hemophilia A mutations
A deficiency in fVIII clotting activity leads to a common bleeding disorder, hemophilia A, which affects 1 in 5000 males worldwide.46 Hemophilia A is an X-linked disorder of variable severity that is due to mutations in the fVIII gene, which is 187 kb long and contains 26 exons. The genetic lesions resulting in hemophilia A include deletions, exon inversions and translocations, nonsense frame shifts, premature stops, and a large number of missense point mutations, all of which can cause defects in the expression, secretion, and/or half-life of fVIII in circulation.47 Alternatively, some hemophilic mutations can generate stable but dysfunctional fVIII. An international database of point mutations that are associated with hemophilia A lists several hundred unique missense mutations within fVIII that all have been observed in vivo and are associated with variable severity of disease symptoms.48 These mutations are distributed uniformly across the entire peptide chain of fVIII, regardless of either disease severity or structural domain of the protein (Figure 5A).
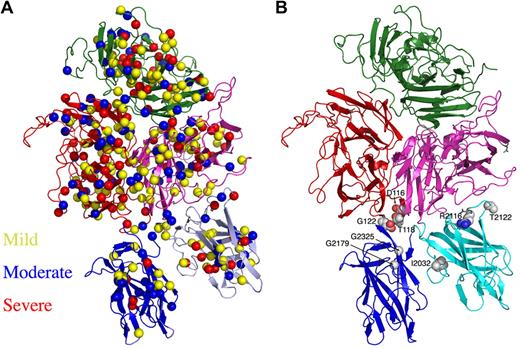
Location of hemophilia A–associated missense mutations across the r-fVIII structure. (A) The disease phenotype/symptom designations of “mild,” “moderate,” and “severe” are taken from the HamSters database48 and correspond to plasma fVIIIa activity levels of 6% to 30%, 1% to 5% and less than 1%, respectively. (B) The presence of mutations in the interfaces of the C domains with each other and with the A3 domain.
Location of hemophilia A–associated missense mutations across the r-fVIII structure. (A) The disease phenotype/symptom designations of “mild,” “moderate,” and “severe” are taken from the HamSters database48 and correspond to plasma fVIIIa activity levels of 6% to 30%, 1% to 5% and less than 1%, respectively. (B) The presence of mutations in the interfaces of the C domains with each other and with the A3 domain.
A variety of studies have mapped hemophilia A mutations to positions across available models of the fVIII structure. For example, upon determination of the crystal structure of the C2 domain and generation of a related homology model of the C1 domain, 57 separate mutations that occur within those regions were analyzed with respect to the correlation between disease severity, effect on circulating levels of r-fVIII, and the position of each residue in the protein fold.29 As a general rule, those residues found in core regions of these folded domains, or among surface epitopes known to be critical for procoagulant-binding activities and functions, were more likely to yield dysfunctional and/or destabilized protein. The structure of the full-length r-fVIII protein provides additional detail for these analyses, particularly for mutations located within interfaces between the C domains and their nearest neighbors in the full-length molecule. For example, 9 separate mutations that yield severe disease symptoms and phenotypes (defined as those that produce less than 1% of normal circulating fVIII activity) are found on the surface of the A1, C1, and C2 domains at positions not known to be involved in binding interactions with membranes, VWF, or fIXa. These residues are found to be located, respectively, in the C1-A3 domain interface (R2116 and T2122), in the A1-C2 interface (T118, E122, and D116) and in the C1-C2 interface (G2026, G2179, G2325, and I2032; Figure 5B). These latter residues are of particular interest, as glycine residues are often of great structural importance for formation of functional protein cores and interfaces due to their ability to assume backbone conformations that are otherwise sterically inaccessible.
Engineered fVIII constructs
Hemophilia A is treated by replacement therapy with concentrated fVIII using typical dosage regiments of 20 to 40 IU/kg 3 times per week, plus prophylactic administration during adverse bleeding episodes.49 A major issue that greatly affects replacement therapy efficacy and cost is the instability and rapid clearance of fVIII. Upon activation, the resulting activated fVIIIa is subject to spontaneous decay of its procoagulant activity, attributable to first-order dissociation of its free A2 subunit.50 In addition, the fVIIIa molecule is cleared by receptor-mediated catabolism, which is mediated by interactions between well-mapped epitopes on the surface of fVIII and 2 receptors from the low-density lipoprotein family (LRP and LDLR; reviewed in Saenko and Pipe49 ). Many investigators have hypothesized that improvement of fVIII stability may allow correction of hemostasis in vivo at lower levels of protein, allowing for longer intervals between therapeutic infusions.
A variety of strategies have been tested for stabilization of fVIII and improvement of its circulating lifetime in replacement therapy.49 Specifically, fVIII has been altered by chemical modification (primarily via “PEGylated” protein formulations), by elimination of the B domain and additional protein modifications that covalently trap the A2 domain (either by elimination of a thrombin-processing site in the fVIII backbone or by introduction of disulfide bonds between A2 and A3 domains), and finally by mutagenesis of proposed receptor-binding residues. The structure of r-fVIII provides insight into the design of engineered fVIII variants, particularly with respect to disulfide cross-linked constructs (Figure 4B). Two such constructs have previously been described in some detail: the first containing a pair of cysteines linking residue 664 of the A2 domain to residue 1826 of the A3 domain (Y664C/T1826C), and the second, located in the same region, linking residues 662 and 1828 (M662C/D1828C).51 In the crystal structure, these residues pairs are positioned within 2 adjacent loops and display appropriate distances for S-S formation (distance between C-alphas is approximately 7 Å and 13 Å, respectively). The use of structural information may be of utility for the creation of new generations of improved fVIII constructs, including those that are stabilized through resculpting the A2 interface solely through increased complementarity of noncovalent contacts and packing using structure-based computational protein design methods.
Antibody inhibitor epitopes
A significant complication of fVIII replacement therapy is the development of antibody inhibitors.52 The most frequent inhibitor incidence occurs on epitopes within the A2 and C2 domains. Inhibitors to the A3 domain and the acidic region between A1 and A2 have also been observed. fVIII antibody inhibitors can block fVIII function in several ways: (1) by blocking the ability of fVIIIa to bind and activate fIXa and fX; (2) by inhibiting the binding of fVIII to VWF and/or negatively charged phospholipid surfaces; or (3) by hindering the activation of fVIII by thrombin (and/or fXa) or the subsequent release of fVIII from VWF.
The antigenic hotspots identified across the surface of r-fVIII generally correspond to the most mobile regions of the r-fVIII structure (Figure S4). A total of 2 epitopes (residues 351-365 in the A1 domain and residues 1674-1684 in the A3 domain) are disordered in the crystal, while another 2 (residues 484-508 in the A2 domain and residues 1814-1819 in the A3 domain) correspond to some of the most elevated B-factors in the structure. In contrast, the C2 domain, of which at least half has been implicated in inhibitor antibody binding, is internally well ordered; however, the domain as a whole is extensively solvent-exposed and only loosely docked to the remainder of the r-fVIII molecule. The importance of this domain in several critical procoagulant functions may make the existence of corresponding inhibitors particularly significant clinically, facilitating their identification and characterization.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Data were collected at the ALS beamline 5.0.2 with assistance from beamline staff. We thank members of the Strong and Ferre D'Amare laboratories at the Hutchinson Center for invaluable assistance and advice.
Data deposition: The coordinates and structure factor amplitudes for the fVIII structure have been deposited in the Research Collaboratory for Structural Bioinformatics (RCSB)53,54 for immediate release (PDB ID code 2R7E).
This project was funded by National Institutes of Health (NIH) grant R01 HL62570 to B.L.S. P.C.S. was funded by NIH training grant T32 GM08268.
Authorship
Contribution: B.W.S. and P.C.S. accomplished X-ray data collection and crystallographic modeling; Y.-H.K., C.-H.C., J.-W.H., J.-S.L., and J.K. generated purified r-fVIII; B.L.S., B.W.S., and P.C.S. analyzed the structure and prepared the manuscript; and all authors provided final review of the manuscript and many revisions.
Conflict-of-interest disclosure: C.-H.C., J.-W.H., J.-S.L., and J.K. are employed by the Green Cross Corporation; the fVIII construct described in this manuscript is a commercial product of that company. The remaining authors declare no competing financial interests.
Correspondence: Barry L. Stoddard, Program in Molecular Biophysics, Structure and Design, Division of Basic Sciences, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave N, A3-023, Seattle, WA; e-mail: bstoddar@fhcrc.org.
References
Author notes
B.W.S. and P.C.S. contributed equally to this study.