Pathogenic activation of the LMO2 proto-oncogene by an oncoretroviral vector insertion in a clinical trial for X-linked severe combined immunodeficiency (X-SCID) has prompted safety concerns. We used an adeno-associated virus vector to achieve targeted insertion of a γ-retroviral long terminal repeat (LTR) driving a GFP expression cassette with flanking loxP sites in a human T-cell line at the precise location of vector integration in one of the patients with X-SCID. The LTR-GFP cassette was inserted into the first intron of the LMO2 gene, resulting in strong activation of LMO2. Cre-mediated cassette exchange was used to replace the original LTR-GFP cassette with one flanked by insulator elements leading to a several fold reduction in LMO2 expression. The LTR-GFP cassette was also replaced with a globin gene regulatory cassette that failed to activate the LMO2 gene in lymphoid cells. A γ-retroviral vector with 2 intact LTRs resulted in activation of the LMO2 gene when inserted into the first intron, but a self-inactivating lentiviral vector with an internal cellular promoter and flanking insulator elements did not activate the LMO2 gene. Thus, this system is useful for comparing the safety profiles of vector cassettes with various regulatory elements for their potential for proto-oncogene activation.
Introduction
Retroviral vectors are potentially useful for gene therapy of blood disorders because they are capable of permanently integrating a therapeutic gene into stem cells and achieving long-term expression in differentiating hematopoietic cells. The initial clinical trials for severe combined immunodeficiency (SCID) secondary to adenosine deaminase deficiency (ADA) were either targeted to T lymphocytes1 or resulted in too low a frequency of gene transfer into stem cells to produce clinical benefit.2 Improved transduction conditions have now resulted in clinical benefit after stem cell–targeted gene transfer for patients with SCID secondary to ADA deficiency3 or secondary to a deficiency in the common γ chain of interleukin receptors (X-SCID).4,–6 In addition, 2 severely affected patients with chronic granulomatous disease (CGD) achieved clearance of life-threatening infections by gene-corrected granulocytes derived from transduced stem cells.7
In the context of these successful clinical trials, unequivocal evidence of the genotoxicity of retroviral vector integration has emerged in that 3 of the patients with X-SCID in one clinical trial8,9 have developed leukemia. In 2, activation of the proto-oncogene, LMO2, was directly implicated as a genotoxic event.8 In addition, the 2 patients with CGD exhibited striking clonal dominance with most of their gene-corrected cells having insertions into or near one or more of the MDS1-EVI1, PRDM16, or SETBP1 loci. Evidence indicated that these retroviral insertion events had activated the nearby proto-oncogene.7 This experience emphasizes that the genotoxicity of retroviral integration is a relevant factor to be considered in designing vectors for future clinical trials.
The γ-retroviral vectors used in clinical trials to date have intact long terminal repeats (LTRs) derived from murine leukemia viruses that contain a powerful enhancer/promoter element. An alternative is the use of self-inactivating (SIN) vectors in which the enhancer/promoter is removed from the 3′ LTR.10,11 During integration, the 3′ LTR is copied so that the integrated 5′ LTR also lacks the enhancer/promoter. Recent results obtained using an immortalization assay of primary bone marrow cells suggest that the SIN design of γ-retroviral vectors reduces the potential for proto-oncogene activation and outgrowth of cytokine-dependent, immortalized myeloid cells.12 SIN vectors require an internal promoter to drive the therapeutic transgene and thus retain some potential for proto-oncogene activation as shown in this immortalization assay.12 In the future, comparison of different promoter elements with respect to proto-oncogene activation will be relevant to optimizing the safety of particular therapeutic vectors.
Another potentially useful safety modification is the inclusion of an insulator element in the 3′ LTR that, when duplicated during vector integration, results in the therapeutic gene being flanked with insulator elements in both LTRs.13,14 Insulator elements function as a barrier by separating heterochromatin from transcriptionally active chromatin.15 In addition to this barrier function, insulator elements may have enhancer-blocking activities that diminish interactions of enhancers with promoters.15,16 One well-studied insulator, the hypersensitive site 4 of the chicken β-globin locus (cHS4), has enhancer blocking activities that depend on binding of the protein, CTCF. Its chromatin barrier function depends on the binding of other proteins that recruit histone and DNA-modifying complexes to the site.15 The cHS4 insulator has been incorporated into retroviral vectors and shown to enhance transgene expression by diminishing the influence of integration position on the potential for gene silencing or variable expression in the progeny of the cell in which the integration event occurred.13,14,17
A third approach for improving the safety profile of therapeutic vectors is to use only tissue-specific enhancer and promoter elements. Our work has focused on the development of a therapeutic vector for treatment of hemoglobinopathies in which gene expression relies on an erythroid-specific promoter and globin locus control region (LCR) regulatory elements.18 Prior studies suggest that these elements have diminished the potential for transgene expression in lineages other than erythroid cells.19 It is therefore reasonable to propose that such elements will have less risk for proto-oncogene activation in self-renewing cells that are capable of clonal dominance or the evolution to leukemia.
Several cell culture and animal model assays have been developed to evaluate the safety of specific vector designs.20,–22 Those assays assess the consequences of vector insertion by virtue of the biologic consequences of random integration events. In contrast, our focus has been on the development of a strategy to recreate specific integration events so that the direct effects of various regulatory elements on proto-oncogene activation can be determined. To achieve this goal, we have used adeno-associated virus (AAV)–mediated homologous gene targeting23,,–26 to create an initial integration event at the precise chromosomal location of the vector integration in one of the patients with X-SCID who developed leukemia8 and Cre recombinase–mediated cassette exchange27,28 to substitute cassettes carrying different regulatory elements at this position. Different loxP sites, the cis-acting sequences required for Cre-mediated recombination, were placed upstream and downstream of the introduced cassette to facilitate site-specific and directional cassette exchange.29,–31
Our experimental efforts have focused on the LMO2 gene, a well-known proto-oncogene that is involved in chromosomal translocations in human T-cell leukemias and results in perturbations of T lymphopoiesis and a predilection to lymphomas in transgenic mice.32 As noted earlier, integration within or near the LMO2 gene has been implicated as a pathogenic event in 2 patients who developed leukemia after successful gene therapy for X-SCID.8 Our experiments to date have used the human T-lymphocyte cell line (Jurkat), but we believe that this approach can be used successfully in other cellular systems. Our data indicate that this system is useful for comparing vectors and strongly support the prediction that a SIN vector with an internal promoter flanked by insulator elements will have a much better safety profile than vectors used in clinical trials to date.
Methods
Plasmid construction
To construct the plasmid containing the AAV-LMO2 homologous segments, the human LMO2 intron sequences from upstream and downstream of the retroviral insertion site8 were amplified by polymerase chain reaction (PCR) on genomic DNA from Jurkat cells with an oligonucleotide primer set containing BamHI (underlined) and StuI (dotted underline) recognition sequences. The sequences are provided in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). The PCR products were individually cloned into a TOPO-TA vector (Invitrogen, Carlsbad, CA), and the sequences were confirmed by automated sequencing. Both intron sequences were then combined in a single vector to generate a StuI restriction site at the original retroviral vector insertion site with flanking BamHI sites. Next, a gene cassette was designed to have the GFP coding sequences under the control of the retroviral 592-bp (base pair) LTR identical to that used in the vector in the gene therapy trial,4,8 as verified by DNA sequencing, followed by a polyadenylation signal; the entire cassette was flanked by loxP (upstream of retroviral LTR) and loxP511 (downstream of polyadenylation signal) sites (pLTR-GFP). This floxed 1.6-kb (kilobase) LTR-GFP cassette was cloned into the StuI site in a reverse orientation to LMO2 intron segments. The 4.4-kb BamHI fragment was cloned between AAV2 ITRs to yield a plasmid containing the genome for generating the single-stranded targeting AAV vector (AAV-LMO2). The construction of the other plasmids used in these studies is described in Document S1.
AAV vector production and target cell transduction
AAV vector particles were produced by transient transfection using a vector plasmid and plasmids pHGTI-adeno1 (which provides adenoviral helper functions) and LT-RC02 (which encodes serotype 2 rep and serotype 1 capsid).33 After transfection of 293T cells using calcium phosphate, the cells were incubated for 48 hours before harvest, microfluidization, and purification on an iodixanol gradient.34 After the iodixanol step gradient purification, viral particles were further purified on a Sephacryl S-300 column in phosphate-buffered saline. The vector preparations were then concentrated using Millipore Amicon Ultra-15 centrifugal filters and stored at 4°C or frozen at −80°C. The human T-lymphoblastic cell line Jurkat (ATCC, Manassas, VA) was maintained in RPMI 1640 medium (Lonza Baltimore, Baltimore, MD) with 10% fetal bovine serum (HyClone, Logan, UT) supplemented with 2mM l-glutamine, penicillin, and streptomycin (Invitrogen, Carlsbad, CA). For AAV vector transduction, proliferating Jurkat cells were pelleted and resuspended in serum-free Opti-MEM (Invitrogen), and 3 × 105 cells were added to each well of a 6-well plate. AAV-LMO2 vector particles were added to each well to give multiplicities of infection (MOIs) of 4 × 104, 2 × 105, or 4 × 105 vector genome copies per cell and incubated overnight. As a control, scAAV-GFP was used at a MOI of 4 × 104. Serum was added 16 hours after transduction. Transduced Jurkat cells were analyzed for GFP expression by flow cytometry every 3 days.
DNA analysis
AAV-LMO2–transduced Jurkat cells were cultured for 9 days, and GFP-expressing single cells were sorted by fluorescence-activated cell sorting (FACS) into 96-well plates containing irradiated (3 Gy [300 rad]) feeder HeLa cells. After culture for an additional 2 to 3 weeks, each GFP-expressing clone was transferred into 12-well plates, and after proliferation genomic DNA was extracted using the Puregene DNA purification kit (Gentra Systems, Minneapolis, MN). The clones were screened by PCR using a primer set with one primer within the LTR and the other primer on the LMO2 intron outside of the AAV-LMO2 homologous arm. Only clones with a targeted insertion could give the predicted 1.5-kb band. The PCR conditions were as follows: initial denaturation at 94°C for 2 minutes followed by 30-cycle amplification at 94°C for 30 seconds, 61°C for 30 seconds, and 68°C for 100 seconds. Genomic DNA from PCR-positive clones was digested with BglII or DraIII, separated in a 1% agarose gel, and transferred onto a GeneScreen Plus membrane (PerkinElmer, Life and Analytical Sciences, Waltham, MA). The blot was hybridized with either a 32P-labeled GFP probe or a LMO2 intron probe, and the signal was detected using a phosphorimager.
Analysis of gene expression
The relative level of LMO2 expression was measured by quantitative real-time reverse transcriptase-polymerase chain reaction (qRT-PCR) using TaqMan Gene Expression Assay primer/probe sets (Applied Biosystems, Foster City, CA).
For Western blot analysis, Jurkat cells were lysed in M-PER mammalian protein extraction reagent (Pierce Chemical, Rockford, IL). Experimental details are provided in Document S1.
Cre recombinase–mediated cassette exchange
A Jurkat cell clone having a LTR-GFP insertion (G-25) was used for the initial DsRed2 cassette exchange. Cells were seeded in Opti-MEM (Invitrogen) without serum, and scAAV-Cre vector particles were added at a MOI of 4 × 104 and incubated overnight. Serum was added the next day and incubation continued for 8 hours. The scAAV-Cre–transduced cells were then transfected with a plasmid containing the floxed DsRed2 cassette using LipofectAmine 2000 (Invitrogen) according to the manufacturer's protocol and expanded for 7 days. Cells expressing DsRed2 were sorted as single cells using FACS and then expanded an additional 2 to 3 weeks. Clones were subjected to a PCR screen followed by Southern blot analysis to evaluate for cassette exchange. A DsRed exchanged clone (R-6) was used for the following cassette exchange experiments. Either a 1.2-kb insulator fragment16 or a 2 × 250-bp core insulator fragment was inserted so that each flanked the original LTR-GFP expression cassette. A γ-retroviral vector flanked by the loxP and the loxP511 sites with 2 intact LTRs and the GFP coding sequences and a SIN lentiviral vector also flanked by the loxP sites with a 1.2-kb insulator fragment in both LTRs and an internal EF1α promoter GFP cassette were also assembled and used in the Cre recombinase–mediated cassette exchange reaction. After AAV-Cre transduction and floxed cassette transfection of R-6, cells were expanded for 4 days, and a GFP-positive population was recovered by FACS. The sorted cell population was cultured 2 more weeks, and single cell clones were isolated by FACS. For the globin cassette, the globin LCR and β-globin promoter driving GFP were removed from a lentiviral vector18 and placed between the loxP and loxP511 sites and used for the exchange reaction. When the initial effort yielded a globin exchange clone lacking GFP expression, additional clones were sorted randomly and screened by PCR (Table S1).
Results
Targeted insertion of a retroviral LTR into the LMO2 intron by the AAV-LMO2 vector
A retroviral insertion into the first intron of the LMO2 gene in a reverse orientation resulted in activation of this proto-oncogene in one of the patients with X-SCID.8 Our initial goal was to recreate this specific insertion in a context in which we could (1) replace the original inserted LTR with the same LTR flanked by insulator elements or (2) substitute a globin gene cassette developed for gene therapy of hemoglobin disorders. The targeting vector, AAV-LMO2, contained a total of 2.8 kb (1.4 kb upstream and downstream) of sequences from the LMO2 first intron flanking an expression cassette (Figure 1A). The expression cassette, composed of the retroviral LTR used in the X-SCID trial4,5 driving GFP followed by a SV40 polyadenylation site, was placed in the middle of the LMO2 intron homologous segment in reverse orientation. Because of the packaging constraints of AAV vectors (up to 4.7 kb), we inserted this simple expression cassette rather than the complete retroviral vector genome.
Insertion of a retroviral LTR expression cassette into the first intron of LMO2. (A) The AAV-LMO2 vector consists of a single retroviral LTR-GFP cassette and 1.4 kb of upstream and 1.4 kb of downstream intron sequences. The LTR-GFP cassette was flanked by loxP (▵) and loxP511 (▴) sites, and the entire assembly was cloned between AAV-ITRs. The restriction enzyme sites (D, DraIII; B, BglII) and the expected band sizes on cassette integration along with probes used in Southern blot hybridization are indicated. (B) Jurkat cells were transduced with scAAV-GFP or AAV-LMO2 at the indicated MOI, and GFP expression was periodically monitored by FACS analysis. (C) BglII-digested DNA (left) or DraIII-digested DNA (right) was probed with GFP (top) or LMO2 (bottom) intron sequences in a Southern blot analysis.
Insertion of a retroviral LTR expression cassette into the first intron of LMO2. (A) The AAV-LMO2 vector consists of a single retroviral LTR-GFP cassette and 1.4 kb of upstream and 1.4 kb of downstream intron sequences. The LTR-GFP cassette was flanked by loxP (▵) and loxP511 (▴) sites, and the entire assembly was cloned between AAV-ITRs. The restriction enzyme sites (D, DraIII; B, BglII) and the expected band sizes on cassette integration along with probes used in Southern blot hybridization are indicated. (B) Jurkat cells were transduced with scAAV-GFP or AAV-LMO2 at the indicated MOI, and GFP expression was periodically monitored by FACS analysis. (C) BglII-digested DNA (left) or DraIII-digested DNA (right) was probed with GFP (top) or LMO2 (bottom) intron sequences in a Southern blot analysis.
Human CD3+ lymphoblastoid cells (Jurkat) were transduced with the AAV-LMO2 vector at 3 different MOIs. As a control, a self-complementary (sc) AAV-GFP vector was used at a MOI of 4 × 104 to estimate the maximal frequency of random integration by AAV vectors in Jurkat cells. GFP expression was periodically monitored (Figure 1B). Although almost all of the cells were initially transduced after 3 days by the scAAV-GFP vector, only a very low percentage of cells (0.2%) remained GFP positive after 21 days, reflecting rare random integration. Similarly, cells expressing GFP were present at maximal frequency 3 days after transduction with AAV-LMO2, and the frequency gradually decreased (Figure 1B). GFP-expressing cells were sorted as single cells 9 days after transduction, when approximately 40% were GFP positive, and then expanded. Only 2.2% of the clones remained GFP positive on further passage. The initial screen for targeted insertion was done by PCR on genomic DNA with a primer located within the retroviral LTR and the other primer in the LMO2 intron outside of the AAV-LMO2 vector homology so that only clones with a true recombination could give a PCR product (Figure 1A arrows). To confirm targeting without rearrangement, genomic DNA from PCR-positive clones was digested with BglII and DraIII and analyzed by Southern blot probed with a fragment containing GFP coding sequences or sequences from the LMO2 intron (Figure 1C). The LMO2 probe showed 2 bands of the predicted sizes for both an LTR-inserted allele and a wild-type allele in 4 clones. The GFP probe detected only the modified allele, as expected (Figure 1C). Nine of the 62 GFP-positive clones at more than 3 weeks were PCR positive, and 4 of these were confirmed as unrearranged by Southern blot analysis and lacking any random genome integrations. Thus, approximately 0.25% of the cells initially transduced by AAV-LMO2 and 6.5% of clones that exhibited stable GFP expression at more than 3 weeks exhibited homologous targeting (Table S2). A second experiment in which sorting of GFP-positive clones was done after 3 weeks also yielded 6.3% targeted clones (Table S2).
Single LTR is sufficient to activate LMO2
The effect of the retroviral LTR insertion on LMO2 transcription was initially estimated by qRT-PCR using primers in exons 4 and 5 (Figure 2A). Untransduced Jurkat cells had low levels of LMO2 expression. Human erythroleukemia cells (K562) that constitutively express LMO2 were used as a positive control. All 4 clones having a targeted insertion into the LMO2 locus showed an increase to 17% to 53% the level of LMO2 expression in control K562 cells compared with less than 0.1% in untransduced Jurkat cells. Control single cell clones lacking the AAV-LMO2 vector genome had similarly low levels of LMO2 expression as the untransduced Jurkat cell population (Figure 2A, M-3 and M-7 clones). Western blot analysis confirmed the increase in LMO2 protein expression consistent with the qRT-PCR results (Figure 2B). LMO2 activation in targeted clones did not convey an obvious growth advantage to the already transformed Jurkat cells in competitive mixing experiments (data not shown).
LTR mediated LMO2 activation. (A) The relative LMO2 expression was compared by real-time qRT-PCR. All targeted clones showed a significant increase in LMO2 transcript over control Jurkat cells. M-3 and M-7 are single cell clones of untransduced Jurkat cells. K562 erythroleukemia cells in which LMO2 is constitutively expressed were used as the positive control. Values are expressed as percentages relative to the K562 control. (B) Expression of LMO2 protein was confirmed by Western blot analysis. Consistent with the qRT-PCR results, LMO2 protein was abundant in targeted clones but not in untransduced Jurkat cells. The upper portion of the Western filter was probed with an antibody to β-actin to provide a lane-to-lane loading control.
LTR mediated LMO2 activation. (A) The relative LMO2 expression was compared by real-time qRT-PCR. All targeted clones showed a significant increase in LMO2 transcript over control Jurkat cells. M-3 and M-7 are single cell clones of untransduced Jurkat cells. K562 erythroleukemia cells in which LMO2 is constitutively expressed were used as the positive control. Values are expressed as percentages relative to the K562 control. (B) Expression of LMO2 protein was confirmed by Western blot analysis. Consistent with the qRT-PCR results, LMO2 protein was abundant in targeted clones but not in untransduced Jurkat cells. The upper portion of the Western filter was probed with an antibody to β-actin to provide a lane-to-lane loading control.
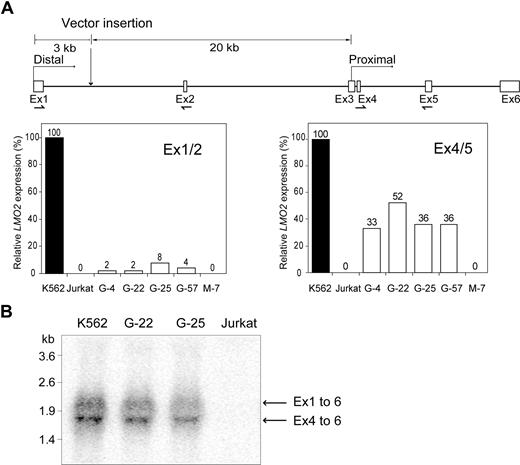
The LMO2 gene has 2 promoters, the distal promoter upstream from exon 1 and a proximal promoter overlapping with exon 3.35,–37 Both transcripts encode an identical protein because the reading frame begins in exon 4 (Figure 3A). To determine which promoter was activated on retroviral LTR insertion, qRT-PCR with a primer pair in exons 1 and 2 (transcript from distal promoter) was compared with results with a primer pair in exons 4 and 5 (total transcript). These data along with the results of Northern blot analysis (Figure 3B) indicate that most of the transcript originates from the proximal promoter. qRT-PCR was also used to quantify expression of nearby genes flanking the LMO2 locus. Neither the GPIAP1 gene (GPI-anchored membrane protein 1), which is 165 kb upstream, nor the FBXO3 gene (F box protein 3), which is 110 kb downstream, were significantly activated (data not shown). Thus, precise targeting of the LTR-GFP cassette into the LMO2 locus resulted in selective activation of this proto-oncogene, thereby reproducing the transcriptional effects of the retroviral vector integration in one of the patients with X-SCID with leukemia.8
Activation of the distal and proximal LMO2 promoters by the LTR. (A) The origin of the 2 transcripts from human LMO2, both of which encode the same protein, is indicated along with the relative distances of the retroviral insertion site from the 2 transcriptional start sites. The relative LMO2 expression from the 2 promoters was established in several clones using TaqMan Gene Expression Assays. The location of each primer pair is indicated on the diagram. Results are expressed as the percentage of the K562 cell control. (B) Northern blot analysis of RNA from 2 clones having an LTR-GFP insertion (G-22 and G-25) and positive control (K562) and negative control (Jurkat) cells was performed. The transcript from the proximal promoter was more abundant that that from the distal promoter in both clones, consistent with the results obtained with the qRT-PCR analysis in panel A.
Activation of the distal and proximal LMO2 promoters by the LTR. (A) The origin of the 2 transcripts from human LMO2, both of which encode the same protein, is indicated along with the relative distances of the retroviral insertion site from the 2 transcriptional start sites. The relative LMO2 expression from the 2 promoters was established in several clones using TaqMan Gene Expression Assays. The location of each primer pair is indicated on the diagram. Results are expressed as the percentage of the K562 cell control. (B) Northern blot analysis of RNA from 2 clones having an LTR-GFP insertion (G-22 and G-25) and positive control (K562) and negative control (Jurkat) cells was performed. The transcript from the proximal promoter was more abundant that that from the distal promoter in both clones, consistent with the results obtained with the qRT-PCR analysis in panel A.
Cre/loxP-mediated cassette exchange
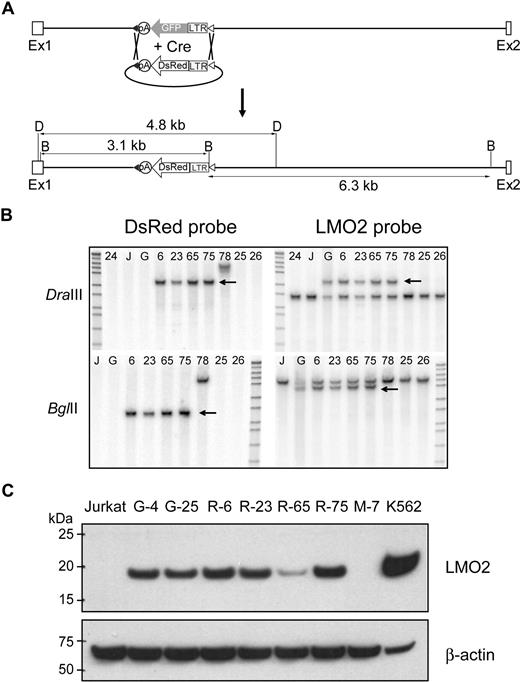
LoxP and loxP511 sites flanked the LTR-GFP cassette, thereby creating the opportunity for directional, Cre-mediated cassette exchange.31 As a test of our system, we assembled an LTR-DsRed2 expression cassette (Figure 4A) that had an identical design to the original LTR-GFP cassette except the GFP coding sequences were replaced with those of DsRed2. Because Jurkat cells are not easily transfected with a conventional transfection protocol (efficiency ranging from 10% to 15% using LipofectAmine2000), a scAAV-Cre vector was used to obtain high-level Cre expression. One of the original targeted clones (G-25) was transduced with scAAV-Cre and transfected with a plasmid containing the LTR-DsRed2 cassette. Seven days later, the cells were initially sorted based on DsRed2 expression; 6 clones that were GFP negative but DsRed positive were isolated, and 4 clones were confirmed by Southern blot analysis to represent authentic cassette exchange (Figure 4B). We estimate that 4% to 8% of the clones initially sorted based on DsRed expression had undergone cassette exchange (Table S1). The efficiency was probably compromised because the dimness of the DsRed signal made it difficult to separate positive and negative cells. The level of LMO2 protein expression in Red clones was similar, with the exception of R-65, to that found in the original GFP clones (Figure 4C). The R-6 clone was used for subsequent cassette exchanges.
Cre recombinase mediated cassette exchange. (A) Shown is a diagram of the predicted exchange reaction and the size of the resulting restriction enzyme fragments. (B) Southern blot analysis was performed to verify that the cassette-mediated exchange reaction had occurred. Shown are the results of Southern blot analysis of selected clones of DNA digested with DraIII (top) or BglII (bottom) and probed with the DsRed probe (left) or the LMO2 probe (right). The arrow marks the position of the predicted band with each probe. G is the original LTR-GFP clone, and 24, 25, and 26 are clones in which the LTR-GFP has been excised without insertion of the LTR-DsRed cassette. (C) Western blot analysis of proteins extracted from selected clones containing the LTR-GFP cassette (G-4 and G-25) or the DsRed cassette (R-6, R-23, R-65, and R-75). An untransduced Jurkat cell clone (M-7) and K562 cells served as negative and positive controls, respectively.
Cre recombinase mediated cassette exchange. (A) Shown is a diagram of the predicted exchange reaction and the size of the resulting restriction enzyme fragments. (B) Southern blot analysis was performed to verify that the cassette-mediated exchange reaction had occurred. Shown are the results of Southern blot analysis of selected clones of DNA digested with DraIII (top) or BglII (bottom) and probed with the DsRed probe (left) or the LMO2 probe (right). The arrow marks the position of the predicted band with each probe. G is the original LTR-GFP clone, and 24, 25, and 26 are clones in which the LTR-GFP has been excised without insertion of the LTR-DsRed cassette. (C) Western blot analysis of proteins extracted from selected clones containing the LTR-GFP cassette (G-4 and G-25) or the DsRed cassette (R-6, R-23, R-65, and R-75). An untransduced Jurkat cell clone (M-7) and K562 cells served as negative and positive controls, respectively.
Flanking chromatin insulator reduced but did not eliminate LMO2 activation
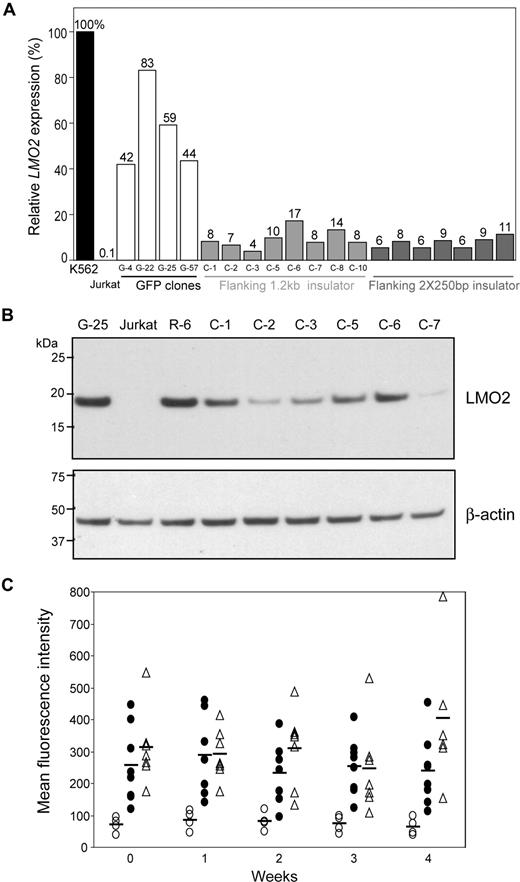
To investigate the effect of cHS4 insulator on LMO2 activation by the retroviral LTR, the original floxed LTR-GFP cassette was modified to incorporate either a 1.2-kb cHS4 or 2 copies of a 250-bp core insulator (2 × 250 bp) fragment flanking the LTR-GFP cassette. After Cre-mediated cassette exchange using the R-6 clone, 10 (1.2 kb) or 9 (2 × 250 bp) clones were shown to have cassette exchange verified by PCR and Southern blot analysis (Figures S1 and S2). Strongly GFP-positive clones were readily isolated, and the exchange efficiency in these clones was 88% or 23%, respectively, for the 2 cassettes. Addition of either insulator element reduced LMO2 transcript levels by 3- to 15-fold compared with the original LTR-GFP cassette (Figure 5A). All values obtained by qRT-PCR were normalized to the K562 value that was set at 100%. The mean of the percentage of K562 values, SD, and P values are as follows: (1) 1.2 kb versus the LTR-GFP control, 9.4% (± 3.9%) versus 56.9% (± 16.5%), P = .014, and (2) 2 × 250 bp versus LTR-GFP control, 7.7% (± 2.1%) versus 56.9% (± 16.5%), P = .014. The LMO2 protein in 6 clones in which the 1.2-kb cHS4 insulator flanked the LTR-GFP cassette was reduced compared with control clones with only the LTR-GFP cassette (Figure 5B) with some variability from clone to clone probably reflecting the semiquantitative nature of Western blot analysis. The relative GFP expression, measured by mean fluorescence intensity (MFI), in clones having the insulated LTR-GFP cassette was approximately 3- to 5-fold higher than the average MFI in clones having the original LTR-GFP cassette (Figure 5C). The mean, SD, and P values for comparison to the control for week 3 are as follows: (1) 1.2 kb versus control; 253 (± 88) versus 72 (± 16), P < .001 and (2) 2 × 250 bp versus control, 246 (± 139) versus 72 (± 16), P = .016. All other weekly comparisons were also statistically different. These data indicate that addition of the cHS4 insulator element to the LTRs of a SIN vector increases transgene expression, as has been reported previously,13,14,17 and diminishes but does not eliminate the potential for an internal oncoretroviral LTR to activate a proto-oncogene.
Attenuation of LMO2 activation by a chromatin insulator (cHS4). (A) The R-6 clone was transduced with scAAV-Cre and transfected with a plasmid containing the LTR-GFP cassette flanked by 1.2-kb cHS4 or 2 × 250-bp core insulator fragments, and single cell clones were identified in which cassette exchange had occurred. The relative LMO2 expression in those clones was compared by qRT-PCR. Both flanking insulators reduced LMO2 expression 3- to 15-fold over that observed in clones having the noninsulated LTR-GFP cassette. (B) Western blot confirmed reduction of LMO2 protein expression by flanking insulator fragments in 6 clones in which the 1.2-kb cHS4 insulator flanked the LTR-GFP cassette relative to the G-25 and R-6 clones. Jurkat cells served as a negative control. (C) The relative GFP expression in clones with or without insulator elements flanking the LTR-GFP cassette was compared by FACS each week for 4 weeks. The average MFI of GFP expression in each clone set is represented by a horizontal bar. LTR-GFP cassette (○), LTR-GFP cassette flanked by a 1.2-kb insulator fragment (●), LTR-GFP cassette flanked by 2 × 250-bp core insulator fragments (▵).
Attenuation of LMO2 activation by a chromatin insulator (cHS4). (A) The R-6 clone was transduced with scAAV-Cre and transfected with a plasmid containing the LTR-GFP cassette flanked by 1.2-kb cHS4 or 2 × 250-bp core insulator fragments, and single cell clones were identified in which cassette exchange had occurred. The relative LMO2 expression in those clones was compared by qRT-PCR. Both flanking insulators reduced LMO2 expression 3- to 15-fold over that observed in clones having the noninsulated LTR-GFP cassette. (B) Western blot confirmed reduction of LMO2 protein expression by flanking insulator fragments in 6 clones in which the 1.2-kb cHS4 insulator flanked the LTR-GFP cassette relative to the G-25 and R-6 clones. Jurkat cells served as a negative control. (C) The relative GFP expression in clones with or without insulator elements flanking the LTR-GFP cassette was compared by FACS each week for 4 weeks. The average MFI of GFP expression in each clone set is represented by a horizontal bar. LTR-GFP cassette (○), LTR-GFP cassette flanked by a 1.2-kb insulator fragment (●), LTR-GFP cassette flanked by 2 × 250-bp core insulator fragments (▵).
Insulator elements can block gene activation by an enhancer when they are placed between it and a promoter.15,16 To test whether this is the case for the LTR insertion in LMO2, single insulator (1.2 kb) constructs were also investigated. Successful exchange was achieved as verified by Southern blot analysis (Figures S3 and S4). When placed between the LTR and the distal promoter, the insulator had little effect on LMO2 expression, whereas a significant reduction occurred when the insulator was placed between the LTR and the proximal promoter (Figure S5). Note that clones having the insulator between the distal promoter and the expression cassette exhibited an increase in GFP MFI over the noninsulated LTR-GFP cassette (294 ± 66 vs 81 ± 24, P = .028), whereas those clones with the insulator element between the proximal promoter and the expression cassette had a lower MFI (25 ± 1, P = .035) than control, suggesting that epigenetic mechanisms that tended to reduce transgene expression originated 5′ to the insertion site (Figure S5).
Globin regulatory elements do not activate LMO2 in lymphoid cells
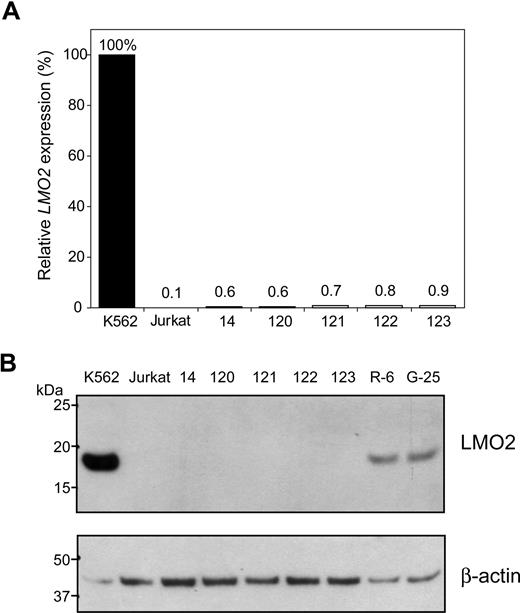
Our laboratory has developed a globin gene lentiviral vector in which the γ-globin coding sequences are under the control of the minimal β-globin promoter and linked to elements from the β-globin LCR.18 A cassette was constructed in which the globin regulatory elements from this vector (mLARβΔγV5) driving GFP coding sequences were flanked by loxP and loxP511 sites. Directional cassette exchange was accomplished in the R-6 clone. In an early experiment, single cells initially expressing GFP were sorted, and a total of 225 clones were recovered. Only one clone showed the expected band in PCR screening and it was GFP negative. In subsequent experiments, transfected cells were randomly sorted. Four additional clones were identified by PCR screening, and Southern blot analysis confirmed that all 5 clones had undergone cassette exchange (Figure S6). Overall, the process of targeted clone identification was relatively inefficient (0.5%-1.4%) because clones had to be screened randomly rather than based on marker expression. The globin regulatory elements failed to activate the LMO2 gene at the mRNA level (Figure 6A) or protein level (Figure 6B). None of these clones expressed GFP by flow analysis (data not shown). These results are consistent with the lineage specificity of globin LCR19 and imply that a lentiviral vector containing a therapeutic globin gene cassette with a SIN design will have a more favorable safety profile with respect to proto-oncogene activation in lymphoid cells than the oncoretroviral vectors with intact LTRs used in the X-SCID4,–6 and CGD7 trials.
LMO2 is not activated by globin regulatory elements. A cassette containing the GFP coding sequences under the control of β-globin gene promoter along with the human β-globin LCR was used in the cassette exchange reaction in R-6 cells. Five clones were confirmed as having undergone the predicted exchange. Activation of the LMO2 gene in all 5 clones was minimal, if any, as determined by qRT-PCR (A) or Western blot analysis (B).
LMO2 is not activated by globin regulatory elements. A cassette containing the GFP coding sequences under the control of β-globin gene promoter along with the human β-globin LCR was used in the cassette exchange reaction in R-6 cells. Five clones were confirmed as having undergone the predicted exchange. Activation of the LMO2 gene in all 5 clones was minimal, if any, as determined by qRT-PCR (A) or Western blot analysis (B).
Vector comparison
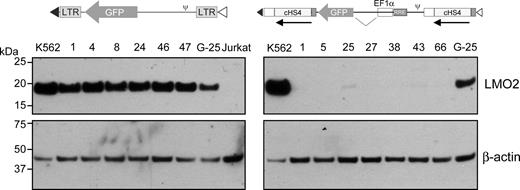
Our experimental system was designed to facilitate the comparison of various vectors with respect to proto-oncogene activation. Accordingly, we compared a conventional γ-retroviral vector having 2 LTRs, similar to that used in clinical trials to date, to a vector engineered to enhance safety by use of a SIN design with insulator fragments in the LTRs and an internal cellular promoter driving the transgene. The 2 LTR γ-retroviral vector activated LMO2 to 87.5% (± 29.9%, n = 5) the level of expression of the gene in K562 cells, whereas the lentiviral vector with insulators gave a value of 1.7% (± 0.7%, n = 7), which is at the level of sensitivity of the qRT-PCR assay. The Western blot analysis shown in Figure 7 confirms strong LMO2 activation by the 2 LTR vectors and minimal activation in only 2 of the clones by the SIN vector with insulators.
Comparison of a conventional γ-retroviral vector to a SIN lentiviral vector with insulator elements in the LTRs and an internal cellular promoter. The organization of the 2 vectors is shown. Each of the 6 clones having the conventional γ-retroviral vector inserted into the LMO2 gene gave a strong signal on Western blot analysis, whereas only 2 of the 7 clones having the insulated lentiviral vector gave a weak signal on Western blot analysis.
Comparison of a conventional γ-retroviral vector to a SIN lentiviral vector with insulator elements in the LTRs and an internal cellular promoter. The organization of the 2 vectors is shown. Each of the 6 clones having the conventional γ-retroviral vector inserted into the LMO2 gene gave a strong signal on Western blot analysis, whereas only 2 of the 7 clones having the insulated lentiviral vector gave a weak signal on Western blot analysis.
Discussion
The focus of our work has been the development of a system capable of recreating specific retroviral insertions into or near proto-oncogenes to evaluate the potential for specific regulatory elements to activate the targeted gene. We have used AAV vector–mediated homologous targeting to introduce an expression cassette24 followed by Cre recombinase–mediated, directional cassette exchange31 to evaluate the potential of specific regulatory elements to attenuate or eliminate activation of the LMO2 proto-oncogene. We targeted LMO2 because it has been implicated in the pathogenesis of leukemia in 2 patients with X-SCID as a consequence of insertional mutagenesis by the therapeutic transgene vector.8 Our results indicate that a single LTR can activate LMO2 while driving expression of the GFP marker gene. Flanking insulator (cHS4) elements attenuated, but did not eliminate, LMO2 activation while simultaneously increasing expression of the marker gene. Of interest with respect to the development of gene therapy for hemoglobin disorders,18 an expression cassette containing elements from the globin LCR and the β-globin promoter was unable to activate LMO2 in lymphoid cells. Furthermore, we showed that a lentiviral vector with a design intended to enhance safety did not activate the LMO2 gene.
The cHS4 insulator has been well characterized both for its enhancer blocking and barrier function activities.16 Although these activities have been predominantly defined in erythroid cells, we observed evidence of both activities in human lymphoid cells. However, neither the full 1.2-kb insulator fragment or tandem repeats of the 250-bp core fragment were sufficient to completely block the enhancer function of the oncoretroviral LTR. The single insulator element may be more useful in vector design because the 2 × 250-bp cassette is prone to rearrange during reverse transcription (P. Hargrove and D.A.P., unpublished observations, November 2006). The clone-to-clone variability in LMO2 activity (Figure 5) is consistent with the inherent variability of epigenetic mechanisms by which insulators act.15 These results are also consistent with our recently published data which indicate that flanking cHS4 insulator elements of a SIN lentiviral vector having an internal oncoretroviral LTR diminish but do not eliminate clonal dominance during short-term culture of transduced Jurkat cells.38 Our new system provides an assay to evaluate the enhancer-blocking activities of the multiple insulators that have been identified throughout the human genome39 with the goal of identifying the most potent for eliminating proto-oncogene activation by a gene therapy vector.
Lentiviral vectors containing globin regulatory elements are likely to be safer for gene therapy applications because the LTRs of SIN vectors lack viral regulatory elements and the globin regulatory elements necessary for high-level globin transgene expression are demonstrably tissue specific.19 Indeed, the globin lentiviral vector currently in a clinical trial, which also includes the 2 × 250-bp cHS4 insulator element in the LTRs, did not accelerate tumorigenesis in a tumor-prone mouse model after transplantation of transduced stem cells.40 Our results are consistent with this observation in that globin regulatory elements failed to activate the LMO2 proto-oncogene in lymphoid cells. However, we have found evidence of distant gene activation by integrated globin vectors in clonal erythroid spleen colonies.41
The use of a SIN design whereby both LTRs of the integrated vector genome lack regulatory elements with an internal promoter has been proposed as a strategy to improve the safety profile of oncoretroviral and lentiviral vectors. Indeed, recently published data using a modified primary bone marrow immortalization assay suggest a significant reduction in immortalization potential by use of a SIN vector having a single internal LTR identical to the 2 LTRs of the conventional control vector.12,41 However, immortalization was still observed with the SIN vector which is consistent with our observation that a single LTR is capable of proto-oncogene activation. Of potential relevance to this issue is the demonstration that a lentiviral vector with an internal cellular phosphoglycerol kinase promoter did not enhance the rate of tumorigenesis in a tumor-prone mouse strain, whereas a conventional γ-retroviral vector accelerated the rate of tumor formation as a function of age.42 The results could be interpreted to suggest that a cellular promoter has a lower potential to activate proto-oncogenes in the context of a SIN vector compared with an oncoretroviral LTR.
We have now tested this prediction in our experimental system. As shown by the results in Figure 7, the newly designed lentiviral vector with features intended to enhance safety had a dramatically lower potential for LMO2 activation than a 2 LTR γ-retroviral vector analogous to that used in all clinical trials to date. Although limited to a single insertion into one of many possible proto-oncogenes, these results lead us to predict that the use of vectors having these safety features in future clinical trials will significantly diminish the risk of insertional mutagenesis, clonal dominance, and leukemic transformation.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the Vector Core at St Jude Children's Research Hospital for providing AAV vectors. We also thank Janice Riberdy (PhD, Hematology) for help in sorting Jurkat cells and Pat Streich for manuscript preparation.
This work was supported in part by The Assisi Foundation of Memphis, by the US National Heart, Lung, and Blood Institute (NHLBI) Program (project grant P01 HL53749; A.W. N.), and by the American Lebanese Syrian Associated Charities. B.Y.R. is a recipient of the Sumara Fellowship from St Jude Children's Research Hospital. The plasmid, pMC-CreN, which encodes Cre-recombinase, was a gift from Fred Alt (Boston Children's Hospital, Boston, MA).
National Institutes of Health
Authorship
Contribution: B.Y.R., D.M.B., D.A.P., and A.W.N. designed the studies; B.Y.R., M.V.E.-G., and A.W.N. conducted experiments; J.T.G. provided essential reagents; and B.Y.R. and A.W.N. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Arthur W. Nienhuis, St Jude Children's Research Hospital, 332 N Lauderdale Street, Memphis, TN 38105; e-mail: arthur.nienhuis@stjude.org.