Small nuclear U1-RNAs (snRNAs), the spliceosome components selectively recognizing donor splice sites (5′ss), were engineered to restore correct mRNA processing in a cellular model of severe coagulation factor VII (FVII) deficiency, caused by the IVS7 9726 + 5g/a change. Three U1-snRNAs, complementary to the mutated 5′ss (U1 + 5a) or to neighboring sequences were expressed with FVII minigenes in a hepatoma cell line. The U1-snRNAs reduced from 80% to 40% the exon 7 skipping, thus increasing exon definition. The U1 + 5a construct also dramatically increased recognition of the correct 5′ss over the 37-bp downstream cryptic site preferentially activated by the mutation, thus inducing appreciable synthesis of normal transcripts (from barely detectable to 50%). This effect, which was dose-dependent, clearly demonstrated that impaired recognition by the U1-snRNA was the mechanism responsible for FVII deficiency. These findings suggest compensatory U1-snRNAs as therapeutic tools in coagulation factor deficiencies caused by mutations at 5′ss, a frequent cause of severe defects.
Introduction
Changes affecting mRNA processing represent a frequent cause of severe coagulation factor defects1,,,,–6 and of all inherited human diseases.7,–9 Different from gene therapy approaches inserting exogenous sequences that drive the expression of the missing factor, the specific correction of the mRNA processing would maintain the proper transcriptional control at the natural chromosomal environment and restore gene expression.
Modified small nuclear RNAs (snRNAs) have been shown to promote changes in mRNA splicing in cellular and animal models of human diseases. However, these approaches were mainly aimed at inducing skipping of defective exons,10,11 an approach that would not produce functional coagulation proteins, or masking newly generated cryptic exons,12,–14 an uncommon event in coagulation factor defects. Few attempts15,16 have been made to redirect recognition of mutated donor splice site (5′ss) consensus sequences, the most frequent target in human disease genes.8 On the other hand, bleeding disorders would significantly benefit even from low production of the correct mRNA and protein.
As a model to exploit snRNAs for restoration of correct splicing in coagulation factor deficiencies, we chose the 9726 + 5g/a mutation17 in FVII occurring in the donor splice site of intron 7 (IVS7) at the + 5 position, whose substitution has been frequently found to be associated with human diseases.7,–9 The homozygous patients experienced life-threatening hemorrhagic symptoms and require replacement therapy. Interestingly, coinheritance of the FVLeiden allele in a 9726 + 5g/a homozygote produced a small increase in factor Xa and thrombin generation, resulting in a mild bleeding phenotype.18
Through expression of minigenes and use of U1-snRNAs designed to selectively target the mutated site, we provided evidence for partial restoration of factor VII (FVII) mRNA processing impaired by the 9726 + 5g/a change.
Methods
Construction of plasmids
The FVII gene region spanning exon 6 through 8 (nt 8870-10 920)19 was amplified from DNA of a normal subjects (carrying 6 IVS7 repeats as the mutant allele) and from a IVS7 + 5g/a homozygote20 using the 5′-GCATCTTTCTGACTTTTGTT-3′ (forward) and 5′-ATCCGAGTAGCCGGCACAGAACATGTACTC-3′ (reverse) primers. On TA cloning (pGEM-T system, Promega, Madison, WI), the fragments were inserted into the pCDNA3 (Invitrogen, Carlsbad, CA) using EcoRI sites.
U1-snRNAs were prepared as previously described.15 The sequence between BclI and BglII sites of the parental U1-snRNA (WT-U1) was replaced with the following oligonucleotides: 5′-GATCTCAATATCTACCTGGCAGGGGAGATACCAT-3′ and 5′-GATCATGGTATCTCCCCTGCCAGGTAGATATGA-3′ for pU1 + 5a, 5′-GATCTCATCAGCACCGCGCAGGGGAGATACCAT-3′ and 5′-GATCATGGTATCTCCCCTGCGCGGTGCTGAT-GA-3′ for pU1Mut1, and 5′-GATCTCATTCGGACAGGGCAGGGGAGA TACCAT-3′ and 5′-GATCATGGTATCTCCCCTGCCCTGTCCGAATGA-3′ for pU1Mut2.
Transfection and reverse transcription–polymerase chain reaction
Hep3B (human hepatoma) and COS-1 (African green monkey kidney fibroblasts) cells were transiently transfected into 30-mm plates15,20 with3 μg of each minigene vector and, in complementation assays, with 0.5×, 1×, 1.5×, 2×, and 2.5× molar excess of pU1-snRNA vectors by exploiting Lipofectamine 2000 (Invitrogen).
RNA isolation and reverse transcription (RT) were conducted as previously described15,20 using a reverse primer 5′-GCCCTCTAGATGCATGCTCGAGCGG-3′ (8R) in the vector. The following primers were used for polymerase chain reaction (PCR): 5′-AAACCCCAAGGCCGA ATT-3′ (6F), 5′-FAM-cattcaAGGTCCTGTTGTTGGTGAAtG-3′ (7FF, fluorescent, Invitrogen), 5′-ACCCTGATCAACACCATCTGG-3′ (7F), and 5′-GCGATGTCGTGGTTGGT GGT-3′ (8R′) (Figure 1A).
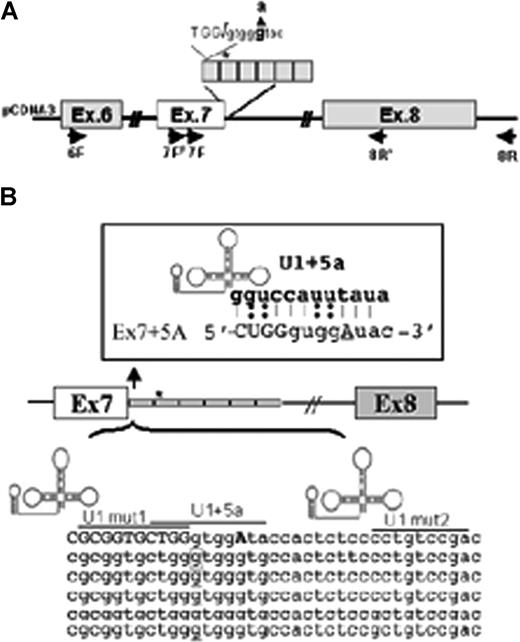
FVII minigene construct and modified U1-snRNAs. (A) Schematic representation of the expression cassette cloned into pCDNA3. The entire region was sequenced to exclude undesired sequence variations. Primers for RT-PCR (6F, 7FF, 7F, 8R′, 8R) are indicated by arrows. The 37bp IVS7 repeats are boxed, and the asterisk represents the 5′ss cryptic site in the first repeat. The 5′ss consensus sequence and the 9726 + 5g/a change (bold) is shown at the top. (B) Schematic representation of the IVS7 repeats and of the U1-snRNA secondary structure. The 5′ tail of U1-snRNAs was engineered to bind to different regions at or near the mutated IVS7 5′ss, and their sequence complementarity is underlined. Complementarity of U1 + 5a is shown at the top. The asterisk and the corresponding boxed nucleotide in the first repeat represent the 5′ss cryptic site.
FVII minigene construct and modified U1-snRNAs. (A) Schematic representation of the expression cassette cloned into pCDNA3. The entire region was sequenced to exclude undesired sequence variations. Primers for RT-PCR (6F, 7FF, 7F, 8R′, 8R) are indicated by arrows. The 37bp IVS7 repeats are boxed, and the asterisk represents the 5′ss cryptic site in the first repeat. The 5′ss consensus sequence and the 9726 + 5g/a change (bold) is shown at the top. (B) Schematic representation of the IVS7 repeats and of the U1-snRNA secondary structure. The 5′ tail of U1-snRNAs was engineered to bind to different regions at or near the mutated IVS7 5′ss, and their sequence complementarity is underlined. Complementarity of U1 + 5a is shown at the top. The asterisk and the corresponding boxed nucleotide in the first repeat represent the 5′ss cryptic site.
Three independent experiments were conducted both in Hep3B and COS-1 cells.
Results and discussion
The 9726 + 5g/a mutation occurs at the +5 position of the consensus sequence of the IVS7 donor splice site (Figure 1A) and maintains the canonical GT dinucleotide, a fundamental feature of eukaryotic introns.21 The IVS7 donor splice site is located in the first of a variable number of highly homologous 37-bp repeats22 containing several potential 5′ss cryptic splice sites, which complicates the selection of the authentic 5′ss by the spliceosome.23 In normal conditions, only the most upstream 5′ss is used, whereas the pseudo-sites remain silent.24
Using a 1-intron minigene system, we previously reported20 that the 9726 + 5g/a substitution induces the selection of the cryptic 5′ss located in the second IVS7 repeat (Figure 1 asterisk), in accordance with the oriented scanning mechanism.24
To explore the effects of the mutation in a more physiologic gene context, we prepared an extended construct with the entire FVII region spanning exon 6 through 8 (Figure 1A). Being FVII of liver origin, the expression experiments were conducted in a human liver cell line (Hep3B) to mimic the proper cell environment. COS-1 cells were also exploited to replicate major findings.
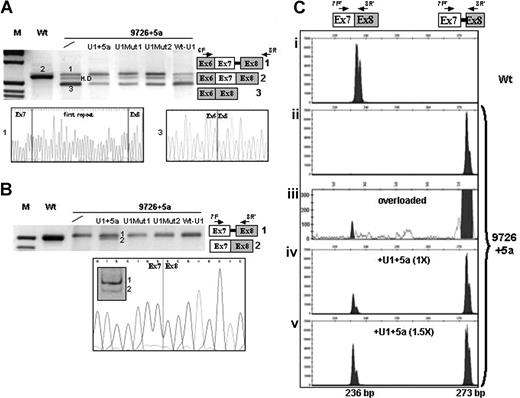
In both cell lines, the 9726 + 5g/a mutation induced exon 7 skipping (Figure 2A transcript 3) and usage of the cryptic 5′ss (transcript 1), predicting FVII mRNA frameshift and premature translation termination. Exon 7 skipping, not detectable with the previous approach,20 accounted for the majority of transcripts (80%), as estimated through semiquantitative analysis of bands. The RT-PCR with primers 7F and 8R′ did not reveal the presence of normal transcripts (Figure 2B). Reduced complementarity and inefficient recognition between the mutated IVS7 5′ss and the 5′-tail of the U1-snRNA belonging to the U1-snRNP complex, a crucial step in exon definition,25 could explain the aberrant splicing induced by the +5g/a change.
Rescue of FVII splicing by modified U1-snRNAs in Hep3B cells. (A,B) Electrophoretic separation on 3% agarose gel, and direct sequencing, of RT-PCR products obtained from total RNA of cells transfected with the expression vectors for normal (Wt) or mutated (9726 + 5a) minigenes, and equimolar concentrations of plasmids encoding for the modified U1-snRNAs. The separation on 12% polyacrylamide gel of the amplified transcripts on U1 + 5a expression and the sequence of the correctly spliced form are also shown in the lower part of panel B. The scheme of transcripts (1, 2, 3) and the primers used (arrows) are depicted on the right side. The additional band observed for the 9726 + 5a mutant (A) corresponded to heteroduplex (H.D), as demonstrated by sequencing. Fragment sizes in panel A were: 637 bp (1), 600 bp (2), and 480 bp (3). Fragment sizes in panel B were: 228 bp (1) and 191 bp (2). M indicates molecular weight marker. (C) Separation on a denaturing capillary system (automated ABI-3100) of fluorescently labeled RT-PCR products obtained from total RNA of cells expressing the Wt (i) or the 9726 + 5a minigene without (ii,iii) and on (iv,v) overexpression of the U1 + 5a snRNA. The scheme of transcripts, and of primers used (arrows), is depicted on the top. Separation of 1 μL of 1:100 diluted RT-PCR reaction is shown, with the exception of panel Ciii) in which 1 μL of the 1:20 dilution was loaded (overloaded). The peaks, which might suggest the presence of a doublet, were referred to as single bands by the automated ABI-3100 system. As expected, the fragment sizes of the normal and aberrant transcripts were 236 bp and 273 bp, respectively.
Rescue of FVII splicing by modified U1-snRNAs in Hep3B cells. (A,B) Electrophoretic separation on 3% agarose gel, and direct sequencing, of RT-PCR products obtained from total RNA of cells transfected with the expression vectors for normal (Wt) or mutated (9726 + 5a) minigenes, and equimolar concentrations of plasmids encoding for the modified U1-snRNAs. The separation on 12% polyacrylamide gel of the amplified transcripts on U1 + 5a expression and the sequence of the correctly spliced form are also shown in the lower part of panel B. The scheme of transcripts (1, 2, 3) and the primers used (arrows) are depicted on the right side. The additional band observed for the 9726 + 5a mutant (A) corresponded to heteroduplex (H.D), as demonstrated by sequencing. Fragment sizes in panel A were: 637 bp (1), 600 bp (2), and 480 bp (3). Fragment sizes in panel B were: 228 bp (1) and 191 bp (2). M indicates molecular weight marker. (C) Separation on a denaturing capillary system (automated ABI-3100) of fluorescently labeled RT-PCR products obtained from total RNA of cells expressing the Wt (i) or the 9726 + 5a minigene without (ii,iii) and on (iv,v) overexpression of the U1 + 5a snRNA. The scheme of transcripts, and of primers used (arrows), is depicted on the top. Separation of 1 μL of 1:100 diluted RT-PCR reaction is shown, with the exception of panel Ciii) in which 1 μL of the 1:20 dilution was loaded (overloaded). The peaks, which might suggest the presence of a doublet, were referred to as single bands by the automated ABI-3100 system. As expected, the fragment sizes of the normal and aberrant transcripts were 236 bp and 273 bp, respectively.
Compensatory U1-snRNAs designed to suppress the effects of 5′ss mutations have been so far exploited only for the TCIRG1-dependent osteoporosis15 and for NF1,16 with variable efficiency.
To restore exon 7 inclusion and selection of the correct 5′ss, we performed complementation experiments with modified U1-snRNAs (Figure 1B). Their 5′-tails were engineered to bind the mutated 5′ss (U1 + 5a) or multiple sites in the IVS7 repeats (U1Mut1, U1Mut2), to get insights on the causative mechanisms of the mutation.
Evaluation of bands revealed in both Hep3B and in COS-1 cells that all mutated U1-snRNAs reduced exon skipping (> 50% reduction) to indicate that their binding at or near the 5′ss favored exon 7 definition (Figure 2A). However, only the U1 + 5a was able to redirect the selection at the correct 5′ss and induce the generation of normal transcripts, whose nature was demonstrated by restriction and sequencing (Figure 2B transcript 2).
The high sequence complementarity19 between transcripts 1 and 2 renders virtually impossible the specific quantification of the rescued form by real-time PCR. This prompted us to evaluate the efficacy of the U1 + 5a and its ability to favor recognition of the authentic over the cryptic 5′ss, by denaturing capillary electrophoresis of fluorescently labeled RT-PCR products (primers 7FF and 8R′).
This methodologic approach revealed traces of normal transcripts (∼0.2%; Figure 2Ciii) in the mutant and further highlighted the U1 + 5a-mediated rescue of correct splicing, which resulted in dose-dependency. In particular, at equimolar concentrations of p9726 + 5a and pU1 + 5a, the normal transcripts in Hep3B were 25% plus or minus 5% of the aberrant form, and reached 53% plus or minus 7% with an excess (1.5×) of pU1 + 5a (Figure 2Civ,v). Evaluation of normal transcript at lower PCR cycle number (20) confirmed the robust correction (40% at 1.5× of pU1 + 5a). Further overexpression of the U1 + 5a did not produce additional effects, and a comparable correction efficiency was observed in COS-1.
These observations were not the result of increased levels of cellular U1-snRNA, as demonstrated by inability of the WT-U1 to rescue splicing (Figure 2A,B). Specificity of U1 + 5a was further suggested by the observation that it did not alter the splicing of the Wt minigene and was not associated with cell culture death. The in vivo application of this strategy will require the evaluation in animal models of hepatocyte gene expression profile modifications as well as of effects on FVII protein and on coagulation.
Taken together, these findings demonstrated that impaired recognition by the U1-snRNA was the mechanism responsible for severe FVII deficiency. The compensatory U1-snRNAs might represent a therapeutic strategy in coagulation factor deficiencies caused by mutations at 5′ss, which account for a large fraction of the severe cases.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dott.ssa Iva Maestri, Department of Experimental and Diagnostic Medicine of the Ferrara University, for her assistance in capillary electrophoresis.
This work was supported by grants from Telethon (Grant General Project 05214), Ministero dell'Università e della Ricerca (MIUR)-Progetti di Ricerca di Interesse Nazionale (PRIN), and the University of Ferrara (M.P., L.R., D.B., N.C., G.M., and F.B.), and by grants from the Associazione Italiana per la Ricerca sul Cancro (AIRC) and the Italian Cystic Fibrosis Foundation (F.P., M.A.L.).
Authorship
Contribution: M.P., F.B., and F.P. conceived the study, analyzed and interpreted data, and wrote the manuscript; L.R., D.B., and N.C. created FVII minigenes and performed expression studies; M.A.L. created modified U1-snRNA for complementation assays; G.M. helped with the analysis and interpretation of results and revised the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mirko Pinotti, Department of Biochemistry and Molecular Biology, University of Ferrara, Via Fossato di Mortara 74, 44100 Ferrara; e-mail: pnm@unife.it; or Dr Franco Pagani, Human Molecular Genetics, ICGEB, Padriciano 99, 34012 Trieste, Italy; e-mail: pagani@icgeb.org.
References
Author notes
M.P. and L.R. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal