We have studied the plasma membrane protein phenotype of human culture-amplified and native bone marrow mesenchymal stem cells (BM MSCs). We have found, using microarrays and flow cytometry, that cultured cells express specifically 113 transcripts and 17 proteins that were not detected in hematopoietic cells. These antigens define a lineage-homogenous cell population of mesenchymal cells, clearly distinct from the hematopoietic lineages, and distinguishable from other cultured skeletal mesenchymal cells (periosteal cells and synovial fibroblasts). Among the specific membrane proteins present on cultured MSCs, 9 allowed the isolation from BM mononuclear cells of a minute population of native MSCs. The enrichment in colony-forming units–fibroblasts was low for CD49b, CD90, and CD105, but high for CD73, CD130, CD146, CD200, and integrin alphaV/beta5. In addition, the expression of CD73, CD146, and CD200 was down-regulated in differentiated cells. The new marker CD200, because of its specificity and immunomodulatory properties, deserves further in-depth studies.
Introduction
Bone marrow mesenchymal stem cells (BM MSCs) are defined as adherent culture-amplified cells giving rise, when cultured in appropriate conditions, to adipocytes, osteoblasts, and chondrocytes.1 The phenotype of cultured MSCs remains limited in spite of several decades of study.2,,–5 In addition, there has been only scattered data on the phenotype of the native BM MSCs because different markers have been used for MSC isolation in independent studies.6,,,,,,,,,,–17 In this work we have studied the plasma membrane protein phenotype of cultured cells, screening for molecules not detected in hematopoietic cells. Hierarchical and principal components analyses of the transcripts coding for membrane proteins and distribution of the membrane antigens indicate lineage-homogeneity. Nine of the proteins detected specifically on cultured mesenchymal cells have proven useful to sort native MSCs from BM mononuclear cells (MNCs). This study should help select the optimal marker for BM MSCs and help discriminate MSCs from other stem/progenitor cells present in the bone marrow (hematopoietic stem cells, multipotent adult progenitor cells,18 SSEA-1+ cells,19 etc).
Methods
Approval for these studies was given by the Comité Consultatif de Protection des Personnes participant à la Recherche Biomédicale (CPPRB) for the University Hospital in Tours. Informed consent was obtained in accordance with the Declaration of Helsinki. Details of materials and methods used are in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
MSC culture and in vitro differentiation
Culture of synovial fibroblasts and periosteal cells
Synovial tissues were collected postmortem from normal joints. Periosteal autografts were harvested from mastoids of patients undergoing mastoidectomy. Tissues were digested with collagenase, cells were cultured as described in Document S1.
Flow cytometry
Cells were incubated with conjugated monoclonal antibodies (mAbs) or with purified mAbs (Table S7). Acquisitions were performed on a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA).
Cell sorting and CFU-F assays
CD235a+, CD45+, and CD11b+ hematopoietic fractions were isolated from BM samples using magnetic-assisted cell sorting (MACS; Miltenyi Biotec, Auburn, CA). The remaining cells were labeled with phycoerythrin (PE)-conjugated mAbs and sorted using a MoFlo cell sorter (Dako, Glostrup, Denmark). For fibroblast colony-forming unit (CFU-F) assays, mononuclear cells were seeded at 2000 to 40 000 cells/cm2, and sorted cells at density 100 to 1000 cells/cm2. Colonies were counted after 10 days.
Microarrays
Microarray data are available in the Gene Expression Omnibus (GEO)21 at http://www.ncbi.nlm.nih.gov/geo/ with accession number GSE9894.
RNA was extracted from 6 P1 MSC samples and 3 samples each of CD235a+, CD45+, and CD11b+ cells. Hybridization on HG-U133 Plus 2.0 microarrays (Affymetrix, Santa Clara, CA) was performed according to standards supplied by Affymetrix. Affymetrix GCOS 1.2 software was used to generate DAT, CEL, and EXP files and to process raw data for signal calculation and pairwise chip comparison. Group comparison and gene retrieval was performed using the online database SiPaGene.22 Hierarchical clustering and principal components analyses were performed using Genesis software (http://www.genesis-softwareonline.com/).
In vivo study of bone formation
Micro-macroporous biphasic calcium phosphate (MBCP) ceramic discs loaded with CD200+ culture-amplifed MSCs were implanted subcutaneously in the backs of nude mice.23 Implants were harvested at 4 weeks and examined by histology (Goldner trichrome) and by scanning electron microscopy (back-scattered electrons mode).
Results and discussion
The pattern of membrane protein expression in cultured MSCs
We determined the transcripts encoding outer plasma membrane proteins (excluding Golgi, mitochondriae, etc.) using Affymetrix microarrays in 6 P1 samples. Of 1624 inventoried molecules, we detected 464 transcripts including 98 CDs (Tables S1,S3). Among these transcripts were 118 channel/transporter proteins, 102 cell-cell or cell-matrix adhesion receptors, 57 cytokine receptors and 20 junction molecules (tight, gap, etc.). Most receptors specific for immune cells (T cells, B cells, NK lymphocytes, dendritic cells, and monocytes/macrophages), and receptors for chemokines, hormones, neuromediators, and neuropeptides were not detected (Tables S2 and S4).
Of 114 membrane proteins studied by flow cytometry, we detected 51 proteins (Figure 1A,B, Tables S6,S7), among which were adhesion receptors (integrins, immunoglobulin superfamily members, tetraspanins, etc), diverse receptor tyrosine kinases, and HLA-ABC (but not HLA-DR). Remarkably, antigens expressed at high (relative mean fluorescence intensity [rMFI] ≥ 100), moderate (10 ≤ rMFI < 100) or low (2 ≤ rMFI < 10) levels remained at the same level from one sample to the other.
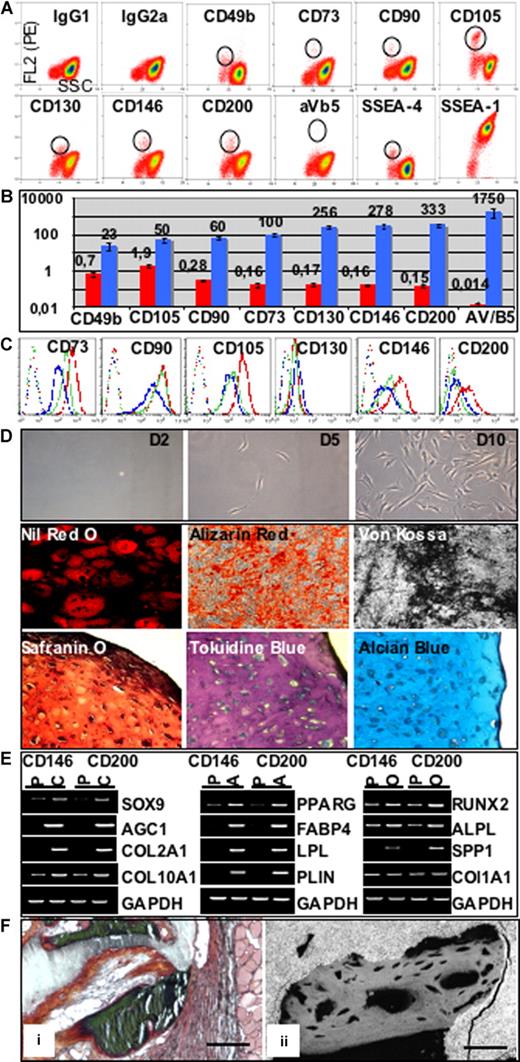
Cultured BM MSCs. (A) Phenotype of P1 cells: positive markers detected using PE-conjugated mAbs. Cells were collected at the end of passage 1 (P1, day 36), after trypsinization of confluent layers. Cells were incubated with PE-conjugated mAbs added at saturating concentration, as indicated in Document S1. (B) Phenotype of P1 cells: positive markers detected using purified unconjugated mAbs. For indirect staining, cells were incubated with the primary mouse antihuman mAbs, then with biotinylated goat anti–mouse Ig Ab, and finally with R-phycoerythrin (RPE)-conjugated streptavidin, as indicated in Document S1. (C) Hierarchical clustering. Hierarchical clustering was performed using a panel of 147 CD transcripts (genes identified as “absent” by GCOS 1.2 in all samples were excluded), on 6 independent samples of P1 MSCs, 3 samples each of CD45+ (CD45), CD11b+ (CD11b), and CD235a+ (GlyA) hematopoietic cells, 3 samples of periosteal cells (POC), and 4 samples of synovial fibroblasts (SFb). Clusters: i: lymphoid cluster including CD2, CD3E, CD3Z, CD8A, CD117, CD122, CD132, CD160, and CD197; ii: erythroid cluster including CD234, CD235a, CD240, CD241, and CD338; iiic: MSC cluster including CD49b, CD51, CD90, CD73, CD105, CD130, CD140a, CD140b, CD146, CD151, CD202b, CD266, CD295, CD325, and CD332; iv: SFb cluster including CD9, CD34, CD42b, CD62P, CD66d, and CD227. (D) Principal components analysis. Analysis was carried out using the same CD transcripts and on the same samples as for hierarchical clustering. Each plotted data point represents a single profile. (E) Specific CD transcripts. Among the 113 transcripts (given in Table S5) specifically expressed in cultured MSCs, 20 are CD membrane antigens. The line plot indicates the mean of signal intensities for the 6 MSC samples [S(MSC)] and the 3 samples each of CD45+ [S (45)], CD11b+ [S(11b)], and CD235a+ [S (235)] cells.
Cultured BM MSCs. (A) Phenotype of P1 cells: positive markers detected using PE-conjugated mAbs. Cells were collected at the end of passage 1 (P1, day 36), after trypsinization of confluent layers. Cells were incubated with PE-conjugated mAbs added at saturating concentration, as indicated in Document S1. (B) Phenotype of P1 cells: positive markers detected using purified unconjugated mAbs. For indirect staining, cells were incubated with the primary mouse antihuman mAbs, then with biotinylated goat anti–mouse Ig Ab, and finally with R-phycoerythrin (RPE)-conjugated streptavidin, as indicated in Document S1. (C) Hierarchical clustering. Hierarchical clustering was performed using a panel of 147 CD transcripts (genes identified as “absent” by GCOS 1.2 in all samples were excluded), on 6 independent samples of P1 MSCs, 3 samples each of CD45+ (CD45), CD11b+ (CD11b), and CD235a+ (GlyA) hematopoietic cells, 3 samples of periosteal cells (POC), and 4 samples of synovial fibroblasts (SFb). Clusters: i: lymphoid cluster including CD2, CD3E, CD3Z, CD8A, CD117, CD122, CD132, CD160, and CD197; ii: erythroid cluster including CD234, CD235a, CD240, CD241, and CD338; iiic: MSC cluster including CD49b, CD51, CD90, CD73, CD105, CD130, CD140a, CD140b, CD146, CD151, CD202b, CD266, CD295, CD325, and CD332; iv: SFb cluster including CD9, CD34, CD42b, CD62P, CD66d, and CD227. (D) Principal components analysis. Analysis was carried out using the same CD transcripts and on the same samples as for hierarchical clustering. Each plotted data point represents a single profile. (E) Specific CD transcripts. Among the 113 transcripts (given in Table S5) specifically expressed in cultured MSCs, 20 are CD membrane antigens. The line plot indicates the mean of signal intensities for the 6 MSC samples [S(MSC)] and the 3 samples each of CD45+ [S (45)], CD11b+ [S(11b)], and CD235a+ [S (235)] cells.
We then investigated whether levels of detected mRNAs were correlated with those of proteins by plotting the rMFI given by flow cytometry studies versus the signal intensity (S) determined by microarrays. The highly significant (r2 = 0.453, P < .001) regression plot of log(rMFI) versus log(S) indicated a parabolic relationship.
The population of cultured BM MSCs is clearly distinct from those of hematopoietic cells
A panel of 147 CDs was selected (genes identified as “absent” by GCOS 1.2 in all samples were excluded). Hierarchical clustering indicated that the population of cultured MSCs belonged to a cluster clearly distinct from populations of hematopoietic cells (Figure 1C). Moreover, the population of cultured MSCs did not contain discernible cell subsets and was distinguishable from other skeletal, but non–BM-derived, cultured mesenchymal cells, that is, periosteal cells (POCs) and synovial fibroblasts (SFbs). The first 3 components of the principal components analysis (PCA) confirmed the distinctions (Figure 1D): hematopoietic cells were discriminated from mesenchymal cells according to the x-axis, while y- and z-axes allowed us to distinguish the different hematopoietic (CD45+, CD11b+, and CD235a+) and mesenchymal (MSC, POC, and SFb) populations.
Of the 464 transcripts (including 98 CDs) expressed in cultured MSCs, the 113 (including 20 CDs) that were not detected in hematopoietic cells (except for 4 transcripts detected at low level) were defined as specific (Table S5). Of the 20 specific CDs (Figure 1E), 17 could be studied at the protein level and were expressed at the plasma membrane: CD49b, CD90, CD73, CD105, CD130, CD140a, CD140b, CD146, CD151, CD200, CD202b, CD266, CD295, CD325, CD332, and integrin alphaV(CD51)/beta5(ITGB5). Five of these (CD146, CD200, CD295, CD325, CD332) discriminated also at the transcript level MSCs from POCs and SFbs.
Our data demonstrate that cultured cells constitute a lineage-homogenous cell population. Lineage specificity is defined by a set of markers estimated on the MSC population as a whole. The mesenchymal lineage was clearly distinct from the hematopoietic lineage because (1) 24.5% (113/464) of the transcripts for membrane protein antigens were detected specifically on cultured MSCs, and (2) hierarchical clustering and PCA showed that the different hematopoietic cell populations segregated apart from cultured MSCs and showed no major gene expression variability from one MSC sample to the other. Moreover, clustering also discriminated MSCs from other non–BM-derived, cultured skeletal mesenchymal cells. Lineage-homogeneity is a population characteristic that does not preclude clonal heterogeneity within the population. Clonal heterogeneity has been evidenced by the study of CFU-Fs whose differentiation potential, albeit variable, remains restricted to the mesenchymal lineages.1,24,25
For cell therapy it is essential to transplant a well-defined and lineage-homogenous cell population, as obtained in our study. Our standardized protocol has been upscaled to provide sufficient cells with known phenotype and differentiation potential for therapeutic administration. Using this protocol, a randomized trial for prevention of acute graft-versus-host disease is in progress, including 12 patients thus far.
Analysis of membrane proteins on bone marrow mononuclear cells allows definition of the cell population of origin
We hypothesized that some of the specific membrane antigens present on cultured MSCs would define the BM mesenchymal cell population of origin containing the CFU-Fs. Of the 17 CD markers specific for cultured MSCs, 9 (53%) reproducibly allowed us to determine a minute population of BM mononuclear cells containing the CFU-Fs: CD49b, CD73, CD90, CD105, CD130, CD146, CD200, and integrin alphaV/beta5 (Figure 2A). The percentage of BM cells recovered varied from 1.9% (CD105) to 0.014% (alphaV/beta5) of the total MNCs, indicating that some of the antibodies, such as CD49b and CD105, selected a population encompassing, but not being restricted to, native MSCs (Figure 2B). The enrichment in CFU-Fs was low (23- to 60-fold) for CD49b, CD105, and CD90; high (100- to 333-fold) for CD73, CD130, CD146, and CD200; and very high (1750) but also highly variable for alphaV/beta 5 (Figure 2B).
Native BM MSCs. (A) Flow cytometric sorting of native cells. BM mononuclear cells were prepared and labeled as indicated in Document S1. The sorting gate was determined on the FL2 vs SSC as shown. For SSEA-1, no specific cell subset corresponding to the sorting gate was identified. The percentages (mean ± SEM, n = 7) of BM cells recovered from the total MNCs was (highest to lowest) 1.9% (± 0,4%; CD105), 0.7% (± 0.2%; CD49b), 0.28% (± 0.03%; CD90), 0.17% (± 0.025%; CD130), 0.16% (± 0.035%; CD73), 0.16% (± 0.015%; CD146), 0.15% (± 0.04%; CD200), and 0.014% (± 0.004%; integrin alphaV/beta5). (B) Cell recovery and enrichment in CFU-Fs in the sorted fractions. Red bars indicate cell recovery (percentage of cells recovered in the sorted fraction). Blue bars indicate enrichment in CFU-Fs (cloning efficiency in sorted cells related to that in total mononuclear cells before sorting). Mean values are indicated on top of each bar; error bars represent SEM (n = 4). AV/B5 indicates integrin alphaV/beta5. (C) Protein expression after differentiation. The pattern of protein expression was studied by flow cytometry before differentiation of culture-amplified BM MSCs at passage 1 (continuous red line) and 10 days after induction in osteogenic (continuous green line) and adipogenic (continuous blue line) media (1 representative experiment of 3). Discontinuous lines indicate irrelevant isotype controls. Notice the decrease in expression for CD73, CD105, CD146 and CD200; for CD90 the expression declined only after adipogenic induction; for CD130 there was no decrease. (D) CFU-Fs from CD200+ cells. CD200+ sorted cells (n = 6) were cultured in alpha-MEM plus 10% FCS plus 1 ng/mL bFGF plus supplements as indicated in Document S1. Cultures were screened at days 2, 5, and 10. CFU-Fs were counted at day 10. CFU-Fs could not be grown from CD200− cells. Similar results were obtained for the different sortings using antibodies indicated in panel A. (E) In vitro adipocytic, osteoblastic, and chondrocytic differentiation of CD200+ cells: histochemical markers. P1 confluent layers obtained from CD200+ cells were trypsinized and cells were seeded in differentiation media as indicated in Document S1. Adipocytic differentiation was assessed after 14 days by revealing the presence of cells containing large Nile Red O+ intracytoplasmic vesicles. Osteoblastic differentiation was assessed after 21 days by revealing the presence of von Kossa+ and Alizarin Red+ mineralized areas. Chondrocytic differentiation was assessed after 21 days by revealing the presence in the micropellets of cartilage-specific glycosaminoglycans stained by Safranin O, Toluidine Blue and Alcian Blue. Similar results were obtained for CD146+ cells (data not shown). Experiments were performed in duplicate. Micrographs were acquired with a Leica Microsystems microscope fitted with 10×/0.22 or 20×/0.30 objectives, a Nikon digital camera (DMX1200F; Nikon, Champigny-sur-Marne, France), and Nikon AXT-1 acquisition software (v2.63). (F) In vitro adipocytic, osteoblastic and chondrocytic differentiation of CD200+ cells: molecular markers. RNA was extracted from CD200+ cells cultured in proliferation medium (P) or differentiated into adipocytes (A), osteoblasts (O) and chondrocytes (C). Experiments were performed in parallel on CD200+ and CD146+ cells. RT-PCRs were performed using primers specific for C: transcription factor SOX-9 (SOX9), aggrecan core protein (AGC1), collagen 2, alpha1 chain (COL2A1), collagen 10, alpha1 chain (COL10A1), A: peroxisome proliferator-activated receptor gamma (PPARG), fatty acid-binding protein (FABP4), lipoprotein lipase (LPL), perilipin (PLIN), O: Runt-related transcription factor 2 (RUNX2), alkaline phosphatase (ALPL), osteopontin (SPP1), collagen 1, alpha1 chain (COL1A1). Housekeeping gene analyzed was glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Experiments were performed in duplicate. (G) In vivo ectopic bone formation by CD200+ cells. CD200+ BM MSCs were cultured in proliferation medium (Passage 1) before loading on MBCP ceramic discs that were implanted subcutaneously in nude mice. Mice were killed after 4 weeks. Ceramic discs implanted subcutaneously without cells served as negative controls. (i) Histology picture (Goldner trichrome stain). Bone is stained green; ceramic has a shadowy white appearance. Bar = 50 μm. (ii) Back-scattered electrons mode (BSEM) picture. Mineralized bone is gray with typical osteocyte lacunae, ceramic is white and nonmineralized tissue is black. Bar = 50 μm. Micrograph i was acquired with a Zeiss Axioplan 2 light microscope (Carl Zeiss, Oberkochen, Germany) fitted with a 40× objective, a Kappa OX-40 CDD camera, and Kappa imageBase software (Kappa Opto-electronics, Gleichen, Germany). Micrograph ii was acquired with a scanning microscope with backscattered electron mode (SEM, LEO1450VP, Germany).
Native BM MSCs. (A) Flow cytometric sorting of native cells. BM mononuclear cells were prepared and labeled as indicated in Document S1. The sorting gate was determined on the FL2 vs SSC as shown. For SSEA-1, no specific cell subset corresponding to the sorting gate was identified. The percentages (mean ± SEM, n = 7) of BM cells recovered from the total MNCs was (highest to lowest) 1.9% (± 0,4%; CD105), 0.7% (± 0.2%; CD49b), 0.28% (± 0.03%; CD90), 0.17% (± 0.025%; CD130), 0.16% (± 0.035%; CD73), 0.16% (± 0.015%; CD146), 0.15% (± 0.04%; CD200), and 0.014% (± 0.004%; integrin alphaV/beta5). (B) Cell recovery and enrichment in CFU-Fs in the sorted fractions. Red bars indicate cell recovery (percentage of cells recovered in the sorted fraction). Blue bars indicate enrichment in CFU-Fs (cloning efficiency in sorted cells related to that in total mononuclear cells before sorting). Mean values are indicated on top of each bar; error bars represent SEM (n = 4). AV/B5 indicates integrin alphaV/beta5. (C) Protein expression after differentiation. The pattern of protein expression was studied by flow cytometry before differentiation of culture-amplified BM MSCs at passage 1 (continuous red line) and 10 days after induction in osteogenic (continuous green line) and adipogenic (continuous blue line) media (1 representative experiment of 3). Discontinuous lines indicate irrelevant isotype controls. Notice the decrease in expression for CD73, CD105, CD146 and CD200; for CD90 the expression declined only after adipogenic induction; for CD130 there was no decrease. (D) CFU-Fs from CD200+ cells. CD200+ sorted cells (n = 6) were cultured in alpha-MEM plus 10% FCS plus 1 ng/mL bFGF plus supplements as indicated in Document S1. Cultures were screened at days 2, 5, and 10. CFU-Fs were counted at day 10. CFU-Fs could not be grown from CD200− cells. Similar results were obtained for the different sortings using antibodies indicated in panel A. (E) In vitro adipocytic, osteoblastic, and chondrocytic differentiation of CD200+ cells: histochemical markers. P1 confluent layers obtained from CD200+ cells were trypsinized and cells were seeded in differentiation media as indicated in Document S1. Adipocytic differentiation was assessed after 14 days by revealing the presence of cells containing large Nile Red O+ intracytoplasmic vesicles. Osteoblastic differentiation was assessed after 21 days by revealing the presence of von Kossa+ and Alizarin Red+ mineralized areas. Chondrocytic differentiation was assessed after 21 days by revealing the presence in the micropellets of cartilage-specific glycosaminoglycans stained by Safranin O, Toluidine Blue and Alcian Blue. Similar results were obtained for CD146+ cells (data not shown). Experiments were performed in duplicate. Micrographs were acquired with a Leica Microsystems microscope fitted with 10×/0.22 or 20×/0.30 objectives, a Nikon digital camera (DMX1200F; Nikon, Champigny-sur-Marne, France), and Nikon AXT-1 acquisition software (v2.63). (F) In vitro adipocytic, osteoblastic and chondrocytic differentiation of CD200+ cells: molecular markers. RNA was extracted from CD200+ cells cultured in proliferation medium (P) or differentiated into adipocytes (A), osteoblasts (O) and chondrocytes (C). Experiments were performed in parallel on CD200+ and CD146+ cells. RT-PCRs were performed using primers specific for C: transcription factor SOX-9 (SOX9), aggrecan core protein (AGC1), collagen 2, alpha1 chain (COL2A1), collagen 10, alpha1 chain (COL10A1), A: peroxisome proliferator-activated receptor gamma (PPARG), fatty acid-binding protein (FABP4), lipoprotein lipase (LPL), perilipin (PLIN), O: Runt-related transcription factor 2 (RUNX2), alkaline phosphatase (ALPL), osteopontin (SPP1), collagen 1, alpha1 chain (COL1A1). Housekeeping gene analyzed was glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Experiments were performed in duplicate. (G) In vivo ectopic bone formation by CD200+ cells. CD200+ BM MSCs were cultured in proliferation medium (Passage 1) before loading on MBCP ceramic discs that were implanted subcutaneously in nude mice. Mice were killed after 4 weeks. Ceramic discs implanted subcutaneously without cells served as negative controls. (i) Histology picture (Goldner trichrome stain). Bone is stained green; ceramic has a shadowy white appearance. Bar = 50 μm. (ii) Back-scattered electrons mode (BSEM) picture. Mineralized bone is gray with typical osteocyte lacunae, ceramic is white and nonmineralized tissue is black. Bar = 50 μm. Micrograph i was acquired with a Zeiss Axioplan 2 light microscope (Carl Zeiss, Oberkochen, Germany) fitted with a 40× objective, a Kappa OX-40 CDD camera, and Kappa imageBase software (Kappa Opto-electronics, Gleichen, Germany). Micrograph ii was acquired with a scanning microscope with backscattered electron mode (SEM, LEO1450VP, Germany).
Finally, after adipogenic and osteogenic induction, the protein expression of CD49b, CD73, CD105, CD146, and CD200 was decreased, if not collapsed (Figure 2C). After chondrogenic induction, the mRNA expression (evaluated by quantitative RT-PCR) of these markers was also clearly decreased and was completely abolished for CD200 (not shown).
According to these results, CD200, a new marker for MSCs not expressed on bone marrow hematopoietic cells in healthy individuals (this report and Moreaux et al26 ), appeared to be one of the most efficient markers to reproducibly purify native MSCs. The in vitro adipogenic, osteogenic, and chondrogenic potential of CD200+ cells was similar to that observed for cells separated by adherence or according to CD146 expression (Figure 2D,E). CD200+ MSCs also generated ectopic bone in vivo in nude mice (Figure 2F). Whether MSCs exert their immunosuppressive activity through CD200, a known immunomodulatory molecule, has to be studied.
In conclusion, we have established the extensive and specific membrane phenotype of culture-amplified BM MSCs and started to identify several specific surface markers to sort native MSCs. Many other specific candidates remain to be tested (Table S5). Nonprotein markers such as SSEA-4 or GD214,17 have also to be included. For transplantation, the advantage of native MSCs might reside not so much in better differentiation and proliferation potential as in improved homing capacity to different tissues.27
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to Jorge Domenenech and members of the laboratory for their helpful discussions, Julien Gaillard and Anja Wachtel for technical assistance, and Christelle Gauthier for secretarial help. We thank BioRetis (Berlin, Germany) for technical support in data analysis and sharing.
This work was supported by the European Community (Key action 1.2.4-3 Integrated Project Genostem, contract No. 503161), by a grant from Inserm (Contract ProA No. A04069FS), and by the national genome research network in Germany (German Federal Ministry of Education and Research, grant 01GS0413).
Authorship
Contribution: B.D. devised the experiments and carried out the cell cultures and flow cytometric analyses with the help of N.G. and J.R. T.H. devised and carried out the microarray experiments. Y.L.V. and D.K. performed the flow cytometry sorting experiments. P.L. performed the experiments for ectopic bone formation in vivo. C.J. contributed to the overall methodology. P.R. and L.S. provided the bone marrow samples and contributed to the overall methodology. P.C. analyzed the microarray data and directed the work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Bruno Delorme, Laboratoire d'Hématopoièse, Faculté de médecine, Batiment Dutrochet, 10 Bvd Tonnellé, Tours 37032, France, e-mail: delorme@med.univ-tours.fr.

![Figure 1. Cultured BM MSCs. (A) Phenotype of P1 cells: positive markers detected using PE-conjugated mAbs. Cells were collected at the end of passage 1 (P1, day 36), after trypsinization of confluent layers. Cells were incubated with PE-conjugated mAbs added at saturating concentration, as indicated in Document S1. (B) Phenotype of P1 cells: positive markers detected using purified unconjugated mAbs. For indirect staining, cells were incubated with the primary mouse antihuman mAbs, then with biotinylated goat anti–mouse Ig Ab, and finally with R-phycoerythrin (RPE)-conjugated streptavidin, as indicated in Document S1. (C) Hierarchical clustering. Hierarchical clustering was performed using a panel of 147 CD transcripts (genes identified as “absent” by GCOS 1.2 in all samples were excluded), on 6 independent samples of P1 MSCs, 3 samples each of CD45+ (CD45), CD11b+ (CD11b), and CD235a+ (GlyA) hematopoietic cells, 3 samples of periosteal cells (POC), and 4 samples of synovial fibroblasts (SFb). Clusters: i: lymphoid cluster including CD2, CD3E, CD3Z, CD8A, CD117, CD122, CD132, CD160, and CD197; ii: erythroid cluster including CD234, CD235a, CD240, CD241, and CD338; iiic: MSC cluster including CD49b, CD51, CD90, CD73, CD105, CD130, CD140a, CD140b, CD146, CD151, CD202b, CD266, CD295, CD325, and CD332; iv: SFb cluster including CD9, CD34, CD42b, CD62P, CD66d, and CD227. (D) Principal components analysis. Analysis was carried out using the same CD transcripts and on the same samples as for hierarchical clustering. Each plotted data point represents a single profile. (E) Specific CD transcripts. Among the 113 transcripts (given in Table S5) specifically expressed in cultured MSCs, 20 are CD membrane antigens. The line plot indicates the mean of signal intensities for the 6 MSC samples [S(MSC)] and the 3 samples each of CD45+ [S (45)], CD11b+ [S(11b)], and CD235a+ [S (235)] cells.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/111/5/10.1182_blood-2007-07-099622/2/m_zh80050815680001.jpeg?Expires=1769090450&Signature=K16PgIdW9AuKhp~mr4vYQXJZF7alSXFPIasClXgQtiUOmCU-oy5tX7XBachYHytgPek~LQjD-lE~yn2t2LB-hsgf-O-eLNg0bbNcBKcm7EickSNcpWjCnXr-nQUI7xj5YEzl3Mms8L6f33OtvZ7yJA-VTZd-9S4RY4Jv77VC0eQULaTnzhpxyg9mOJs4IemFtwaJweLA8zG1lPwWy-AxZJMGyfKb9n7cxqBSFyQ4ND3sDf0vk6WQTriEZ-TNyVaYbrpuL2cZVuc~dlOMHvbilfEls3TPdqZ9SLBMvPWTUPlXChLZUfthn7Oxp0ODQ5YZWSQ04RGlaq4ZzDautTgaGw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)