Abstract
Small-molecule αIIbβ3 antagonists competitively block ligand binding by spanning between the D224 in αIIb and the MIDAS metal ion in β3. They variably induce conformational changes in the receptor, which may have undesirable consequences. To identify αIIbβ3 antagonists with novel structures, we tested 33 264 small molecules for their ability to inhibit the adhesion of washed platelets to immobilized fibrinogen at 16 μM. A total of 102 compounds demonstrated 50% or more inhibition, and one of these (compound 1, 265 g/mol) inhibited ADP-induced platelet aggregation (IC50: 13± 5 μM), the binding of soluble fibrinogen to platelets induced by mAb AP5, and the binding of soluble fibrinogen and a cyclic RGD peptide to purified αIIbβ3. Compound 1 did not affect the function of GPIb, α2β1, or the other β3 family receptor αVβ3. Molecular docking simulations suggest that compound 1 interacts with αIIb but not β3. Compound 1 induced partial exposure of an αIIb ligand-induced binding site (LIBS), but did not induce exposure of 2 β3 LIBS. Transient exposure of purified αIIbβ3 to eptifibatide, but not compound 1, enhanced fibrinogen binding (“priming”). Compound 1 provides a prototype for small molecule selective inhibition of αIIbβ3, without receptor priming, via targeting αIIb.
Introduction
The platelet αIIbβ3 integrin plays a central role in platelet adhesion and aggregation.1-3 Thus, it can support platelet adhesion to immobilized fibrinogen even in the absence of exogenous activators.4,5 Moreover, when activated, the αIIbβ3 heterodimer can bind soluble ligands, including fibrinogen and von Willebrand factor, which can span between platelets to form aggregates.1,3,6,7 Loss of the receptor or its function on an inherited basis results in the hemorrhagic diathesis Glanzmann thrombasthenia,8 and inhibitors of the receptor have proven effective in the prevention and treatment of coronary artery thrombosis.9,10 Biochemical, molecular biologic, and crystallographic evidence indicate that ligands bind to a groove in αIIbβ3 that is at the intersection of the αIIb β propeller domain and the β3 βA (I-like) domain.11 Fibrinogen binds to αIIbβ3 via a carboxyl-terminal dodecapeptide sequence in its γ chain that contains both a positively charged Lys and a negatively charged Asp (HHLGGAKQAGDV).12-14 The integrin also binds ligands containing the sequence Arg-Gly-Asp (RGD) or Lys-Gly-Asp (KGD), including von Willebrand factor6,15 and snake venom–derived disintegrins.16 The drugs eptifibatide and tirofiban, which are patterned after the KGD and RGD sequences, respectively, span the αIIbβ3 ligand binding groove with orientations similar to that of an RGD-containing peptide (cilengitide) in the related receptor αVβ317 ; thus, their positively charged groups interact with αIIb Asp224 and their negatively charged carboxyl groups contribute to the coordination of the metal ion in the β3 metal ion–dependent adhesion site (MIDAS).11
Conformational changes in αIIbβ3 occur upon receptor activation, and additional changes occur after the binding of ligand to the receptor, leading to the exposure of ligand-induced binding sites (LIBS) that can be detected by LIBS-specific monoclonal antibodies (mAbs).18-21 The binding of RGD peptides and both eptifibatide and tirofiban increase the binding of LIBS-specific mAbs.22 Since αIIbβ3 may remain in its high-affinity conformation after dissociation of the competitive inhibitors, transient interactions of these compounds with the receptor may actually facilitate ligand binding by “priming” the receptor.23 It has been postulated that this effect may have contributed to the increased mortality observed during treatment with orally active inhibitors of αIIbβ3 that were administered on a chronic basis.24-29 Moreover, the conformational changes induced by all of the antagonists may contribute to the thrombocytopenia observed with these agents.30
To identify novel small molecules capable of inhibiting the interaction of fibrinogen with αIIbβ3, we used high-throughput screening of several libraries of small molecules, testing the ability of the compounds to inhibit platelet adhesion to fibrinogen. We identified one compound with unique features that provide insights into αIIbβ3 structure and function.
Methods
Monoclonal antibodies and cell lines
Monoclonal antibodies (mAbs) 6D131 (anti-GPIb), 6F132 (anti-α2β1), 7H233 (anti-αIIbβ3 and αVβ3), 7E334 (anti-αIIbβ3 and αVβ3), and 10E535 (anti-αIIbβ3) were produced at the National Cell Culture Center (Minneapolis, MN). The mAb AP521 was generously provided by Peter Newman (Blood Center of Southeastern Wisconsin). The mAbs PMI-136 and LIBS-119 were the generous gift of Dr Mark H. Ginsberg (University of California). HEK293 cells stably expressing normal human αIIbβ3 were prepared as previously described.34 CS1 cells stably expressing normal human αV were a generous gift of Dr David Cheresh (University of California, San Diego), and were transfected with cDNA encoding normal human β3 as previously described.37
Platelet preparation for primary screen
Platelet concentrates (1500 × 109 to 3000 × 109 platelets/L, ADVIA 120; Bayer, Tarrytown, NY), obtained from the New York Blood Center, were divided into 5-mL aliquots and then 5 mL HEPES-modified Tyrode buffer (HBMT; 138 mM NaCl, 12 mM NaHCO3, 10 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid], 2.7 mM KCl, 0.4 mM NaH2PO4, 0.1% glucose, 0.35% BSA, pH 7.4) was added, and the suspensions were treated with prostaglandin E1 (PGE1, final concentration: 1 μM; Sigma-Aldrich, St Louis, MO). Samples were centrifuged at 1200g for 8 minutes at 22°C and resuspended in HBMT. Platelets were fluorescently labeled by incubation with calcein-acetoxymethyl ester (7 μM; Invitrogen, Carlsbad, CA) for 30 minutes at 22°C in the dark, and washed with HBMT/PGE1. Platelet pellets were then resuspended in HBMT containing 2 mM CaCl2 and 1 mM MgCl2, and the platelet counts were adjusted.
Platelet adhesion assay
Human fibrinogen (50 μg/mL in 100 mM NaCl, 50 mM Tris/HCl, pH 7.4 [Tris/saline]; American Diagnostica, Stamford, CT) was added to black-walled, clear-bottomed, untreated polystyrene, nonsterile 384-well microtiter plate wells (Corning no. 3711; Acton, MA) using a peristaltic microplate dispenser (WellMate; Matrix, Hudson, NH). After incubating at 22°C for 1 hour, plates were washed 4 times with Tris/saline using an automated plate washer (ELX405; Bio-Tek, Winooski, VT), and wells were then blocked with HBMT for at least 1 hour. After 2 additional washes, 15 μL HBMT was allowed to remain in each well. Compounds were added to the wells with a liquid handling robot (MiniTrak V; PerkinElmer, Wellesley, MA) using a pin tool (VP Scientific, San Diego, CA) that dispensed 0.1-μL aliquots of 5-mM solutions of compounds in DMSO. Calcein-labeled platelets were then added (15 μL, 500 × 109/L; final platelet count: 250 × 109/L, final compound concentration: 16 μM). After 1 hour of adhesion, wells were washed 4 times with HBMT-CaCl2/MgCl2. Positive controls consisted of wells containing platelets without compounds. Negative controls were wells containing platelets and known inhibitors of αIIbβ3, including mAbs 7E3 and 10E5, and EDTA (ethylenediaminetetraacetic acid). The relative number of adherent platelets was measured by exciting calcein with 490 nm light and reading fluorescence intensity at 515 nm using an automated plate reader (Envision; PerkinElmer). The reliability of the assay was assessed by calculating the Z' factor, which is determined from the separation between and the standard deviations of the positive and negative controls on a plate-by-plate basis.38 Every plate tested had a Z' value more than 0.65; values more than 0.5 indicate excellent assay quality.
Platelet preparation from peripheral blood
This study was approved by the Rockefeller University Institutional Review Board. Blood was obtained with consent from healthy volunteers who had not taken any medication known to inhibit platelet function for at least 7 days using a 19-gauge needle and anticoagulated with either a 0.10 volume of 3.8% sodium citrate or a 0.15 volume of acid-citrate-dextrose (ACD; 74.8 mM sodium citrate, 38 mM citric acid, 124 mM dextrose). Platelet-rich plasma (PRP) was prepared by centrifugation at 650g for 4 minutes at 22°C. Platelet-poor plasma (PPP) was prepared by further centrifugation of the remaining blood at 1200g for 8 minutes at 22°C. Washed platelets (WPs) were prepared by treating ACD-anticoagulated PRP with 1 μM PGE1, adding 0.10 vol ACD, and centrifuging at 1200g for 8 minutes at 22°C. The platelet pellet was resuspended in HBMT.
Platelet aggregation in 96-well microtiter plates
As modified from Fratantoni and Poindexter,39 untreated 96-well polystyrene plates (Nunc, Rochester, NY) were blocked for at least 1 hour with HBMT, and then compounds were added to the wells (33 μL of 50 μM in HBMT), along with a 67-μL sample of citrated PRP (final compound concentration: 16 μM). After plates equilibrated at 37°C in the plate reader (SpectraMax; Molecular Devices, Sunnyvale, CA) for 10 minutes, aggregation was initiated by adding ADP (final concentration: 5 μM; Chrono-Log Corp, Havertown, PA). Absorbance (Abs) was measured at 563 nm every 10 seconds with 3 seconds of shaking between determinations for 8 minutes. Percent aggregation was calculated by the equation (AbsPRP − Abssample)/(AbsPRP − AbsPPP).
Platelet aggregation in aggregometer
Washed platelets (250 × 109/L in HBMT containing 100 μM CaCl2, 50 μM MgCl2, and 200 μg/mL fibrinogen) or citrated PRP in aggregometer cuvettes was incubated with compounds or controls for 15 minutes at 37°C. After 30 seconds in the aggregometer (Kowa AG-10E; Tokyo, Japan) at 37°C with stirring, agonists were added, and light transmittance was measured for 8 minutes. Agonists included ADP, collagen (native type 1 collagen fibrils), ristocetin, arachidonic acid (all from Chrono-Log), and thrombin receptor activating peptide (TRAP [SFLLRN]; prepared at the State University of New York at Stony Brook).
mAb binding
WPs were prepared in HBMT containing 2 mM CaCl2/1 mM MgCl2. Platelet count was adjusted to 250 × 109/L and compound 1 (100 μM), tirofiban (100 μM), EDTA (10 mM), or mAb 10E5 (20 μg/mL) was added to samples (30 μL) and incubated for 15 minutes at 37°C. Fluorescently labeled monoclonal antibodies (PMI-1, LIBS-1, AP5, 7H2, 7E3, or 10E5; Alexa-488 conjugated; Invitrogen) were then added (20 μL, final concentration: 5 μg/mL) and incubated for 30 minutes at 22°C in the dark, after which samples were diluted 1:10 in HBMT with CaCl2/MgCl2 for analysis by flow cytometry (FACSCalibur; Becton Dickinson, Franklin Lakes, NJ). Antibody binding was reported as the geometric mean fluorescence intensity; nonspecific binding was determined by adding a 50-fold excess of unlabeled antibody before adding the labeled antibody.
Platelet and cell adhesion
Washed platelets prepared as per the primary screening assay, CS1 cells stably expressing normal human αVβ3, or HEK293 cells stably expressing normal human αIIbβ3 were suspended in HBMT containing either 1 mM MgCl2 (CS1 cells and platelets for adhesion to collagen) or 2 mM CaCl2/1 mM MgCl2 (HEK293 cells and platelets for adhesion to fibrinogen), and the cell counts were adjusted to 2 × 106/mL (CS1 cells), 106/mL (HEK293 cells), or 250 × 109/L (platelets). Polystyrene 96-well microtiter plates (Nunc) were coated with either fibrinogen (50 μg/mL), vitronectin (5 μg/mL; generously provided by Dr Deane F. Mosher, University of Wisconsin), or collagen (33 μg/mL, rat tail type 1; Becton Dickinson) for 1 hour, and blocked with HBMT for at least 1 hour. Platelets and cells were treated with compounds or control solutions for 15 minutes at 37°C before being added to the microtiter wells. After adhering for 1 hour at either 22°C (platelets) or 37°C (cells), nonadherent platelets or cells were removed by washing 3 times with HBMT containing the same ion composition as the buffer used for adhesion. Adherent platelets or cells were quantified by their endogenous acid phosphatase activity on p-nitrophenyl phosphate (pNPP) as previously described40 (1 mg/mL in 0.1 M sodium citrate, 0.1% Triton X-100, pH 5.4). In other experiments, 8-chambered glass coverslips (Nunc) were coated with collagen (33 μg/mL) for 1 hour at 22°C. Washed platelets were allowed to adhere for 1 hour at 22°C and the coverslips were stained with the Alexa-488–conjugated β3-specific mAb 7H2. Adherent platelets were imaged using a Zeiss LSM-510 confocal system with Axiovert 200 microscope (Carl Zeiss, Heidelberg, Germany) using a Plan-Apochromat 100×/1.4 NA oil DIC objective.
Fluorescent fibrinogen binding
Washed platelets (250 × 109/L) or HEK293 cells expressing αIIbβ3 (1.0 × 105/mL), both in HBMT with 2 mM CaCl2/1 mM MgCl2, were incubated with compounds or control solutions and Alexa-488–conjugated fibrinogen (200 μg/mL; Invitrogen), with or without the αIIbβ3-activating mAb AP5 (60 μg/mL), for 30 minutes at 22°C in the dark. Unbound fibrinogen was removed by centrifugation at 1800g (platelets) in the presence of 1 μM PGE1 or 300g (cells) for 3 minutes at 22°C and resuspending in HBMT containing ions before flow cytometric analysis.
Fibrinogen binding to purified αIIbβ3
The procedure for measuring the binding of fibrinogen to purified αIIbβ3 was adapted from the method of Kouns et al.41 The anti-β3 mAb 7H2 (10 μg/mL) was adsorbed to polystyrene microtiter plate wells overnight at 4°C and wells were then blocked with 3.5% BSA for 1 hour at 22°C. Purified αIIbβ3 (Enzyme Research Laboratories, South Bend, IN) was diluted in buffer A (50 mM Tris/HCl, 100 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, 1% BSA, 0.0035% Triton X-100) to 10 μg/mL, added to the wells, and captured by the immobilized mAb 7H2 for 2 hours at 37°C. Wells were then washed 3 times with buffer A. Fibrinogen, prepared in buffer A (20 μg/mL) in the presence or absence of mAb AP5 (60 μg/mL), was incubated in plate wells for 2 hours at 37°C with or without compounds or control solutions. Wells were washed 3 times with buffer A and then incubated with a horseradish peroxidase–conjugated polyclonal antifibrinogen antibody (1:1000 in buffer A; DAKO, Glostrup, Denmark) for 1 hour at 22°C. Wells were washed 3 times, 200 μL of a peroxidase substrate [3,3′,5,5′-tetramethylbenzidine (TMB, Sigma-Aldrich) was added, the reaction was terminated after 30 minutes by acidification with 50 μL 0.5 M H2SO4, and absorbance was determined at 450 nm. For priming experiments, the procedure was the same except (1) αIIbβ3 was allowed to adhere in the presence of either eptifibatide (10 μM), RGDS (10 μM; Rockefeller University Proteomics Resource Center), compound 1 (100 μM), or DMSO (1%); (2) wells were washed 10 times after αIIbβ3 adhesion; and (3) absorbance was determined at 655 nm after 10 minutes.
Platelet adhesion under shear stress
A cone and plate system (Diamed, Cressier, Switzerland) was used to assess the effect of compound 1 on platelet deposition under shear stress as described by Varon et al.42 Briefly, 0.25 mL of citrated whole blood was incubated with nothing, DMSO (0.5%), compound 1 (50 μM), or tirofiban (10 μM) for 15 minutes at 22°C and placed in polystyrene wells (16-mm diameter; Nunc). A constant uniform shear force of 1800 s−1 was applied across the well using a Teflon cone for 2 minutes at 22°C. Wells were washed with buffer, fixed with 1% paraformaldehyde, stained with Wright stain (Sigma-Aldrich), and analyzed for platelet adhesion using a 20× objective in an inverted microscope (IX51; Olympus, Center Valley, PA). Platelet adhesion was assessed by measuring the area occupied by platelets in each of 4 uniformly selected viewing areas in duplicate wells using image analysis software (SlideBook; Intelligent Imaging Innovations, Denver, CO).
Fluorescent RGD peptide binding to αIIbβ3
A fluorescent cyclic RGD-containing pentapeptide (5(6)-carboxyfluorescein-c[KRGDf]; generously provided by Dr Wei Wang, University of Texas, M. D. Anderson Cancer Center) was used as described by Wang et al.43 Briefly, 300 nM αIIbβ3 was prepared in 0.15 M NaCl, 50 mM Tris/HCl containing 1 mM MnCl2, and buffer, compound 1, a functionally inactive derivative of compound 1, DMSO, or tirofiban was added and after 10 minutes at 22°C, fluorescence polarization was measured using a fluorescence polarization microplate reader (Envision; PerkinElmer). Polarization was expressed in millipolarization units (mP). Values obtained in the presence of tirofiban were taken as background, and all other values were expressed as a percentage of the millipolarization value observed in the presence of buffer alone (no inhibitor).
Molecular docking
The automated docking protocol Flexcdock44 was used to dock compound 1 into the ligand binding sites of αIIbβ311 (PDB ID: ITY6, 1TY5) and αVβ317 (PDB ID: 1L5G). This algorithm carries out an exhaustive translational and rotational search of the ligand within the binding site, while considering the protein to be rigid. The nonbonded interaction energies based on the AMBER force field using an all-atom model are used to model the ligand-receptor interaction (van der Waals and Coulombic term) as the scoring energy function. To account for the flexibility of the ligand, the protocol takes into account its torsional degrees of freedom, which are discretized into coarse rotameric states (36 rotamers generated for compound 1). The ligand library was then docked into the ligand-binding sites of the integrins and the 20 best-scored solutions were selected for further analysis. An average-linkage hierarchic clustering of the different solutions was carried out based on the pair-wise root mean square deviations (RMSD) of the ligand heavy atoms, using CLUSBAS.45 As controls, eptifibatide (αIIbβ3), tirofiban (αIIbβ3), and the RGD peptide cilengitide (αVβ3) were docked back into their native crystal structures (PDB entries: 1TY6, 1TY5, and 1L5G, respectively). For these redocking studies, the ligands were considered to have the conformations observed in their crystal structures.
Results
Screening
A total of 33 264 compounds were analyzed in the platelet adhesion to fibrinogen assay (primary screen), and a total of 102 compounds were identified that inhibited platelet adhesion by more than 50% on 2 separate days (0.31% hit rate). Of these 102 compounds, 32 inhibited platelet adhesion by more than 60%, and 9 inhibited adhesion by more than 70%.
Compound 1 inhibits platelet aggregation and platelet deposition under shear
All 102 compounds identified in the primary screen were next tested for their ability to inhibit the initial slope of ADP (5 μM)–induced platelet aggregation in PRP in microtiter plates. Only 2 compounds demonstrated significant inhibitory activity in this assay, with compound 1 (Figure 1) reducing the initial slope of aggregation by 84%, and compound 2 inhibiting it by 72%. Subsequent studies demonstrated that compound 2's effects were not due to direct inhibition of αIIbβ3 (eg, it induced cell death in 74% ± 21% of CS1 cells [mean ± SD, n = 3] after incubation for 2 hours at 100 μM [as judged by trypan blue exclusion46 ], compared with 26% [± 5%] of cells treated with vehicle [1% DMSO] or 14% [± 13%] of cells treated with compound 1 [100 μM]; it induced washed platelets to adopt a spherical shape when incubated at concentrations of 30 μM or greater in a time-dependent manner; and it failed to inhibit fibrinogen binding to purified αIIbβ3), and thus it was not considered further.
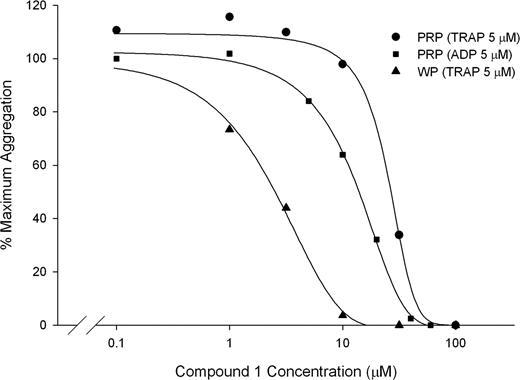
Compound 1 also inhibited the aggregation of platelets in citrated PRP induced by 5 μM ADP, 5 μM TRAP, and 1 mM arachidonic acid with IC50s of 13 μM (± 5 μM), 29 μM (± 6 μM), and 8.9 μM (± 5.7μM), respectively, and inhibited the aggregation of washed platelets induced by 5 μM TRAP with an IC50 of 3.4 μM (± 0.4 μM; n = 3; Figure 2). Compound 1 (100 μM), also completely inhibited platelet aggregation in PRP induced by collagen (2 μg/mL). Like the αIIbβ3-specific mAb 10E5, compound 1 did not inhibit the initial slope of ristocetin-induced (1.8 mg/mL) agglutination/aggregation of PRP, but did inhibit the later phase of aggregation (data not shown). In contrast, the GPIb-specific mAb 6D1 (20 μg/mL) completely abolished the initial wave of ristocetin-induced agglutination.
Compound 1 inhibits aggregation of PRP induced by 5 μM ADP (IC50: 13 ± 5 μM) and 5 μM TRAP (IC50: 29 ± 6 μM) and inhibits the aggregation of washed platelets induced by 5 μM TRAP (IC50: 3.4 ± 0.4). Citrated PRP or washed platelets (250 × 109/L, 100 μM CaCl2, 50 μM MgCl2, 200 μg/mL fibrinogen) was treated with compound 1 for 15 minutes at 37°C before initiating aggregation with 5 μM ADP or 5 μM TRAP (PRP and washed platelets). Aggregation was calculated as the percentage of the maximum initial slope of the change in light transmittance from 15 to 90 seconds. Results from 1 representative experiment from each condition are shown, of a total of 3. Note that data are from different donors tested on different days.
Compound 1 inhibits aggregation of PRP induced by 5 μM ADP (IC50: 13 ± 5 μM) and 5 μM TRAP (IC50: 29 ± 6 μM) and inhibits the aggregation of washed platelets induced by 5 μM TRAP (IC50: 3.4 ± 0.4). Citrated PRP or washed platelets (250 × 109/L, 100 μM CaCl2, 50 μM MgCl2, 200 μg/mL fibrinogen) was treated with compound 1 for 15 minutes at 37°C before initiating aggregation with 5 μM ADP or 5 μM TRAP (PRP and washed platelets). Aggregation was calculated as the percentage of the maximum initial slope of the change in light transmittance from 15 to 90 seconds. Results from 1 representative experiment from each condition are shown, of a total of 3. Note that data are from different donors tested on different days.
Untreated platelets and platelets treated with DMSO (0.5%) adhered to polystyrene wells when subjected to shear. Compared with the area occupied by platelets in samples treated with DMSO, compound 1 (50 μM) inhibited the deposition by 87% (± 10%; P < .025) and tirofiban (10 μM) inhibited the deposition by 99% (± 1%; P < .015, n = 3).
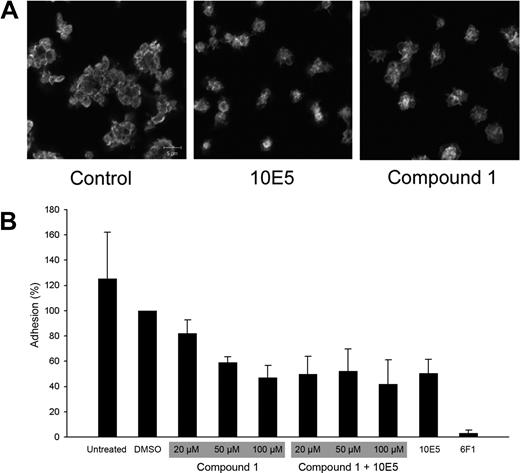
Compound 1 inhibits αIIbβ3-mediated platelet recruitment to platelets adherent to collagen
Compound 1 was tested for its effect on platelet adhesion to immobilized collagen. Confocal microscopy of adherent platelets in the presence of either no compound or DMSO revealed clusters of platelets, which has previously been shown to reflect both α2β1-mediated platelet adhesion to collagen and subsequent αIIbβ3-dependent recruitment of additional platelets to the adherent ones47 (Figure 3). Platelet adhesion was nearly abolished by an anti-α2β1 mAb (6F1, 20 μg/mL). In contrast, the anti-αIIbβ3 mAb (10E5, 20-40 μg/mL) did not affect the initial interaction of platelets with collagen, but nearly abolished the recruitment of additional platelets to the adherent platelets. In the presence of compound 1 (50-250 μM) platelets adhered to collagen, but like samples treated with mAb 10E5, there was no recruitment of additional platelets to the adherent platelets. To quantify these observations, the assay was conducted in microtiter plates (Figure 3). The mAb 6F1 nearly abolished adhesion, whereas compound 1 (100 μM) reduced the total number of adherent platelet by approximately 50%, a value similar to that obtained for mAb 10E5. Adding both compound 1 and mAb 10E5 did not result in greater inhibition of platelet adhesion than with either agent alone, supporting the hypothesis that they operate through similar mechanisms.
Compound 1 does not prevent platelet adhesion to collagen, but does prevent recruitment of additional platelets to adherent platelets; compound 1 and the anti-αIIbβ3 mAb 10E5 only partially inhibit platelet deposition on immobilized collagen, and their effects are not additive. (A) Washed platelets were allowed to adhere to collagen on a glass slide for 1 hour, and were then labeled with Alexa-488–conjugated anti-β3 mAb 7H2. Confocal microscopy images were obtained with a 100× objective as described in “Platelet and cell adhesion.” (B) Washed platelets were left untreated or treated with DMSO (1%); compound 1 at 20, 50, or 100 μM, with or without mAb 10E5 (20 μg/mL); mAb 10E5; or mAb 6F1 (anti-α2β1, 20 μg/mL) for 15 minutes at 37°C before being added to microtiter wells coated with collagen. After 1 hour, nonadherent platelets were removed by washing and the number of adherent platelets was determined by pNPP assay as described in “Platelet and cell adhesion.” Adhesion is expressed as the percentage of adhesion of control, DMSO (1%)–treated platelets. The mean and SD of 4 separate experiments is depicted for each condition.
Compound 1 does not prevent platelet adhesion to collagen, but does prevent recruitment of additional platelets to adherent platelets; compound 1 and the anti-αIIbβ3 mAb 10E5 only partially inhibit platelet deposition on immobilized collagen, and their effects are not additive. (A) Washed platelets were allowed to adhere to collagen on a glass slide for 1 hour, and were then labeled with Alexa-488–conjugated anti-β3 mAb 7H2. Confocal microscopy images were obtained with a 100× objective as described in “Platelet and cell adhesion.” (B) Washed platelets were left untreated or treated with DMSO (1%); compound 1 at 20, 50, or 100 μM, with or without mAb 10E5 (20 μg/mL); mAb 10E5; or mAb 6F1 (anti-α2β1, 20 μg/mL) for 15 minutes at 37°C before being added to microtiter wells coated with collagen. After 1 hour, nonadherent platelets were removed by washing and the number of adherent platelets was determined by pNPP assay as described in “Platelet and cell adhesion.” Adhesion is expressed as the percentage of adhesion of control, DMSO (1%)–treated platelets. The mean and SD of 4 separate experiments is depicted for each condition.
Compound 1 partially exposes an αIIb LIBS but neither of 2 β3 LIBS
Compounds were tested for their effect on the binding of various mAbs specific for αIIb, β3, or the αIIbβ3-complex (Table 1). As expected, the LIBS mAbs PMI-1 (anti-αIIb), LIBS-1 (anti-β3), and AP5 (anti-β3) bound poorly to untreated platelets, and binding was increased in the presence of the ligand mimetic antagonist tirofiban (∼ 10-fold increases for PMI-1 and LIBS-1 and a ∼ 5-fold increase for AP5; data not shown). The binding of the non-LIBS mAbs (7H2 and 10E5) was not affected by either compound 1 or tirofiban (data not shown). Compound 1 increased the binding of the αIIb LIBS-specific mAb PMI-1, but the increase was less than one half the increase produced by tirofiban (Table 1). The binding of the β3 LIBS-specific mAbs LIBS-1 and AP5 was not significantly affected by compound 1, although there was a small increase in AP5 binding.
Compound 1 does not increase the binding of β 3 LIBS antibodies and only partially increases the binding of an α IIb LIBS antibody to washed platelets
| mAb . | Target . | mAb binding, % value with tirofiban . | |
|---|---|---|---|
| Untreated platelets . | Compound 1 . | ||
| PMI-1 | αIIb | 11 ± 4.1 | 44 ± 8.7* |
| LIBS-1 | β3 | 9.0 ± 3.1 | 5.7 ± 6.3 |
| AP5 | β3 | 19 ± 6.2 | 26 ± 5.9 |
| mAb . | Target . | mAb binding, % value with tirofiban . | |
|---|---|---|---|
| Untreated platelets . | Compound 1 . | ||
| PMI-1 | αIIb | 11 ± 4.1 | 44 ± 8.7* |
| LIBS-1 | β3 | 9.0 ± 3.1 | 5.7 ± 6.3 |
| AP5 | β3 | 19 ± 6.2 | 26 ± 5.9 |
Washed platelets were treated with compound 1 (100 μM), DMSO (1%), or tirofiban (100 μM) for 15 minutes at 37°C before labeling with mAbs for 30 minutes at 22°C in the dark. Samples were diluted 1:10 and analyzed by flow cytometry. Binding was calculated as the percentage of binding to platelets treated with 100 μM tirofiban. Data are presented as mean plus or minus SD values (n = 5).
P < .001 versus PMI-1 binding to untreated platelets.
Compound 1 inhibits fibrinogen binding to platelets, recombinant cells, and purified αIIbβ3 and has no “priming” activity
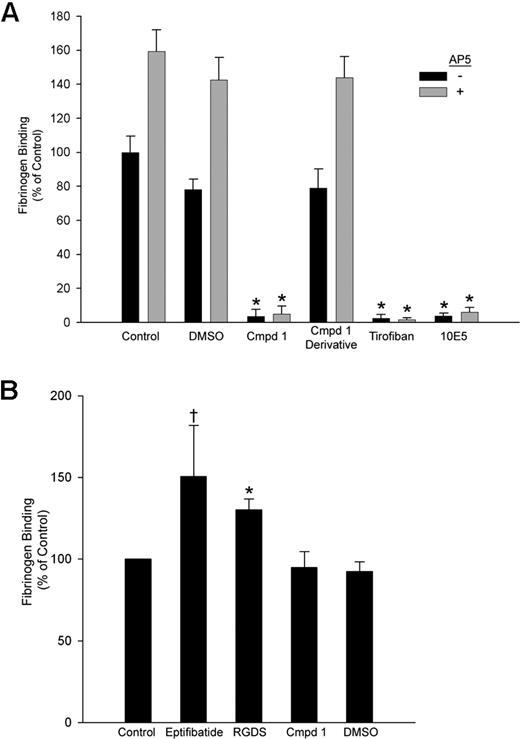
Compound 1 inhibited fibrinogen binding to platelets induced by the activating LIBS mAb AP5 with an IC50 of 29 ± 2 μM (n = 3), a value similar to its IC50 for inhibiting AP5-induced fibrinogen binding to HEK293 cells (21 ± 0.3 μM; n = 3). Fibrinogen bound to purified αIIbβ3 in microtiter plates in both the presence and the absence of 1% DMSO or a functionally inactive derivative of compound 1 in which the piperazine group was altered (Figure 4A). The binding was blocked by compound 1 (IC50: 1.6 ± 0.4 μM, n = 3), tirofiban, and mAb 10E5. The αIIbβ3-activating mAb AP5 increased fibrinogen binding to purified αIIbβ3 in the presence of buffer, DMSO, or the compound 1 derivative, but mAb AP5 did not increase fibrinogen binding to purified αIIbβ3 in the presence of compound 1, tirofiban, or mAb 10E5.
Compound 1 inhibits fibrinogen binding to purified αIIbβ3; priming with eptifibatide or RGDS peptide, but not compound 1, enhances fibrinogen binding. (A) Purified αIIbβ3 was allowed to adhere to wells coated with the anti-β3 mAb 7H2 for 2 hours at 37°C. Fibrinogen binding to αIIbβ3 took place in the presence of buffer alone; DMSO (1%); compound 1 (Cmpd 1, 100 μM); an analog of compound 1 in which the piperazine group is disrupted (100 μM); tirofiban (10 μM); or mAb 10E5 (20 μg/mL), with or without AP5 (60 μg/mL) for 2 hours at 37°C. The extent of fibrinogen binding was determined using HRP-conjugated antifibrinogen polyclonal antibodies and a peroxidase substrate as described in “Fibrinogen binding to purified αIIbβ3.” (B) For priming experiments, the same procedure was followed with the following exceptions: priming agents were added along with αIIbβ3, and wells were washed 10 times following fibrinogen addition. Binding is presented as mean plus or minus SD for 4 separate experiments, calculated as the percentage of “control” (fibrinogen binding in the presence of buffer alone without AP5) (*P < .001, †P < .02 both vs control).
Compound 1 inhibits fibrinogen binding to purified αIIbβ3; priming with eptifibatide or RGDS peptide, but not compound 1, enhances fibrinogen binding. (A) Purified αIIbβ3 was allowed to adhere to wells coated with the anti-β3 mAb 7H2 for 2 hours at 37°C. Fibrinogen binding to αIIbβ3 took place in the presence of buffer alone; DMSO (1%); compound 1 (Cmpd 1, 100 μM); an analog of compound 1 in which the piperazine group is disrupted (100 μM); tirofiban (10 μM); or mAb 10E5 (20 μg/mL), with or without AP5 (60 μg/mL) for 2 hours at 37°C. The extent of fibrinogen binding was determined using HRP-conjugated antifibrinogen polyclonal antibodies and a peroxidase substrate as described in “Fibrinogen binding to purified αIIbβ3.” (B) For priming experiments, the same procedure was followed with the following exceptions: priming agents were added along with αIIbβ3, and wells were washed 10 times following fibrinogen addition. Binding is presented as mean plus or minus SD for 4 separate experiments, calculated as the percentage of “control” (fibrinogen binding in the presence of buffer alone without AP5) (*P < .001, †P < .02 both vs control).
In experiments designed to assess the ability of different compounds to enhance fibrinogen binding to purified αIIbβ3 (priming), transient exposure of αIIbβ3 to eptifibatide or RGDS peptide resulted in increased fibrinogen binding (150% ± 31% [P < .02] and 130% ± 7% [P < .001], respectively) compared with untreated αIIbβ3 (Figure 4B). In sharp contrast, pretreatment with DMSO or compound 1 did not increase fibrinogen binding.
Compound 1 inhibits binding of c(KRGDf) to purified αIIbβ3
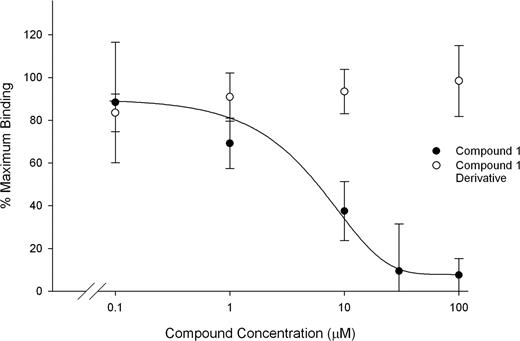
The fluorescent cyclic RGD-containing pentapeptide bound to purified αIIbβ3 as judged by fluorescence polarization, and the binding was inhibited by tirofiban (1 μM). Neither DMSO nor an inactive derivative of compound 1 inhibited peptide binding, whereas compound 1 inhibited peptide binding with an IC50 of 6.6 μM (± 2.2 μM; Figure 5).
Compound 1 inhibits binding of a fluorescent cyclic RGD peptide to αIIbβ3. Purified αIIbβ3 (300 nM) was incubated with buffer alone, DMSO, tirofiban (1 μM), compound 1, or a functionally inactive derivative of compound 1. FITC-labeled c(KRGDf) was added at 10 nM and after 10 minutes at 22°C fluorescence polarization was assessed. Binding is presented as mean plus or minus SD for 3 separate experiments, calculated as the percentage of the maximum mP value observed in the presence of buffer alone. The mP observed with tirofiban treatment was taken as background.
Compound 1 inhibits binding of a fluorescent cyclic RGD peptide to αIIbβ3. Purified αIIbβ3 (300 nM) was incubated with buffer alone, DMSO, tirofiban (1 μM), compound 1, or a functionally inactive derivative of compound 1. FITC-labeled c(KRGDf) was added at 10 nM and after 10 minutes at 22°C fluorescence polarization was assessed. Binding is presented as mean plus or minus SD for 3 separate experiments, calculated as the percentage of the maximum mP value observed in the presence of buffer alone. The mP observed with tirofiban treatment was taken as background.
Compound 1 inhibits αIIbβ3-mediated cell adhesion but not αVβ3-mediated cell adhesion
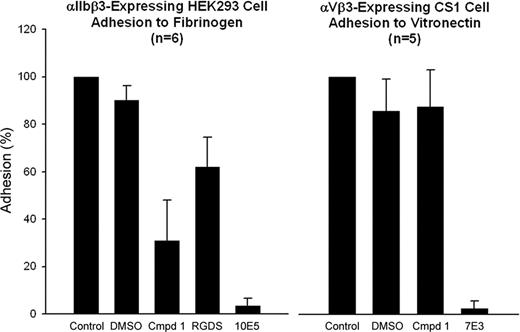
HEK293 cells expressing αIIbβ3 adhered to immobilized fibrinogen in microtiter plate wells in the presence of 2 mM CaCl2/1 mM MgCl2 with or without treatment with 1% DMSO (Figure 6). Compound 1 at 100 μM inhibited adhesion by 69% (± 17%; n = 6, P < .001), whereas mAb 10E5 (20 μg/mL) inhibited cell adhesion by 97% (± 3%; n = 6, P < .001). Untreated αVβ3-expressing CS1 cells adhered to immobilized vitronectin in the presence of 1 mM MgCl2, and adhesion was not significantly inhibited by 1% DMSO or the αIIbβ3-specific mAb 10E5 (Figure 6). Adhesion was nearly abolished by the mAb 7E3, which binds to both αIIbβ3 and αVβ3. Compound 1 did not inhibit CS1 cell adhesion.
Compound 1 inhibits adhesion of cells expressing αIIbβ3 to fibrinogen, but does not inhibit adhesion of cells expressing αVβ3 to vitronectin. αIIbβ3-Expressing HEK293 cells or αVβ3-expressing CS1 cells were treated with DMSO (1%), compound 1 (100 μM), RGDS (1 mM), mAb 10E5 (20 μg/mL), or mAb 7E3 (20 μg/mL) for 15 minutes at 37°C. Cells were allowed to adhere to either immobilized fibrinogen or vitronectin, respectively, for 1 hour at 37°C. Nonadherent cells were removed by washing and the extent of cell adhesion was determined by pNPP assay as described in “Platelet and cell adhesion.” Adhesion is expressed as the percentage of cells adhering relative to the untreated cells. Means and SD are shown.
Compound 1 inhibits adhesion of cells expressing αIIbβ3 to fibrinogen, but does not inhibit adhesion of cells expressing αVβ3 to vitronectin. αIIbβ3-Expressing HEK293 cells or αVβ3-expressing CS1 cells were treated with DMSO (1%), compound 1 (100 μM), RGDS (1 mM), mAb 10E5 (20 μg/mL), or mAb 7E3 (20 μg/mL) for 15 minutes at 37°C. Cells were allowed to adhere to either immobilized fibrinogen or vitronectin, respectively, for 1 hour at 37°C. Nonadherent cells were removed by washing and the extent of cell adhesion was determined by pNPP assay as described in “Platelet and cell adhesion.” Adhesion is expressed as the percentage of cells adhering relative to the untreated cells. Means and SD are shown.
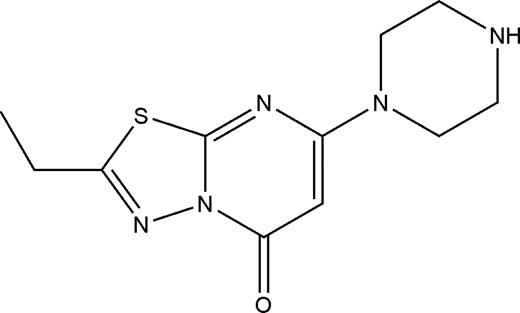
Structure of compound 1
Compound 1 (265 g/mol) contains a positively charged piperazine group attached to a nitrogen- and sulfur-containing heterocyclic core (Figure 1). Of note, 4 compounds containing the same cyclic core scaffold, but with aromatic groups attached to the piperazine group, were also tested, but none of these compounds inhibited platelet adhesion to fibrinogen.
Molecular docking studies
Docking results are presented in Table 2 and Figure 7. Compound 1 was docked into 2 different conformations of αIIbβ3, corresponding to the bound structures of this receptor with the cyclic peptide eptifibatide (PDB entry: 1TY6) or the non–peptide inhibitor tirofiban (PDB entry: 1TY5). Support for the reliability of the docking program was obtained by docking eptifibatide and tirofiban back into their corresponding crystal structures, yielding RMSDs of heavy atom coordinates of 0.3 Å and 0.4 Å, respectively. The preferred orientation of compound 1 in the ligand-binding cavity of αIIbβ3 was similar in both receptor conformations. The clustering of the 20 best-scored solutions obtained in both structures based on RMSD-similarity criteria identified a predominant binding solution comprising 28 of the 40 solutions analyzed (13 of the 20 in the eptifibatide-bound crystal and 15 of the 20 in the tirofiban-bound structure). In this preferred orientation, the basic piperazinyl nitrogen of compound 1 interacts with the carboxyl side chain of D224, as do the basic groups of both eptifibatide and tirofiban (Figure 7A-C). Unlike these latter compounds, however, compound 1 is not long enough to reach the MIDAS metal ion, nor does it contain a negatively charged group capable of coordinating the MIDAS. The distance from the end of compound 1 to the MIDAS (8 Å) also makes it unlikely that a water molecule bridges between compound 1 and the MIDAS metal ion. The ring structure of compound 1 interacts with the aromatic pocket formed by the αIIb residues F160, Y190, and F231. The computed docking energy, that is, the protein-ligand interaction energy, of the minimum energy member of the most populated solution (ranked 1 in the tirofiban crystal and 2 in the eptifibatide crystal) was approximately −92 kcal/mol.
Compound 1 docks into αIIbβ3 with a more favorable interaction energy than into αVβ3
| Receptor-antagonist complex (PDBID)* . | Compound 1 docking energies . | Percentage of solutions with similar orientation‖ . | ||
|---|---|---|---|---|
| Total† . | van der Waals‡ . | Electrostatic§ . | ||
| αIIbβ3-tirofiban (1TY5) | −91.6 | −31.5 | −60.1 | 75 |
| αIIbβ3-eptifibatide (1TY6) | −92.1 | −31.4 | −60.7 | 65 |
| αVβ3-cilengitide (cyclic RGD, 1L5G) | −73.0 | −12.2 | −60.9 | 55 |
| Receptor-antagonist complex (PDBID)* . | Compound 1 docking energies . | Percentage of solutions with similar orientation‖ . | ||
|---|---|---|---|---|
| Total† . | van der Waals‡ . | Electrostatic§ . | ||
| αIIbβ3-tirofiban (1TY5) | −91.6 | −31.5 | −60.1 | 75 |
| αIIbβ3-eptifibatide (1TY6) | −92.1 | −31.4 | −60.7 | 65 |
| αVβ3-cilengitide (cyclic RGD, 1L5G) | −73.0 | −12.2 | −60.9 | 55 |
Receptor-antagonist complex used in the docking experiment and its corresponding Protein Data Bank entry.
Total energy of interaction (kcal/mol) of compound 1 in the best-scored docking orientation. This is the sum of van der Waals and electrostatic forces.
Van der Waals contributions to the docking energy (kcal/mol).
Electrostatic contribution to the docking energy (kcal/mol).
Percentage of solutions (of 20 best-scored) belonging to the most populated group of solutions (cutoff 2 Å RMSD).
Compound 1 docks into αIIbβ3 with an orientation similar to that of eptifibatide and tirofiban, but interacts only with αIIb; compound 1 docks into αVβ3 with a different and less favorable orientation compared with αIIbβ3. (A) Close-up view of the αIIbβ3 ligand binding site corresponding to the crystal structure of αIIbβ3 with bound eptifibatide (PDB entry 1TY6). (B) Most favorable docking mode of compound 1 in the same crystal structure. (C) Close-up view of the αIIbβ3 ligand binding site corresponding to the crystal structure of αIIbβ3 with bound tirofiban (PDB entry 1TY5). (D) The surface representation (1.4 Å probe radius) for the ligand binding site of αIIbβ3 with docked compound 1 is shown. Residue character is indicated by the following color designations: blue (basic), red (acidic), white (aliphatic), cyan (aromatic), and green (polar, mainly residues containing hydroxyl groups). (E) Surface representation of the ligand binding site of αVβ3 with docked compound 1. Visual Molecular Dynamics48 (VMD) was used for all molecular graphics. (See Videos S1,S2 [available on the Blood website; see the Supplemental Materials link at the top of the online article] for additional visual representations of compound 1 docking into αIIbβ3 and αVβ3, respectively.)
Compound 1 docks into αIIbβ3 with an orientation similar to that of eptifibatide and tirofiban, but interacts only with αIIb; compound 1 docks into αVβ3 with a different and less favorable orientation compared with αIIbβ3. (A) Close-up view of the αIIbβ3 ligand binding site corresponding to the crystal structure of αIIbβ3 with bound eptifibatide (PDB entry 1TY6). (B) Most favorable docking mode of compound 1 in the same crystal structure. (C) Close-up view of the αIIbβ3 ligand binding site corresponding to the crystal structure of αIIbβ3 with bound tirofiban (PDB entry 1TY5). (D) The surface representation (1.4 Å probe radius) for the ligand binding site of αIIbβ3 with docked compound 1 is shown. Residue character is indicated by the following color designations: blue (basic), red (acidic), white (aliphatic), cyan (aromatic), and green (polar, mainly residues containing hydroxyl groups). (E) Surface representation of the ligand binding site of αVβ3 with docked compound 1. Visual Molecular Dynamics48 (VMD) was used for all molecular graphics. (See Videos S1,S2 [available on the Blood website; see the Supplemental Materials link at the top of the online article] for additional visual representations of compound 1 docking into αIIbβ3 and αVβ3, respectively.)
Docking of compound 1 into αVβ3 also yielded a predominant binding mode (11 of 20 solutions), but it was very different from the one obtained in αIIbβ3 and yielded a less favorable interaction energy (−73 kcal/mol; Figure 7D,E). Differences in αIIb and αV in this region provide a possible explanation for the differences in docking patterns. Since there is no equivalent of the αIIb D224 in αV, compound 1's basic piperazinyl nitrogen atom interacts with the positively charged D218, which does not exist in αIIb. Moreover, there are fewer opportunities for hydrophobic interactions of compound 1 in αV because αV does not have an aromatic group equivalent to F160 in αIIb (it resides within an inserted loop in αIIb11 ), and the position occupied by F231 in αIIb is occupied by D218 in αV.11,17 These differences orient compound 1 away from β3 in αVβ3 in its preferred docking solution (Figure 7E), rather than toward β3 as in αIIbβ3 (Figure 7D). Thus, both the difference in the location of the negatively charged carboxyl group that interacts with the piperazinal nitrogen and the reduced hydrophobicity of the αV pocket appear to contribute to the selectivity of compound 1 for αIIbβ3 compared with αVβ3. This interpretation is supported by the calculated components of the interaction energies since the electrostatic components are very similar (∼ −60 kcal/mol) in both docking solutions, whereas the hydrophobic van der Waals component is more favorable in αIIbβ3 (∼ −31 kcal/mol) than in αVβ3 (∼ −12 kcal/mol).
Discussion
We have applied high-throughput screening to identify compounds that can inhibit platelet adhesion to fibrinogen, and from more than 30 000 compounds tested we have identified one compound that appears to interact specifically with αIIbβ3. Compound 1 inhibits αIIbβ3-mediated platelet adhesion under static and shear conditions, inhibits αIIbβ3-mediated platelet aggregation, inhibits the binding of soluble fibrinogen and a fluorescent cyclic RGD peptide to purified αIIbβ3, and nearly abolishes fibrinogen binding to platelets induced by the LIBS mAb AP5. Thus, compound 1's mechanism of action appears to be direct inhibition of fibrinogen binding to αIIbβ3, rather than inhibition of platelet signaling. In fact, preliminary data indicate that compound 1 does not inhibit the increase in platelet Ca2+ induced by TRAP (5 or 10 μM), whereas PGE1 consistently does inhibit the increase in Ca2+.
Compound 1 also does not affect the initial slope of ristocetin-induced platelet agglutination or adhesion of washed platelets to collagen, indicating a lack of effect on GPIb and α2β1, respectively. In both cases, however, compound 1 did inhibit the subsequent αIIbβ3-mediated contribution to these phenomena, as did a mAb specific for αIIbβ3. Compound 1, with IC50 values in the low micromolar range for inhibition of ADP-induced platelet aggregation and AP5-induced fibrinogen binding to platelets is, however, a less potent inhibitor of platelet aggregation and platelet adhesion than known inhibitors such as mAb 10E5 or tirofiban. Of particular note, compound 1 did not inhibit αVβ3-mediated cell adhesion to vitronectin, indicating its specificity for αIIbβ3 over αVβ3, despite these receptors sharing the β3 subunit and the high homology between αIIb and αV.49 We conclude that compound 1 shows specificity for αIIbβ3 relative to 3 other receptors, but we cannot exclude reactivity with other receptors.
To assess the potential mechanism of compound 1's inhibition of αIIbβ3, we performed docking studies of compound 1 in the ligand binding sites of αIIbβ3 and αVβ3. We found that compound 1 could dock into the ligand binding site of αIIbβ3 with an orientation similar to those of eptifibatide and tirofiban, but unlike these compounds, it interacted only with αIIb. The positively charged piperazine group in compound 1 interacted with the negatively charged D224 in αIIb, and the cyclic core of compound 1 interacted with the aromatic residues F160, Y190, and F231 in the αIIb binding pocket. Preliminary support for the importance of the positively charged piperazine group comes from studies of 4 derivatives of compound 1 in which the basic piperazine group was disrupted by attaching various aromatic groups, since none of these compounds inhibited platelet adhesion to fibrinogen. A more comprehensive exploration of the chemical space of compound 1, however, is required to formally assess the importance of the piperazine moiety. The docking studies also suggest a potential mechanism to explain the selectivity of compound 1 for αIIbβ3 compared with αVβ3, since compound 1 oriented differently in αVβ3 and had a less favorable interaction energy. This results from the absence of D224 in αVβ3, the presence of a different negatively charged group in αV (D218) with which compound 1's piperazine group interacts, and the reduced aromatic character of the αV binding pocket.
A series of compounds similar to compound 1 in structure have been reported to inhibit platelet aggregation, but mechanistic studies indicated effects on platelet phosphodiesterase 3 and/or enhanced NO production.50-53 The drug pentamidine (340 g/mol) has been reported to inhibit αIIbβ3, and it shows some similarity to compound 1, with a positively charged amidine group connected to an aromatic ring.54 In contrast to compound 1, however, it contains 2 benzamidine groups and its extended conformation is approximately twice the length of compound 1.
The binding of LIBS-specific mAbs is dependent on the conformation of αIIbβ3, and ligand binding results in increased LIBS mAb binding.18-21 In the presence of tirofiban, a ligand mimetic antagonist of αIIbβ3, platelet binding of the LIBS mAbs AP5, LIBS-1, and PMI-1 all increased dramatically. Treatment with compound 1 induced a significant increase in the binding of the αIIb LIBS mAb PMI-1, but the increase was less than one-half that produced by tirofiban. Moreover, compound 1 did not significantly increase the binding of 2 different β3 LIBS antibodies, consistent with the suggestion from the docking experiments that compound 1 interacts with αIIb and not β3. These data are of note in view of the studies of Honda et al, who analyzed 6 αIIbβ3 antagonists for their ability to induce LIBS in αIIb (PMI-1) or β3 (AP5). The researchers found that all 6 induced αIIb LIBS, but only 4 induced β3 LIBS. Only the 4 that induced β3 LIBS also induced outside-in signaling in thrombin-activated platelets, including thromboxane production and an increase in intracellular calcium.55 These data are also consistent with the evidence that the cytoplasmic domain of β3 is important in outside-in signaling.3
To assess whether compound 1's reduced ability to induce LIBS exposure is associated with reduced ability to induce ligand binding (priming), we studied the effect of transient exposure of purified αIIbβ3 to compound 1 on fibrinogen binding. Unlike eptifibatide and RGDS, pretreatment of purified αIIbβ3 with compound 1 did not enhance fibrinogen binding. Compound 1's lack of αIIbβ3 priming may result from its inability to interact with the MIDAS metal ion since ligand-induced reorganization of the β3 metal ions is associated with the development of the open (swing-out) configuration of β3, which is presumed to have higher affinity for ligand.11 We are currently preparing derivatives of compound 1 to test this hypothesis and to identify compounds with higher affinity for the αIIb binding region. It should be noted, however, that the priming assay is not intended to predict clinical outcomes; the compounds that prime purified αIIbβ3 may not have the same effect on αIIbβ3 expressed on the surface of platelets56 ; and eptifibatide is used only as a ligand-mimetic inducer of conformational changes in αIIbβ3.
In conclusion, we have identified a novel low-molecular-weight compound that selectively inhibits ligand binding to αIIbβ3 compared with αVβ3 with a micromolar IC50. Molecular docking studies suggest that its primary mechanism of action involves interacting with αIIb D224 and the aromatic amino acids that line the αIIb portion of the ligand binding pocket. Unlike ligand-mimetic antagonists of αIIbβ3, it does not contain a carboxyl group capable of coordinating the MIDAS metal ion, and it is not long enough to span between the αIIb D224 and the MIDAS metal ion in β3. Moreover, it does not increase the binding of β3 LIBS antibodies to αIIbβ3, and it is much less potent than tirofiban in its ability to increase the binding of an αIIb LIBS antibody. Finally, transient exposure of purified αIIbβ3 to compound 1 does not result in enhanced fibrinogen binding. Since the induction of conformational changes in αIIbβ3 by currently available antagonists has been postulated to contribute to the development of thrombocytopenia in some patients,30 and since activation of the αIIbβ3 receptors by orally active αIIbβ3 antagonists has been postulated to contribute to the increased mortality associated with these agents,24-29 there may be benefit in developing new antagonists that have a diminished capacity to induce conformational changes in the receptor. It will, however, require compounds with higher affinity than compound 1 and human studies to test these hypotheses.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported in part by grant 19278 from the National Heart, Lung, and Blood Institute; by a General Clinical Research Center grant (M01-RR00102) and a Clinical and Translational Science Award (UL1-RR024143) from the National Center for Research Resources at the NIH; and by funds from Stony Brook University.
RGDS peptide synthesis was carried out in the Proteomics Resource Center of the Rockefeller University. The confocal microscopy was performed at the Bio-Imaging Resource Center at Rockefeller University with help from Dr Alison J. North. Excellent technical assistance in fluorescence polarization and fluid shear experiments was provided by Tatiana Deveney.
Authorship
Contribution: R.B. designed and performed research, analyzed data, and wrote the paper; M.M. performed docking experiments and helped write the paper; C.K. helped design the screening assay and fluorescence polarization experiments; M.J. helped design research and analyze data; B.S.C. designed research, analyzed data, and helped write the paper.
Conflict-of-interest disclosure: A patent application for “Compound 1,” described in the manuscript, has been filed under the auspices of Rockefeller University. B.S.C. and R.B. are named as inventors. The other authors declare no competing financial interests.
Correspondence: Barry S. Coller, The Rockefeller University, 1230 York Ave, Box 309, New York, NY 10021; e-mail: collerb@rockefeller.edu.

![Figure 7. Compound 1 docks into αIIbβ3 with an orientation similar to that of eptifibatide and tirofiban, but interacts only with αIIb; compound 1 docks into αVβ3 with a different and less favorable orientation compared with αIIbβ3. (A) Close-up view of the αIIbβ3 ligand binding site corresponding to the crystal structure of αIIbβ3 with bound eptifibatide (PDB entry 1TY6). (B) Most favorable docking mode of compound 1 in the same crystal structure. (C) Close-up view of the αIIbβ3 ligand binding site corresponding to the crystal structure of αIIbβ3 with bound tirofiban (PDB entry 1TY5). (D) The surface representation (1.4 Å probe radius) for the ligand binding site of αIIbβ3 with docked compound 1 is shown. Residue character is indicated by the following color designations: blue (basic), red (acidic), white (aliphatic), cyan (aromatic), and green (polar, mainly residues containing hydroxyl groups). (E) Surface representation of the ligand binding site of αVβ3 with docked compound 1. Visual Molecular Dynamics48 (VMD) was used for all molecular graphics. (See Videos S1,S2 [available on the Blood website; see the Supplemental Materials link at the top of the online article] for additional visual representations of compound 1 docking into αIIbβ3 and αVβ3, respectively.)](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/111/3/10.1182_blood-2007-08-105544/6/m_zh80050813380007.jpeg?Expires=1765921664&Signature=qTbyfTnwrjewMG5NY8Ct8IxulR1cgnqKBXb3cIUkZ1IEexzYRSxzg0c2V07z~Z6eg60lE9MzA79FdcQhwJ0Hyhk8iiGsUCQXfkSI3YUPiJRqBM2k6sjRnDnpnmheat-NFCAhBYPOWEucVAUQ4P8lXFbbmzj5UUnM~5jTkQ11Zju9DrJG75nf5cD3qUj1PnJuSsLuoWq0LJMbPOlPd0R7wMXiRAoCLE~BfAbdVinRlL-cMS48gW~OMAmxl3XWAOnR4ObL~Mk4U6rHyYg4AVYJdixIiamMSW5r2bdzqfmv69oiT3NIpUNPJ8vl57AKKOxDy8H7Q04fi-lXBjf7AqkCsQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal