Differentiating human embryonic stem cells (HESCs) represent an experimental platform for establishing the relationships between the earliest lineages that emerge during human development. Here we report the targeted insertion in HESCs of sequences encoding green fluorescent protein (GFP) into the locus of MIXL1, a gene transiently expressed in the primitive streak during embryogenesis.1,2 GFP fluorescence in MIXL1GFP/w HESCs differentiated in the presence of BMP4 reported the expression of MIXL1, permitting the identification of viable human primitive streak-like cells. The use of GFP as a reporter for MIXL1 combined with cell surface staining for platelet-derived growth factor receptor alpha (PDGFRα) enabled the isolation of a cell population that was highly enriched in primitive hematopoietic precursors, the earliest derivatives of the primitive streak. These experiments demonstrate the utility of MIXL1GFP/w HESCs for analyzing the previously inaccessible events surrounding the development of human primitive streak-like cells and their subsequent commitment to hematopoiesis.
Introduction
In vertebrate species, a prerequisite for the development of the primary germ layers is the commitment of primitive ectoderm (epiblast) cells to gastrulation.3,,–6 In mammalian embryos, this process is accompanied by the formation of the primitive streak, a morphologic structure initiating at the prospective embryonic posterior.6,–8 In the mouse epiblast, cells ingressing through the streak emerge as either definitive endoderm or mesoderm, the latter including the progenitors of the hematopoietic system.9
In the mouse, primitive streak cells are marked by expression of the transcription factor Mixl11,2 and mouse embryos deficient in Mixl1 display multiple defects in the formation of mesodermal and endodermal derived structures.10 Consistent with this, more recent studies have confirmed that Mixl1 expression marks precursors of both mesoderm11 and endoderm.12 These latter studies took advantage of embryonic stem cells (ESCs) or mice in which one Mixl1 allele had been replaced by sequences encoding green fluorescent protein (GFP), facilitating the identification and isolation of viable GFP+ (Mixl1+) primitive streak-like cells. Analysis of Mixl1GFP/w mouse ESCs showed that a GFP+ (Mixl1+) population present at differentiation days 3 and 4 contained hematopoietic precursors,11 supporting previous data indicating that, in mouse embryos, such precursors arise directly from the primitive streak.13 The majority of progenitors at this time were hemangioblasts, precursors with both hematopoietic and endothelial potential which, in the embryo, contribute to the primary vascular plexus and primitive erythropoiesis of the yolk sac. Thus, these progenitors as well as lineage restricted primitive erythroid precursors represent the first differentiated mesodermal derivatives that arise after the onset of gastrulation at embryonic day (E) 6.5.9
Because of the scarcity of examples, events surrounding gastrulation in the human have largely been inferred from comparative embryology,8 a situation that has led to uncertainty surrounding the relationship between the first mesodermal like cells (mesoblasts) documented from postovulation day 13 onward, the appearance of hematopoietic cells, and the overt manifestation of the primitive streak, a structure that is first visible in embryos representing embryonic day 15 (E15).8,14,–16
Differentiating human embryonic stem cells (HESCs) represent an experimental platform for dissecting the relationship between specific lineages and the early differentiation events surrounding formation of the primary germ layers. To examine the correlation between mesoderm formation in the human and the emergence of hematopoietic precursors, we targeted sequences encoding GFP to the MIXL1 locus using homologous recombination. We demonstrate that GFP fluorescence faithfully reported expression of the endogenous MIXL1 gene and that a mesodermal cell population defined by coexpression of GFP (MIXL1) and the platelet-derived growth factor receptor alpha (PDGFRα) was highly enriched in primitive hematopoietic precursors, the earliest derivatives of the primitive streak.
Methods
Generation and identification of targeted MIXL1GFP/w HESCs
The MIXL1 targeting vector comprised a 9.4 kb 5′ homology arm, GFP, loxP flanked PGK-promoter-neomycin resistance gene and a 1.9 kb 3′ homology arm. The homology arms were derived from previously described genomic clones of the human MIXL1 locus2 and spanned sequences from a PacI site situated 9466 bp 5′ of the ATG to an HpaI site located 2242 bp 3′ of the ATG. The vector was digested with the restriction enzymes PacI and NotI before electroporation into HESCs as described elsewhere.17 HESC clones with a putative targeted MIXL1 allele were identified using a polymerase chain reaction (PCR)-based screening strategy using the primer, Neo4, in conjunction with MIXL1 ScreenRev (primer b in Figure 1A), a primer located immediately 3′ of the genomic sequences encompassed by the targeting vector (Table S1, primer details, available on the Blood website; see the Supplemental Materials link at the top of the online article). Using this criterion, several clones were identified in which the vector appeared to be correctly integrated into the MIXL1 locus. Two independent HES3-derived MIXL1GFPNeoR/w clones (clone 7 and clone 10) were expanded and transiently transfected with a pEFBOS-cre-IRESPuro vector using Fugene 6 transfection reagent according to the manufacturer's instructions (Roche, Mannheim, Germany). This vector was designed to express a single transcript encoding cre recombinase and puromycin resistance, the latter translated from an internal ribosomal entry site (IRES). At 24 to 32 hours after transfection, cells were selected in 2 μg/mL puromycin for 2 days and subsequently allowed to form colonies for a further 7 days. Several colonies representing each primary clone were picked and screened for the loss of the neomycin resistance cassette and for the absence of the cre expression plasmid using a PCR based approach (Table S1, primer details and PCR conditions). Southern blot analysis was performed as described elsewhere.18 The 5′ external DNA probe included a mixture of fragments corresponding to human genomic sequences flanked by primer pairs listed in Table S1. The GFP probe used to verify the presence of a single integration event encompassed the coding sequences of EGFP (Invitrogen). The DNA fragment generated by PCR using the primers GFP1 (primer a in Figure 1A) and MIXL1 3′ probe #1 was cloned and sequenced to establish that the 3′ arm of the targeting vector had correctly integrated into the locus.
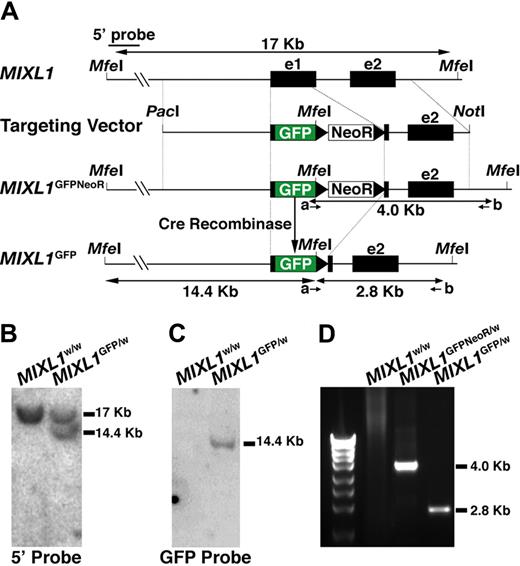
Targeting of GFP to the MIXL1 locus in HESCs. (A) Structure of the gene targeting vector used to insert sequences encoding GFP into exon 1 of the MIXL1 locus using homologous recombination. PacI and NotI are restriction enzyme sites used to linearize the vector before electroporation. NeoR is the PGKNeo cassette encoding G418 resistance, flanked by loxP sites (black triangles). The positions of MfeI sites used to map the structure of the modified locus are shown, as are the position of primers (a, b) used to identify correctly targeted clones. (B) Southern blot analysis of MfeI digested genomic DNA shows that a 5′ external probe detects a fragment of 17 kb representing the endogenous locus from both parental HES3 cells and HESCs with a targeted MIXL1 locus. An additional fragment of 14.4 kb is also detected in the genetically modified cells, representing the distance from the 5′ external MfeI site to the 3′ end of GFP. (C) Probing this same DNA with GFP sequences indicates that these cells contain a single copy of the GFP gene consistent with a single genetic modification at the MIXL1 locus. (D) The integrity of sequences 3′ of the GFP gene was validated using a PCR-based approach (with primers a and b) to amplify DNA representing the junction of the targeting vector with the chromosomal DNA. Sequence analysis of this fragment confirmed that the relationship between the vector DNA and adjacent chromosomal sequences were as expected (data not shown).
Targeting of GFP to the MIXL1 locus in HESCs. (A) Structure of the gene targeting vector used to insert sequences encoding GFP into exon 1 of the MIXL1 locus using homologous recombination. PacI and NotI are restriction enzyme sites used to linearize the vector before electroporation. NeoR is the PGKNeo cassette encoding G418 resistance, flanked by loxP sites (black triangles). The positions of MfeI sites used to map the structure of the modified locus are shown, as are the position of primers (a, b) used to identify correctly targeted clones. (B) Southern blot analysis of MfeI digested genomic DNA shows that a 5′ external probe detects a fragment of 17 kb representing the endogenous locus from both parental HES3 cells and HESCs with a targeted MIXL1 locus. An additional fragment of 14.4 kb is also detected in the genetically modified cells, representing the distance from the 5′ external MfeI site to the 3′ end of GFP. (C) Probing this same DNA with GFP sequences indicates that these cells contain a single copy of the GFP gene consistent with a single genetic modification at the MIXL1 locus. (D) The integrity of sequences 3′ of the GFP gene was validated using a PCR-based approach (with primers a and b) to amplify DNA representing the junction of the targeting vector with the chromosomal DNA. Sequence analysis of this fragment confirmed that the relationship between the vector DNA and adjacent chromosomal sequences were as expected (data not shown).
Cell culture and differentiation
HESC lines were passaged as reported elsewhere17,19 and differentiated as spin EBs according to previously established protocols.20 Serum free differentiation medium (SFM),20 containing recombinant human albumin and 0.05% to 0.25% polyvinylalcohol in some experiments was supplemented with the following growth factors at the concentrations indicated: 10 to 100 ng/mL BMP4, 50 ng/mL Activin A (R&D Systems, Minneapolis, MN), 50 to 100 ng/mL FGF2, 10 to 50 ng/mL VEGF, 20 to 100 ng/mL SCF, 30 ng/mL IL3, 30 ng/mL IL-6, 30 ng/mL Tpo, 3 U/mL erythropoietin (PeproTech, Haifa, Israel). EBs were dissociated using either 0.25% w/v Trypsin-EDTA (Invitrogen, Carlsbad, CA) or TrypLE Select (Invitrogen). Preparation and analysis of methylcellulose cultures and cytocentrifuge preparations were conducted according to Ng et al.20 Karyotype analysis and teratoma assays were performed as described previously.19
Flow cytometric analysis
Intracellular flow cytometry with anti-Mixl1 and anti-Oct4 antibodies was performed as described previously.21 For analysis and sorting of live cells, HESCs were dissociated to give a single-cell suspension and labeled with antibodies as described previously.20 The antibodies used in this study were phycoerythrin (PE)-conjugated mouse anti–human CD34 (BD Biosciences), mouse anti–human E-CADHERIN (Zymed, South San Francisco, CA), mouse anti–human PDGFRα (BD Biosciences), mouse anti–human CD43 (BD Biosciences), PE-conjugated mouse anti–human CD45 (BD Biosciences), allophycocyanin (APC)-conjugated mouse anti–human glycophorin A (BD Biosciences), and mouse anti–human Tra-1-60 (Chemicon, Temecula, CA). Unconjugated primary antibodies were detected with either PE or APC-conjugated goat anti–mouse IgG (BD Biosciences). Flow cytometry gates were set using control cells (HES3) and MIXL1GFP/w HESCs labeled with the appropriate isotype control antibody. Alternatively, gates were set relative to MIXL1GFP/w HESCs differentiated in FGF2, which do not express MIXL1. Single-cell cloning was performed using the single-cell deposition function of a FACSaria FACS station to place single-cells into each well of 10 96-well trays preseeded with irradiated primary mouse embryonic fibroblasts (PMEFs) and containing HESC culture media.19 For reaggregation/reculture experiments, cells obtained from flow cytometric sorting were aggregated using the spin EB protocol (104/well), in SFM supplemented with 30 ng/mL BMP4, 30 ng/mL VEGF, and 40 ng/mL SCF.
Immunofluorescence analysis
Dissociated cells from day (d)4 EBs were resuspended in SFM and allowed to adhere to a poly-L-lysine–coated glass slide for 20 minutes. The cells were fixed and permeabilized with Cytofix/Cytoperm (BD Biosciences) and labeled with the anti-MIXL1 antibody, 6-G2,21 or a rat IgG control antibody. Primary antibodies were detected with a Texas Red conjugated goat anti–rat immunoglobulin G (IgG; Jackson ImmunoResearch, West Grove, PA) and confocal images were captured using an Olympus FluoView 1000 (Olympus, Tokyo, Japan).
Gene-expression analysis
RNA was prepared using RNAeasy according to manufacturer's instructions (QIAGEN, Valencia, CA). RNA samples were reverse transcribed and normalized as described previously.20 PCR was performed under standard conditions (30 cycles of 94°C, 20 seconds; 60°C, 30 seconds; 68°C, 60 seconds) using the primer sets listed in Table S2. For PCR with PAX6 specific primers, an annealing temperature of 55°C was used. Real-time PCR was performed using Taqman gene expression probes and expression levels calculated as described previously.22
Results
The MIXL1-GFP targeting vector (Figure 1A) was electroporated into HESCs and G418 resistant colonies isolated as described elsewhere.17 Correctly targeted clones were identified using a PCR-based strategy with the primers indicated (Table S1). After removal of the G418 resistance cassette (see “Methods”), the structural integrity of the targeted locus was verified by Southern blot analysis (Figure 1B,C) and sequencing of the PCR product representing the 3′ junction between the vector and flanking genomic DNA (Figure 1D and data not shown). In addition, one MIXL1GFP/w HESC line was cloned by single-cell deposition using a flow cytometer into 96-well trays (cloning efficiency of ∼ 5%). The parental line and subclones were phenotypically indistinguishable (data not shown). MIXL1GFP/w HESCs had normal karyotypes, formed teratomas, and expressed markers of undifferentiated HESCs (Figure S1 and data not shown).
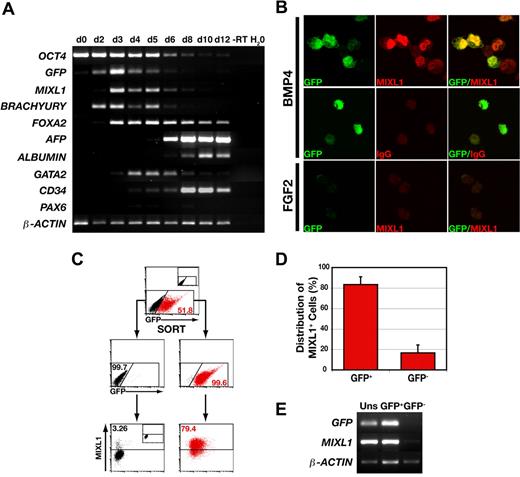
To examine the temporal association between expression of GFP and the endogenous MIXL1 allele, the gene expression profile of MIXL1GFP/w HESCs differentiated in response to BMP4 was analyzed over a 12-day period using RT-PCR (Figure 2A). The expression profile of GFPtranscripts mirrored that of MIXL1, and as previously reported,20 MIXL1 expression was contemporaneous with that of BRACHYURY, a transcription factor also present in the primitive streak. The decline in the level of expression of these primitive streak genes between days 6 to 8, overlapped with the expression of mesodermal (GATA2, CD34) and endodermal (FOXA2, ALPHA FETOPROTEIN, ALBUMIN) genes. This transient wave of MIXL1 expression was consistent with our previous data demonstrating the kinetics of differentiation using the “spin embryoid body” (spin EB) system in SFM supplemented with BMP4.20 Generally, these kinetics paralleled those reported by others for BMP4 dependent HESC differentiation in serum free media, with transient expression of BRACHYURY and a gradual diminution in the levels of OCT4.23 Also, in agreement with the studies of Kennedy et al,23 substantial CD34 expression was not observed until after day 4 (Figure 2A). As evidenced by the weak expression of PAX6, BMP4 did not promote neurectodermal differentiation.
GFP marks MIXL1+ cells during the early stages of HESC differentiation. (A) PCR analysis indicates that GFP expression mirrors the wave of expression of endogenous MIXL1. This analysis also shows the progressive down-regulation of the stem-cell marker, OCT4, the transient expression of the primitive streak genes, MIXL1 and BRACHYURY, and activation of genes expressed in endodermal (FOXA2, AFP-alpha fetoprotein, ALBUMIN) and mesodermal (GATA2, CD34) cell types. – RT indicates − reverse transcriptase. This panel is a composite of images for individual ethidium bromide-stained agarose gels for each set of genes. (B) Immunofluorescent images of day 4 MIXL1GFP/w HESCs differentiated in SFM in the presence of either 30 ng/mL BMP4 (top and middle panels) or 100 ng/mL FGF2 (bottom panel). The bottom panel shows that MIXL1GFP/w HESCs differentiated in FGF2 expressed neither GFP nor MIXL1. (C) Sorting and reanalysis experiments examining the relationship between expression of GFP and MIXL1 protein by flow cytometric analysis. The top panel shows the profile of GFP expressing cells in day 4 MIXL1GFP/w EBs. The division between GFP+ (red) and GFP− (black) fractions was based on gates set using MIXL1w/w (HES3) control EBs (inset). The middle panel shows the reanalysis of the sorted populations with the distribution of GFP+ cells and GFP− cells indicated. Endogenous MIXL1 protein (bottom panels), as determined by intracellular flow cytometry with an anti-MIXL1 antibody, is largely restricted to GFP+ cells and excluded from the GFP− cells. The position of gates for intracellular flow cytometry were set with MIXL1GFP/w day 4 EBs differentiated in SFM containing 100 ng/mL FGF2 and stained with the anti-MIXL1 antibody (inset, bottom panel). (D) Graphic representation showing the distribution of MIXL1+ cells between the GFP+ and GFP− sorted populations from 4 separate experiments using 2 independent MIXL1GFP/w HESC lines (P < .001). Error bars represent the SEM. Calculations are shown in Table S3A-C. (E) PCR analysis of the GFP-sorted fractions from C showing both GFP and MIXL1 transcripts are essentially restricted to the GFP+ population.
GFP marks MIXL1+ cells during the early stages of HESC differentiation. (A) PCR analysis indicates that GFP expression mirrors the wave of expression of endogenous MIXL1. This analysis also shows the progressive down-regulation of the stem-cell marker, OCT4, the transient expression of the primitive streak genes, MIXL1 and BRACHYURY, and activation of genes expressed in endodermal (FOXA2, AFP-alpha fetoprotein, ALBUMIN) and mesodermal (GATA2, CD34) cell types. – RT indicates − reverse transcriptase. This panel is a composite of images for individual ethidium bromide-stained agarose gels for each set of genes. (B) Immunofluorescent images of day 4 MIXL1GFP/w HESCs differentiated in SFM in the presence of either 30 ng/mL BMP4 (top and middle panels) or 100 ng/mL FGF2 (bottom panel). The bottom panel shows that MIXL1GFP/w HESCs differentiated in FGF2 expressed neither GFP nor MIXL1. (C) Sorting and reanalysis experiments examining the relationship between expression of GFP and MIXL1 protein by flow cytometric analysis. The top panel shows the profile of GFP expressing cells in day 4 MIXL1GFP/w EBs. The division between GFP+ (red) and GFP− (black) fractions was based on gates set using MIXL1w/w (HES3) control EBs (inset). The middle panel shows the reanalysis of the sorted populations with the distribution of GFP+ cells and GFP− cells indicated. Endogenous MIXL1 protein (bottom panels), as determined by intracellular flow cytometry with an anti-MIXL1 antibody, is largely restricted to GFP+ cells and excluded from the GFP− cells. The position of gates for intracellular flow cytometry were set with MIXL1GFP/w day 4 EBs differentiated in SFM containing 100 ng/mL FGF2 and stained with the anti-MIXL1 antibody (inset, bottom panel). (D) Graphic representation showing the distribution of MIXL1+ cells between the GFP+ and GFP− sorted populations from 4 separate experiments using 2 independent MIXL1GFP/w HESC lines (P < .001). Error bars represent the SEM. Calculations are shown in Table S3A-C. (E) PCR analysis of the GFP-sorted fractions from C showing both GFP and MIXL1 transcripts are essentially restricted to the GFP+ population.
Immunofluorescence analysis of MIXL1GFP/w HESCs differentiated for 4 days in BMP4 revealed a correlation between GFP expression and MIXLl protein. In the example shown, 5 of 6 intact cells coexpress MIXL1 and GFP (Figure 2B top panels). Specific localized staining was not observed in GFP+ cells labeled with an isotype control antibody (middle panels), nor in the uniformly GFP− cells derived from FGF2 differentiated cultures that were labeled with anti-MIXL1 antibodies (bottom panels).
To further document the relationship between MIXL1 expression and GFP, MIXL1GFP/w HESCs were differentiated for 4 days and flow cytometrically purified GFP+ and GFP− fractions analyzed by intracellular flow cytometry using MIXL1 antibodies (Figure 2C). This analysis demonstrated that, at day 4, cells expressing MIXL1 protein were enriched in the GFP+ fraction and that MIXL1 was essentially absent from the GFP− fraction. The concordance between presence of MIXL1 protein and GFP expression at day 4 or day 5 was such that, on average, 83.3 (± 7.7, mean ± SEM, n = 4) of the MIXL1+ cells resided in the GFP+ fraction (Table S3, primary data and explanation of frequency calculations). This correlation was also reflected in the distribution of MIXL1 and GFP transcripts between the sorted populations (Figures 2E, S2A). At later times, GFP expression persisted beyond the point when MIXL1 protein levels had substantially diminished (Figure S2B), suggesting that the half-life of GFP, which is greater than 20 hours,24 exceeds that of MIXL1. In this regard, expression of GFP also functioned as a lineage tracer, identifying cells that had previously passed through a stage of being MIXL1+.
Consistent with previous studies where the expression of MIXL1 has been analyzed,22 GFP expression was absolutely dependent on the inclusion of an inducing growth factor, in this case BMP4 (Figure 3A). This analysis revealed a wave of GFP expression that mirrored that of MIXL1 and GFP RNA (Figure 2A). In this experiment, the frequency of GFP+ cells was maximal at days 4 to 6 (∼ 50%) and declined gradually thereafter, becoming negligible by d12. Although the precise time point of peak GFP induction varied between individual experiments (related to the concentration or activity of the BMP4 preparation used as an inducer), the transient nature of this expression was a common feature of all differentiations performed with both independently derived MIXL1GFP/w HESC lines (Figure S3).
BMP4 induces a wave of GFP expression in differentiating MIXL1GFP/w HESCs. (A) Time course of GFP expression determined by flow cytometric analysis of differentiating MIXL1GFP/w HESCs shows the transient appearance of mesendodermal progenitors in response to 50 ng/mL BMP4. Note the absence of GFP+ cells in cultures differentiated in SFM alone (top panel). The proportion of GFP+ cells for each time point is indicated. (B) GFP expression is also induced in day 5 MIXL1GFP/w EBs formed in SFM supplemented with either 100 ng/mL BMP4 or 50 ng/mL Activin A (Act A) but not with 100 ng/mL FGF2. BF indicates bright field. (C) Flow cytometric analysis substantiates the capacity of BMP4 and Activin A, but not FGF2, to induce GFP expression in MIXL1GFP/w EBs (left panels). Intracellular flow cytometric analysis of endogenous MIXL1 protein shows that GFP mirrors MIXL1 expression (right panels). (D) Time course analysis of GFP (MIXL1), E-CAD, and PDGFRα expression in MIXL1GFP/w HESCs differentiated in SFM containing BMP4, VEGF, and SCF, shows the transit of cells from undifferentiated E-CAD+GFP−PDGFRα− HESCs toward GFP+PDGFRα+ mesoderm. This latter population gives rise to CD34+ cells (bottom panel). As expected, cells differentiated in FGF2 did not express GFP or PDGFRα. Region statistics relating to each population were calculated as described in Figure S4A. GFP+ cells are shown in red in all plots. The proportion of the population expressing E-CAD or PDGFRα is shown above the line, and percentages of negative cells are shown below the line. In all instances, the proportion of cells expressing GFP is shown in red. CD34+ cells are boxed with the GFP+ and GFP− portions indicated with red and black type, respectively.
BMP4 induces a wave of GFP expression in differentiating MIXL1GFP/w HESCs. (A) Time course of GFP expression determined by flow cytometric analysis of differentiating MIXL1GFP/w HESCs shows the transient appearance of mesendodermal progenitors in response to 50 ng/mL BMP4. Note the absence of GFP+ cells in cultures differentiated in SFM alone (top panel). The proportion of GFP+ cells for each time point is indicated. (B) GFP expression is also induced in day 5 MIXL1GFP/w EBs formed in SFM supplemented with either 100 ng/mL BMP4 or 50 ng/mL Activin A (Act A) but not with 100 ng/mL FGF2. BF indicates bright field. (C) Flow cytometric analysis substantiates the capacity of BMP4 and Activin A, but not FGF2, to induce GFP expression in MIXL1GFP/w EBs (left panels). Intracellular flow cytometric analysis of endogenous MIXL1 protein shows that GFP mirrors MIXL1 expression (right panels). (D) Time course analysis of GFP (MIXL1), E-CAD, and PDGFRα expression in MIXL1GFP/w HESCs differentiated in SFM containing BMP4, VEGF, and SCF, shows the transit of cells from undifferentiated E-CAD+GFP−PDGFRα− HESCs toward GFP+PDGFRα+ mesoderm. This latter population gives rise to CD34+ cells (bottom panel). As expected, cells differentiated in FGF2 did not express GFP or PDGFRα. Region statistics relating to each population were calculated as described in Figure S4A. GFP+ cells are shown in red in all plots. The proportion of the population expressing E-CAD or PDGFRα is shown above the line, and percentages of negative cells are shown below the line. In all instances, the proportion of cells expressing GFP is shown in red. CD34+ cells are boxed with the GFP+ and GFP− portions indicated with red and black type, respectively.
Apart from the more prolonged kinetics of differentiation, these results are analogous to those we obtained with mouse Mixl1GFP/w ESCs differentiated under similar conditions.11 The enhanced viability associated with BMP4 treatment seen in differentiating mouse ESCs11 also occurred with HESCs (data not shown). In addition to BMP4, GFP expression was also induced by activin A (Figure 3B), consistent with previous studies showing that MIXL1 expression is up-regulated during activin-induced endodermal differentiation.25 Similarly, in line with its known role in blocking HESC differentiation,26 FGF2 failed to induce GFP expression (Figure 3B). In all cases, the induction of GFP correlated with the expression of MIXL1 protein, as determined by intracellular flow cytometry (Figure 3C). The results of experiments in which MIXL1GFP/w HESCs were differentiated in the presence of BMP4, activin A, or FGF2 paralleled those obtained with mouse Mixl1GFP/w ESCs.11 The similar behavior of mouse and human ESCs differentiated under comparable conditions suggests that the signaling pathways underlying MIXL1 induction are probably to be conserved between these 2 species.
During mouse embryogenesis, epiblast cells entering the primitive streak retain E-cadherin (E-cad) before passing through a transition during which E-cad expression is down regulated and expression of early mesodermal genes, including the receptors for vascular endothelial growth factor (Flk1) and platelet-derived growth factor (PDRFRα) are increased.27,28 We have previously shown that, in differentiating mouse Mixl1GFP/w ESCs, GFP expression spanned the interval during which cells transit from E-cad+ epiblast to E-cad− Flk1+ mesoderm.11 We sought to determine whether a similar transition period could be discerned during the course of HESC differentiation. However, because we and others have observed that the human homologue of Flk1, KDR, is expressed on undifferentiated HESCs23 (E.S.N., E.G.S., and A.G.E., unpublished data, January 2004), we instead examined the relationship between the expression of GFP, E-CAD, and PDGFRα (Figure 3D). Flow cytometric analysis of MIXL1GFP/w HESCs differentiated in BMP4, VEGF, and SCF (BVS) showed that the GFP+ cells were regularly seen from day 3. These cells were E-CAD+, and some already expressed PDGFRα. By day 5, the proportion of E-CAD+ cells started to fall while the frequency of cells expressing PDGFRα had increased to approximately 40% (Figure S4A, details of how percentages were calculated). In this experiment, the peak in the frequency of GFP+ cells occurred at day 7 (33%), and the majority of these were PDGFRα+. At day 10, the proportion of E-CAD+ cells had fallen to below 40% and almost all of the GFP+ cells were also PDRFRα+. Previous studies in our laboratory showed that cell-surface expression of the hematopoietic and endothelial marker, CD34, was virtually absent prior to differentiation at day 4 (Figure S4B).22 Examination of differentiating MIXL1GFP/w HESCs from day 8 onwards revealed a population of CD34+ cells, which appeared to derive from the preexisting GFP+ PDGFRα+ fraction, as evidenced by the coexpression of these markers on a proportion of CD34+ cells (Figure 3D and data not shown).
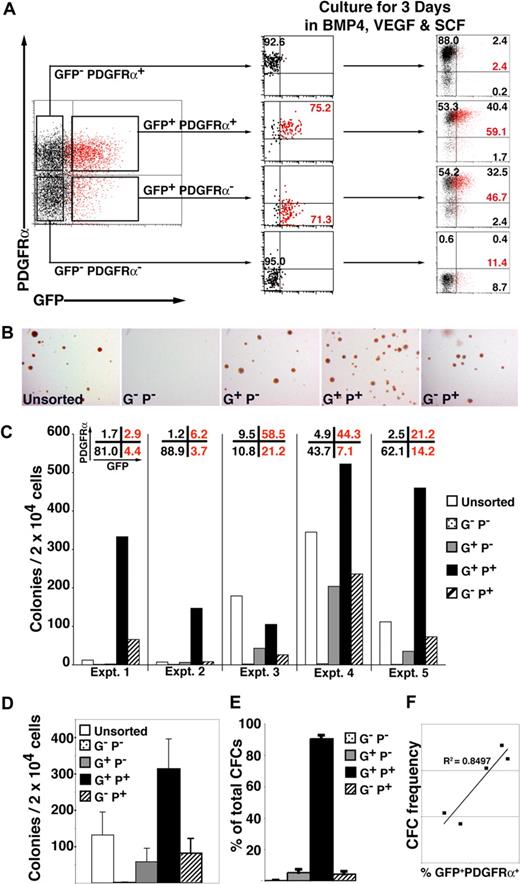
To examine the relationship between GFP and PDGFRα expression, we isolated cells expressing combinations of these markers by flow cytometric sorting and recultured each fraction for 3 days in SFM supplemented with BVS (Figures 4A, S5, gating strategy). This analysis demonstrated that GFP+ PDGFRα+ cells arose from the GFP+ PDGFRα− population and implied that cells sequentially acquired the expression of GFP (MIXL1), PDGFRα, and subsequently CD34, reflecting the sequential commitment to primitive streak, mesoderm, and hematopoietic development. PDGFRα expression was quite stable, as evidenced by the high proportion of cells (∼ 85%-95%) from both GFP− PDGFRα+ and GFP+ PDGFRα+ populations that retained PDGFRα expression 3 days after sorting and reculturing. Conversely, GFP expression was only retained in approximately 40% to 60% of GFP+ PDGFRα− or GFP+ PDGFRα+ cells over this same time period. These experiments also showed that GFP− PDGFRα+ cells did not give rise to GFP+ cells and that little new GFP or PDGFRα expression was induced after day 4 from GFP− PDGFRα− (Figure 4A).
Hematopoietic progenitors are enriched in the MIXL1+PDGFRα+ fraction of differentiating HESCs. (A) Cell sorting and reculture experiment showing that day 4 GFP+ PDGFRα− cells give rise to GFP+PDGFRα+ cells when cultured in SFM supplemented with BVS. Some GFP+ cells can still develop from the GFP− PDGFRα− fraction, but they remained PDGFRα− at the time points examined. The fraction of MIXL1+ (red) cells is indicated, as is the proportion of cells in each quadrant (black text). (B) Low-power images of methylcellulose cultures showing that compared with the GFP+ PDGFRα+ fraction, approximately 5-fold fewer CFCs were present in cell populations expressing only GFP (G+P−) or PDGFRα (G−P+) (original magnification, ×12). (C) Results from 5 independent experiments (experiments 1–5) confirmed that the frequency of blast (Bl)-CFCs was highest in the GFP+(MIXL1+) PDGFRα+ fraction. The proportion of each subpopulation present at the time of sorting is shown across the top of the panel. (D) Summary of data in panel C showing that on average the GFP+ PDGFRα+ fraction contained approximately 300 Bl-CFCs/20 000 cells plated. (E) Summary of the blast colony distribution based on the data in panel C showing that approximately 90% of Bl-CFCs are present in the GFP+ PDGFRα+ fraction. (F) Graph showing that the frequency Bl-CFCs at day 4 correlates with proportion of cells that are GFP+ PDGFRα+. Error bars represent SEM. G−P−, GFP− PDGFRα−; G+P−, GFP+PDGFRα−; G+P+, GFP+ PDGFRα+; G−P+, GFP− PDGFRα+.
Hematopoietic progenitors are enriched in the MIXL1+PDGFRα+ fraction of differentiating HESCs. (A) Cell sorting and reculture experiment showing that day 4 GFP+ PDGFRα− cells give rise to GFP+PDGFRα+ cells when cultured in SFM supplemented with BVS. Some GFP+ cells can still develop from the GFP− PDGFRα− fraction, but they remained PDGFRα− at the time points examined. The fraction of MIXL1+ (red) cells is indicated, as is the proportion of cells in each quadrant (black text). (B) Low-power images of methylcellulose cultures showing that compared with the GFP+ PDGFRα+ fraction, approximately 5-fold fewer CFCs were present in cell populations expressing only GFP (G+P−) or PDGFRα (G−P+) (original magnification, ×12). (C) Results from 5 independent experiments (experiments 1–5) confirmed that the frequency of blast (Bl)-CFCs was highest in the GFP+(MIXL1+) PDGFRα+ fraction. The proportion of each subpopulation present at the time of sorting is shown across the top of the panel. (D) Summary of data in panel C showing that on average the GFP+ PDGFRα+ fraction contained approximately 300 Bl-CFCs/20 000 cells plated. (E) Summary of the blast colony distribution based on the data in panel C showing that approximately 90% of Bl-CFCs are present in the GFP+ PDGFRα+ fraction. (F) Graph showing that the frequency Bl-CFCs at day 4 correlates with proportion of cells that are GFP+ PDGFRα+. Error bars represent SEM. G−P−, GFP− PDGFRα−; G+P−, GFP+PDGFRα−; G+P+, GFP+ PDGFRα+; G−P+, GFP− PDGFRα+.
Quantitative PCR analysis confirmed that the mesendodermal and hematopoietic markers BRACHYURY, GSC, FOXA2, GATA2, and RUNX1 were expressed in day 4 EBs (Figure S6A). The distribution of transcripts representing these markers varied between the different GFP and PDGFRα subfractions, perhaps reflecting difficulties in analyzing dynamic populations. Nevertheless, it was generally found that expression of these markers was higher in the GFP+ PDGFR+ than in the GFP− PDGFR− fractions (Figure S6B), as would be predicted from the hypothesis that GFP+ PDGFR+ cells represent primitive streak and nascent mesoderm while the GFP− PDGFR− cells include less differentiated cells or ectodermal precursors.
We and others have observed that the earliest hematopoietic precursors that develop from HESCs, termed blast colony forming cells (Bl-CFCs), are seen after 3 to 4 days of differentiation23,29 (E.S.N., E.G.S., and A.G.E., unpublished data, May 2006). Experiments with HESCs indicated that some Bl-CFCs have the capacity to form both hematopoietic and endothelial lineages,23,29 indicating this population contains hemangioblasts similar to those identified during the early phases of mouse ESC differentiation and mouse development.13,30 In this regard, Bl-CFCs most probably correspond to progenitors that give rise to hematopoietic cells that have been documented in the yolk sac of the human embryos at approximately embryonic day 15.8,14
In view of previous data showing that Bl-CFCs arose during the early phases of HESC differentiation, we examined the methylcellulose colony forming ability of day 4 subpopulations isolated on the basis of their GFP and PDGFRα expression. In the 5 consecutive experiments shown in Figure 4B,C, the first 2 used a batch of BMP4 with a 3- to 5-fold lower specific activity (batch 1) than the batch used in the last 3 experiments (batch 2). This led to a lower percentage of GFP+ and PDGFRα+ expressing cells in the first 2 experiments that could be correlated with the low frequency of Bl-CFCs in the unsorted day 4 EBs. For example, 7 to 12 Bl-CFCs per 2 × 104 cells were observed in the day 4 EBs differentiated in BMP4 from batch 1, while 112 to 349 Bl-CFCs per 2 × 104 cells were seen in the day 4 EBs cultured in BMP4 from batch 2. Despite these differences, hematopoietic Bl-CFCs were highly enriched in the GFP+ PDGFRα+ fraction in all 5 experiments (Figure 4B-D), demonstrating that, as in the mouse, the earliest human hematopoietic progenitors arose within the primitive streak and nascent mesoderm.13 Although the GFP+ PDGFRα− and GFP− PDGFRα+ populations also contained hematopoietic CFCs, 90.5% plus or minus 2.2% of CFCs were present in the GFP+ PDGFRα+ fraction (Figure 4E; Tables S4Table S5. Sum of calculated colony distribution in each sorted fraction (PDF, 46.6 KB)Table S6. Progenitor yield from sorted fractions compared to CFC yield from unsorted cells (PDF, 13.9 KB)–S7). This conclusion is reinforced by the strong correlation (R2 = 0.8497) between the frequency of Bl-CFCs and the percentage of GFP+ PDGFRα+ cells (Figure 4F). Bl-CFCs were essentially absent from the GFP− PDGFRα− populations.
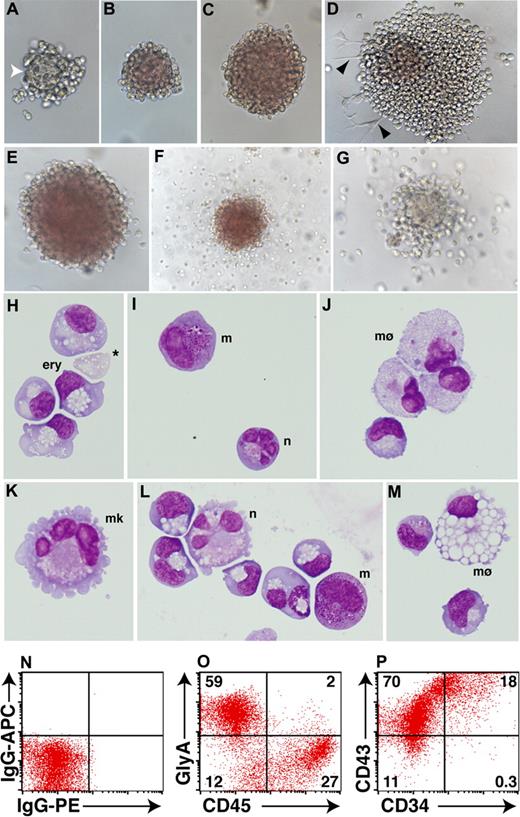
Although the frequency of hematopoietic colonies varied between the different sorted populations, a similar spectrum of colony morphologies was detected in each case. At early stages of blast colony formation, blood cells emerged from a dense core of cells that was morphologically similar to the mesodermal core observed in mouse hemangioblast colonies31 (Figure 5A). In the presence of erythropoietin, developing blast colonies became overtly hemoglobinized (Figure 5B,C). Where colonies contacted the plate surface, hematopoietic cells developed in association with adherent cells with a morphology resembling hemangioblast derived endothelial cells recently reported23,29 (Figure 5D). Although erythroid colonies, occasionally with a halo of migrating myeloid cells, comprised the most frequent colony types (∼ 95%) (Figure 5E,F and data not shown), colonies wholly composed of migrating myeloid cells were also routinely observed (∼ 5%) (Figure 5G).
Blast colonies derived from day EBs contain primitive erythroid and myeloid cells. (A-D) After 8 days of methylcellulose culture, day 4 MIXl1+PDGFRα+ cells gave rise to hematopoietic colonies representing different stages of blast colony maturation. (A) Early stage colonies often contained a dense central core (white arrowhead) with a morphology distinct from the surrounding hematopoietic cells. (B,C) In more mature colonies, this feature was lost as cells within the colony underwent hemoglobinization. (D) Some colonies also contained adherent cells (black arrowheads). (E-G) Colonies arising from the day 4 MIXL1+PDGFRα+ fraction after 11 days of methylcellulose culture displayed phenotypes indicative of erythroid, myeloid, and bipotential progenitors. (H-M) Cytocentrifuge preparations of day 4 colonies after 13 days of methylcellulose confirmed the presence of nucleated primitive erythroid and myeloid cells. Enucleated erythroid cells were also observed (* in panel H) as well as cells with the morphologic appearance of neutrophils (n), megakaryocytes (mk), macrophages (mø), and mast cells (m). Panels K-M are derived from a cytocentrifuge preparation of a single erythroid colony similar to that shown in E. (N-P) Flow cytometric analysis of 15-day methylcellulose cultures showing that the majority of cells express the erythroid marker glycophorin A (GLYA) or CD45. Approximately 90% of cells also express the pan-hematopoietic marker CD43 and, of these, approximately 20% also express CD34.
Blast colonies derived from day EBs contain primitive erythroid and myeloid cells. (A-D) After 8 days of methylcellulose culture, day 4 MIXl1+PDGFRα+ cells gave rise to hematopoietic colonies representing different stages of blast colony maturation. (A) Early stage colonies often contained a dense central core (white arrowhead) with a morphology distinct from the surrounding hematopoietic cells. (B,C) In more mature colonies, this feature was lost as cells within the colony underwent hemoglobinization. (D) Some colonies also contained adherent cells (black arrowheads). (E-G) Colonies arising from the day 4 MIXL1+PDGFRα+ fraction after 11 days of methylcellulose culture displayed phenotypes indicative of erythroid, myeloid, and bipotential progenitors. (H-M) Cytocentrifuge preparations of day 4 colonies after 13 days of methylcellulose confirmed the presence of nucleated primitive erythroid and myeloid cells. Enucleated erythroid cells were also observed (* in panel H) as well as cells with the morphologic appearance of neutrophils (n), megakaryocytes (mk), macrophages (mø), and mast cells (m). Panels K-M are derived from a cytocentrifuge preparation of a single erythroid colony similar to that shown in E. (N-P) Flow cytometric analysis of 15-day methylcellulose cultures showing that the majority of cells express the erythroid marker glycophorin A (GLYA) or CD45. Approximately 90% of cells also express the pan-hematopoietic marker CD43 and, of these, approximately 20% also express CD34.
Examination of May-Grünwald-Giemsa stained cytospin preparations revealed that most colonies were composed of primitive nucleated erythrocytes, although the presence of small numbers of enucleated erythrocytes was frequently observed (Figure 5H). Myeloid colonies contained macrophages, often in combination with mast cells or neutrophils (Figure 5I,J). Many erythroid colonies, even those without an obvious myeloid component (such as Figure 5E), contained macrophages, mast cells, neutrophils, and megakaryocytes at a low frequency, indicating that these CFCs were multipotent (Figure 5K-M). The proportions of cells representing the erythroid and myeloid lineages that were present in day 15 methylcellulose cultures was determined by flow cytometric analysis. As expected, the majority of cells were glycophorin A+ (GLYA+), and a substantial proportion were CD45+ (Figure 5O). Expression of CD45 and GlyA was essentially mutually exclusive, consistent with down-regulation of the former as cells committed to erythroid differentiation (GLYA+). The hematopoietic phenotype of cells present in these methylcellulose cultures was further confirmed by the high frequency of cells that expressed CD43,32 an antigen found on the surface of cells belonging to the erythroid, myeloid, and lymphoid lineages (Figure 5P). A sizable fraction of cells also expressed CD34, suggesting, as well as mature cells representing the erythroid and myeloid lineages, these cultures also contained hematopoietic progenitors (Figure 5P).
Discussion
The in vitro analysis of lineage commitment from differentiating mouse ESCs has been greatly facilitated by the availability of ESC lines containing reporter genes inserted into loci whose expression marks critical developmental milestones. The most reliable method of generating genetically tagged lines is by targeting the reporter gene to the chosen locus using homologous recombination. Descriptions of gene targeting in HESCs have paved the way for the application of this approach to genetic tagging experiments in the human system.33,34 However, these previous reports have used a promoter trapping approach that takes advantage of expression from the target locus,33 or methods that rely on drug resistance resulting from disruption of the targeted gene.34 Because most genes are not amenable to targeting by such approaches, we developed a generic strategy using conventional gene targeting in which the selectable marker is driven from a promoter within the vector and that does not require expression of the target locus in undifferentiated ESCs.17 Using this approach, we have obtained several HESC lines in which sequences encoding a fluorescent protein have been targeted to developmentally significant loci17 (current study; E.G.S. and A.G.E., unpublished results). In a similar fashion to genetically tagged mouse ESCs, these lines are likely to prove useful in dissecting relationships between cell lineages that emerge during the course of HESC differentiation.
In this study, we have used gene targeting to insert sequences encoding GFP into the locus of MIXL1, the human ortholog of a gene expressed in primitive streak cells of other vertebrate species. During the early phases of HESC differentiation, GFP reliably identified cells that expressed MIXL1 protein, indicating that this reporter could be used to isolate cells corresponding to a primitive streak-like stage of human embryogenesis. Because of the long half-life of GFP,24 at later time points GFP fluorescence persisted in cells that had lost MIXL1 expression, enabling a proportion of CD34+ cells to be identified as direct descendants of a preexisting MIXL1+ PDGFRα+ population. This latter property of GFP may prove useful in analyzing the lineage relationships between MIXL1+ cells and cells representing other mesodermal and endodermal derivatives.
Using these genetically tagged MIXL1GFP/w HESCs, we examined the relationship between primitive streak-like MIXL1+ cells and the earliest hematopoietic progenitors, blast colony forming cells (Bl-CFCs).23,29 The results of this analysis provided a concrete example of how fundamental aspects of early ESC differentiation are conserved between mouse and human. In both species, SFM supplemented by BMP4 induces Mixl1+ cells that give rise to a mesoderm-committed subpopulation that harbors progenitors of primitive hematopoiesis.11
In the mouse, Mixl1 expression is localized to the primitive streak and emerging mesendoderm, providing a molecular marker of this process, which spans approximately 3 days, beginning at E6.5 and ending with the generation of mesodermal derivatives within the tail bud of the E9.5 embryo.1,2 Similarly, in differentiating mouse ESCs, Mixl1 expression also spans a 3-day period, from its onset at approximately day 3 and extinction by day 6. Thus, in the mouse, the time interval between the blastocyst stage (E3.5) or undifferentiated ESC (day 0) stages to the onset of Mixl1 expression is similar in the in vivo and in vitro systems. In the human, although a primitive streak has been documented as early as E15, the appearance of extraembryonic mesoderm from E12 suggests that gastrulation may have begun well before it is apparent morphologically. Analysis of early human embryos suggest that mesoderm continues to be generated at least until E19, suggesting that human gastrulation may span up to 7 days.8 Indeed, depending on the growth milieu, the duration of MIXL1 expression in differentiating HESCS, from day 2 up to day 8, translated into GFP expression between day 3 and day 12, and is consistent with the protracted gastrulation stage in humans relative to mice. What is surprising is the relatively brief period between the initiation of differentiation and the onset of MIXL1 expression observed in this study and by others.20,25 If HESCs correspond to the inner cell mass of an E6 embryo, then expression of gastrulation markers would not be expected for around 6 days after the initiation of HESC differentiation. The brevity of this interval may suggest that HESCs represent a cell type that more closely resembles the epiblast than the inner cell mass. Alternatively, our results might suggest that, like mouse, human gastrulation begins around 3 days after implantation. Such a time line would then provide ample opportunity for generation and migration of mesodermal like cells observed in the few examples of early stage human embryos that have been examined.8 Future comparative studies with the human and mouse MIXL1GFP/w ESCs should enable a better understanding of the events surrounding this inaccessible but critical period of human development.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Robyn Mayberry and Kathy Koutsis for provision of HESCs and Andrew Fryga and Darren Ellemor for flow cytometric sorting.
This work was supported by the Australian Stem Cell Center, the Juvenile Diabetes Research Foundation, and the National Health and Medical Research Council of Australia.
Authorship
Contribution: R.P.D., E.S.N., M.C., A.K.M., and K.S. performed the research and analyzed data; R.P.D., A.G.E., and E.G.S. designed the research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Edouard G. Stanley, Monash Immunology and Stem Cell Laboratories, Level 3, Building 75, Monash University, Clayton, Victoria, 3800, Australia; e-mail: ed.stanley@med.monash.edu.au.
References
Supplemental data
Data is shown as CFC/2 × 104 cells plated. The value for each fraction was derived by multiplying the CFC number by the % of cells for each fraction (data in Table S3). The sum of the sorted fractions for each experiment is shown in the first data column.
The sum of the CFC in the sorted fractions (sum of sorted fractions, see Table S4) was compared with the number of CFC/2 × 104 cells from the unsorted cells to calculate the CFC recovery after flow cytometric sorting.
The distribution of CFC found within each sorted fraction was calculated from data shown in Table S4 by dividing the CFC found in each sorted fraction by the sum of CFC in the sorted fractions and expressing the result as a percentage.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal