Abstract
Using metaphase cytogenetics (MC), chromosomal abnormalities are found in only a proportion of patients with myelodysplastic syndrome (MDS). We hypothesized that with new precise methods more cryptic karyotypic lesions can be uncovered that may show important clinical implications. We have applied 250K single nucleotide polymorphisms (SNP) arrays (SNP-A) to study chromosomal lesions in samples from 174 patients (94 MDS, 33 secondary acute myeloid leukemia [sAML], and 47 myelodysplastic/myeloproliferative disease [MDS/MPD]) and 76 controls. Using SNP-A, aberrations were found in around three-fourths of MDS, MDS/MPD, and sAML (vs 59%, 37%, 53% by MC; in 8% of patients MC was unsuccessful). Previously unrecognized lesions were detected in patients with normal MC and in those with known lesions. Moreover, segmental uniparental disomy (UPD) was found in 20% of MDS, 23% of sAML, and 35% of MDS/MPD patients, a lesion resulting in copy-neutral loss of heterozygosity undetectable by MC. The potential clinical significance of abnormalities detected by SNP-A, but not seen on MC, was demonstrated by their impact on overall survival. UPD involving chromosomes frequently affected by deletions may have prognostic implications similar to the deletions visible by MC. SNP-A–based karyotyping shows superior resolution for chromosomal defects, including UPD. This technique further complements MC to improve clinical prognosis and targeted therapies.
Introduction
The myelodysplastic syndromes (MDS) are a heterogeneous group of clonal hematopoietic stem-cell disorders characterized by cytopenias and frequent leukemic progression. MDS constitutes a prototype of age-related malignancy, with a prevalence in the United States that may be more than 100 000.1 Its incidence in the United States, estimated to be more than 10 000 yearly, is likely to further increase due to the greater life expectancy of the general population (http://www.census.gov/).
Chromosomal aberrations can be detected by metaphase cytogenetics (MC) in approximately 50% of MDS patients and are responsible for some of the observed clinical diversity. Based on the experience that certain chromosomal lesions have a major impact on survival in MDS,2-5 cytogenetic results were included in The International Prognostic Scoring System (IPSS), the most commonly applied prognostic algorithm for MDS. Moreover, recent studies demonstrate that MDS patients with certain cytogenetic abnormalities may be candidates for targeted therapies. For example, lenalidomide results in a high remission rate in MDS patients with 5q- abnormalities.6,7
High-resolution single nucleotide polymorphisms arrays (SNP-A) can be applied in karyotypic analysis. SNP-A–based karyotyping does not depend upon the availability of live, dividing cells, and consequently can yield results when routine MC is not informative. Moreover, due to the higher resolution of SNP-A as compared with MC, smaller, previously cryptic deletions and duplications can be detected. A major advantage of this technology over MC is its ability to identify loss of heterozygosity (LOH) that occurs without concurrent changes in the gene copy number (CN). Such defects are consistent with acquired uniparental disomy (UPD) and can be attributed to errors in mitotic recombination occurring in somatic cells. Acquired segmental UPD is being increasingly recognized in a variety of neoplasms.8,9 UPD has been described in chronic lymphocytic leukemia10 and polycythemia vera as a mechanism leading to homozygosity for the Jak2 mutation.11 Recently, an extensive study of acute lymphoblastic leukemia using SNP-A revealed chromosomal deletions and amplifications, many of them involving genes encoding principal regulators of B-lymphocyte development.12 SNP-A also has been used for detecting genomic lesions in smaller case series of myeloma,13 leukemias,14-16 and lymphoma.17 Initially using 50K arrays, we have demonstrated the potential diagnostic value of this technology, in a smaller cohort of myelodysplastic syndrome (MDS) patients.18 This preliminary study demonstrated frequent detection of UPD in MDS. Subsequent larger studies limited to low-risk MDS showed similar results.19
MDS is a particularly suitable target for demonstrating the use of SNP-A, as acquired cytogenetic abnormalities are relatively frequent and mostly unbalanced.20 Using this disease as a model, we tested the hypothesis that high-density SNP-A can complement routine MC and enhance its diagnostic resolution and prognostic value. We studied a large cohort of patients with MDS using MC and 250K SNP-A to validate the diagnostic use of this technology in MDS.
Methods
Patients
Bone marrow and blood aspirates were collected from 174 patients (mean age, 68; range, 17-88) seen between 2002 and 2007 who were grouped according to the World Health Organization (WHO) classification system21 and the IPSS2 (Table 1). Informed consent was obtained according to protocols approved by the Cleveland Clinic International Review Board. Aspirates and blood obtained from 76 healthy individuals (mean age, 44; range, 16-76) were used as controls. Clinical data analyzed included blood counts, marrow morphology, blast counts, time to leukemic progression, length of follow-up, and overall survival. Median follow-up for low-risk MDS, advanced MDS, myelodysplastic/myeloproliferative disease (MDS/MPD), and secondary acute myeloid leukemia (sAML) was 15, 11, 12, and 13 months, respectively. There were 4 cases of MDS secondary to aplastic anemia. Treatment-related MDS cases were not included.
General characteristics of patients (N = 174)
| Characteristics . | Sex (M/F) . | Age, y (SE) . | No. (%) . | IPSS, mean . |
|---|---|---|---|---|
| WHO classification | ||||
| MDS, n = 94 | ||||
| Low grade, n = 66 | ||||
| RA/RCMD (20) | 14/6 | 57 ± 15 | 11.5 | 0.55Ψ |
| RARS/RCMD-RS (29) | 20/9 | 69 ± 11 | 16.7 | 0.55 |
| 5q- syndrome (2) | 0/2 | 70 ± 16 | 1.1 | 0 |
| MDS-U (15) | 9/6 | 71 ± 8 | 8.6 | 0.63Ψ |
| Advanced grade, n = 28: RAEB1/2 | 16/13 | 62 ± 14 | 16.1 | 1.66 |
| MDS/MPD, n = 47 | ||||
| RARS-T (11) | 5/6 | 70 ± 7 | 6.3 | 0.05 |
| MDS/MPD-U (12) | 7/5 | 67 ± 7 | 6.9 | 0.5 |
| CMML1/2 (24) | 16/8 | 66 ± 9 | 13.8 | 0.94 |
| sAML†, n = 33 | 17/16 | 68 ± 11 | 19 | 2.75* |
| Controls, n = 76 | 32/44 | 44 ± 19 | — | — |
| Characteristics . | Sex (M/F) . | Age, y (SE) . | No. (%) . | IPSS, mean . |
|---|---|---|---|---|
| WHO classification | ||||
| MDS, n = 94 | ||||
| Low grade, n = 66 | ||||
| RA/RCMD (20) | 14/6 | 57 ± 15 | 11.5 | 0.55Ψ |
| RARS/RCMD-RS (29) | 20/9 | 69 ± 11 | 16.7 | 0.55 |
| 5q- syndrome (2) | 0/2 | 70 ± 16 | 1.1 | 0 |
| MDS-U (15) | 9/6 | 71 ± 8 | 8.6 | 0.63Ψ |
| Advanced grade, n = 28: RAEB1/2 | 16/13 | 62 ± 14 | 16.1 | 1.66 |
| MDS/MPD, n = 47 | ||||
| RARS-T (11) | 5/6 | 70 ± 7 | 6.3 | 0.05 |
| MDS/MPD-U (12) | 7/5 | 67 ± 7 | 6.9 | 0.5 |
| CMML1/2 (24) | 16/8 | 66 ± 9 | 13.8 | 0.94 |
| sAML†, n = 33 | 17/16 | 68 ± 11 | 19 | 2.75* |
| Controls, n = 76 | 32/44 | 44 ± 19 | — | — |
— indicates not applicable.
RAEB-T (FAB) accounts for 15 of 33 secondary AML (WHO).
Has patients that cannot be analyzed for IPSS.
Only patients with sAML who fulfilled FAB criteria for RAEB-T were included in IPSS analysis.
Cytogenetic analysis
Cytogenetic analysis was performed on marrow aspirates according to standard methods. Chromosome preparations were G-banded using trypsin and Giemsa (GTG) and karyotypes were described according to the International System for Human Cytogenetic Nomenclature (ISCN).22
DNA extraction
DNA was extracted using the Puregene DNA Purification Kit (Gentra, Minneapolis, MN) from whole blood and bone marrow. Total bone marrow cells were compared with lymphocytes serving as germ line control. In some experiments we also used density gradient-separated granulocytes to distinguish somatic from germ-line defects. T lymphocytes (CD3+) were isolated using immunomagnetic beads (Miltenyi Biotech, Auburn, CA) and used for SNP-A and microsatellite analysis.
SNP-A analysis
Gene Chip Mapping 250K Assay Kit (Affymetrix, Santa Clara, CA) was used. Following Nsp I digestion, fragmented DNA was ligated to adaptor followed by polymerase chain reaction (PCR) amplification. The PCR product was hybridized to the GeneChip Mapping 250K Nsp Array, processed with the Fluidic Station and the Gene Chip Scanner 3000 (Affymetrix). We were able to perform bioinformatic evaluation of DNA samples with a call rate of more than 90%, as we noted that such a call rate does not impede karyotyping using CNAG software.
Microsatellite and gene CN analysis
Regions of LOH were confirmed by microsatellite (MS) polymorphism analysis. Primer sequences were obtained from the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov). Forward primers were modified at the 5′ end with carboxyfluorescein (FAM) fluorescent dye. DNA was amplified and amplicons analyzed using ABI Prism 310 Genetic Analyzer (Applied Biosystems, Foster City, CA). CN analysis was performed by microsatellite analysis using a Real-Time TaqMan chemistry protocol.23 The probe for detection of CA repeats was designed as a 21-bp oligomer containing GT repeats with FAM and Black Hole Quencher modifications on 5′ and 3′ ends, respectively. All reactions were performed in triplicate using the D12S1699 amplicon as an endogenous control.
Biostatistical evaluation
Signal intensity was analyzed and SNP calls determined using Gene Chip Genotyping Analysis Software Version 4.0 (GTYPE). CN was investigated using a Hidden Markov Model and CN Analyzer for Affymetrix GeneChip Mapping 250K arrays (CNAG 2.0)24 using log2 signal intensity as a parameter (compared with 3-10 best fit references with the lowest SD). Segmental LOH was identified by a statistical assessment of the likelihood that consecutive SNP loci would exhibit heterozygosity given the corresponding allelic frequency of particular SNP in the normal population (CNAG). The 2-sided Fisher exact test was used to analyze the difference between the distribution of dichotomized variables among the groups.
Results
Principles of cytogenetic analysis using SNP microarrays
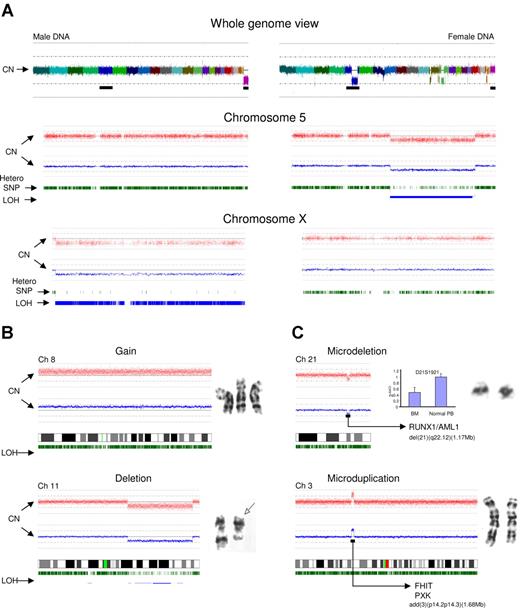
SNP-A can be used for genotyping and CN determination to detect unbalanced chromosomal defects as well as CN-neutral LOH. SNP probes positioned across the human genome allow for precise identification of hypoploidy and frequency of heterozygous SNPs (Figure 1A). Chromosome idiograms that map hybridization signals corresponding to individual SNPs allow for the analysis of the whole genome or analysis by individual chromosomes (as demonstrated using chromosomes X and 5 as examples, Figure 1A). The idiograms display SNP CN as well as the frequency and chromosomal location of heterozygous SNPs. Deletion of a chromosomal segment results in a decrease in the intensity of hybridization and loss of heterozygous calls along this region, indicating segmental LOH. Of note is that the remaining heterozygous calls are due to contamination with cells bearing an intact chromosome 5. Overall, SNP-A–based karyotyping allowed for confirmation of approximately 82% of unbalanced aberrations identified by MC (Figure 1B). The remaining defects seen by MC and “missed” by SNP arrays included lesions present in more than 2/20 but less than 8/20 metaphases and numerical aberrations of chromosome Y (which was not included in the SNP probe coverage of the 250K array).
Analysis approach and type of lesions detected by SNP-A. (A) Normal male (left portion) and abnormal female karyotype with multiple lesions, including del(5q) (right portion). Whole genome scan is shown (top panel); each color represents copy number of a different chromosome (chromosome Y is not included on the array). Chromosome 5 is presented (middle panel) for both patients. Red dots depict single SNP signal intensity, while blue lines present an average value of SNP signal intensity. Green vertical bars represent heterozygous SNP loci, while blue bars show areas of LOH. In comparison to the normal chromosome 5 (left portion), the deletion 5q can be easily observed as a reduction in copy number and area of homozygous SNP loci (right portion). Idiogram of chromosome X is shown (bottom panel) demonstrating the haploid copy number with homozygous SNP loci (male DNA, left portion) and diploid DNA copy number with characteristic distribution of heterozygous SNP loci (female DNA, right portion). MC-detected chromosomal abnormalities (karyograms included) confirmed by SNP-A (B). (C) SNP-A–detected microdeletion of chromosome 21 and microduplication of chromosome 3. Copy number confirmation using Taq-Man Real-Time PCR shown as blue bars and compared with normal T cells (CD3+) obtained from the same patient. Microsatellite ID marker is displayed above the bars. Y axis (2−ΔΔCt) presents the relative quantification scale, where 1 equals diploid DNA copy number. Examples of deleted and duplicated genes are included. Error bars represent SD.
Analysis approach and type of lesions detected by SNP-A. (A) Normal male (left portion) and abnormal female karyotype with multiple lesions, including del(5q) (right portion). Whole genome scan is shown (top panel); each color represents copy number of a different chromosome (chromosome Y is not included on the array). Chromosome 5 is presented (middle panel) for both patients. Red dots depict single SNP signal intensity, while blue lines present an average value of SNP signal intensity. Green vertical bars represent heterozygous SNP loci, while blue bars show areas of LOH. In comparison to the normal chromosome 5 (left portion), the deletion 5q can be easily observed as a reduction in copy number and area of homozygous SNP loci (right portion). Idiogram of chromosome X is shown (bottom panel) demonstrating the haploid copy number with homozygous SNP loci (male DNA, left portion) and diploid DNA copy number with characteristic distribution of heterozygous SNP loci (female DNA, right portion). MC-detected chromosomal abnormalities (karyograms included) confirmed by SNP-A (B). (C) SNP-A–detected microdeletion of chromosome 21 and microduplication of chromosome 3. Copy number confirmation using Taq-Man Real-Time PCR shown as blue bars and compared with normal T cells (CD3+) obtained from the same patient. Microsatellite ID marker is displayed above the bars. Y axis (2−ΔΔCt) presents the relative quantification scale, where 1 equals diploid DNA copy number. Examples of deleted and duplicated genes are included. Error bars represent SD.
To determine the minimal clonal size that can be detected by SNP-A, dilution studies of trisomy 21 DNA with normal diploid DNA were performed. Clonal populations compromising 25% to 50% of total cells can be detected (not shown). Repetitive testing of the same sample (n = 3) yielded identical results. We also have examined simultaneously obtained whole blood and marrow for 6 patients. In all cases we obtained concordant results (Figure 2A). When serial bone marrow samples were analyzed for 6 patients, stable chromosomal configurations concordant with MC results were seen in 4. In the 2 remaining patients, in addition to previously seen defects, new aberrations (del(4q) and del(7q) in one patient and new monosomy 7 in the second) were detected. Of note is that in the latter patient the initial MC was unsuccessful due to lack of evaluable metaphases (Figure 2B).
SNP-A karyotyping results obtained through simultaneous testing blood and marrow and serial bone marrow exams. (A) Comparison of SNP-A results for both marrow and blood. (B) Results of serial testing performed during the disease course.
SNP-A karyotyping results obtained through simultaneous testing blood and marrow and serial bone marrow exams. (A) Comparison of SNP-A results for both marrow and blood. (B) Results of serial testing performed during the disease course.
Bone marrow and blood from healthy controls (N = 76) were studied using SNP-A to determine if any lesions detected in MDS patients also might be present in controls. In controls, we observed expected differences in the signal intensity in regions affected by known CN polymorphisms.25 In addition, deletion and gains present in multiple controls were considered as germ line–encoded CN polymorphisms. The remaining unique areas of copy number variation were randomly tested using either MS analysis of nonclonal lymphoid cells (n = 4) or human androgen receptor assay (HUMARA) for clonal X chromosome inactivation in informative women (n = 2). In all instances, copy number changes were found to be nonclonal. Consequently, for the analysis of MDS patients, these changes were not deemed pathologic and were not included in the evaluation.
New lesions discovered by SNP-A karyotyping, including UPD
We have analyzed 174 patients with MDS, MDS/MPD, or MDS-derived AML (Table 1 and Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). In addition to the larger defects, smaller previously unknown cryptic chromosomal aberrations were found using SNP-A (Figure 1C). These defects appeared in bone marrow cells (as opposed to nonclonal lymphocytes) and in some instances were limited to one gene; they therefore likely represented defects with pathologic significance. The results of confirmatory TaqMan PCR studies (CN-neutral LOH) and MS analysis (n = 7) provided independent validation of the SNP-A findings (Figures 1C, 3A). Confirmation of the somatic nature of SNP-A–detected lesions was performed using paired SNP-A analysis applied to both bone marrow and sorted nonclonal CD3+ cells (n = 5); confirmation included upd (6)(p21.2-pter), upd (11)(q13.5-qter), upd (4)(q23-qter), upd (7)(q11.23-qter), del (7)(q22.1) (Figure S1). Such segmental UPD was most commonly found in chromosomal regions frequently identified as abnormal in MDS by MC and was not present in nonclonal lymphocytes, suggesting a somatic origin of SNP-A–detected lesions. Types of lesions and clinical features of the patients are included in Table S1. UPD, deletions, or duplications involved several important chromosomal regions affected in multiple patients (Figure 4).
Improved mapping of commonly detected lesions in MDS using SNP-A
SNP-A facilitates precise mapping of lesions; LOH affecting common regions due to deletion and UPD can be grouped to better define boundaries of minimal overlapping regions (Figures 3B, 4). As demonstrated, in addition to larger lesions and UPD, small defects also can be found in these regions. Analysis of chromosomal lesions and their locations showed areas of overlap.
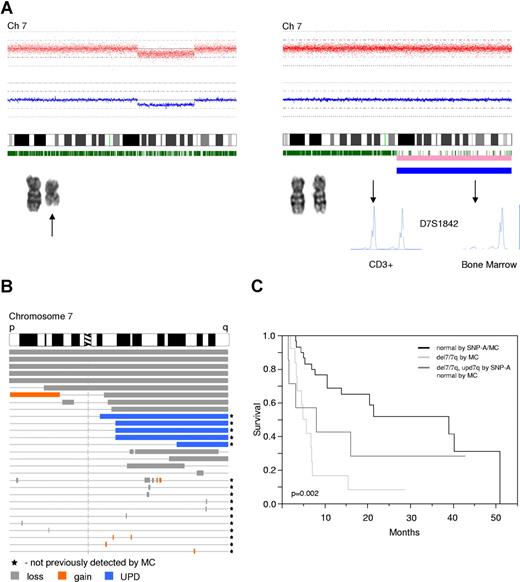
Detection and comparison of different types of lesions, including UPD, that affect chromosome 7 and influence the survival. (A) Two types of lesions resulting in LOH. Left portion demonstrates a deletion spanning part of the long arm of chromosome 7, shown here as reduction of copy number detected by SNP-A, which is concordant with MC finding (black arrow). Right portion demonstrates copy number neutral LOH (UPD), shown here as normal copy number (SNP-A) and normal karyogram by MC. Pink and blue bars below the idiogram indicate areas of LOH, with the thicker the blue bar the higher the probability of LOH. LOH was confirmed by MS genotyping using CD3+ cells as a nonclonal control. MS marker ID is displayed. Arrow indicates the allelic discrepancies. Location and type of lesions (loss-gray, gain-orange, upd-blue) affecting chromosome 7 are shown (B). Lesions previously not found by MC are marked with a black star. Kaplan-Meier analysis of the survival of patients (irrespective of the treatment received) with −7/del(7q) by MC and with new lesions affecting chromosome (del(7q)/upd(7q)) compared with patients with normal SNP-A analysis (C).
Detection and comparison of different types of lesions, including UPD, that affect chromosome 7 and influence the survival. (A) Two types of lesions resulting in LOH. Left portion demonstrates a deletion spanning part of the long arm of chromosome 7, shown here as reduction of copy number detected by SNP-A, which is concordant with MC finding (black arrow). Right portion demonstrates copy number neutral LOH (UPD), shown here as normal copy number (SNP-A) and normal karyogram by MC. Pink and blue bars below the idiogram indicate areas of LOH, with the thicker the blue bar the higher the probability of LOH. LOH was confirmed by MS genotyping using CD3+ cells as a nonclonal control. MS marker ID is displayed. Arrow indicates the allelic discrepancies. Location and type of lesions (loss-gray, gain-orange, upd-blue) affecting chromosome 7 are shown (B). Lesions previously not found by MC are marked with a black star. Kaplan-Meier analysis of the survival of patients (irrespective of the treatment received) with −7/del(7q) by MC and with new lesions affecting chromosome (del(7q)/upd(7q)) compared with patients with normal SNP-A analysis (C).
Chromosomal distribution of the lesions detected by SNP-A. Location and the size of the lesions are indicated next to the chromosomes' idiograms.
Chromosomal distribution of the lesions detected by SNP-A. Location and the size of the lesions are indicated next to the chromosomes' idiograms.
For demonstration purposes, we focused here on specific lesions on chromosome 7 and their prognostic significance. When we compared the survival of 3 groups of patients—one with normal SNP-A karyotype, another with previously known deletion 7/7q, and another with normal MC and new cryptic lesions (including deletions and UPD) on chromosome 7—those with previously known and new abnormalities showed a comparably poor prognosis, distinct from those with a normal karyotype by both MC and SNP-A (median survival 6 vs 8 vs 39 months, respectively, P = .002, Figure 3C). When the group of patients with UPD7q (3 patients with normal MC, one with +13 and one with −Y) was analyzed separately, shorter survival was observed (median survival 3 vs 39 months, P < .001), indicating the clinical relevance of SNP-A and UPD7.
Comparison between SNP-A–based karyotyping and MC
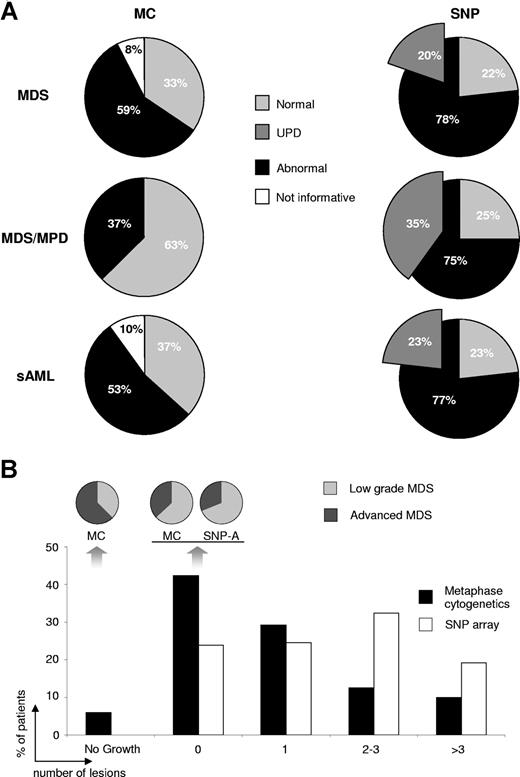
Overall, SNP-A allowed for identification of chromosomal defects in 78% of MDS patients, as compared with 59% by MC. Similarly, a higher detection rate of lesions was found in patients with MDS/MPD and sAML at 75% versus 37%, and 77% versus 53% for SNP-A and MC, respectively. SNP-A does not rely on cell growth, and SNP-A testing was successful in all patients (Figure 5A). UPD as the sole or a concurrent defect was detected in 20%, 35%, and 23% of patients with MDS, MDS/MPD, and sAML, respectively. More chromosomal aberrations were revealed by SNP-A as compared with MC, with a greater proportion of patients showing more than 1 defect (P < .001, Figure 5B). As a result, when patient groups with normal MC or SNP-A results were compared, fewer patients with low-grade disease were found if the karyotype was assessed to be normal using the new technology, consistent with an overall negative clinical impact of newly identified lesions (Figure 5B). In general, MDS patients in whom multiple lesions were detected were more likely to have a more advanced disease and show a more rapid increase in blasts. For example, in a patient with MDS/MPD-U, evolution to sAML was associated with occurrence of additional lesions such as UPD6p (not shown; for other examples see Figure 2B). Separate analysis of patients with normal or noninformative MC showed that SNP-A identified cryptic chromosomal aberrations in 62% and 44% of these patients, respectively (Figure 6A).
Type, frequency, and number of lesions detected by MC and SNP-A. (A) The frequency of chromosomal aberrations and noninformative results as detected by both MC and SNP-A. The insert in the SNP-A pie chart shows the portion of acquired UPD, either as the sole change or in addition to other abnormalities. (B) The percentage of patients with 0, 1, 2-3, and more than 3 lesions, respectively, as detected by MC (■) and SNP-A (□). Pie charts demonstrate distribution of low versus advanced stage of MDS within noninformative and normal karyotypes detected by MC and SNP-A.
Type, frequency, and number of lesions detected by MC and SNP-A. (A) The frequency of chromosomal aberrations and noninformative results as detected by both MC and SNP-A. The insert in the SNP-A pie chart shows the portion of acquired UPD, either as the sole change or in addition to other abnormalities. (B) The percentage of patients with 0, 1, 2-3, and more than 3 lesions, respectively, as detected by MC (■) and SNP-A (□). Pie charts demonstrate distribution of low versus advanced stage of MDS within noninformative and normal karyotypes detected by MC and SNP-A.
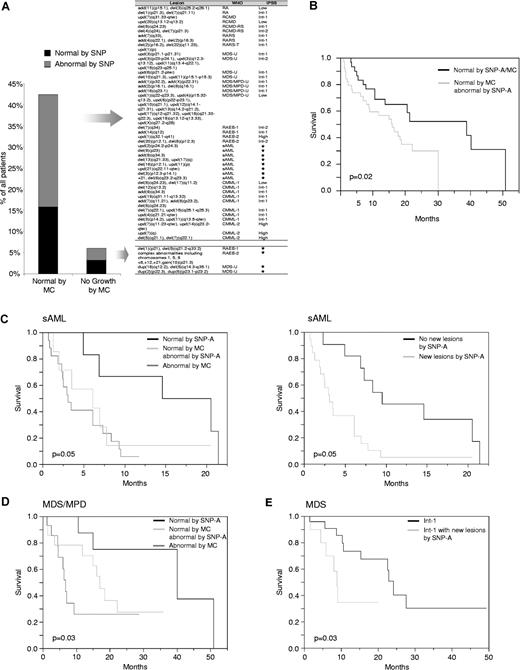
Types and prognostic impact of new lesions found in patients with noninformative or normal MC. (A) The percentage of patients with normal or noninformative MC in whom new chromosomal lesions were found by SNP-A (left). Type and location of abnormalities are displayed in Table S1 and Figure 4. Stars mark the patients with sAML and noninformative MC in whom IPSS risk category cannot be determined. (B) Kaplan-Meier analysis of survival of patients (irrespective of the treatment received) with normal MC in whom new defects were identified using SNP-A as compared with these with a normal karyotype as evidenced by both negative SNP-A and MC. (C) Survival curves of sAML patients with normal SNP-A result versus patients with abnormal MC and those with normal MC in whom new lesions were found by SNP-A (left). Survival of sAML patients grouped into those with and without additional defects identified by SNP-A (right). (D) Survival curves of patients with MDS/MPD subgrouped into those with normal SNP-A, normal MC, but additional lesions by SNP-A, and patients with abnormal MC results. (E) Survival curves of patients with MDS and IPSS int-1 subgrouped based on the presence or absence of new lesions found by SNP-A irrespective of the MC result.
Types and prognostic impact of new lesions found in patients with noninformative or normal MC. (A) The percentage of patients with normal or noninformative MC in whom new chromosomal lesions were found by SNP-A (left). Type and location of abnormalities are displayed in Table S1 and Figure 4. Stars mark the patients with sAML and noninformative MC in whom IPSS risk category cannot be determined. (B) Kaplan-Meier analysis of survival of patients (irrespective of the treatment received) with normal MC in whom new defects were identified using SNP-A as compared with these with a normal karyotype as evidenced by both negative SNP-A and MC. (C) Survival curves of sAML patients with normal SNP-A result versus patients with abnormal MC and those with normal MC in whom new lesions were found by SNP-A (left). Survival of sAML patients grouped into those with and without additional defects identified by SNP-A (right). (D) Survival curves of patients with MDS/MPD subgrouped into those with normal SNP-A, normal MC, but additional lesions by SNP-A, and patients with abnormal MC results. (E) Survival curves of patients with MDS and IPSS int-1 subgrouped based on the presence or absence of new lesions found by SNP-A irrespective of the MC result.
Prognostic value of combined traditional karyotyping, SNP-based cytogenetics, and clinical information
The clinical relevance of individual new recurrent lesions (Figure 4) remains to be established, but it is clear that these lesions contribute to clinical diversity. We analyzed the survival of MDS, MDS/MPD, and sAML patients (irrespective of the type and duration of the therapy received) based on MC and SNP-A findings. We stratified the patients with normal MC into 2 groups: one with a normal karyotype according to both MC and SNP-A, and the other comprised of patients with normal MC in whom new lesions were identified using SNP-A (Figure 6B). The latter group showed a reduced overall survival compared with those with normal MC and SNP-A karyotype (median survival 16 vs 39 months, P = .02).
We also have studied the impact of SNP-A karyotyping results on survival within MDS, MDS/MPD, and sAML patients treated with diverse regimens separately (Figure 6C-E). While in MDS the survival was not significantly affected, most likely due to short follow-up time (in comparison to the average survival length of MDS patients), in patients with MDS/MPD and sAML newly detected lesions were associated with worse survival (Figure 6C,D). In particular, in sAML newly detected lesions in patients with normal MC conveyed worse prognosis, as did additional lesions in those with already established abnormal MC.
In MDS, we also have tested whether the newly identified lesions have a potential to modify the currently accepted IPSS system (excluding sAML, as this diagnosis is now classified separately, though these patients would have been classified per the French-American-British (FAB) classification as RAEBt and given an IPSS score). MDS patients were grouped only based on their risk category including IPSS-low, Int-1, Int-2, and high-risk groups. For each risk group, patients were subdivided into those in whom SNP-A did not detect additional aberrations and those in whom new lesions were found. The survival curves clearly diverged for patients originally classified as IPSS Int-1, suggesting that SNP-A conveys additional information allowing for better prognostic resolution (median survival 28 vs 9 months, P = .03) (Figure 6E). Due to a short observation period and variable impact of individual lesions, the survival within the low-risk group did not differ between SNP-A–confirmed karyotypes and those in whom MC was modified.
Discussion
The prognostic value of cytogenetic abnormalities identified by MC is well established in many malignancies.26-29 In MDS the IPSS algorithm, a widely recognized prognostic scheme, includes cytogenetic defects because some of the nonrandom chromosomal aberrations have a significant clinical impact.30 For example, the presence of monosomy 7 indicates poor risk AML or MDS,31 while other defects may have a more favorable prognosis.32-34 Despite these known features of certain abnormalities, a great deal of heterogeneity remains even in cases with identical lesions. In a significant proportion of MDS patients who show normal MC, this technique does not allow for further prognostic resolution. In all likelihood, clonal defects are present in most of the patients, but their detection is precluded by the low resolution of routine techniques.
MDS is closely related to MDS/MPD and sAML (treated as separate entities according to WHO classification). Therefore, our investigations included these conditions, allowing for analysis of chromosomal abnormalities across morphologic boundaries, but within what is likely a pathophysiologic continuum. Of note is that AML (as defined by WHO) that evolved from MDS often shows a much distinct course from that seen in typical AML.
Previously, in a smaller cohort of patients and using 50K SNP-A, we have demonstrated the use of this technology platform to detect chromosomal abnormalities in MDS and found frequent UPD even at a lower resolution level.18 Our results demonstrate that, using SNP-A, detection of karyotypic defects can be improved in MDS and likely also in other hematologic conditions. We show that this technology allows for detection of cryptic lesions in patients with both normal and abnormal karyotypes. Consequently, the proportion of MDS, MDS/MPD, and sAML patients without obvious chromosomal lesions decreases when compared with routine MC. Since SNP-A karyotyping is a DNA-based technique that does not require live, dividing cells, it permits detection of aberrations in patients with noninformative cytogenetic studies.
In addition to duplications and deletions, SNP-A can identify CN-neutral LOH, a lesion that cannot be detected by MC nor fluorescence in situ hybridization. Segmental acquired UPD, occurring as a result of mitotic recombination, appears to be a common event in MDS, present in 20% of MDS and 23% of sAML patients as either the sole abnormality or as a concurrent defect. Remarkably, 35% of patients with MDS/MPD, in particular chronic myelomonocytic leukemia (CMML), showed the presence of UPD on various chromosomes, suggesting that this type of abnormality is not limited to the UPD9p seen in Jak2 mutation–positive cases. It is possible that UPD represents an attempt to repair chromosomal deletions using the remaining chromosome copy as a template,35,36 but results in LOH with consequent homozygosity for somatic or germ-line mutations. The clinical significance of UPD9p has been demonstrated recently in myeloproliferative syndromes with JAK2 V617F mutation.11,37
Of utmost significance for the future diagnostic application of SNP-A is the impact of newly detected lesions on clinical outcomes, not only in MDS, but also in MDS/MPD and sAML. Several lines of evidence presented here suggest that SNP-A will be clinically relevant. First, previously cryptic lesions found in patients with normal MC (across all subentities studied) were associated with reduced overall survival as demonstrated for sAML, MDS/MPD, and selected subtypes of MDS; as a result, patients with truly normal karyotypes as determined by both SNP-A and MC have a more favorable prognosis. This could have a significant impact on the IPSS scores assigned to patients, and thus on selecting the most appropriate therapies (for low-risk vs high-risk disease). Of note is that due to the relatively long natural history of MDS (in particular in lower-risk categories), the follow-up time was insufficient to determine survival endpoints, as occurs in most clinical trials within MDS. As these data mature, many of the SNP-A–detected defects may be proven to have a negative impact on survival. One example of such a defect may be UPD7q. Our results also demonstrate that UPD affecting regions altered by common deletions such as chromosome 7q has clinical significance. Second, factoring in new lesions identified by SNP-A allowed for further stratification within prognostic groups defined by the IPSS. It is anticipated that when similar data are available for each type of defect based on larger studies, SNP-A will greatly improve prediction of clinical outcomes. Finally, the additional lesions found by this novel technology result in more complex karyotypes, which coincide with morphologic subtypes of MDS associated with advanced disease or sAML, a result expected based on the experience with MC. Other types of newly detected lesions include microdeletions often located on chromosomes affected by large lesions. These defects have to be distinguished from CN polymorphisms and likely point toward pathogenic defects, in particular when present in multiple patients and coinciding with specific genes implicated in malignant transformation. One example among many new recurrent/overlapping lesions is the RUNX1(AML1) locus, for which we have found a microdeletion and UPD. Apart from the obvious clinical applications, mapping UPD and LOH that occur in regions shared between patients may help to refine minimal deleted chromosome segments containing causative tumor suppressor genes or allow for definition of specific disease phenotypes.
Precision and robustness are major advantages of SNP karyotyping. One possible criticism of any new technology with a high resolution (sensitivity for smaller lesions) is that such a method also will have a high rate of false positives, and patients with good or intermediate risk will inappropriately be classified as having adverse risk cytogenetics. Our data belie this claim, in that, for example, patients who were reclassified as having adverse-risk cytogenetics did in fact have a worse survival, and thus were appropriately reclassified. Using control bone marrows and polymorphism maps, we excluded lesions likely reflecting nonpathogenic germ-line variations. The study of the X chromosome showed only a low false call rate, and identical results were obtained on serial testing. However, it is important to also note inherent limitations that include an inability to distinguish one clone with several defects from several distinct clones, and an inability to detect balanced rearrangements. While much more precise (detection of smaller lesions), SNP-A appears to have a comparable sensitivity (proportion of abnormal cells) to MC; as a result, only large clinically relevant clones can be detected.
Establishment of the prognostic value of individual recurrent lesions detected by SNP-A and, in general, the clinical use of the SNP-A–based karyotyping platform will require significant follow-up. Most patients receive multiple sequential treatment regimens, which may or may not have an impact on survival. Recurrent lesions may occur in the context of additional defects and various morphologic subtypes, making analysis of global outcome data very difficult. Even for MC, the prognostic value of some less frequent aberrations is not established.
In summary, our investigation represents a systematic application of SNP-based karyotyping for the detection of chromosomal defects in MDS, sAML, and MPD/MDS. Our results suggest that this technology will have clinical use for detection of new defects with clinical relevance not only in MDS, but also in other malignancies, and will complement traditional MC. In particular, it allows for detection of UPD, a form of LOH that appears to occur frequently in these conditions, and may have important clinical implications.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by NIH R01 HL082983 (J.P.M.), U54 RR019391 (J.P.M., M.A.S.), K24 HL077522 (J.P.M.), and a charitable donation from the Robert Duggan Cancer Research Fund.
Authorship
Contribution: L.P.G. performed experiments, analyzed the data, and wrote the manuscript. K.S.T. performed and helped to interpret cytogenetic results. R.T. performed clinical analysis of patients and collected samples. C.L.O participated in experiments and helped prepare the manuscript. M.A.S. recruited patients, analyzed data, and helped write the manuscript. J.P.M. conceived the idea, designed the experiment, provided financial and administrative support, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jaroslaw P. Maciejewski, Experimental Hematology and Hematopoiesis Section, Taussig Cancer Center R-40, Cleveland Clinic, 9500 Euclid Ave, Cleveland, OH; e-mail: maciejj@ccf.org.