An internal tandem duplication in the fms-like tyrosine kinase 3 gene (FLT3/ITD) is associated with poor prognosis in acute myeloid leukemia (AML), but the impact of mutant level, size, and interaction with nucleophosmin 1 (NPM1) mutations remains controversial. We evaluated these characteristics in a large cohort of young adult AML patients. There was a highly significant trend for worsening in relapse risk (RR) and overall survival (OS) with increasing FLT3/ITD mutant level (P < .001 for both), and even in the low level mutant group (1%-24% of total FLT3 alleles), RR was significantly worse than in the FLT3 wild-type (WT) group (P < .001). In multivariate analysis, mutant level was the most powerful prognostic factor for RR. Mutant size and number had no significant impact on outcome. The beneficial impact of an NPM1 mutation on RR and OS was seen in FLT3/ITD+ as well as FLT3/WT patients; both markers were highly significant independent predictors of outcome (P < .001). Stratification using both markers identified 3 prognostic groups: good (FLT3/ITD−NPM1+), intermediate (FLT3/ITD−NPM1− or FLT3/ITD+NPM1+), and poor (FLT3/ITD+NPM1−). Patients with high FLT3/ITD mutant level (greater than 50%) or FLT3/ITD+ in the absence of an NPM1 mutation may be good candidates for more experimental therapeutic approaches.
Introduction
In recent years there have been considerable advances in the identification of acquired molecular markers that are of prognostic significance in patients with acute myeloid leukemia (AML) and may add additional information to the currently used stratification according to cytogenetic risk group. An internal tandem duplication (ITD) in the juxtamembrane domain of the fms-like tyrosine kinase-3 (FLT3) gene was one of the earliest markers detected, first reported by Nakao et al in 1996,1 and is a common finding in approximately 25% of younger adult AML patients.2 The length of duplicated DNA varies between 3 and more than 400 base pairs (bp), and its exact position differs between cases, but it is always in frame and therefore expected to produce a functional protein. In vitro studies have shown that such insertions lead to a constitutively activated receptor,3,4 and it is now widely accepted that in AML patients FLT3/ITDs are associated with leukocytosis, a high percentage of bone marrow blast cells, an increased risk of relapse from complete remission (CR), and reduced survival.2,5,6
Despite the evident poor outcome associated with an FLT3/ITD, it is as yet unclear whether this can be explained simply by the presence or absence of the abnormality, or whether there are other factors that influence its prognostic impact. For example, the relative mutant level varies widely, and several studies have suggested that it is only patients with a higher mutant level that have a poor outcome. In 2 studies examining younger adults (younger than 60 years of age) with either intermediate cytogenetics or a normal karyotype, a poor prognosis compared with wild-type (WT) cases was observed only in patients with a high level of FLT3/ITD that was indicative of loss of WT alleles.7,8 In a study of pediatric patients, those with a mutant/WT allelic ratio greater than 0.4 (equivalent to more than 29% FLT3/ITD) had a significantly poorer outcome than patients with either a lower mutant level or WT FLT3.9 The number of FLT3/ITDs in an individual patient may also vary, and up to 5 different mutants of varying size and relative level have been reported in a proportion of patients.8,10,11 In our earlier study this was observed in 23% of FLT3/ITD-positive (FLT3/ITD+) patients, and there was a suggestion that this was associated with a significantly worse overall survival (OS) at 5 years.10 Recent studies also have suggested that the size of the inserted DNA may be of prognostic significance, although the results are contradictory, with reports of a worse overall prognosis in cases with both longer and shorter FLT3/ITDs and of no prognostic significance according to FLT3/ITD size.12,–14
All these factors relate to characteristics of the FLT3/ITD per se, but the potential interaction of an FLT3/ITD with other molecular abnormalities also is likely to be important when considering prognostic significance. For example, acquired mutations in exon 12 of the nucleophosmin 1 gene (NPM1) have recently been reported, particularly in patients with a normal karyotype, and a significant proportion of patients carry both an FLT3/ITD and an NPM1 mutation.15 Overall, NPM1 mutations generally are associated with a trend for improved OS and significantly better event-free or relapse-free survival, and in multivariate analysis they are an independent good prognostic factor for outcome.16,,,–20 However, these studies indicated that this benefit is lost in the presence of an FLT3/ITD, with a favorable outcome only in FLT3/WT NPM1 mutant-positive (NPM1+) patients. Furthermore, in normal karyotype patients they reported that the presence of an FLT3/ITD has little prognostic impact on patients who do not have an NPM1 mutation, although this may be due to the relatively small numbers of NPM1−FLT3/ITD+patients in these cohorts.16,,,–20
Patients with an FLT3/ITD may derive particular benefit from the addition of FLT3 inhibitors to conventional chemotherapy, as blast cells containing an FLT3/ITD are more sensitive to these inhibitors in vitro.21,22 Furthermore, it has been suggested that as an FLT3/ITD is a marker of poor prognosis, it should be used as an indication for transplantation or other experimental therapies.23,24 FLT3/ITD+ AML is, however, clearly heterogeneous from a genetic and biological standpoint, and it is important to understand those factors that modify prognosis. To do this, we have investigated the impact of FLT3/ITD mutant level, number and size of mutants, and interaction with NPM1 mutations in a large cohort of young adult AML patients, excluding acute promyelocytic leukemia (APL), uniformly treated in the United Kingdom Medical Research Council (MRC) AML 10 and 12 trials and with a long clinical follow-up.
Methods
Patients
DNA was available from peripheral blood (PB) or bone marrow (BM) from 1425 (38%) of 3803 young adult AML patients, excluding APL, who were entered into either the UK MRC AML10 (n = 548) or AML12 (n = 877) trials between 1988 and 2002. The majority of patients (1307 of 1425, 92%) had de novo AML. Median age at trial entry was 43 years of age; all patients were 15 years of age or older, and only 35 were over 60 years of age. Details of randomization and therapy for patients entered into these trials have been published elsewhere.25 Ethical approval for the trials and tissue collection for research was obtained from the Multi-Centre Research Ethics Committee of Wales and local research ethics committees as appropriate, and informed consent was obtained in accordance with the Declaration of Helsinki. Trial MRC AML12 is registered at http://controlled-trials.com as #55678797.
Endpoints
CR was defined as a normocellular BM containing fewer than 5% blasts and showing evidence of normal maturation of other marrow elements. PB regeneration was not a requirement, but 97% of cases defined as CR achieved a neutrophil count of 109/L and a platelet count of 100 × 109/L. Remission failures were classified by the clinicians as either partial remission (PR, defined as 5%-15% blasts or fewer than 5% blasts but a hypocellular BM), resistant disease (RD, more than 15% blasts in the BM), or induction death (ID; ie, related to treatment or hypoplasia). Where the clinician's evaluation was not available, deaths within 30 days of entry were classified as ID and deaths later than 30 days after entry as RD. OS was defined as the time from randomization to death. For patients achieving CR, disease-free survival (DFS) was the time from the date of first CR to an event (death in first CR or relapse) and relapse risk (RR), the cumulative probability of relapse censoring at death in CR.
FLT3/ITD and NPM1 exon 12 mutation analysis
Exons 14 and 15 and the intervening intron of the FLT3 gene were amplified from DNA using the polymerase chain reaction (PCR) as previously described.10 Any patient with an additional higher molecular weight band was considered to be FLT3/ITD+. To obtain the size and relative level of mutant(s), PCR was performed using a fluorescently labeled primer and only 25 cycles of amplification. Products were analyzed by fragment analysis on a CEQ8000 DNA Genetic Analysis System (Beckman Coulter UK, High Wycombe, United Kingdom). To screen for presence of an insertion in exon 12 of the NPM1 gene, approximately 50 ng genomic DNA was amplified in a total volume of 20 μL containing 1× manufacturer's buffer, 1 mM MgCl2, 200 μM dNTPs, 10 pmol each of primers NPMex12/F (CTTAACCACATTTCTTTTTTTTTTTTTCCAG), and fluorescently labeled NPMex12/R (GGACAACATTTATCAAACACGGTAG), with 1 unit of BIOTAQ DNA polymerase (Bioline, London, United Kingdom). Either 25 or 28 cycles were performed, each 45 seconds at 95°C, 45 seconds at 62°C, 45 seconds at 72°C, with a final extension of 15 minutes at 72°C, and PCR products were analyzed by fragment analysis on the CEQ8000 system. WT product size was 198 bp. For both FLT3/ITDs and NPM1 mutants, relative mutant level was calculated using the area under the peak and expressed as a percentage of total FLT3 or NPM1 alleles.
X-chromosome inactivation patterns
X-chromosome inactivation pattern (XCIP) analysis was performed using the human androgen receptor gene assay (HUMARA) as previously described26 except with a fluorescently labeled primer and analysis on the CEQ8000 system. Results were expressed as the relative percentage of the shorter and longer X-chromosome alleles (A%:B%), respectively.
Statistical methods
The Wilcoxon 2 sample test (for continuous data), Mantel-Haenszel test for trend (for ordinal data), and χ2 tests were used to test for differences in clinical and demographic data by FLT3/ITD and NPM1 mutant positivity. Unstratified analyses of remission were performed using the Mantel-Haenszel test. Kaplan-Meier life-tables were constructed for survival data and were compared by means of the log rank test, with surviving patients being censored March 22, 2006. Follow-up was up-to-date for more than 95% of patients, and the small numbers of patients lost to follow-up are censored at the date they were last known to be alive. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated from the observed minus expected number of events (O − E) and its variance, and forest plots were used to illustrate differences in the relative effect of FLT3/ITD between prognostic factor subgroups. Multivariate logistic regression analysis was used to find the factors most closely associated with CR rate, and multivariate Cox models were used to analyze OS, DFS, and RR. Models were fitted using forward selection, with variables added to the model if they had a P value less than .05.
Results
Response to therapy and clinical outcome according to FLT3/ITD mutant level
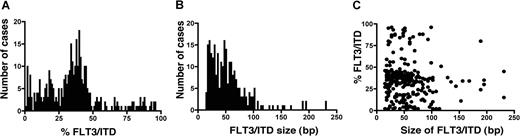
FLT3 results were obtained from the total cohort of 1425 cases; 354 (26%) were FLT3/ITD+. Details of these patients are in Table 1 Screening on 1135 of these patients was reported in a previous study.25 Survival data were available on the whole cohort, with a median follow-up of 9.3 years (range, 25-215 months). FLT3/ITD quantification and sizing was feasible in all FLT3/ITD+ cases. Median total mutant level for all patients was 35% of total FLT3, but the variation in mutant level was wide, ranging from 1% to 96%. The distribution of FLT3/ITD+ cases according to total mutant level suggested 3 different groups: those with less than 25% mutant, 25% to 50% mutant, and greater than 50% mutant (Figure 1A), which henceforward will be called low, intermediate, and high mutant level. The proportion of patients in each of these 3 groups was 29%, 56%, and 15%, respectively, which represented 7%, 14%, and 4% of the total cohort of cases. Median mutant level in each group was 14%, 38%, and 77%. The 50% cutoff value is consistent with the fact that levels above this are indicative of the presence of biallelic disease, at least in some cells. In the high-level group of 53 cases, 49 (92%) had a single mutant comprising greater than 50% of total FLT3 alleles; only 4 cases (8%) had more than one mutant. The relative mutant level was highly correlated with presenting white blood cell count (WBC), median WBC 16.6 × 109/L for WT cases; 23.9, 57.5, and 75.0 × 109/L for low, intermediate, and high mutant cases, respectively (P < .001).
Clinical and demographic characteristics of 1425 non-APL AML patients in the total cohort
| . | Total . | FLT3 . | P . | NPM1 . | P . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| WT . | ITD low . | ITD int. . | ITD high . | Total . | WT . | Mutant . | ||||
| Total, no. (%) | 1425 | 1071 (75) | 102 (7) | 199 (14) | 53 (4) | .4* | 1217 | 714 (59) | 503 (41) | — |
| Type AML, no. (%) | .02* | .003 | ||||||||
| De novo | 1307 | 977 (75) | 89 (7) | 191 (14) | 52 (4) | — | 1124 | 646 (57) | 478 (43) | — |
| Secondary | 116 | 94 (81) | 13 (11) | 8 (7) | 1 (1) | — | 93 | 68 (73) | 25 (27) | — |
| FAB type, no. (%) | — | — | ||||||||
| M0 | 48 | 44 (92) | 1 (2) | 3 (6) | 0 | — | 37 | 36 (97) | 1 (3) | — |
| M1 | 269 | 191 (71) | 15 (6) | 52 (19) | 11 (4) | — | 238 | 137 (58) | 101 (42) | — |
| M2 | 422 | 325 (77) | 40 (10) | 47 (11) | 10 (2) | — | 358 | 233 (65) | 125 (35) | — |
| M4 | 358 | 251 (70) | 24 (7) | 61 (17) | 22 (6) | — | 316 | 156 (49) | 160 (51) | — |
| M5 | 166 | 117 (71) | 11 (7) | 29 (17) | 9 (5) | — | 143 | 73 (51) | 70 (49) | — |
| M6 | 42 | 40 (96) | 1 (2) | 1 (2) | 0 | — | 33 | 21 (64) | 12 (36) | — |
| M7 | 21 | 20 (95) | 1 (5) | 0 | 0 | — | 16 | 12 (75) | 4 (25) | — |
| RAEB-t | 21 | 20 (95) | 1 (5) | 0 | 0 | — | 18 | 10 (56) | 8 (44) | — |
| Unknown | 78 | 63 (81) | 8 (10) | 6 (8) | 1 (1) | — | 58 | 36 (62) | 22 (38) | — |
| Sex, no. (%) | .15* | <.001 | ||||||||
| Female | 714 | 528 (74) | 51 (7) | 102 (14) | 33 (5) | — | 620 | 328 (53) | 292 (47) | — |
| Male | 711 | 543 (76) | 51 (7) | 97 (14) | 20 (3) | — | 597 | 386 (65) | 211 (35) | — |
| Median age, y | 43 | 42 | 43 | 43 | 44 | .10* | 43 | 41 | 46 | <.001* |
| Median WBC, × 109/L | 21.9 | 16.6 | 23.9 | 57.5 | 75.0 | <.001* | 24.4 | 18.5 | 35.4 | <.001* |
| Cytogenetics, no. (%) | .9* | .04* | ||||||||
| Favorable | 176 | 157 (89) | 10 (6) | 8 (5) | 1 (<1) | — | 123 | 119 (97) | 4 (3) | — |
| Intermediate (total) | 849 | 593 (70) | 69 (8) | 144 (17) | 43 (5) | <.001 | 766 | 373 (49) | 393 (51) | <.001 |
| Normal karyotype | 574 | 379 (66) | 51 (9) | 114 (20) | 30 (5) | — | 550 | 210 (38) | 340 (62) | — |
| Adverse | 139 | 130 (93) | 4 (3) | 5 (4) | 0 | — | 88 | 84 (95) | 4 (5) | — |
| Unknown | 261 | 191 (73) | 19 (7) | 42 (16) | 9 (3) | — | 240 | 138 (58) | 102 (42) | — |
| FLT3 status | <.001* | |||||||||
| WT | — | — | — | — | — | — | 872 | 577 (66) | 295 (34) | — |
| Low-level ITD | — | — | — | — | — | — | 99 | 48 (48) | 51 (51) | — |
| Intermediate-level ITD | — | — | — | — | — | — | 194 | 79 (41) | 115 (59) | — |
| High-level ITD | — | — | — | — | — | — | 52 | 10 (19) | 42 (81) | — |
| . | Total . | FLT3 . | P . | NPM1 . | P . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| WT . | ITD low . | ITD int. . | ITD high . | Total . | WT . | Mutant . | ||||
| Total, no. (%) | 1425 | 1071 (75) | 102 (7) | 199 (14) | 53 (4) | .4* | 1217 | 714 (59) | 503 (41) | — |
| Type AML, no. (%) | .02* | .003 | ||||||||
| De novo | 1307 | 977 (75) | 89 (7) | 191 (14) | 52 (4) | — | 1124 | 646 (57) | 478 (43) | — |
| Secondary | 116 | 94 (81) | 13 (11) | 8 (7) | 1 (1) | — | 93 | 68 (73) | 25 (27) | — |
| FAB type, no. (%) | — | — | ||||||||
| M0 | 48 | 44 (92) | 1 (2) | 3 (6) | 0 | — | 37 | 36 (97) | 1 (3) | — |
| M1 | 269 | 191 (71) | 15 (6) | 52 (19) | 11 (4) | — | 238 | 137 (58) | 101 (42) | — |
| M2 | 422 | 325 (77) | 40 (10) | 47 (11) | 10 (2) | — | 358 | 233 (65) | 125 (35) | — |
| M4 | 358 | 251 (70) | 24 (7) | 61 (17) | 22 (6) | — | 316 | 156 (49) | 160 (51) | — |
| M5 | 166 | 117 (71) | 11 (7) | 29 (17) | 9 (5) | — | 143 | 73 (51) | 70 (49) | — |
| M6 | 42 | 40 (96) | 1 (2) | 1 (2) | 0 | — | 33 | 21 (64) | 12 (36) | — |
| M7 | 21 | 20 (95) | 1 (5) | 0 | 0 | — | 16 | 12 (75) | 4 (25) | — |
| RAEB-t | 21 | 20 (95) | 1 (5) | 0 | 0 | — | 18 | 10 (56) | 8 (44) | — |
| Unknown | 78 | 63 (81) | 8 (10) | 6 (8) | 1 (1) | — | 58 | 36 (62) | 22 (38) | — |
| Sex, no. (%) | .15* | <.001 | ||||||||
| Female | 714 | 528 (74) | 51 (7) | 102 (14) | 33 (5) | — | 620 | 328 (53) | 292 (47) | — |
| Male | 711 | 543 (76) | 51 (7) | 97 (14) | 20 (3) | — | 597 | 386 (65) | 211 (35) | — |
| Median age, y | 43 | 42 | 43 | 43 | 44 | .10* | 43 | 41 | 46 | <.001* |
| Median WBC, × 109/L | 21.9 | 16.6 | 23.9 | 57.5 | 75.0 | <.001* | 24.4 | 18.5 | 35.4 | <.001* |
| Cytogenetics, no. (%) | .9* | .04* | ||||||||
| Favorable | 176 | 157 (89) | 10 (6) | 8 (5) | 1 (<1) | — | 123 | 119 (97) | 4 (3) | — |
| Intermediate (total) | 849 | 593 (70) | 69 (8) | 144 (17) | 43 (5) | <.001 | 766 | 373 (49) | 393 (51) | <.001 |
| Normal karyotype | 574 | 379 (66) | 51 (9) | 114 (20) | 30 (5) | — | 550 | 210 (38) | 340 (62) | — |
| Adverse | 139 | 130 (93) | 4 (3) | 5 (4) | 0 | — | 88 | 84 (95) | 4 (5) | — |
| Unknown | 261 | 191 (73) | 19 (7) | 42 (16) | 9 (3) | — | 240 | 138 (58) | 102 (42) | — |
| FLT3 status | <.001* | |||||||||
| WT | — | — | — | — | — | — | 872 | 577 (66) | 295 (34) | — |
| Low-level ITD | — | — | — | — | — | — | 99 | 48 (48) | 51 (51) | — |
| Intermediate-level ITD | — | — | — | — | — | — | 194 | 79 (41) | 115 (59) | — |
| High-level ITD | — | — | — | — | — | — | 52 | 10 (19) | 42 (81) | — |
P values are for heterogeneity (Chi-squared test), or * for trend (Mantel-Haenszel test).
WBC indicates white blood cell count; WT, wild type; int., intermediate; and —, not applicable.
FLT3/ITD mutant level and size. (A) Total FLT3/ITD mutant level in 354 FLT3/ITD+ patients. (B) FLT3/ITD size in 260 cases with a single mutant. (C) Correlation between FLT3/ITD level and size.
FLT3/ITD mutant level and size. (A) Total FLT3/ITD mutant level in 354 FLT3/ITD+ patients. (B) FLT3/ITD size in 260 cases with a single mutant. (C) Correlation between FLT3/ITD level and size.
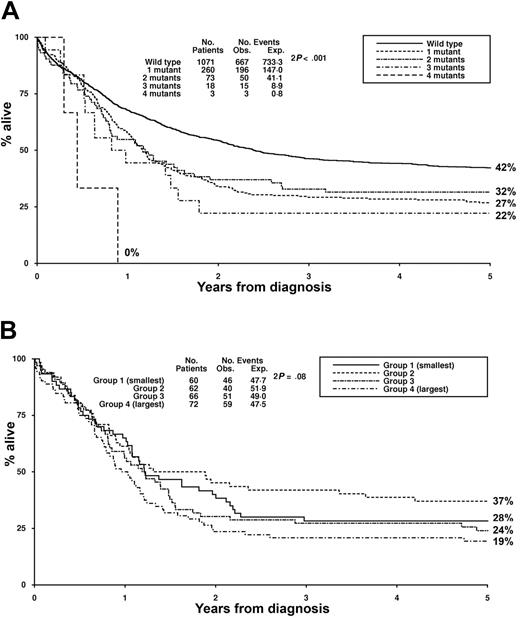
There was no difference in the remission rate between the 4 groups of patients stratified according to mutant level (Table 2). The frequency of RD and ID in the different groups also was similar. In univariate analysis, there was a highly significant trend across the 4 groups for a worsening in RR, DFS, and OS with increasing mutant level (Table 2). RR at 5 years was 49%, 69%, 64%, and 82% for the WT, low, intermediate, and high mutant level groups, respectively (OR 1.35, CI 1.24-1.47, P < .001) (Figure 2A). It is noteworthy that, in the high mutant level group, most of the patients who would ultimately relapse had done so within one year of achieving remission. Similar results were obtained for OS, with a significantly worse OS at 5 years with increasing mutant level across the 4 groups (Figure 2B, Table 2).
Outcome data according to FLT3/ITD mutant level
| FLT3 . | WT, % . | ITD low, % . | ITD int., % . | ITD high, % . | OR (CI) . | P . |
|---|---|---|---|---|---|---|
| CR | 84 | 82 | 83 | 85 | 1.00 (0.85-1.17) | 1.0 |
| RD | 10 | 11 | 10 | 8 | 0.97 (0.79-1.19) | .7 |
| ID | 6 | 7 | 7 | 8 | 1.04 (0.82-1.33) | .7 |
| OS at 5 y | 42 | 31 | 28 | 15 | 1.24 (1.16-1.33) | <.001 |
| DFS at 5 y | 41 | 26 | 27 | 16 | 1.28 (1.19-1.38) | <.001 |
| RR at 5 y | 49 | 69 | 64 | 82 | 1.35 (1.24-1.47) | <.001 |
| FLT3 . | WT, % . | ITD low, % . | ITD int., % . | ITD high, % . | OR (CI) . | P . |
|---|---|---|---|---|---|---|
| CR | 84 | 82 | 83 | 85 | 1.00 (0.85-1.17) | 1.0 |
| RD | 10 | 11 | 10 | 8 | 0.97 (0.79-1.19) | .7 |
| ID | 6 | 7 | 7 | 8 | 1.04 (0.82-1.33) | .7 |
| OS at 5 y | 42 | 31 | 28 | 15 | 1.24 (1.16-1.33) | <.001 |
| DFS at 5 y | 41 | 26 | 27 | 16 | 1.28 (1.19-1.38) | <.001 |
| RR at 5 y | 49 | 69 | 64 | 82 | 1.35 (1.24-1.47) | <.001 |
For FLT3 levels, odds ratios (OR) and 95% confidence intervals (CI) were determined by logistic regression.
Int. indicates intermediate mutant level.
Clinical outcome stratified according to total FLT3/ITD level. (A) Relapse rate. (B) Overall survival. Low level indicates 1%-24%; intermediate, 25%-50%; and high level, greater than 50% FLT3/ITD, respectively.
Clinical outcome stratified according to total FLT3/ITD level. (A) Relapse rate. (B) Overall survival. Low level indicates 1%-24%; intermediate, 25%-50%; and high level, greater than 50% FLT3/ITD, respectively.
The relative FLT3/ITD mutant level remained a significant adverse prognostic factor for both RR and OS in a Cox multivariate analysis, the variables considered being age, cytogenetics, de novo or secondary AML, presentation WBC, performance status, FLT3/ITD mutant level as a 3-group variable (low, intermediate, or high), and NPM1 mutation status. For RR, FLT3/ITD mutant level was the most significant factor, entering the model above cytogenetics (Table 3). However, for OS, cytogenetics was the more powerful prognostic factor because of its additional relationship to remission rate. In a Cox regression analysis to test presence or absence of an FLT3/ITD against FLT3/ITD mutant level, the mutant level was more significant for both RR and OS (χ2 for RR: 43.5 for presence or absence of an FLT3/ITD, 47.2 for FLT3/ITD mutant level, P < .001; for OS: 28.4 and 36.5, respectively, P < .001). In analysis of individual groups, RR was significantly worse in the high-level compared with intermediate-level group (P = .005). Even in the low-level mutant group, RR was significantly worse than FLT3/WT (P < .001). For the low- and intermediate-level groups, the value of 25% as a cutoff between them was derived from the distribution data and was somewhat arbitrary. Nevertheless, stepwise threshold analysis did not reveal a specific cutoff level in these groups at which a major prognostic change occurred.
Results of multivariate analysis for CR, RR, and OS
| Outcome, variable . | OR (CI) . | P . |
|---|---|---|
| CR | ||
| Cytogenetics | 4.69 (3.03-7.26) | <.001 |
| NPM1 status | 0.27 (0.17-0.43) | <.001 |
| WBC | 1.007 (1.004-1.009) | <.001 |
| Age | 1.04 (1.02-1.06) | <.001 |
| WHO Performance status | 1.43 (1.19-1.71) | <.001 |
| De novo/secondary | 2.39 (1.33-4.29) | .003 |
| RR | ||
| FLT3/ITD level | 1.55 (1.40-1.72) | <.001 |
| NPM1 status | 0.43 (0.35-0.54) | <.001 |
| Cytogenetics | 1.95 (1.56-2.43) | <.001 |
| Age | 1.01 (1.01-1.02) | <.001 |
| OS | ||
| Cytogenetics | 1.89 (1.59-2.25) | <.001 |
| NPM1 status | 0.47 (0.39-0.56) | <.001 |
| FLT3/ITD level | 1.34 (1.23-1.47) | <.001 |
| Age | 1.02 (1.01-1.03) | <.001 |
| WBC | 1.002 (1.001-1.003) | <.001 |
| WHO Performance status | 1.13 (1.05-1.22) | .003 |
| De novo/secondary | 1.37 (1.04-1.80) | .02 |
| Outcome, variable . | OR (CI) . | P . |
|---|---|---|
| CR | ||
| Cytogenetics | 4.69 (3.03-7.26) | <.001 |
| NPM1 status | 0.27 (0.17-0.43) | <.001 |
| WBC | 1.007 (1.004-1.009) | <.001 |
| Age | 1.04 (1.02-1.06) | <.001 |
| WHO Performance status | 1.43 (1.19-1.71) | <.001 |
| De novo/secondary | 2.39 (1.33-4.29) | .003 |
| RR | ||
| FLT3/ITD level | 1.55 (1.40-1.72) | <.001 |
| NPM1 status | 0.43 (0.35-0.54) | <.001 |
| Cytogenetics | 1.95 (1.56-2.43) | <.001 |
| Age | 1.01 (1.01-1.02) | <.001 |
| OS | ||
| Cytogenetics | 1.89 (1.59-2.25) | <.001 |
| NPM1 status | 0.47 (0.39-0.56) | <.001 |
| FLT3/ITD level | 1.34 (1.23-1.47) | <.001 |
| Age | 1.02 (1.01-1.03) | <.001 |
| WBC | 1.002 (1.001-1.003) | <.001 |
| WHO Performance status | 1.13 (1.05-1.22) | .003 |
| De novo/secondary | 1.37 (1.04-1.80) | .02 |
WBC indicates white blood cell count; and WHO, World Health Organization.
Clonality analyses in cases with low-level FLT3/ITDs
To examine whether the low mutant level obtained in 102 FLT3/ITD+ cases could be due to the presence of contaminating nonleukemic cells, other markers of clonality were studied. An NPM1 exon 12 mutation was detected in 51 (50%) of the 102 cases (see “Outcome according to FLT3/ITD and NPM1 mutant status”). The median NPM1 mutant level was 43% (range, 19%-72%), and only 2 cases had mutant levels that were less than 25% of total NPM1 alleles (19% and 20%, respectively). These results indicate that the majority of cells in these cases were clonal. Further analysis was not possible in 40 of the remaining 51 cases. None had another known molecular marker, 32 were male, and 8 of the 19 female cases were either not informative for the trinucleotide repeat in the human androgen receptor gene or no DNA was available. XCIP analysis using the HUMARA was therefore performed in 11 cases. In each case there was greater than 75% expression of one X-allele (Table 4), which is in accord with the presence of a predominantly clonal population, although T-cell DNA was not available to exclude the possibility of a constitutively skewed XCIP in individual cases. Taken together, these results suggest that in most cases a low FLT3/ITD mutant level is not because the leukemic cells comprise only a small proportion of the total cells analyzed but rather that the mutation is in a subclone of the leukemic cells.
XCIPs in 11 female patients with low mutant level FLT3/ITDs
| Patient . | FLT3/ITD, % . | XCIP, A%:B% . |
|---|---|---|
| 1 | 2 | 4:96 |
| 2 | 4 | 3:97 |
| 3 | 5 | 3:97 |
| 4 | 9 | 0:100 |
| 5 | 11 | 88:12 |
| 6 | 11 | 81:19 |
| 7 | 14 | 5:95 |
| 8 | 14 | 8:92 |
| 9 | 19 | 100:0 |
| 10 | 21 | 0:100 |
| 11 | 23 | 4:96 |
| Patient . | FLT3/ITD, % . | XCIP, A%:B% . |
|---|---|---|
| 1 | 2 | 4:96 |
| 2 | 4 | 3:97 |
| 3 | 5 | 3:97 |
| 4 | 9 | 0:100 |
| 5 | 11 | 88:12 |
| 6 | 11 | 81:19 |
| 7 | 14 | 5:95 |
| 8 | 14 | 8:92 |
| 9 | 19 | 100:0 |
| 10 | 21 | 0:100 |
| 11 | 23 | 4:96 |
Outcome according to number of FLT3/ITD mutants
The majority (73%) of FLT3/ITD+ cases had a single mutant (260 of 354), however, 21% (73 cases) had 2 mutants, 5% (18 cases) had 3 mutants, and 1% (3 cases) had 4 mutants. The median total mutant level did not differ according to the number of mutants and varied between 35% and 37% for the 4 groups. In the 94 cases with more than 1 mutant, 63 (67%) had one predominant mutant and one or more minor mutants; in 31 (33%) cases the relative level of the mutants was approximately equal.
There was an apparent decrease in the remission rate with an increasing number of FLT3/ITD mutants: CR 84%, 86%, 78%, 67%, and 67% for the cases with WT FLT3, 1, 2, 3 and 4 mutants, respectively, but this difference was not significant (P = .17 for trend). There was no significant difference according to mutant number for either RR or OS (Figure 3A).
Clinical outcome stratified according to mutant number and size. (A) Overall survival in patients with up to 4 FLT3/ITD mutants. (B) Overall survival in patients with a single FLT3/ITD grouped in quartiles according to size. Group 1 (smallest) indicates 15 to 27 bp; group 2, 30 to 45 bp; group 3, 48 to 63 bp; and group 4 (largest), 66 to 213 bp, respectively.
Clinical outcome stratified according to mutant number and size. (A) Overall survival in patients with up to 4 FLT3/ITD mutants. (B) Overall survival in patients with a single FLT3/ITD grouped in quartiles according to size. Group 1 (smallest) indicates 15 to 27 bp; group 2, 30 to 45 bp; group 3, 48 to 63 bp; and group 4 (largest), 66 to 213 bp, respectively.
Impact of FLT3/ITD mutant size
Analysis of the impact of FLT3/ITD size on outcome was limited to the 260 cases with only a single mutant. Median mutant size was 48 bp (range, 15 to 231 bp). No obvious groupings were evident according to the distribution of mutant size (Figure 1B), and therefore the cohort was split into quartiles for the analysis, with the groups containing cases of 15 to 27 bp, 30 to 45 bp, 48 to 63 bp, and 66 to 213 bp, respectively. There was no difference between the groups with respect to type of leukemia, French-American-British (FAB) type, age, presentation WBC, or karyotype (data not shown). No correlation between mutant size and level was observed (Figure 1C), and the median size for the low, intermediate, and high mutant level groups was 54 bp, 45 bp, and 48 bp, respectively.
Increasing mutant size appeared to affect CR rate, which was 90%, 92%, 88%, and 76% for the smallest to largest mutant size groups respectively (OR 1.51, CI 1.08-2.11, P = .02 for trend). There was no significant difference according to mutant size in either RR or OS (Figure 3B).
Outcome according to FLT3/ITD and NPM1 mutant status
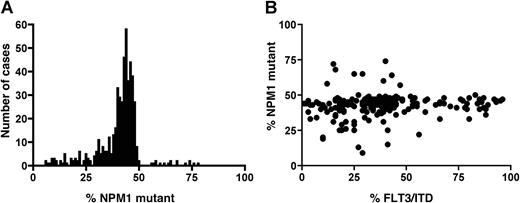
DNA was available only from 1217 of the original cohort of 1425 patients to screen for NPM1 exon 12 mutations, as this test was performed at a later date when some of the samples had been used up: 714 (59%) were WT (NPM1−) and 503 (41%) positive (NPM1+). Patient details are given in Table 1. The mutation was more common in de novo than in secondary AML (43% and 27%, respectively, P = .003) and in patients with FAB subtypes M4 and M5 (51% and 49%, respectively) but uncommon in FAB type M0 (3%). It also was associated with increasing age and leukocytosis (P < .001 for both). All mutant-positive patients had a 4-bp insertion. The median mutant NPM1 level was 43%, and although the range in level varied between 6% and 78%, the distribution was much tighter than that observed for FLT3/ITD mutant level (Figure 4A). Only 39 cases (8% of mutant-positive patients) had less than 25% mutant NPM1, and 16 cases (3%) had greater than 50% mutant, indicative of loss of WT NPM1. None of the latter cases were known to have cytogenetic evidence for loss of material on chromosome 5q, the location of the NPM1 gene.
NPM1 mutant level. (A) Total NPM1 mutant level in 503 NPM1+ patients. (B) Correlation of FLT3/ITD and NPM1 mutant levels in 208 patients positive for both mutations.
NPM1 mutant level. (A) Total NPM1 mutant level in 503 NPM1+ patients. (B) Correlation of FLT3/ITD and NPM1 mutant levels in 208 patients positive for both mutations.
Of the 1217 patients, 345 (28%) had an FLT3/ITD, which is slightly higher than in the total cohort of 1425 patients and probably reflects the higher proportion of patients with a normal karyotype in this cohort (40% of the total cohort, 45% of the patients screened for an NPM1 mutation). Overall, 47% of patients (577 of 1217) were WT for both mutations (FLT3/ITD−NPM1−), 11% (137 cases) had only an FLT3/ITD (FLT3/ITD+NPM1−), 24% (295 cases) had only an NPM1 mutation (FLT3/ITD−NPM1+), and 17% (208 cases) had both mutations (FLT3/ITD+NPM1+). There was a significant correlation between the presence of both mutations (P < .001). There was no correlation, however, between the FLT3/ITD and NPM1 mutant levels in the 208 cases positive for both (Figure 4B), although the presence of an NPM1 mutation did increase with relative FLT3/ITD mutant level: 51%, 52%, 59%, and 81% of the FLT3/WT, low, intermediate, and high FLT3/ITD mutant level groups, respectively, had an NPM1 mutation (P < .001, Table 1).
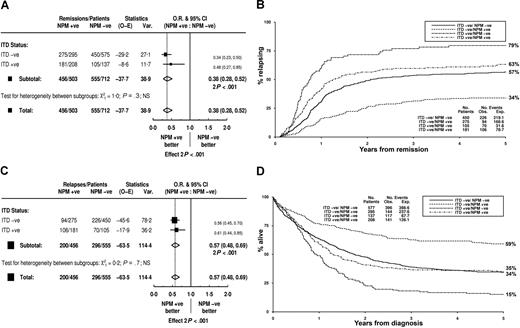
The presence of an NPM1 mutation had a beneficial effect on the remission rate that was probably due to a lower rate of RD (Table 5). There was no difference in ID. This benefit was not influenced by the presence or absence of an FLT3/ITD (Figure 5A, Table 5). NPM1+ patients had a significantly reduced RR that was observed in patients both without and with an FLT3/ITD (Figure 5B,C, Table 5). Likewise, OS was significantly better in NPM1+ patients, and this was seen in both FLT3/ITD+ and FLT3/WT patients (Figure 5D, Table 5). Stratifying according to both molecular markers defined 3 clear prognostic groups. Patients with just an NPM1 mutation had a good prognosis; those with only an FLT3/ITD, a poor prognosis; and those either lacking or positive for both were intermediate (Figure 5B,D).
Outcome data according to NPM1 and FLT3/ITD mutant status in the total cohort of 1217 patients
| . | NPM1−, % . | NPM1+, % . | OR (CI) . | P . | NPM1−, ITD−, % . | NPM1−, ITD+, % . | NPM1+, ITD−, % . | NPM1+, ITD+, % . | OR (CI) for NPM1 stratified by ITD status . | P . | Het. . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CR | 78 | 91 | 0.40 (0.30-0.55) | <.001 | 78 | 77 | 94 | 87 | 0.38 (0.28-0.52) | <.001 | 0.3 |
| RD | 15 | 3 | 0.28 (0.19-0.41) | <.001 | 15 | 17 | 2 | 6 | 0.26 (0.18-0.38) | <.001 | 0.6 |
| ID | 7 | 6 | 0.86 (0.54-1.36) | .5 | 7 | 7 | 5 | 7 | 0.83 (0.52-1.34) | .4 | 0.4 |
| OS at 5 yr | 31 | 49 | 0.67 (0.58-0.77) | <.001 | 34 | 15 | 59 | 35 | 0.60 (0.52-0.69) | <.001 | 0.6 |
| DFS at 5 yr | 31 | 45 | 0.72 (0.62-0.84) | <.001 | 34 | 15 | 55 | 31 | 0.62 (0.53-0.73) | <.001 | 0.7 |
| RR at 5 yr | 61 | 46 | 0.69 (0.58-0.82) | <.001 | 57 | 79 | 34 | 63 | 0.57 (0.48-0.69) | <.001 | 0.6 |
| . | NPM1−, % . | NPM1+, % . | OR (CI) . | P . | NPM1−, ITD−, % . | NPM1−, ITD+, % . | NPM1+, ITD−, % . | NPM1+, ITD+, % . | OR (CI) for NPM1 stratified by ITD status . | P . | Het. . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CR | 78 | 91 | 0.40 (0.30-0.55) | <.001 | 78 | 77 | 94 | 87 | 0.38 (0.28-0.52) | <.001 | 0.3 |
| RD | 15 | 3 | 0.28 (0.19-0.41) | <.001 | 15 | 17 | 2 | 6 | 0.26 (0.18-0.38) | <.001 | 0.6 |
| ID | 7 | 6 | 0.86 (0.54-1.36) | .5 | 7 | 7 | 5 | 7 | 0.83 (0.52-1.34) | .4 | 0.4 |
| OS at 5 yr | 31 | 49 | 0.67 (0.58-0.77) | <.001 | 34 | 15 | 59 | 35 | 0.60 (0.52-0.69) | <.001 | 0.6 |
| DFS at 5 yr | 31 | 45 | 0.72 (0.62-0.84) | <.001 | 34 | 15 | 55 | 31 | 0.62 (0.53-0.73) | <.001 | 0.7 |
| RR at 5 yr | 61 | 46 | 0.69 (0.58-0.82) | <.001 | 57 | 79 | 34 | 63 | 0.57 (0.48-0.69) | <.001 | 0.6 |
Het. indicates testing for heterogeneity (P value for difference in effect of an NPM1 mutation in FLT3/ITD− and ITD+ patients).
Clinical outcome stratified according to FLT3/ITD and NPM1 mutant status in total cohort. (A) Impact on remission rate of an NPM1 mutation in FLT3/ITD+ and ITD− patients. (B) Kaplan-Meier curves for relapse risk. (C) Impact on relapse rate of an NPM1 mutation in FLT3/ITD+ and ITD− patients. (D) Kaplan-Meier curves for overall survival. O-E indicates observed minus expected; Var, variance; and NS, not significant.
Clinical outcome stratified according to FLT3/ITD and NPM1 mutant status in total cohort. (A) Impact on remission rate of an NPM1 mutation in FLT3/ITD+ and ITD− patients. (B) Kaplan-Meier curves for relapse risk. (C) Impact on relapse rate of an NPM1 mutation in FLT3/ITD+ and ITD− patients. (D) Kaplan-Meier curves for overall survival. O-E indicates observed minus expected; Var, variance; and NS, not significant.
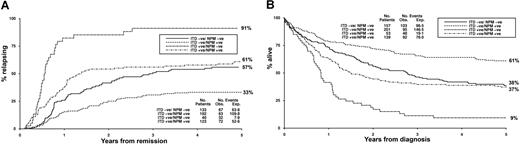
Cytogenetics were available in 977 patients, 401 (41%) of whom had an NPM1 mutation (Table 1). The majority of NPM1+ patients had a normal karyotype (340 of 401, 85%); conversely, 62% (340 of 550) of patients with a normal karyotype had an NPM1 mutation. The mutations were uncommon in patients with either a favorable (4 of 123, 3%) or adverse (4 of 88, 5%) karyotype. Since FLT3/ITDs and NPM1 mutations were both common in patients with a normal karyotype, clinical outcome according to the presence of the 2 mutations was evaluated in this subgroup of 550 patients. Overall, 28% were FLT3/ITD−NPM1−, 10% FLT3/ITD+NPM1−, 37% FLT3/ITD−NPM1+, and 25% FLT3/ITD+NPM1+. The results were not different from those observed in the total cohort. RR for FLT3/ITD−NPM1−, FLT3/ITD+NPM1−, FLT3/ITD−NPM1+, and FLT3/ITD+NPM1+ patients was 57%, 91%, 33%, and 61%, respectively, and OS 38%, 9%, 61%, and 37% (Figure 6A,B). Tests for heterogeneity showed that the impact of FLT3/ITD and NPM1 mutant status on outcome did not differ between the different cytogenetic subgroups (data not shown).
Clinical outcome in 550 patients with a normal karyotype. (A) Relapse rate. (B) Overall survival.
Clinical outcome in 550 patients with a normal karyotype. (A) Relapse rate. (B) Overall survival.
In multivariate analysis considering both the presence of an NPM1 mutation and FLT3/ITD mutant level, the favorable outcome associated with an NPM1 mutation remained an independent prognostic factor for CR and was more powerful than presentation WBC but less predictive than cytogenetics. FLT3/ITD mutant level did not enter the model (Table 3). NPM1 positivity also remained highly significant as an independent good prognostic factor for RR, but FLT3/ITD mutant level was the more powerful predictive factor. Conversely, for OS, presence of an NPM1 mutation was a better prognostic indicator than FLT3/ITD mutant level, although cytogenetics was the most powerful factor (Table 3).
Discussion
In this study we have examined the impact of mutant level, size, and number on clinical outcome in a large cohort of 354 young adult FLT3/ITD+ patients. The relative level of FLT3/ITD(s) as a proportion of total FLT3 alleles had no impact on the ability to achieve remission but was a strong predictor of survival, with a highly significant worsening in both RR and OS with increasing mutant level across the 4 groups analyzed, which was still evident in a multivariate analysis (P < .001 for both). Indeed, for RR the FLT3/ITD mutant level was the most significant predictor of outcome.
A high total mutant level (more than 50% FLT3/ITD), which is consistent with the loss of WT FLT3, was found in 15% of mutant-positive cases. Both RR and OS were significantly worse in this group than in patients with intermediate-level (25%-50%) mutant. These results are in keeping with other studies that only found a significantly adverse outcome in patients with high-level FLT3/ITD.7,8 Presence of such a high level of mutant in these cases would suggest uniparental disomy (UPD) of all or part of chromosome 13 in at least a proportion of cells.27,28 Acquisition of a biallelic mutation could occur as an early event in leukemogenesis, or it may reflect disease progression due to mitotic recombination in a cell carrying a heterozygous mutation. In keeping with the latter, several studies of paired presentation-relapse samples have reported increased levels of an FLT3/ITD at relapse.29,–31 To be selected in this way, the biallelic mutation must provide some advantage to the cell that is greater than that of a single mutated allele. For example, in our cohort there was a significantly higher WBC with increasing FLT3/ITD level, suggesting that it may lead to a greater proliferative drive. Alternatively, a biallelic mutation may provide a greater degree of chemoresistance to a cell. Presence of UPD also may reflect more widespread underlying genomic damage, with abnormalities in other genes of pathological significance leading to clonal selection. Intermediate mutant levels could represent either a heterozygous mutation in the majority of cells, with a new event required to increase mutant level, or UPD of a mutated allele in up to half of the cells that will progressively outgrow heterozygous mutant cells with time. Further studies will be required to examine whether the latter is a frequent occurrence.
In our cohort we also found that patients with a low FLT3/ITD mutant level (1%-24%) had a significantly worse RR than patients with WT FLT3, although this was less significant for OS. A low level of mutant could be due to the presence of contaminating normal (WT) cells or may indicate acquisition of an FLT3/ITD as a later, secondary event present in only a subclone of cells. Although we cannot exclude the former possibility in all cases, half of the 102 cases with a low-level FLT3/ITD were also NPM1 mutant-positive, with a median NPM1 mutant level of 43%, indicating that the majority of cells in these patients were clonal. In addition, the XCIP analysis in 11 cases further suggests that the majority of cases of low-level FLT3/ITDs are not due to nonleukemic cell contamination. Our results are in contrast to other studies, where no significant difference in outcome was found between lower level mutants and WT FLT3, although the exact cutoffs varied.7,–9 The reasons for this difference are not clear but may relate to heterogeneity of patient inclusion criteria, therapy received, and length of follow-up in other series, or the play of chance owing to the smaller number of patients in the low mutant level category. Our results do, however, indicate that presence of lower-level FLT3/ITDs cannot be excluded altogether from prognostic risk stratification. If the presence of an FLT3/ITD provides chemoresistance, for example, by enhancing DNA repair and salvage of damaged cells,32 then mutant-carrying cells, irrespective of their relative proportion in the total population, will have a survival advantage on treatment with conventional chemotherapy, and this is reflected in the higher mutant levels at relapse.29,–31 Alternatively, or in addition, FLT3/ITDs may serve as markers of the level of genomic damage in a cell and reflect other (unknown) abnormalities that lead to increased chemoresistance.
Unlike the impact of mutant level, we were unable to find a significant correlation between FLT3/ITD mutant number and clinical outcome. More than one FLT3/ITD could indicate a higher degree of susceptibility to genomic damage, and in a smaller cohort we originally reported that the number of mutants was an independent variable predicting for OS, although this may have been a chance finding due to a significantly higher number of deaths in remission in patients with more than one FLT3/ITD.10 In the current larger cohort, however, mutant number had no impact on RR or OS.
We also were unable to find a significant correlation between FLT3/ITD size and clinical outcome. The inserted sequence is thought to lead to constitutive activation of the kinase domain by disrupting the auto-inhibitory interaction between the juxtamembrane domain and activation loop that normally stabilizes the kinase in its inactive conformation and protects the ATP binding pocket.33,34 An increasing length of additional sequence may therefore influence the ability of this interaction to either partially form or be completely abolished, and could lead to progressively increasing levels of activation. However, studies of the impact of ITD length on outcome have produced contradictory results, with worse survival in patients with longer length ITDs in one study12 that was not confirmed by others.13,14 This may be due to the heterogeneity of the patients included in these studies, with a wide age range, relatively few mutant-positive patients, variation of cutoff sizes from 33 to 70 bp, and inclusion of APL patients in one study. In our large cohort of younger adult non-APL patients, where only patients with a single FLT3/ITD mutant were considered and the cohort was divided into quartiles according to length, we found no difference in RR with increasing FLT3/ITD size.
Multiple hits are thought to be required for leukemogenesis and, therefore, the impact of any one abnormality may depend to some degree on its interaction with the other cytogenetic/molecular abnormalities that are present in the same cell. In keeping with other studies, we observed a highly significant clinical benefit associated with an NPM1 mutation, and a correlation between the presence of NPM1 mutations and FLT3/ITDs.16,,,–20 However, unlike these cohorts, where the favorable effect of an NPM1 mutation was not seen in the presence of an FLT3/ITD, we found no evidence of interaction between the 2 mutations. In multivariate analysis, both were highly significant independent predictors of outcome. Of note, the intermediate prognosis in FLT3/ITD−NPM1− patients was quite distinct from the poor prognosis of FLT3/ITD+NPM1− patients, and this has implications for risk stratification and therapeutic options.
These results raise interesting questions about the underlying genetic cause, the target cell of transformation, and the biologic interaction of the 2 mutations. Roles attributed to WT NPM1 include stabilization of the critical tumor suppressors p53 and p19ARF, maintenance of genomic stability, and control of DNA repair.35 However, although the correlation between NPM1 and FLT3/ITD mutations could imply a susceptibility to genomic instability, this is not consistent with the preponderance of NPM1 mutations in patients with a normal karyotype. The relative mutant levels would suggest that, in at least a proportion of double-positive patients, the NPM1 mutation is acquired before the FLT3/ITD, which has been observed by others,19 although we found no evidence that the improved outcome with an NPM1 mutation differed between the groups with either WT or increasing FLT3/ITD mutant levels. A recent study demonstrated that the mutant NPM1 can act as an oncogene in classical in vitro transformation assays only if its pro-senescence activity is evaded, which was not achieved by cooperation with an FLT3/ITD.36 Although these assays were not performed in hematopoietic cells, it is clear that further studies are required to elucidate the biologic consequences of an NPM1 mutation.
With the development of compounds and antibodies targeting specific leukemic abnormalities, current therapy is moving toward a more individual-tailored therapeutic approach, which puts increasing importance on determining which dia-gnostic markers are relevant for risk stratification and which patients will most benefit from specific therapies. This is challenging due to the heterogeneity of AML and the potential diversity of combinations of markers and their impact on clinical outcome, and means that large cohorts of patients must be studied in order to significantly determine prognostic usefulness. In our study patients could be stratified into 3 prognostic groups using their FLT3 and NPM1 mutant status, either good (FLT3/ITD−NPM1+), intermediate (FLT3/ITD−NPM1− and FLT3/ITD+NPM1+), or poor (FLT3/ITD+NPM1−). In particular, 2 groups of patients were identified as being at particularly high risk of early relapse within of one year of achieving remission: those with either a high FLT3/ITD mutant level consistent with the loss of WT alleles or those carrying an FLT3/ITD in the absence of an NPM1 mutation. These patients may be good candidates for more experimental therapeutic approaches. However, the poor outcome associated with FLT3/ITD+ patients in our cohort would suggest that all patients carrying an FLT3/ITD may benefit from FLT3-targeted therapy, irrespective of mutant level.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to the clinical investigators who entered and managed patients in these 2 trials. The work was undertaken at University College London Hospital/University College London, which received a proportion of funding from the Department of Health's NIHR Biomedical Research Centres funding scheme.
This work was supported by the Leukaemia Research Fund of Great Britain.
A complete list of participants in the Medical Research Council Adult Leukaemia Working Party may be found in Document S1, available on the Blood website; see the Supplemental Materials link at the top of the online article.
Authorship
Contribution: Experimental analysis was performed by R.E.G., C.G., and C.A.; data analysis by R.K.H., R.E.G., D.C.L., and A.J.M. designed the study. A.K.B. is the chief trial investigator. R.E.G. and D.C.L. wrote the paper with contributions from R.K.H. and A.J.M.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Rosemary E. Gale, Department of Haematology, Royal Free and University College Medical School, University College London, 98 Chenies Mews, London WC1E 6HX, United Kingdom; e-mail: rosemary.gale@ucl.ac.uk.