Abstract
Aberrant inhibition of B-cell receptor (BCR)-induced programmed cell death pathways is frequently associated with the development of human auto-reactive B-cell lymphomas. Here, we integrated loss-of-function, genomic, and bioinformatics approaches for the identification of oncogenic mechanisms linked to the inhibition of BCR-induced clonal deletion pathways in human B-cell lymphomas. Lentiviral (HIV)-based RNA interference screen identified MCL1 as a key survival molecule linked to BCR signaling. Loss of MCL1 by RNA interference rendered human B-cell lymphomas sensitive to BCR-induced programmed cell death. Conversely, MCL1 overexpression blocked programmed cell death on BCR stimulation. To get insight into the mechanisms of MCL1-induced survival and transformation, we screened 41 000 human genes in a genome-wide gene expression profile analysis of MCL1-overexpressing B-cell lymphomas. Bioinformatic gene network reconstruction illustrated reprogramming of relevant oncoproteins within β-catenin–T-cell factor signaling pathways induced by enforced MCL1 expression. Overall, our findings not only illustrate MCL1 as an aberrantly expressed reprogramming oncoprotein in follicular lymphomas but also highlight MCL1 as key therapeutic target.
Introduction
Reciprocal chromosomal translocations involving one of the IG loci and a proto-oncogene are hallmarks of many types of human B-cell non-Hodgkin lymphomas (B-NHL).1 As a consequence of such translocations, several proto-oncogenes are placed under the transcriptional control of the active IG locus, causing constitutive expression of the oncogenes with subsequent B-cell transformation.2 During B-cell tumor progression, IG chromosomal translocations are thought to be early events in the pathogenesis of these neoplasias and result in critical deregulation of multiple oncogenes such as BCL2 t(14;18)(q32;q21) in follicular lymphomas, MYC t(8;14)(q24;q32) in Burkitt lymphomas, CCND1 t(11;14)(q13;q32) in mantle cell lymphomas, MALT1 t(1;14)(p21;q32) in extranodal MALT lymphomas, or BCL6 t(3;V)(q27;V) in diffuse large cell lymphomas.3
In addition to these chromosomal aberrations, several observations suggest that B-cell receptor (BCR) signaling supplies important tonic survival signals that might be required for the maintenance of B-cell lymphomas.4 Considering the allelic exclusion model, chromosomal translocation events should occur at equal frequency on the expressed IGH allele and the nonexpressed IGH allele. Because translocations of proto-oncogenes into the IGH-loci are always found on the nonproductively rearranged IGH loci, with few exceptions5 ; these findings suggest that survival of B-cell lymphoma clones is incompatible with translocations of proto-oncogenes into the productively rearranged IGH loci because this type of translocation would induce inability to express immunoglobulin (Ig),2 and suggests the dependency of Ig expression for B-cell lymphoma survival. Moreover, taking into account that the somatic hypermutation machinery induces 2 types of destructive somatic mutations (nonsense mutations and duplications causing reading-frame shifts); these events would lead to the generation of B-cell lymphoma clones that lack Ig expression under nonselective conditions.6 Because that treatment of patients with anti-idiotypic antibodies (a selective condition) did not result in the selection of Ig-negative B-cell lymphoma clones,7 this observation also suggests the dependency of Ig expression for the survival of many B-cell lymphomas.2
In support of this hypothesis, several molecules integrated in the BCR regulatory network appear to act as oncoproteins, such as SYK,8 ZAP70,9 BLK,10 and MALT1,11 whereas other BCR regulatory molecules surprisingly promote programmed cell death, such as LYN,12 IP3R,13 calpains,14 SLP65,15 and BTK.16 These observations exemplify both positive and negative regulatory roles for BCR in signaling and raise the question of how the BCR signal transduction pathway can generate such distinct outcomes and what is the contribution of BCR signaling in B-cell transformation.
B-cell antigen receptors specific for self-molecules are normally produced as a consequence of the random VDJ recombination. B cells that recognize a self-antigen undergo programmed cell death serving thus as a mechanism to remove self-reactive B cells.17 A theoretical collapse in human B-cell tolerance by intrinsic inactivation of the BCR-induced programmed cell death pathway would lead to an autoimmune lymphoproliferative syndrome, characterized by the enrichment of auto-reactive B-cell clones secreting auto-antibodies.18 Some studies have demonstrated the presence of auto-reactive B-cell lymphomas in most individuals with follicular lymphomas.19 These auto-reactive B-cell lymphomas are characterized by the expression of B-cell antigen receptors with restricted IgVH/IgVL repertoires, showing strong IgVH-CDR3 motifs recognizing rheumatoid factors.20 Thus, all these observations lead to the general idea that an intrinsic inactivation of BCR-induced clonal deletion pathways is commonly coupled to the generation of human autoimmune syndromes and B-cell neoplasias.
These observations prompted us to ask about inactivation mechanisms of BCR-induced clonal deletion in human B-cell lymphomas. We integrated here loss-of-function, genomic, and bioinformatics approaches for the identification of relevant BCR-linked survival molecules in human B-cell lymphomas. We engineered a small hairpin RNA interference (shRNAi) mini-library to interrogate survival pathways on BCR signaling in B-NHL cells. Our library is composed of 277 sequence-verified shRNAi clones that permit shRNAi cassettes to be packaged in lentiviruses. Using this multiple loss-of-function approach, we identified MCL1 as a key survival molecule linked to BCR signaling networks. MCL1 ablation by shRNAi induced sensitivity to BCR-induced programmed cell death, whereas MCL1 overexpression blocked programmed cell death activated by BCR stimulation. To understand the mechanisms of MCL1-induced survival, we performed a genome-wide gene expression profiling experiments revealing MCL1 as a key reprogramming factor that transcriptionally regulates multiple oncoproteins integrated in β-catenin–T-cell factor (TCF) and PPAR-RXRA signaling pathways. We also demonstrated that MCL1 is aberrantly coexpressed with a substantial number of MCL1 targets in follicular lymphomas. Thus, all these data together indicate that MCL1 deregulation in human B-cell lymphomas provides both survival and reprogramming signals that contribute to B-cell transformation.
Methods
This study was approved by the institutional review board at CNIO, Madrid, Spain.
B-cell non-Hodgkin lymphomas (B-NHL)
B-NHL cells included DOHH2 (DSMZ) [immortalized human follicular lymphoma (BCL2+P53−) bearing chromosomal translocation t(14;18)21 and biallelic loss of TP53 [del(17p)]22 ], Z-138 [immortalized human mantle cell lymphoma (BCL1+) bearing chromosomal translocation t(11;14)23 and homozygous loss of BIM [del2q13]24 ], REC-1 (DSMZ) [immortalized human mantle cell lymphoma (BCL1+) bearing chromosomal translocation t(11;14)25 ], JEKO-1 (DSMZ) [immortalized human mantle cell lymphoma (BCL1+) showing highly rearranged hypertriploid karyotype including chromosomal translocation t(11;14)26,27 ], SU-DHL-6 (DSMZ) [immortalized human diffuse large B-cell lymphoma (BCL2+) bearing chromosomal translocation t(14;18)28 ], RAMOS RA-1 (Promochem-ATCC) [immortalized Burkitt lymphoma showing a complex karyotype involving chromosomal translocation t(8;14), gains of regions at 2p, 3q, 13q, and 16q, and losses at 3p, 4q, and 17p29 ], MHH-PREB-1 (DSMZ) [immortalized human lymphoblastic lymphoma bearing a hyperdiploid karyotype including t(8;14)] and MEC-1 (DSMZ) [immortalized human B-cell chronic lymphocytic leukemia Epstein-Barr virus positive (EBV+)30 ].
Lentiviral-mediated shRNAi transfer
Lentiviral shRNAi transfer was performed using high-titer lentiviral stocks generated by transient cotransfection using Fugene HD (Roche Diagnostics, Indianapolis, IN) in the 293-T packaging cell line (American Type Culture Collection, Manassas, VA). DNA plasmids (∼0.5 μg/μL) were produced using TOP10 Escherichia coli (Invitrogen, Carlsbad, CA) cultured in 3 mL terrific broth. DNA plasmids were further purified using a Wizard Plus SV Miniprep DNA purification system (Promega, Madison, WI) and resuspended in 50 μL of H2O (Promega). Before cotransfection, DNA concentration and quality were estimated (per duplicate) using NanoDrop (Nanodrop Technologies, Wilmington, DE). For cotransfection, 1 μg plasmid (pCMVdeltaR8.91 derived from pCMVR8.931 ) containing HIV-1 gag, polymerase (RT) and accessory proteins Tat and Rev; 1 μg plasmid containing the envelope glycoprotein VSV-G (pMD.G), and 1 μg lentiviral vector pA179. Helix containing the shRNAi sequences (Figure 1A) were gently mixed in Eppendorf tubes and then incubated for 10 minutes. The DNA mixture was then gently applied in 100 μL Optimen medium (Invitrogen) and gently vortexed by hand. The resulting mixture was further incubated with 7 μL Fugene HD and then gently vortexed by hand; samples were subsequently incubated for 30 minutes. 293-T packaging cells cultured in 60-mm-diameter dishes (Falcon, BD Biosciences Discovery Labware, Bedford, MA) at 70% confluents in 2 mL fresh warm DMEM medium were cotransfected by dropping the entire Fugene-DNA mixture into the dishes. After this, dishes were slowly shaken. Levels of transfection in 293-T cells were analyzed after 24 hours by immunofluorescence microscopy for green fluorescent protein (GFP) expression. Lentiviral supernatant was recovered at 48 hours after cotransfection. For viral transduction, RAMOS RA-1 B cells (5 × 105 cells/well in 12-well plates, Falcon) were spun (1500g, 37°C, 1 hour) with 100 μL lentiviral supernatant plus 900 μL RMPI media containing 10 μg/mL polybrene (Sigma-Aldrich, St Louis, MO). RAMOS RA-1 B cells were then incubated with the lentiviral supernatant for 2 hours at 37°C in a humidified atmosphere containing 5% CO2. Then, RAMOS RA-1 B cells were spun down (1500g, 37°C, 5 minutes) and resuspended (5 × 105 cells/well in 6-well plates) in 3 mL fresh RPMI 1640 medium (Sigma-Aldrich) containing additives plus FCS. B cells were incubated at 37°C in a humidified 5% CO2 atmosphere for 72 hours. GFP+ B cells were then analyzed (Figure 2B) by FACS analysis (FACSCalibur, BD Biosciences, San Jose, CA).
shRNAi lentiviral mini-library for targeting B-cell receptor (BCR) regulatory genes. (A) Cloning strategy for the lentiviral vector pA179.Helix. Only the relevant portions of the plasmids are shown. The pA179.Helix vector is a new version of pFUGW vector engineered at the CNIO Genomic Unit (http://www.cnio.es/es/index.asp). The pA179.Helix contains a CMV enhancer substituted for the U3 region of the 5′ LTR to maximize expression of viral RNA genomes during transient transfection.31 The pA179.Helix also contains a deletion in the U3 region of the 3′ LTR that makes the 5′ LTR of the integrated provirus transcriptionally inactive66 as well as the woodchuck hepatitis virus posttranscriptional regulatory element (WRE) inserted downstream of GFP to increase the level of transcription.67 (B) BCR-induced programmed cell death in a group of human B-cell lymphomas. MHH-PREB-1, RAMOS RA-1, SU-DHL-6, Z-138, JEKO, REC-1, MEC-1, and DOHH2 B-NHL cells were cultured (50 000 cells/ 200 μL/ well in 96-well plates) either in medium alone (RPMI plus additives and 10% FCS) or anti-IgM treated [goat F(ab′)2 antihuman IgM (10 μg/mL) antibody]. B cells were collected at 24 hours and programmed cell death was quantified using annexin V-APC staining. Cell death was quantified by FACS analysis, followed by analysis using FlowJo software (mean ± SD). Representative flow cytometric plots are shown. Numbers on plots represent the pecentage of cells in each quadrant over the total number of cells gated. (C) Western blot for human Argonuate-2 and Dicer in total protein extracts (RIPA extraction) from Z-138, DOHH-2, and RAMOS RA-1 B cells.
shRNAi lentiviral mini-library for targeting B-cell receptor (BCR) regulatory genes. (A) Cloning strategy for the lentiviral vector pA179.Helix. Only the relevant portions of the plasmids are shown. The pA179.Helix vector is a new version of pFUGW vector engineered at the CNIO Genomic Unit (http://www.cnio.es/es/index.asp). The pA179.Helix contains a CMV enhancer substituted for the U3 region of the 5′ LTR to maximize expression of viral RNA genomes during transient transfection.31 The pA179.Helix also contains a deletion in the U3 region of the 3′ LTR that makes the 5′ LTR of the integrated provirus transcriptionally inactive66 as well as the woodchuck hepatitis virus posttranscriptional regulatory element (WRE) inserted downstream of GFP to increase the level of transcription.67 (B) BCR-induced programmed cell death in a group of human B-cell lymphomas. MHH-PREB-1, RAMOS RA-1, SU-DHL-6, Z-138, JEKO, REC-1, MEC-1, and DOHH2 B-NHL cells were cultured (50 000 cells/ 200 μL/ well in 96-well plates) either in medium alone (RPMI plus additives and 10% FCS) or anti-IgM treated [goat F(ab′)2 antihuman IgM (10 μg/mL) antibody]. B cells were collected at 24 hours and programmed cell death was quantified using annexin V-APC staining. Cell death was quantified by FACS analysis, followed by analysis using FlowJo software (mean ± SD). Representative flow cytometric plots are shown. Numbers on plots represent the pecentage of cells in each quadrant over the total number of cells gated. (C) Western blot for human Argonuate-2 and Dicer in total protein extracts (RIPA extraction) from Z-138, DOHH-2, and RAMOS RA-1 B cells.
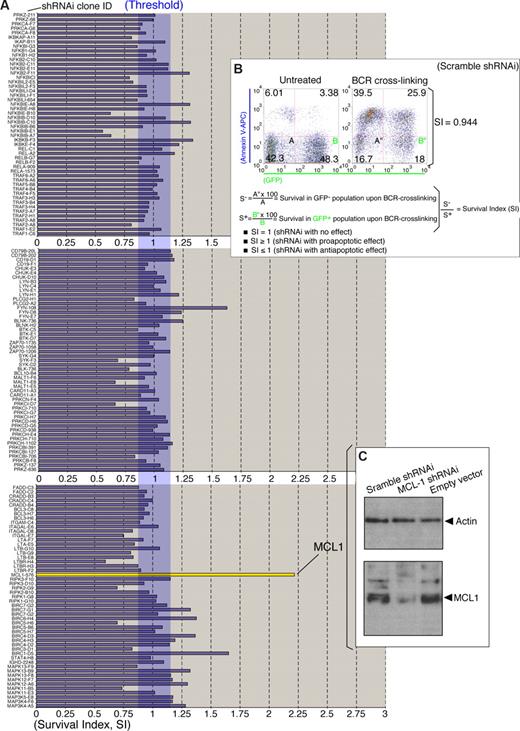
shRNAi lentiviral screen in RAMOS RA-1 B-NHL cells. (A) Lentiviral (HIV)-based screen for BCR regulatory molecules. RAMOS RA-1 B cells were transduced with individual lentiviruses from the shRNAi lentiviral mini-library (Table S1) pseudo-typed with vesicular stomatitis glycoprotein (VSVG) as described in “Lentiviral-mediated shRNAi transfer.” After lentiviral infection, transduced B cells were assayed for BCR-induced programmed cell death as described in Figure 1B. Survival index (SI) for each individual transduction was calculated as specified in Figure 2B. Representative SI value from 150 shRNAi lentiviral clones is shown. (B) Effect of scramble RNAi in the threshold of BCR-induced programmed cell death. RAMOS RA-1 B cells were transduced with lentiviruses containing a scramble shRNAi (negative control). SI value was calculated according to the formula described in the figure. Data for scramble shRNAi lentivirus infections are representative of 5 independent experiments. Scramble sequence included GAGACACCAAAGCATACAA. (C) Western blot for human MCL1 and Actin (loading control) in total protein extracts (RIPA extraction) from GFP+ purified MCL1 shRNAi, scramble shRNAi and empty vector-transduced RAMOS RA-1 B cells. GFP+ B cells were purified using Flow Sorter (FACSAria BD).
shRNAi lentiviral screen in RAMOS RA-1 B-NHL cells. (A) Lentiviral (HIV)-based screen for BCR regulatory molecules. RAMOS RA-1 B cells were transduced with individual lentiviruses from the shRNAi lentiviral mini-library (Table S1) pseudo-typed with vesicular stomatitis glycoprotein (VSVG) as described in “Lentiviral-mediated shRNAi transfer.” After lentiviral infection, transduced B cells were assayed for BCR-induced programmed cell death as described in Figure 1B. Survival index (SI) for each individual transduction was calculated as specified in Figure 2B. Representative SI value from 150 shRNAi lentiviral clones is shown. (B) Effect of scramble RNAi in the threshold of BCR-induced programmed cell death. RAMOS RA-1 B cells were transduced with lentiviruses containing a scramble shRNAi (negative control). SI value was calculated according to the formula described in the figure. Data for scramble shRNAi lentivirus infections are representative of 5 independent experiments. Scramble sequence included GAGACACCAAAGCATACAA. (C) Western blot for human MCL1 and Actin (loading control) in total protein extracts (RIPA extraction) from GFP+ purified MCL1 shRNAi, scramble shRNAi and empty vector-transduced RAMOS RA-1 B cells. GFP+ B cells were purified using Flow Sorter (FACSAria BD).
shRNA mini-library construction
Hairpins were cloned by standard PCR into the pENTR-D TOPO vector (Invitrogen) downstream of the H1 promoter as follows. The complete promoter sequence used was: GAACGCTGACGTCATCAACCCGCTCCAAGGAATCGCGGGCCCAGTGTCACTGGCGGGAACACCCAGCGC-GCGTGCGCCCTGGCAGGAAGATGGCTGTGAGGGACAGGGGAGT-GGCGCCCTGATATTTGCATGTCGCTATGTGTTCTGGGAATCACCA-TAAACGTGAAATGTCTTTGGATTTGGAATCTTATAAGTTCTGTATGA-GACCACTCGTTCCC. Inserts were obtained as PCR expression cassettes as previously described by McManus et al.32 Pfu DNA polymerase (Stratagene, La Jolla, CA) was used in standard conditions. The template providing the H1 promoter was obtained from pSUPER vector (OligoEngine, Seattle, WA), and the primers included H1 forward: GAACGCTGACGTCATCAAC and reverse hairpin: CACCGGAATTCCAAAAA-N19-TCTCTTGAA-N19-GGGAACGAGTGGTCTCATT (Sigma-Genosys, The Woodlands, TX), where the N19 hairpin targets were separately synthesized for each gene as previously described.33 Complete list of the hairpins and target genes can be found in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). PCR expression cassettes were subsequently cloned into the pENTR-D TOPO vector using the TOPO cloning procedure (pENTR Directional TOPO cloning kit, Invitrogen). Selected sequence-verified clones were subcloned into the destination vector pA179.Helix, which contains the CmR-ccdB selection cassette (Figure 1A); shRNAi clones were packaged into pA179.Helix lentiviral vector at the attR1 and attR2 sides using the recombinases Int, Xis, and IHF (Figure 1A). Positive clones were further evaluated by digestion with the Pst-I restriction enzyme.
Genome-wide gene-expression profile (DNA microarray)
Gene expression of follicular lymphoma
The analysis of the follicular lymphomas (FLs) was performed using SOTA algorithms (euclidean squared distances) as implemented in GEPAS (http://gepas.bioinfo.cipf.es/)53 . The Cluster Accuracy Analysis Tool from GEPAS was used for the identification of clusters of coregulated genes. The increased expression of MCL1 in multiple NHL types was also confirmed using Oncomine database.35,36 In all cases, fresh frozen samples provided from CNIO Tumor Bank were used. The cases were selected based on the standard criteria from World Health Organization classification criteria.
Western blot analysis
Total protein extracts were prepared by lysing B cells in RIPA lysis buffer (Sigma-Aldrich) containing protease inhibitor cocktail set III (Calbiochem, San Diego, CA) plus protease inhibitor cocktail set VIII (Calbiochem) for 10 minutes on ice. Cell debris was removed by centrifugation (10 000g, 10 minutes). Protein concentration was measured using Protein assays reagents A, B, and S (BioRad, Hercules, CA) following the manufacturer's instructions. Samples (30 μg protein) were resolved on a 4% to 12% gradient NuPAGE gel, transferred to nitrocellulose membranes, and blocked overnight with 5% non-fat dry milk in TBS buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl). Subsequent antibody incubations and membrane washes were performed in TBS-T buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.2% Tween 20) with 5% non-fat dry milk. Antibodies were detected using enhanced chemoluminescence (Western Lightning; Perkin-Elmer, Waltham, MA). Films were scanned using a HP F380 scanner and HP Scan Pro software. Digital image integrity were maintained in all Western blot pictures presented in this work and were not subjected to digital adjustment including brightness, contrast, color balance, nonlinear adjustment, or any other digital fabrication.
Antibodies
Antibodies for immunoblot analysis included mouse antiactin mAb (clone AC-15, Sigma-Aldrich), polyclonal rabbit antimouse MCL1 (Rockland Immunochemicals, Gilbertsville, PA), and rabbit anti-AGO2 (Upstate Biotechnology, Charlottesville, VA). We also used HRP-conjugated antirabbit and antimouse (Dako, Glostrup, Denmark). Antibody for programmed cell death induction included mouse F(ab′)2 antihuman IgM (μ-chain specific).
Cell viability assay
Cell death was quantitated by annexin-V-APC (BD Pharmingen) staining according to manufacturer's protocol. Briefly, cells were washed in PBS, resuspended in 500 μL binding buffer (BD Biosciences PharMingen, San Diego, CA) containing 0.5 μg/mL annexin V-APC, followed by flow cytometric analysis with a FACSCalibur (BD Biosciences PharMingen). Programmed cell death was quantified by FACS, followed by analysis using FlowJo software (Tree Star, Ashland, OR).
Results
Generation of a lentiviral RNA interference mini-library for the targeting of B-cell receptor regulatory molecules
In mammalian cells, 21- to 23- nucleotide (nt) double-stranded RNAs are normally produced by the RNase III endonuclease activity of the Dicer nuclease.37 Double-stranded RNAs associate with a multiprotein complex formed by Argonaute-2 that triggers the degradation of their complementary mRNA.38 Based on this biologic process, a new powerful tool called RNA interference (RNAi) has been engineered to inactivate specific gene targets.39,40 RNAi provokes the loss of activity of a gene through the specific degradation of its mRNA.39 RNAi is based on single-stranded RNAs that form a 19-nt long duplex with 2-nt 3′ overhangs. The ability of RNAi to silence genes in cultured cells has been rapidly adapted to different expression vectors, such as retroviruses and lentiviruses, to facilitate the delivery of these constructs into mammalian cells.41 Because the BCR signal transduction pathway is comprised of multiple adaptors, kinases, phosphatases, G-proteins, and transcription factors,42 this new technology provides the potential to genetically dissect the role of individual molecules in survival signaling within the BCR regulatory network (Human Protein Reference Database).43,44
To gain an insight into the key molecules controlling BCR-induced survival in B-NHL cells, we engineered a RNAi mini-library for the targeting of BCR regulatory molecules. To design this mini-library, small hairpins located in the open reading frame of the target genes were selected using the SIDE software.45 To avoid off-target gene knock-downs normally associated with the presence of a perfect match into the 3′ untranslated region of nontarget genes,46 hairpins were selected exclusively within the open reading frame of the target genes (Table S1). Individual small hairpins (sh) were cloned by standard PCR into pENTR vector downstream of the H1 promoter generating 277 sequence-verified shRNAi pENTR clones that subsequently were subcloned into the pA179.Helix lentiviral (HIV)-based vector. This lentiviral vector contains the CmR-ccdB selection cassette and is a new version of the pFUGW lentiviral vector, which was originally engineered by Lois et al.31 shRNAi pENTR clones were packaged into the pA179.Helix lentiviral vector at the attR1 and attR2 sides using the Int, Xis, and IHF recombinases (Figure 1A). The lentiviral backbone used was engineered to carry the GFP gene driven by a ubiquitously expressing promoter (UBC; Figure 1A), allowing the GFP+ transduced B cells to be tracked by FACS analysis.
We then addressed the characterization of suitable biologic systems for screen survival molecules within BCR regulatory networks. We first examined the competence of different B-NHL cells to initiate programmed cell death on BCR stimulation (Figure 1B). Programmed cell death in these B-NHL cell lines was activated through BCR crosslinking with a goat F(ab′)2 antihuman IgM (μ-heavy chain specific) antibody. Cell death was further evaluated after staining for annexin V at 24 hours after treatment (Figure 1B). Notably, BCR crosslinking induced programmed cell death in DOHH2, REC-1, Z-138, SU-DHL-6 and RAMOS RA-1 B-NHL cells (Figure 1B). In contrast JEKO, MHH-PREB-1, and MEC-1 B-NHL cells remained refractory to BCR-induced programmed cell death (Figure 1B). RAMOS RA-1 B-NHL cells showed a significant response to BCR-induced programmed cell death, exhibiting a high increase in programmed cell death compared with DOHH2 and Z-138 B-NHL cells. We also examined the expression levels of Argonuate-2 and Dicer in RAMOS RA-1, Z-138, and DOHH2 B-NHL cells (Figure 1C). We therefore decided to use RAMOS RA-1 because (1) this cell line responded properly to BCR-induced cell death, (2) showed good transduction efficiency, (3) showed expression of Dicer and Argonuate, and (4) RAMOS RA1, unlike other B-cell lines such as Z-138, is a model system classically used to study the molecular mechanisms of BCR-induced programmed cell death.47-51 RAMOS RA-1 B cells were therefore deemed to be an appropriate biologic model system to genetically screen survival factors linked to the BCR regulatory signaling network by RNA interference.
Lentiviral RNA interference screen for survival regulators linked to BCR regulatory networks
To better understand the molecular mechanisms of BCR-induced programmed cell death, we checked the effect of shRNAi lentiviral clones in RAMOS RA-1 B cells; shRNAi viruses pseudo-typed with vesicular stomatitis virus glycoproteins (VSVG) were initially examined for their ability to alter the threshold of programmed cell death in RAMOS RA-1 B-NHL cells on BCR stimulation. To this end, 277 independent infections were performed and a survival index (SI) calculated for each individual shRNAi clone (Figure 2A,B; Table S2). We validated our methodology by examining the effect on BCR-induced programmed cell death using scramble shRNAi viruses (Figure 2B). Importantly, SI is a ratio between survival in GFP− (endogenous control) and GFP+ B cells on BCR crosslinking, which helps to normalize data obtained in each individual infection. SI is thereby an indicator of the effect of shRNAi expression on BCR-induced programmed cell death. In this regard, a value of SI = 1 denotes that GFP+ and GFP− B cells die equally on BCR stimulation; a value of SI greater than 1 indicates that GFP+ B cells die more rapidly compared with GFP− B cells, whereas a SI value less than 1 reveals that GFP+ B cells die slower compared with GFP− B cells (Figure 2B).
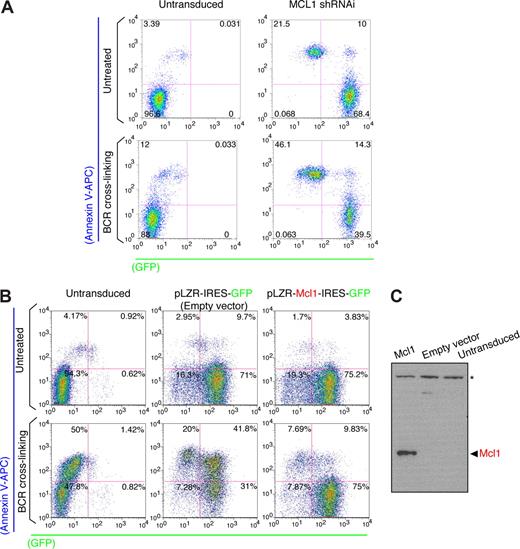
Most of the shRNAi lentiviral clones showed an SI value ranging between 0.75 and 1.25 (Table S2), showing no major impact of the hairpins in BCR-induced programmed cell death. Interestingly, a MCL1 shRNAi clone showed an SI value more than 2 (Figure 2A), revealing MCL1 as a critical survival molecule. We next sought to determine whether MCL1 shRNAi expression leads to MCL1 knock-down. Western blot analysis in RAMOS RA-1 B-NHL cells transduced with MCL1 shRNAi exhibited lower levels of MCL1 compared with cells infected with lentiviruses generated with empty lentivirus vector or scramble shRNAi (Figure 2C). These results thus indicate that MCL1 is an inhibitor of BCR-induced programmed cell death. MCL1 knock-down was also examined in the MEC1 cell line (a cell line resistant to BCR-induced programmed cell death). Our data indicate that MCL1 shRNAi promotes spontaneous cell death in MEC1 B-cell lymphomas and sensitizes to programmed cell death on BCR stimulation (Figure 3A). These results reflect the requirement of MCL1 for cell survival in many B-cell lymphomas. To be certain that unintended silencing of nontargeted genes was not responsible for the observed alteration in programmed cell death by MCL1 shRNAi, a functional rescue experiment was performed in RAMOS-RA B-NHL cells overexpressing murine Mcl1 (Figure 3B,C). FYN-108 and BIRC1-D5 shRNAi clones also promoted programmed cell death on BCR stimulation implying that FYN and BIRC1 may present antiapoptotic functions. Nevertheless, subsequent validation experiments confirmed these clones as false positives. In our screen, we have used 3 distinct FYN shRNAi clones (108, D8, and E7) showing similar Reynolds score,33 the ability of FYN-108 shRNAi to make an impact in cell death may reflect off-target effects of this hairpin. Our results show unequivocally that the final outcome of the BCR response in human B-NHL cells is determined by levels of MCL1 expression. Significantly, our results suggest that MCL1 might block BCR-induced programmed cell death by keeping proapoptotic (BAX and BAK)52 and BH3-only (BIM) proteins in check.18,53
Enforced MCL1 expression blocks BCR-induced programmed cell death in B-NHL cells. (A) Analysis of BCR-induced programmed cell death in MEC1 B cells. MEC1 B cells were transduced with lentiviruses containing MCL1 shRNAi. After infection, GFP+ B cells were purified using Flow Sorter (FACSAria BD), whereas untransduced MEC1 B cells were used as negative controls. Programmed cell death on BCR stimulation was performed and evaluated as described in Figures 1B and 2B. (B) Analysis of BCR-induced programmed cell death in MCL1 overexpressing B-cell lymphomas. RAMOS RA-1 B cells were transduced with retroviruses (using the pLZR-IRES-GFP backbone68 ) containing empty vector or Flag-murine Mcl1 cassette. The Flag-Mcl1 construct was cloned from pCMV-3xFlag-Mcl1 (3xFlag epitope tag cloned in frame with mouse Mcl-1 cDNA including ATG) into the EcoR1 site of the pLZR-IRES-GFP retroviral vector. Retroviruses were pseudo-typed with amphotropic envelopes using the envelope-expressing packaging cell line BING-CAK8 (ATCC). After infection, GFP+ B cells were purified (∼80% of enrichment) using Flow Sorter (FACSAria BD), whereas untransduced RAMOS RA-1 B cells were used as negative controls. Programmed cell death on BCR stimulation was performed and evaluated as described in Figures 1B and 2B. (C) Western blot for murine Mcl1 in total protein extracts (RIPA extraction) from GFP+ purified MCL1 overexpressing and empty vector (negative control) transduced RAMOS RA-1 B cells. Untransduced RAMOS RA-1 B cells were used as negative control as well. *Background band that can be used as loading control.
Enforced MCL1 expression blocks BCR-induced programmed cell death in B-NHL cells. (A) Analysis of BCR-induced programmed cell death in MEC1 B cells. MEC1 B cells were transduced with lentiviruses containing MCL1 shRNAi. After infection, GFP+ B cells were purified using Flow Sorter (FACSAria BD), whereas untransduced MEC1 B cells were used as negative controls. Programmed cell death on BCR stimulation was performed and evaluated as described in Figures 1B and 2B. (B) Analysis of BCR-induced programmed cell death in MCL1 overexpressing B-cell lymphomas. RAMOS RA-1 B cells were transduced with retroviruses (using the pLZR-IRES-GFP backbone68 ) containing empty vector or Flag-murine Mcl1 cassette. The Flag-Mcl1 construct was cloned from pCMV-3xFlag-Mcl1 (3xFlag epitope tag cloned in frame with mouse Mcl-1 cDNA including ATG) into the EcoR1 site of the pLZR-IRES-GFP retroviral vector. Retroviruses were pseudo-typed with amphotropic envelopes using the envelope-expressing packaging cell line BING-CAK8 (ATCC). After infection, GFP+ B cells were purified (∼80% of enrichment) using Flow Sorter (FACSAria BD), whereas untransduced RAMOS RA-1 B cells were used as negative controls. Programmed cell death on BCR stimulation was performed and evaluated as described in Figures 1B and 2B. (C) Western blot for murine Mcl1 in total protein extracts (RIPA extraction) from GFP+ purified MCL1 overexpressing and empty vector (negative control) transduced RAMOS RA-1 B cells. Untransduced RAMOS RA-1 B cells were used as negative control as well. *Background band that can be used as loading control.
Genome-wide gene expression profile analysis of Mcl1 overexpressing B-NHL cells
Our MCL1 loss- and gain-of function data in B-NHL cells are in agreement with: (1) results obtained in Mcl1 conditional mice showing that elimination of Mcl1 during early lymphocyte differentiation increased programmed cell death and arrested the development of pro-B cells,54 and (2) those obtained in Mcl1 transgenic mice, showing that Mcl1 overexpression leads to the generation of FLs, working thus as an oncoprotein.55 In human, MCL1 appears deregulated in high-grade B-cell lymphomas56 and FLs.57 Thus, collectively, these results point to MCL1 as a key oncoprotein, although the biologic functions critical for MCL1-induced B-cell transformation have not been fully elucidated.
These results and our observations prompted us to ask about the mechanisms of MCL1-induced survival and transformation. To understand how MCL1 overload contributes in B-cell neoplastic transformation, 41 000 human genes were screened in a genome-wide gene expression profile analysis of MCL1 overexpressing B-NHL cells. Two-color (Cy5/Cy3) microarray-based gene expression formats presenting high-density oligonucleotide probes printed on glass slides were used (GEO accession number GPL1708; http://www.ncbi.nlm.nih.gov/geo/). This type of microarray increases the sensitivity due to a smaller microarray footprint and increased hybridization concentration. Total RNAs from MCL1 overexpressing RAMOS RA-1 and Z-138 B-NHL cells were labeled with Cy5-CTP, whereas RNAs from references (empty vector/RAMOS RA-1 and empty vector/Z-138 B-NHL cells) were labeled with Cy3-CTP. After competitive hybridization (RNA-Cy5 from MCL1 overexpressing RAMOS RA-1 or MCL1 overexpressing Z-138 vs RNA-Cy3 from either empty vector/ RAMOS RA-1 or empty vector/ Z-138, respectively), gene expression data were pre-processed using Feature Extraction software and GEPAS tools.58 The resulting raw data were deposited at GEO database (accession number GSE8834). Unsupervised analysis of the genes showed 468 human genes differentially expressed in both cell types by MCL1 overexpression (Table S3). Supervised analysis of the genes was performed using BABELOMICS tools59 identifying 209 genes up- or down-regulated by MCL1 overexpression using a Log2 Cy5/ Cy3 cut-off of 1 (Figure 4A). These data demonstrate that acquisition of survival properties in MCL1 overexpressing B-NHL cells is associated with global cellular reprogramming.
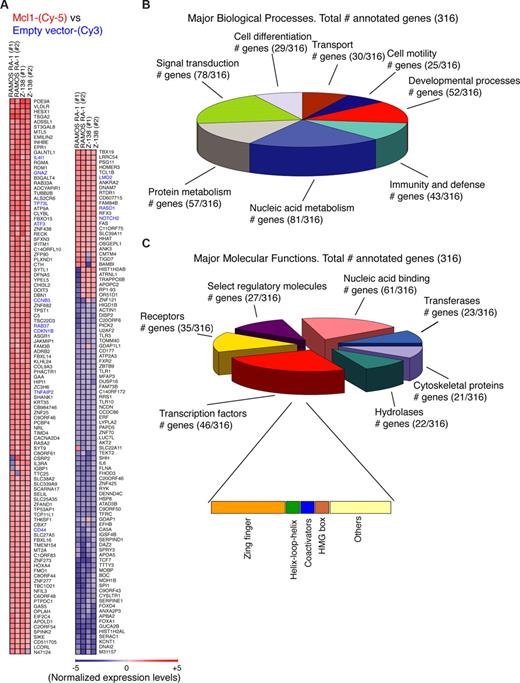
Transcriptome analysis of MCL1 overexpressing B-NHL cells. (A) Gene-expression profile of MCL1-overexpressing B-cell lymphomas. Gene-expression profiling was performed in duplicate for MCL1 overexpressing RAMOS RA-1 vs empty vector RAMOS RA-1, as well as MCL1 overexpressing Z-138 vs empty vector Z-138. The heat map represents expression levels for each sample. A total of 209 top significant genes (Log2 (Cy5/ Cy3) cut-off = 1) were ranked by Euclidean squared functions using GEPAS and BABELOMICS tools (http://babelomics.bioinfo.cipf.es/). Genes in blue denote genes that were found cross-validated in human follicular B-cell lymphomas (Figure 5). (B) Gene ontology analysis for the 316 annotated human genes commonly up- or down-regulated by MCL1 overexpression (Log2 (Cy5/ Cy3) cut-off = 0.5) was performed with the PANTHER program. Categories with more than 25 genes in biologic processes are shown. (C) Gene ontology analysis of the genes regulated by MCL1 overexpression was done with the PANTHER program. Of the 468 human genes showing a Log2 (Cy5/ Cy3) cut-off = 0.5 commonly up- or down-regulated by MCL1 overexpression, 316 were assigned with multiple molecular functions. Proteins for which no molecular function could be assigned were omitted from this display. Categories with more than 20 assigned proteins are shown. Subdivisions of the category transcription factor (46 proteins) is also shown.
Transcriptome analysis of MCL1 overexpressing B-NHL cells. (A) Gene-expression profile of MCL1-overexpressing B-cell lymphomas. Gene-expression profiling was performed in duplicate for MCL1 overexpressing RAMOS RA-1 vs empty vector RAMOS RA-1, as well as MCL1 overexpressing Z-138 vs empty vector Z-138. The heat map represents expression levels for each sample. A total of 209 top significant genes (Log2 (Cy5/ Cy3) cut-off = 1) were ranked by Euclidean squared functions using GEPAS and BABELOMICS tools (http://babelomics.bioinfo.cipf.es/). Genes in blue denote genes that were found cross-validated in human follicular B-cell lymphomas (Figure 5). (B) Gene ontology analysis for the 316 annotated human genes commonly up- or down-regulated by MCL1 overexpression (Log2 (Cy5/ Cy3) cut-off = 0.5) was performed with the PANTHER program. Categories with more than 25 genes in biologic processes are shown. (C) Gene ontology analysis of the genes regulated by MCL1 overexpression was done with the PANTHER program. Of the 468 human genes showing a Log2 (Cy5/ Cy3) cut-off = 0.5 commonly up- or down-regulated by MCL1 overexpression, 316 were assigned with multiple molecular functions. Proteins for which no molecular function could be assigned were omitted from this display. Categories with more than 20 assigned proteins are shown. Subdivisions of the category transcription factor (46 proteins) is also shown.
Genes were also annotated in the gene ontology format (Figure 4B). Of the 468 originally identified genes showing a Log2 Cy5/ Cy3 cut-off of 0.5, only 316 genes were annotated with a biologic process by the PANTHER program60 (Figure 4B). Particularly, this database classifies genes by their functions, using published scientific experimental evidence and evolutionary relationships to predict function even in the absence of direct experimental evidence; thus, proteins are categorized by molecular function and biologic process ontology terms. Among these genes, 81 were assigned to the nucleic acid metabolism category that includes DNA replication, DNA repair, and other categories that involve nucleic acids; 78 were integrated in the signal transduction category and 57 in protein metabolism pathways (Figure 4B). Further subdivision of genes into molecular functions revealed that 61 genes were linked to nucleic acid binding activities, 46 genes were transcription factors, and 35 were receptors (Figure 4C). Another measure of the significance of certain functional categories among these MCL1 targets is their enrichment relative to the total numbers in their respective categories; thus, zinc-finger transcription factors showed a highly significant enrichment (Figure 4C).
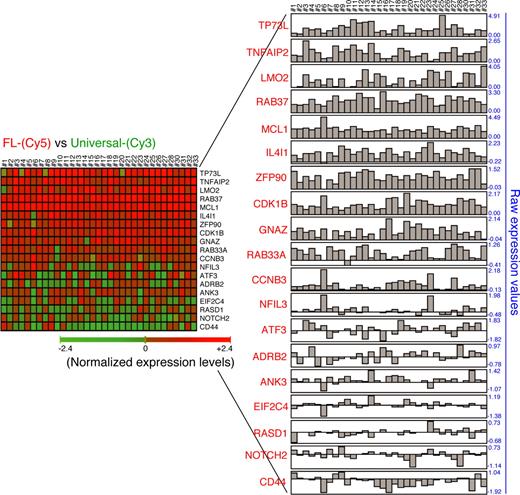
We then asked whether the results obtained from microarray hybridizations agreed with the transcriptome of MCL1-positive human B-cell lymphomas. We therefore assessed the relevance of the MCL1-induced signature in human follicular B-cell lymphomas. Gene cross-validation analysis was performed using CNIO Oncochip microarrays in 33 cases of human primary follicular B-cell lymphomas.61 We surveyed genes considered to be “nearest neighbors” of MCL1 based on the close agreement of their expression profiles. Analysis of the resultant gene expression signatures revealed a significant concordance with recognized MCL1 targets. The majority of cases with high levels of MCL1 expression (33 of 33) essentially showed co-regulation with a substantial number of MCL1 targets, such as TP53L (29 of 33), TNFAIP2 (33 of 33), LMO2 (32 of 33), RAB37 (33 of 33), IL4I1 (32 of 33), ZFP90 (31 of 33), CDK1B (33 of 33), and GNAZ (32 of 33) (Figure 5), indicating that MCL1-induced reprogramming might act in human primary follicular B-cell lymphomas as well. Future experiments are, however, necessary to determine whether the genes that are similarly expressed in all MCL1-positive B-cell lymphomas are part of a universal MCL1-induced signature.
Gene cross-validation in human primary follicular B-cell lymphomas. Correlation in gene expression profile of MCL1-positive primary follicular B-cell lymphomas. Gene expression profiling was performed in 33 cases of follicular B-cell lymphomas using CNIO Oncochip 12K microarrays (ArrayExpress accession number A-MEXP-261). Total RNA was isolated in 2 steps using Trizol (Invitrogen) followed by RNeasy (Qiagen, Valencia, CA) purification. Amplification of RNA was performed from 4 μg of total RNA using the Superscript System for cDNA synthesis (Invitrogen) and the T7 Megascript in vitro transcription kit (Ambion, Austin, TX). Amplified RNA (2.5 μg) was directly labeled with Cy5-conjugated dUTP. RNA (2.5 μg) from the Universal Human Reference RNA (Stratagene) was labeled with Cy3-conjugated dUTP as reference. Raw data (Cy5/Cy3 ratios) for 33 cases of MCL1-positive human follicular B-cell lymphoma are shown.
Gene cross-validation in human primary follicular B-cell lymphomas. Correlation in gene expression profile of MCL1-positive primary follicular B-cell lymphomas. Gene expression profiling was performed in 33 cases of follicular B-cell lymphomas using CNIO Oncochip 12K microarrays (ArrayExpress accession number A-MEXP-261). Total RNA was isolated in 2 steps using Trizol (Invitrogen) followed by RNeasy (Qiagen, Valencia, CA) purification. Amplification of RNA was performed from 4 μg of total RNA using the Superscript System for cDNA synthesis (Invitrogen) and the T7 Megascript in vitro transcription kit (Ambion, Austin, TX). Amplified RNA (2.5 μg) was directly labeled with Cy5-conjugated dUTP. RNA (2.5 μg) from the Universal Human Reference RNA (Stratagene) was labeled with Cy3-conjugated dUTP as reference. Raw data (Cy5/Cy3 ratios) for 33 cases of MCL1-positive human follicular B-cell lymphoma are shown.
Signaling pathways that interface with MCL1 expression
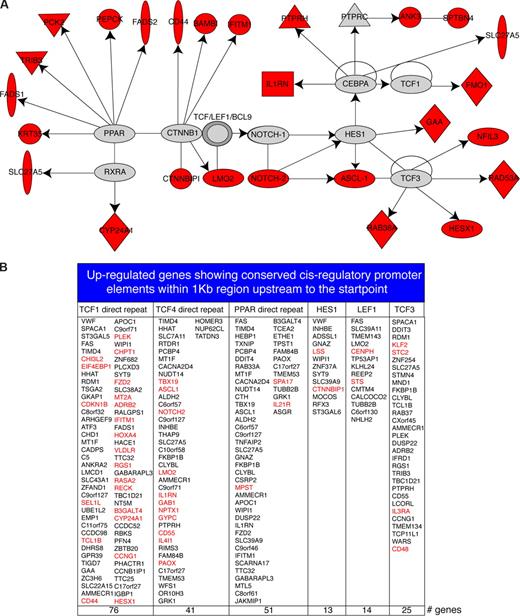
These results encouraged us to ask about the mechanisms of MCL1-induced reprogramming. The genes regulated by MCL1 were thereby analyzed using Ingenuity Systems IPA 5.0 database that provides a high quality knowledge base of biologic networks based on protein–protein interactions and transcriptional regulation. After analysis, an integrated module containing PPAR-RXRA and β-catenin-TCF pathways was identified (Figure 6A), which includes several oncoproteins such as LMO2 (also identified in human primary follicular B-cell lymphomas, Figure 5), NOTCH2, and HESX1 that are up-regulated by enforced MCL1 expression. This integrated module contains direct transcriptional TCF targets including HES1, NOTCH2, NFIL3, RAD53A, HESX1, RAB38A, and FMO1, which are effectors of the WNT signaling pathway62 (Figure 6B). A total of 26 up-regulated molecules were present in this module. This extensive network indicates that the processes of MCL1-induced gene regulation are probably to be heavily regulated by WNT signaling molecules. Moreover, analysis of canonical pathways using Ingenuity database identified canonical WNT signaling molecules up-regulated by MCL1 overexpression such as APC2, FZD2, and CD44 (Tables S4,S5).
Network modeling. (A) Module identified using the Ingenuity program. A solid line indicates direct interaction; a line without arrowhead indicates binding; an arrow from protein A to protein B indicates A transcriptionally activates B. Node shapes are indicative: triangle, kinase; diamond, enzyme; double circle, protein complexes; oval (horizontal), transcription regulator; oval (vertical), transmembrane receptor. Proteins identified in this screen are marked in red. (B) Promoter analysis for the up-regulated genes. Up-regulated genes containing TCF1, TCF4, PPAR, HES1, LEF1, and TCF3 binding sites within 1 kb region upstream of the transcriptional start point. Red denotes genes previously involved in B-cell transformation and cancer progression according to GeneCards database.
Network modeling. (A) Module identified using the Ingenuity program. A solid line indicates direct interaction; a line without arrowhead indicates binding; an arrow from protein A to protein B indicates A transcriptionally activates B. Node shapes are indicative: triangle, kinase; diamond, enzyme; double circle, protein complexes; oval (horizontal), transcription regulator; oval (vertical), transmembrane receptor. Proteins identified in this screen are marked in red. (B) Promoter analysis for the up-regulated genes. Up-regulated genes containing TCF1, TCF4, PPAR, HES1, LEF1, and TCF3 binding sites within 1 kb region upstream of the transcriptional start point. Red denotes genes previously involved in B-cell transformation and cancer progression according to GeneCards database.
To obtain an insight into the mechanisms of MCL1-induced transcriptional activation, conserved cis-regulatory promoter elements within the 1 kb region located upstream of the transcriptional start point were examined in the up-regulated genes using GEPAS tools.58 The data file containing 468 MCL1-regulated genes was clustered using SOTArray,59 using co-relation distance allowing determination of the genes up-regulated by MCL1 expression. Genes were further clustered using Cluster Accuracy Analysis Tool analysis,59 allowing visualization of the gene list using an unsupervised hierarchical clustering algorithm; this approach provides a ranked list of genes for up-regulated and down-regulated genes induced by MCL1 overexpression. Cis-regulatory elements located within 1 kb upstream of the start point were then searched exclusively among the up-regulated gene list using FATISCAN.59
This approach led to the identification of 76 up-regulated genes containing conserved consensus sequences for the transcription factor TCF1, 41 up-regulated genes presenting consensus for TCF4, and 25 up-regulated genes exhibiting TCF3 consensus sequences (Figure 6B). The extent of molecules linked to TCF transcription factors that we observed was unanticipated. For example, among the TCF targets, FMO1, NFIL3, RAD53A, HESX1, RAB38A, CD44, BAMBI, and IFITM1 had been previously identified in experimental conditions (Figure 6A), and these are considered to be well-characterized TCF targets. The quality of this identification is supported by the fact that many TCF targets were found multiple times by specific TCF transcriptional factors (such as TCF1, TCF3, and TCF4) (Figure 6B). Furthermore, other transcription factors linked to WNT signaling such as HES1 and LEF1 exhibited a substantial number of gene targets (14 and 13 genes, respectively) (Figure 6B). In addition, multiple up-regulated oncoproteins were identified as TCF targets, such as TCL1B, CD44, HOXA4, RASA2, HESX1, TBX19, NOTCH2, GAB1, KLF2, or LMO2 (Figure 6B). Taken together these data reveal MCL1 as reprogramming factor that regulates multiple oncoproteins integrated in β-catenin–TCF and PPAR-RXRA signaling pathways.
Discussion
Despite the important accomplishment of molecular biology in the identification of chromosomal translocations in human B-cell lymphomas, many key questions remain unanswered. Because the majority of proteins in the cell mediate their functions together with other proteins, pathologic processes, such as oncogenic transformation, should be considered as an extraordinarily complex network of interconnected proteins. For any disease mechanism, for example, B-cell transformation, we might consider a “systems biology approach” in which the behavior of regulatory networks can be analyzed as a whole.63
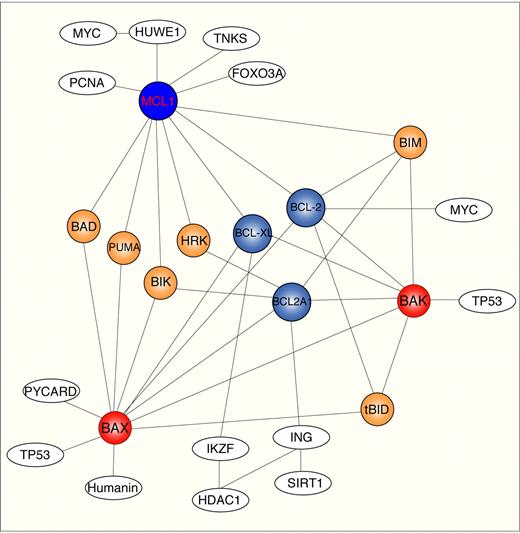
In this study, MCL1 was found not simply to be an aberrantly expressed antiapoptotic molecule linked to BCR signaling in human B-cell follicular lymphomas, but also a provider of transcriptional signaling linked to β-catenin–TCF pathways. These findings indicate that the connection from MCL1 to the BCR regulatory network is surprisingly more complex than previously anticipated. In addition to the antiapoptotic MCL1 function through the sequestration of proapoptotic BAX-BAK and BIM proteins activated after BCR stimulation,18,51 a biochemical interaction of MCL1 with transcription factors (FOXA3), replication factors (PCNA), and telomerase-associated proteins (TNK) (Figure 7) has also been found. How all these potential MCL1 targets are sensed in MCL1-overexpressing B-NHL cells is part of the MCL1 puzzle. Moreover, many canonical pathways appear to have multiple connections with MCL1 (Tables S4,S5). At this point, it is not clear whether these are all MCL1 targets or possibly other secondary molecules activated by intermediary molecules.
MCL1 interactome map. Schematic representation of MCL1 interactome using Ingenuity database, Human Protein Reference Database, and BioGrid. Only a limited number of protein–protein interactions are shown. Solid line indicates direct interaction. Nodo colors are indicatives: white, transcription and chromatin remodeling proteins; red, multidomain proapoptotic proteins; blue, multidomain antiapoptotic proteins; orange, BH3-only proteins.
MCL1 interactome map. Schematic representation of MCL1 interactome using Ingenuity database, Human Protein Reference Database, and BioGrid. Only a limited number of protein–protein interactions are shown. Solid line indicates direct interaction. Nodo colors are indicatives: white, transcription and chromatin remodeling proteins; red, multidomain proapoptotic proteins; blue, multidomain antiapoptotic proteins; orange, BH3-only proteins.
The large number of β-catenin–TCF targets discovered here (Figure 6B) also indicates that MCL1 overload alters WNT signaling pathway. We propose a mechanism for the induction of β-catenin–TCF targets by MCL1 overexpression in human B-cell lymphomas. We identified accumulation of β-catenin in the nucleus of MCL1 overexpressing B-NHL cells (data not shown). Our experiments suggest that MCL1 may disrupt the β-catenin/APC/AXIN/GSK3 protein complex in the cytoplasm.62 Interestingly, MCL1 is a substrate of GSK3-α and GSK3-β,64 suggesting that MCL1 may work as a decoy substrate altering the standard rate of β-catenin degradation induced by GSK3-α and GSK3-β in the cytoplasm and thereby allowing accumulation of β-catenin in the nucleus.
These data illustrate the broad landscape of MCL1 biology, which extends far beyond what was anticipated from previous studies addressing the antiapoptotic functions of BCL2 family members.65 These findings also indicate that MCL1 displays multiple interfaces with other relevant pathways. The elucidation of these novel connections and the roles played in B-cell transformation should provide a solid foundation for a systems biology understanding of MCL1-induced B-cell transformation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank David Livingston (Harvard Medical School, Dana-Farber Cancer Institute ([DFCI], Boston, MA) and Fumitaka Takahashi (University of Tokyo, Tokyo, Japan) for the gift of anti-Dicer antibodies, J.T. Opferman (St. Jude Children's Research Hospital, Memphis, TN) for the gift of the Flag-Mcl1 clone, Scott Armstrong (Harvard Medical School, DFCI) for the gift of pLKO-shRNAi MCL1, Dirk Buscher (Cellerix, Madrid, Spain) for the gift of plasmids containing retroviral amphotropic envelope, Ingrid Derks (Academic Medical Center, Amsterdam, the Netherlands) for the gift of antihuman IgM antibody (MH15), Aime Vazquez (Paul Brousse Hospital, INSERM, Villejuif, France) for the gift of antihuman IgM antibody (DA44), and members of the Centro Nacional de Investigaciones Oncológicas (CNIO) Lymphoma Group (Madrid, Spain) for discussions and the sequencing staff at the Genomics Unit (CNIO).
This work was supported by grants from the Spanish Ministry of Science and Technology (SAF2005-00 221, SAF2004-04 286).
Authorship
Contribution: A.R.-V. and M.A.P. coordinated author contributions and data analyses; A.R.-V., C.T., and B.H. performed cloning in lentiviral (HIV)-based vector; A.R.-V., D.M.-P., and M.A. performed computational analyses; A.R.-V. performed FACS analysis and lentiviral infections; P.d.C. performed microarray processing; A.R.-V. and M.A.P. helped with the overall interpretation of the data; and A.R.-V. wrote the manuscript. O.D. coordinated the high-throughput pENTR cloning performed by staff at the Genomics Unit.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Antonio Ruiz-Vela, Lymphoma Group, Department of Molecular Pathology, Biotechnology Program, Spanish National Cancer Centre, Melchor Fernández Almagro, E-28029, Madrid, Spain; e-mail: antonio.ruizvela@universia.es; or Miguel A. Piris, Lymphoma Group, Department of Molecular Pathology, Biotechnology Program, Spanish National Cancer Centre, Melchor Fernández Almagro, E-28029, Madrid, Spain; e-mail: mapiris@cnio.es.

![Figure 1. shRNAi lentiviral mini-library for targeting B-cell receptor (BCR) regulatory genes. (A) Cloning strategy for the lentiviral vector pA179.Helix. Only the relevant portions of the plasmids are shown. The pA179.Helix vector is a new version of pFUGW vector engineered at the CNIO Genomic Unit (http://www.cnio.es/es/index.asp). The pA179.Helix contains a CMV enhancer substituted for the U3 region of the 5′ LTR to maximize expression of viral RNA genomes during transient transfection.31 The pA179.Helix also contains a deletion in the U3 region of the 3′ LTR that makes the 5′ LTR of the integrated provirus transcriptionally inactive66 as well as the woodchuck hepatitis virus posttranscriptional regulatory element (WRE) inserted downstream of GFP to increase the level of transcription.67 (B) BCR-induced programmed cell death in a group of human B-cell lymphomas. MHH-PREB-1, RAMOS RA-1, SU-DHL-6, Z-138, JEKO, REC-1, MEC-1, and DOHH2 B-NHL cells were cultured (50 000 cells/ 200 μL/ well in 96-well plates) either in medium alone (RPMI plus additives and 10% FCS) or anti-IgM treated [goat F(ab′)2 antihuman IgM (10 μg/mL) antibody]. B cells were collected at 24 hours and programmed cell death was quantified using annexin V-APC staining. Cell death was quantified by FACS analysis, followed by analysis using FlowJo software (mean ± SD). Representative flow cytometric plots are shown. Numbers on plots represent the pecentage of cells in each quadrant over the total number of cells gated. (C) Western blot for human Argonuate-2 and Dicer in total protein extracts (RIPA extraction) from Z-138, DOHH-2, and RAMOS RA-1 B cells.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/111/3/10.1182_blood-2007-09-110601/6/m_zh80030814510001.jpeg?Expires=1769093613&Signature=E477Xdtijo7HytyaNZ8RWsR9cGSIwtlibfvEujAiwPS2WdGyilC3ExTJY0yl~FbeJYkmK5rQH6SnjtTgyR-wIwulbl79D78N-Lozc8sSrutXqOFyeHqupR0Ll4DBtHBxOHzVyOkdQfRjTJ124n~4QOVgxsMDYsWkJ9dFeOVy5bpiu3QGNRzCzqQve~1Ip8IVMXcrosQZtUc34slav~wgJJu5wrcFVPF4TdgAAN8-lS36BQp4bYGd4Frk5UiwBYybg8olrS0lpdOoFXt~3UlaZ~SFhRvmzzm0LRTCAKjkwZCw8URdvnDn2yYjlSDHBtl0t0gmsQJAu09XYMVoe6nMlA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)