Abstract
Acquired molecular abnormalities (mutations or chromosomal translocations) of the RUNX1 transcription factor gene are frequent in acute myeloblastic leukemias (AMLs) and in therapy-related myelodysplastic syndromes, but rarely in acute lymphoblastic leukemias (ALLs) and chronic myelogenous leukemias (CMLs). Among 18 BCR-ABL+ leukemias presenting acquired trisomy of chromosome 21, we report a high frequency (33%) of recurrent point mutations (4 in myeloid blast crisis [BC] CML and one in chronic phase CML) within the DNA-binding region of RUNX1. We did not found any mutation in de novo BCR-ABL+ ALLs or lymphoid BC CML. Emergence of the RUNX1 mutations was detected at diagnosis or before the acquisition of trisomy 21 during disease progression. In addition, we also report a high frequency of cryptic chromosomal RUNX1 translocation to a novel recently described gene partner, PRDM16 on chromosome 1p36, for 3 (21.4%) of 14 investigated patients: 2 myeloid BC CMLs and, for the first time, 1 therapy-related BCR-ABL+ ALL. Two patients presented both RUNX1 mutations and RUNX1-PRDM16 fusion. These events are associated with a short survival and support the concept of a cooperative effect of BCR-ABL with molecular RUNX1 abnormalities on the differentiation arrest phenotype observed during progression of CML and in BCR-ABL+ ALL.
Introduction
BCR-ABL, a constitutively activated tyrosine kinase (TK), is the product of the derived t(9;22)(q34;q11) chromosome 22 Philadelphia (Ph). The chimeric gene is present in virtually all cases of chronic myeloid leukemia (CML) and in 20% of adults with Philadelphia-positive acute lymphoblastic leukemia (ALL). During the clinical course of CML, progression from chronic phase (CP; characterized by the accumulation of apparently normal neutrophils), to a rapidly fatal blast crisis (BC; characterized by clonal expansion of differentiation-arrested myeloid or lymphoid blasts) is observed.1 The disease progression is frequently accompanied by a cytogenetic clonal evolution with the appearance of additional chromosomal aberrations besides the Ph chromosome. Ph+ ALL patients generally have a rapid disease course and a poor overall prognosis.2
Imatinib mesylate (IM) is a potent specific c-ABL TK inhibitor. In CML, even though it successfully controls the disease in CP, an initial lack of response or a recurrence of the disease after a transient initial response is usually seen in patients with advanced phases.3 The addition of IM in the therapeutic arsenal for Ph+ ALL patients has also improved their prognosis, but survival still remains poor.4 Besides the extensively studied point mutations in the BCR-ABL TK domain as the most important clinical cause of resistance in either CML or Ph+ ALL, additional acquired genetic events contributing to resistance/disease progression are not fully understood. Many indications suggest that the enhanced survival and differentiation arrest of the CML-BC cells depends on the cooperation of BCR-ABL with other genes deregulated during disease progression. Indeed, deletions or inactivation of p53, p16, and Rb have been reported to be primarily required for cell proliferation and survival in IM-treated patients.5 To date, the analysis of other relevant candidates that regulate the expression of differentiation-related genes is focused mostly on transcription factors. A recent study has identified the RUNX1/AML1 transcription factor gene as a modulator of the cellular response toward IM in vitro and in vivo in mice, suggesting its possible involvement in disease persistence in IM-resistant CML patients.6 RUNX1 formed a heterodimeric complex with a non–DNA-binding β subunit and the resulting core binding factor (CBF) complex regulates transcription of several genes relevant to both myeloid and lymphoid developments.7 The conserved Runt homology domain of 128 amino acids (exons 3 to 5) at the RUNX1 NH2-terminus is required for DNA binding and CBFβ heterodimerization, whereas the COOH-terminus (exons 6 to 8) contains transcriptional activation and repressor domains.8 Deletions, insertions, or point mutations within the Runt domain of RUNX1 have recently been identified in myeloid disorders.9 These mutations are found at highest incidence in minimally differentiated leukemias (acute myeloid leukemia [AML]–M0: 25%), in secondary myelodysplastic syndromes (MDSs), and in treatment-related AMLs, mainly with acquired trisomy 21.9,10 To date, among 92 CML and 93 ALL patients, only 4 BC CML patients harbored RUNX1 mutations in the Runt domain.10,11 Different chromosomal translocations that involve the RUNX1 gene have also been reported in human acute leukemias.6,9,10 These include, for example, the translocation t(8;21)(q22;q22) resulting in the AML1-ETO fusion gene; the t(12;21)(p12;q22) generating the TEL-AML1 fusion transcript; or less frequently the (3,21)(q26;q22) resulting in the MDS1(EVI1)-RUNX1 transcript.12,13 In all described cases, the fusion proteins mediate their oncogenic activity by dominantly repressing RUNX1 target genes.14-16 Recently, the cryptic t(1;21)(p36;q22) has also been reported as a rare but recurrent translocation associated with 5 therapy-related AML/MDS cases17 and 1 AML-M4.18 In all cases, the resulting fusion involved RUNX1 and an EVI1-like gene, PRDM16. Moreover, even if translocation is a rare event in the progression of CML, Hazourli et al19 described the t(1;21)(p36;q22) in one IM-resistant CML blast crisis, and another similar but complex RUNX1-PRDM16 fusion has been currently identified in our laboratory in an advanced CML case with acquired trisomy 21.
In this study, we report the frequent involvement of RUNX1 abnormalities in a selected Ph+ leukemia cohort of 18 patients presenting an acquired trisomy of chromosome 21. Our data reveal a high frequency of RUNX1 mutations within the Runt domain and/or cryptic translocation t(1;21)(p36;q22). These events are associated with short survival, and may support the concept of an additive effect of RUNX1 alterations in the disease progression and persistence during IM treatment.
Methods
Patients
Retrospectively, 13 advanced CML cases (1 CP-CML, 3 AP-CML, 8 myeloid BC, 1 lymphoid BC), 2 de novo Ph+ ALL-B, 1 de novo AML, and 1 therapy-related Ph+ ALL presenting acquired trisomy 21 were examined (8 females, 10 males; median age at diagnosis of the acquired trisomy 21 was 50 years [range: 35-80], Table 1). It is of note that partial data concerning UPN 18 have previously been reported.11 At time of the trisomy 21 acquisition, 10 patients had previously been treated with IM alone or in association and 10 patients had previously received conventional chemotherapy. Six patients were allografted. Response and resistance to IM therapy was assessed according to the European LeukemiaNet guidelines.20 Screening for BCR-ABL mutations was performed according to the methods described previously.21 Furthermore, 48 patients presenting CML without acquired trisomy 21 were included as control group: 33 CP-CML and 15 advanced CML (including 11 CP-CML and 8 AP-CML patients with additional cytogenetics abnormalities) (data not shown). Informed consent was obtained from all patients in accordance with the Declaration of Helsinki.
Main clinical and biological features and mutation status of patients
| UPN . | Age, y . | Sex . | Disease status at trisomy 21 . | Karyotype . | Treatments since diagnosis . | Response to IM . | BCR-ABL mutation . | RUNX1 mutation . | RUNX1-PRDM16 fusion . | Follow-up since trisomy 21, mo . | Outcome at latest follow-up . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 80 | F | Myeloid BC | 46,XX,t(9;22)(q34;q11) [9]/ 48,idem,+21,+der(22)t(9;22) [22] | Hydroxy-urea | ND | ND | K83Q | ND | 2 | Deceased |
| 2 | 49 | F | Myeloid BC (granulocytic sarcoma) | 47−48, XX, −3, der(5)t(3;5)(p21;q13), +8,t(9;22)(q34;q11),der(17)t(13;17)(q21;q23),−18,+20,+21,+mar [15] | IFN+ ARA-C radiotherapy, allograft, IM | None | ND | WT | ND | 1 | Deceased |
| 3 | 56 | F | Myeloid BC | 49,XX,−7, +8, t(9;22)(q34;q11), +19,+21,+der(22)t(9;22) [10] / 50,idem,+15[20] | IFN+ARA-C, IM, IM+ daunorubicin | CHR | ND | WT | ND | 2 | Deceased |
| 4 | 56 | F | De novo B-ALL | 51,XX,+6,+8,t(9;22)(q34;q11),+13,+21,+der(22)t(9;22) [44] / 46,XX [6] | Daunorubicin+ EDX+VCR + steroids, IM | CHR | ND | WT | ND | 3 | Deceased |
| 5 | 35 | F | Myeloid BC | 50,XX,+8,t(9;22)(q34;q11),+15, der(17)t(1;17)(q22;q22),+21,+der(22)t(9;22) [50] | IFN + ARA-C, IM, IM+ daunorubicin + ARA-C | CHR | WT | R80C | ND | 22 | Deceased |
| 6 | 40 | M | Myeloid BC | 46,XY, t(9;22)(q34;q11)[11]/ 57−60, idem, +1x2,+2,+3,add(4)(q33),+6,+8,+10,+15,+16,+21,+der(22)t(9;22)x2,+mar1,+mar2 [10] | Hydrea IFN autograft | ND | ND | WT | ND | 2 | Deceased |
| 7 | 50 | M | De novo B-ALL | 57,XY,+Xx2,+2,+4,+6,t(9;22)(q34;q11),+14,+16,+20,+21x2,+der(22)t(9;22)[22] / 46, XY[3] | Daunorubicin+ EDX+VCR+steroids, IM | Lost CCyR | ND | WT | ND | 2.5 | Deceased |
| 8 | 70 | M | Myeloid BC | 46,XY, t(9;22)(q34;q11) [8] / 46,XY, idem, i(17)(q10)[1] / 50,XY,+Y,idem,+19,+21,+der(22)t(9;22) [1] | Hydroxy-urea, ARA-C, autograft | ND | ND | WT | ND | 2 | Deceased |
| 9 | 36 | M | CP-CML | 47,XY, t(9;22)(q34;q11), +21[20] | Hydroxy-urea allograft | ND | ND | S114Stop | Yes | 144 | Alive |
| 10 | 67 | M | Myeloid BC | 59,XY,+6,t(9;22)(q34;q11),+10,+11,+13,+14,+17,+21x2,+22,+der(22)t(9;22),+mar1,+mar2,+mar t(?;17)(?;q11) [12] | Hydroxy-urea, ARA-C, idarubicin + ARA-C | ND | ND | WT | ND | 1 | Deceased |
| 11 | 56 | M | Myeloid BC | 51,XY,−6,+8,+9,t(9;22)(q34;q11)x2, add(12)(p11),−17,+19,+21,+der(22)t(9;22),+2 mar[4] / 46,XY [16] | IFN, IM, allograft | CHR | ND | L29S | Yes | NA | NA |
| 12 | 62 | F | Lymphoid BC | 48,XX,t(9;22)(q34;q11),+16,+21[3] / 46,XX [17] | IM+ARA-C; VCR+DXM, dasatinib | Lost CCyR | L384M | WT | ND | 15 | Alive |
| 13 | 50 | M | AP-CML | 52,XY,+6,+8,t(9;22)(q34;q11),+10,+17,+21,+der(22)t(9;22) [10] | IM, dasatinib | PHR | WT | WT | WT | 1 | Deceased |
| 14 | 56 | M | AML after CML myeloid BC | 46,XY,inv(3)(q21;q26),t(9;22)(q34;q11) [19] / 47,XY,idem,+21 [1] | IM, dasatinib, allograft | Lost CCyR | T315I | R80C | WT | 12 | Deceased |
| 15 | 41 | M | AP-CML | 51,XY,+7,+8,t(9;22)(q34;q11),+21, +der(22)t(9;22),+mar [12] | IFN, allograft, dasatinib | ND | ND | WT | ND | 40 | Deceased |
| 16 | 56 | F | De novo AML | 48,XX,+8, ,t(9;22)(q34;q11), add(9)(q22), del(17)(q22), +21 <inc> [4] | Dasatinib + danaurubicin DXM | ND | ND | WT | WT | 1 | Deceased |
| 17 | 40 | F | Th-ALL | 50,XX,+4,i(7)(q10),t(9;22)(q34;q11),del(12)(p11;p13),+20,+21,+der(22)t(9;22) [6] / 46,XX [2] | Dasatinib + DXM | ND | ND | WT | Yes | 3 | Alive |
| 18 | 50 | M | AP-CML | 47,XY, t(9;22)(q34;q11),+21[30] | Hydrea, IFN, ARA-C, IM, allograft | CHR | M244V | R196P | WT | 56 | Alive |
| UPN . | Age, y . | Sex . | Disease status at trisomy 21 . | Karyotype . | Treatments since diagnosis . | Response to IM . | BCR-ABL mutation . | RUNX1 mutation . | RUNX1-PRDM16 fusion . | Follow-up since trisomy 21, mo . | Outcome at latest follow-up . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 80 | F | Myeloid BC | 46,XX,t(9;22)(q34;q11) [9]/ 48,idem,+21,+der(22)t(9;22) [22] | Hydroxy-urea | ND | ND | K83Q | ND | 2 | Deceased |
| 2 | 49 | F | Myeloid BC (granulocytic sarcoma) | 47−48, XX, −3, der(5)t(3;5)(p21;q13), +8,t(9;22)(q34;q11),der(17)t(13;17)(q21;q23),−18,+20,+21,+mar [15] | IFN+ ARA-C radiotherapy, allograft, IM | None | ND | WT | ND | 1 | Deceased |
| 3 | 56 | F | Myeloid BC | 49,XX,−7, +8, t(9;22)(q34;q11), +19,+21,+der(22)t(9;22) [10] / 50,idem,+15[20] | IFN+ARA-C, IM, IM+ daunorubicin | CHR | ND | WT | ND | 2 | Deceased |
| 4 | 56 | F | De novo B-ALL | 51,XX,+6,+8,t(9;22)(q34;q11),+13,+21,+der(22)t(9;22) [44] / 46,XX [6] | Daunorubicin+ EDX+VCR + steroids, IM | CHR | ND | WT | ND | 3 | Deceased |
| 5 | 35 | F | Myeloid BC | 50,XX,+8,t(9;22)(q34;q11),+15, der(17)t(1;17)(q22;q22),+21,+der(22)t(9;22) [50] | IFN + ARA-C, IM, IM+ daunorubicin + ARA-C | CHR | WT | R80C | ND | 22 | Deceased |
| 6 | 40 | M | Myeloid BC | 46,XY, t(9;22)(q34;q11)[11]/ 57−60, idem, +1x2,+2,+3,add(4)(q33),+6,+8,+10,+15,+16,+21,+der(22)t(9;22)x2,+mar1,+mar2 [10] | Hydrea IFN autograft | ND | ND | WT | ND | 2 | Deceased |
| 7 | 50 | M | De novo B-ALL | 57,XY,+Xx2,+2,+4,+6,t(9;22)(q34;q11),+14,+16,+20,+21x2,+der(22)t(9;22)[22] / 46, XY[3] | Daunorubicin+ EDX+VCR+steroids, IM | Lost CCyR | ND | WT | ND | 2.5 | Deceased |
| 8 | 70 | M | Myeloid BC | 46,XY, t(9;22)(q34;q11) [8] / 46,XY, idem, i(17)(q10)[1] / 50,XY,+Y,idem,+19,+21,+der(22)t(9;22) [1] | Hydroxy-urea, ARA-C, autograft | ND | ND | WT | ND | 2 | Deceased |
| 9 | 36 | M | CP-CML | 47,XY, t(9;22)(q34;q11), +21[20] | Hydroxy-urea allograft | ND | ND | S114Stop | Yes | 144 | Alive |
| 10 | 67 | M | Myeloid BC | 59,XY,+6,t(9;22)(q34;q11),+10,+11,+13,+14,+17,+21x2,+22,+der(22)t(9;22),+mar1,+mar2,+mar t(?;17)(?;q11) [12] | Hydroxy-urea, ARA-C, idarubicin + ARA-C | ND | ND | WT | ND | 1 | Deceased |
| 11 | 56 | M | Myeloid BC | 51,XY,−6,+8,+9,t(9;22)(q34;q11)x2, add(12)(p11),−17,+19,+21,+der(22)t(9;22),+2 mar[4] / 46,XY [16] | IFN, IM, allograft | CHR | ND | L29S | Yes | NA | NA |
| 12 | 62 | F | Lymphoid BC | 48,XX,t(9;22)(q34;q11),+16,+21[3] / 46,XX [17] | IM+ARA-C; VCR+DXM, dasatinib | Lost CCyR | L384M | WT | ND | 15 | Alive |
| 13 | 50 | M | AP-CML | 52,XY,+6,+8,t(9;22)(q34;q11),+10,+17,+21,+der(22)t(9;22) [10] | IM, dasatinib | PHR | WT | WT | WT | 1 | Deceased |
| 14 | 56 | M | AML after CML myeloid BC | 46,XY,inv(3)(q21;q26),t(9;22)(q34;q11) [19] / 47,XY,idem,+21 [1] | IM, dasatinib, allograft | Lost CCyR | T315I | R80C | WT | 12 | Deceased |
| 15 | 41 | M | AP-CML | 51,XY,+7,+8,t(9;22)(q34;q11),+21, +der(22)t(9;22),+mar [12] | IFN, allograft, dasatinib | ND | ND | WT | ND | 40 | Deceased |
| 16 | 56 | F | De novo AML | 48,XX,+8, ,t(9;22)(q34;q11), add(9)(q22), del(17)(q22), +21 <inc> [4] | Dasatinib + danaurubicin DXM | ND | ND | WT | WT | 1 | Deceased |
| 17 | 40 | F | Th-ALL | 50,XX,+4,i(7)(q10),t(9;22)(q34;q11),del(12)(p11;p13),+20,+21,+der(22)t(9;22) [6] / 46,XX [2] | Dasatinib + DXM | ND | ND | WT | Yes | 3 | Alive |
| 18 | 50 | M | AP-CML | 47,XY, t(9;22)(q34;q11),+21[30] | Hydrea, IFN, ARA-C, IM, allograft | CHR | M244V | R196P | WT | 56 | Alive |
IM indicates imatinib mesylate; BC, blast crisis; ND, not done; IFN, interferon; ARA-C, cytarabine; WT, wild type; CHR, complete hematologic response; EDX, endoxan; VCR: vincristine; CCyR: complete cytogenetic response; NA, not available; DXM, dexamethasone; Th-ALL, therapy-related ALL; AP, accelerated phase; PHR, partial hematologic response; and UPN, unique patient number.
Karyotypes
For conventional cytogenetic analyses on bone marrow samples, GTG- or RHG-banded chromosomes and classification were performed according to the International System for Cytogenetic Nomenclature (ISCN).22
Identification of RUNX1 mutations
Genomic DNA was extracted from frozen aliquots of 107 peripheral blood leucocytes using QIAmp mini kit (Qiagen, Hilden, Germany). RUNX1 DNA-specific polymerase chain reaction (PCR) products corresponding to the Runt domain and to the COOH-terminal regions (exons 3 to 8) were obtained using previously described flanking intronic forward/reverse primers.23 After purification on QIAquick column (Qiagen), PCR fragments were sequenced on both strands according to standard methods. The sequence of each identified mutation was confirmed on another independent PCR product.
Screening for the PRDM16-RUNX1 cryptic fusion
Total RNA was extracted from frozen aliquots of 107 peripheral blood leucocytes with Trizol reagent (Life Technologies, Paisley, United Kingdom) according to the manufacturer's instructions and cDNA synthesis from 1 μg total RNA and reverse-transcription (RT)–PCR were performed as previously described.18 Briefly, cDNA PCR products corresponding to fusion transcripts were obtained using the RUNX1 exon 5– and PRDM16 exon 2–specific primers. A positive control corresponding to a cloned cDNA fragment encompassing the junction between RUNX1 exon 6 and PRDM16 exon 2 (kindly provided by Dr I. Sakai, Ehime University School of Medicine, Toon, Japan) was added to each experiment. Quality of RT-PCR was controlled for each sample by EF1α cDNA amplification using overlapping adjacent exons EF1α forward 5′-CTGGAGCCAAGTGCTAACATG-3′ and EF1α reverse 5′-CCGGGTTTGAGAACACCAGT-3′ primers.
The presence of the cryptic translocation t(1;21)(p36;q22) was confirmed by sequencing RT-PCR products according to standard procedure, and fluorescent in situ hybridization (FISH) was performed on bone marrow metaphases spreads from RUNX1-PRDM16 RT-PCR–positive patients, according to standard methods and the manufacturer's instructions using either the AML1/ETO or TEL/AML1 dual-color/dual fusion translocation probes (Vysis, Downer's Grove, IL).
RUNX1-PRDM16 mutation status
For the 2 patients presenting both RUNX1 mutation and RUNX1-PRDM16 fusion (UPN 9 and UPN 11), the mutation status of RUNX1 alleles not fused to PRDM16 have been investigated by sequencing the full RUNX1 cDNA using forward primers targeting RUNX1 exon 2, 5′-GCAGGGTCCTAACTCAATCG-3′, and reverse primers targeting RUNX1 exon 7, 5′-GTGAAGGCGCCTGGATAGT-3′. The RT-PCR products were further sequenced on both strands using nested primers forward, 5′-GATGCGTATCCCCGTAGATG-3′ and reverse, 5′-GCTGCGGTAGCATTTCTCA-3′.
Results
High frequency of RUNX1 mutations in advanced CML with acquired trisomy 21
All data are summarized in Table 1. Among the 18 analyzed patients, 6 CML cases (33%) harboring RUNX1 mutations within the runt domain have been found: 4 missense mutations (K83Q [UPN 1]; R80C [UPN 5 and 14]; L29S [UPN 11]; and a previously reported11 R139P [UPN 18]) and 1 stop mutation at codon S114 (UPN 9). All but one RUNX1-mutated patient (UPN 9) were in myeloid BC. All mutations were heterozygous, and electropherogram analysis of our sequencing data revealed a higher 2/3 level of the mutated sequence compared with the wt for 5 of the 6 mutated patients in our study (data not shown, excepted for UPN 11, Figure 3A). For the last one (UPN 14), mutated and wt sequences are at the same 1/2 level. No mutation was found within the COOH-terminal region or among investigated de novo Ph+ ALL or lymphoid BC-CML. Conversely with UPN 18 (for whom the RUNX1 mutation has been detected at diagnosis and before the acquired trisomy 21),11 retrospective sequencing analysis during UPN 14 follow-up revealed the lack of the R80C mutation at diagnosis of CML, but the emergence of the mutation was found at relapse after allografting and preceded the acquisition of trisomy 21 (data not shown).
RUNX1 is also disrupted by cryptic t(1;21)(p36;q22) translocation in advanced CML and Ph+ ALL with acquired trisomy 21
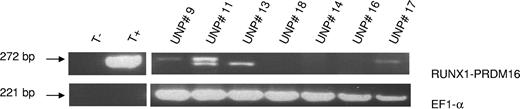
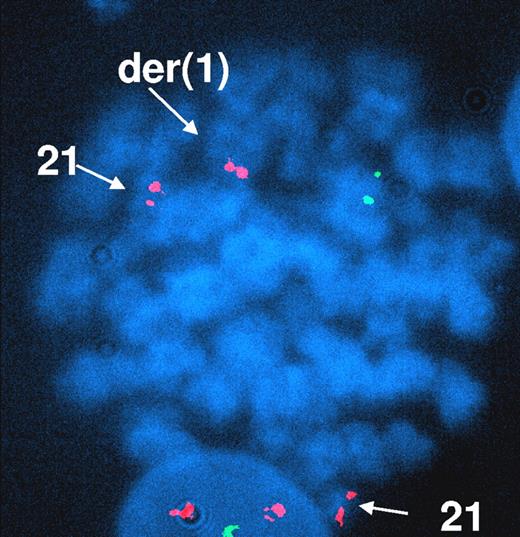
In all 18 analyzed cases, no structural abnormalities of chromosomes 1 and 21 were found by conventional cytogenetic analyses. We investigated the cryptic t(1;21)(p36;q22) by RT-PCR and/or by FISH. Data are summarized in Table 1 and in Figures 1 and 2. After sequencing the 272-bp RT-PCR products, 3 of 7 analyzed patients presenting acquired trisomy 21 (43%; 2 CML and 1 therapy-related Ph+ ALL patients; UPN 9, 11, and 17; Figure 1) were found to harbor a RUNX1 exon 5 and PRDM16 exon 2 fusion by RT-PCR as previously described.19 No RUNX1-PRDM16 fusion has been detected among the control group. FISH analyses on bone marrow metaphases using AML1/ETO or TEL/AML1, a dual fusion probe, revealed a RUNX1 signal to chromosome 1pter for one of these RT-PCR–positive cases (UPN 17), confirming the cryptic rearrangement and RT-PCR data (Figure 2). For UPN 9 and UPN 11, the t(1;21)(p36;q22) was not detected by FISH, but only few metaphases were available for the analysis (ie, for UPN 9 only 8 metaphases revealed trisomy 21 from the 30 analyzed and all revealed a normal FISH pattern, and for UPN 11 only 3 nuclei revealed trisomy 21 of the 197 analyzed). Unfortunately, as no RNA was available for the 11 remaining patients, the presence of a RUNX1-PRDM16 fusion transcript by RT-PCR could not be further investigated, and FISH analysis performed on 7 of these 11 patients revealed the absence of t(1;21)(p16;q22)-positive cells. Nevertheless, our data report the occurrence of this recurrent genomic rearrangement in a subfraction of resistant CML patients in a surprisingly high frequency (3 of 14, 21.4%) and, for the first time, in Ph+ ALL.
Ethidium bromide–stained gel showing expression of RUNX1-PRDM16 fusion transcript (upper RT-PCR product of 272 bp, confirmed by sequencing analysis) for 3 of 7 analyzed patients (UPNs 9, 11, and 17). Sequencing data after purification of the lower band revealed unspecific RT-PCR products (UPN 11 and UPN 13). The EF1α RT-PCR product (221 bp) was used as the endogenous control.
Ethidium bromide–stained gel showing expression of RUNX1-PRDM16 fusion transcript (upper RT-PCR product of 272 bp, confirmed by sequencing analysis) for 3 of 7 analyzed patients (UPNs 9, 11, and 17). Sequencing data after purification of the lower band revealed unspecific RT-PCR products (UPN 11 and UPN 13). The EF1α RT-PCR product (221 bp) was used as the endogenous control.
FISH analysis of UPN 17 using the TEL/AML1 dual-color, dual-fusion translocation probe. Three RUNX1/AML1 signals are observed in red, on 2 normal chromosomes 21 and on the derivative chromosome 1. The residual signal RUNX1/AML1 on the derivative chromosome 21 is lost. The green signal corresponds to the TEL locus 12p12 and the loss of the second green signal confirms the del(12)(p11;p13) observed on karyotyping analysis. Image was acquired with a Leica DMXRA microscope (Leica, Wetzlar, Germany) fitted with a 100×/1.30 numeric aperture oil objective, a Photometrics Sensys CCD camera (Roper Scientific, Tucson, AZ) and QFISH image acquisition software (Leica).
FISH analysis of UPN 17 using the TEL/AML1 dual-color, dual-fusion translocation probe. Three RUNX1/AML1 signals are observed in red, on 2 normal chromosomes 21 and on the derivative chromosome 1. The residual signal RUNX1/AML1 on the derivative chromosome 21 is lost. The green signal corresponds to the TEL locus 12p12 and the loss of the second green signal confirms the del(12)(p11;p13) observed on karyotyping analysis. Image was acquired with a Leica DMXRA microscope (Leica, Wetzlar, Germany) fitted with a 100×/1.30 numeric aperture oil objective, a Photometrics Sensys CCD camera (Roper Scientific, Tucson, AZ) and QFISH image acquisition software (Leica).
Co-occurrence of mutated RUNX1 fused and not fused to PRDM16: the RUNX1-PRDM16 fusion could be a secondary event to the acquired RUNX1 mutation
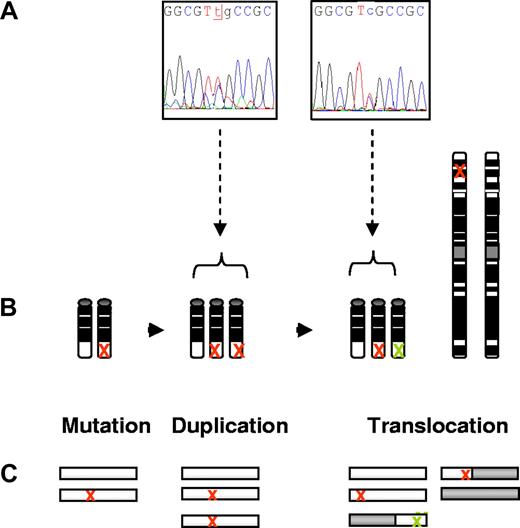
We found 2 CML patients (UPN 9 and UPN 11) harboring both a RUNX1 Runt mutation and a RUNX1-PRDM16 fusion. Sequencing the full (not fused) RUNX1 cDNA revealed after electrophoregram analysis that the previously detected mutations L29S and S144Stop were at heterozygous 1/2 level for both patients (Figure 3A for UPN 11). Because the mutations have been detected after initial DNA sequencing at a 2/3 level, this result revealed the alteration of at least 2 RUNX1 copies on both one fused and one nonfused allele for these 2 patients (see Figure 3A-C, illustrating the UPN 11 data). In fact, if the mutations were on the 2 nonfused RUNX1 alleles, sequencing analysis of the full RUNX1 cDNA would exhibit the mutations at a 2/2 level. Nevertheless, it is of note that electropherograms from sequencing data could provide only partial proofs for the ratio of full RUNX1-mutated and wild-type alleles in cases of potential contamination of nonleukemic cells. But in this study, UPN 11 is a blast crisis CML patient (harboring more than 90% of blast cells in the sample used for the analysis), and bone marrow aspiration from UPN 9 revealed only 3% of lymphocytes and 2% of erythrocytes, suggesting a majority of cells were from the myeloid lineage. Furthermore, despite the chronic phase status of UPN 9, peripheral blood count revealed a significant proportion of immature cells and a BCR-ABL/ABL transcript rate of 120% on the international scale24 (data not shown). It is therefore reasonable to consider the proportion of nonleukemic cells at a very low level in these 2 reported cases. Therefore, as illustrated in Figure 3, it is likely that during the evolution of the disease for UPN 9 and UPN 11, the mutation first arose on 1 of the 2 RUNX1 alleles, followed by its duplication through the acquisition of trisomy 21, and lastly one of the 2 altered RUNX1 copies translocated to chromosome 1p36.
Schematic representation of the different molecular events during progression of CML, supporting the co-occurrence of mutated RUNX1 alleles on one chromosome 21q22 and on the derived chromosome 1p36 harboring the RUNX1-PRDM16 fusion (example of UPN 11). (A) Sequencing data of the L29S mutation. Left: DNA-sequencing electrophoregram revealing the L29S mutation at 2/3 level. Right: RNA-sequencing electrophoregram of the full RUNX1 revealing the L29S mutation at 1/2 level. (B) Scenario of genetic and chromosomal events during the disease progression. Red cross: the mutated RUNX1 allele. Green cross: the remaining 3′ region of truncated RUNX1 gene after t(1;21)(p36;q22). (C) Representation of the corresponding various alleles of RUNX1 (white open bar) and PRDM16 (gray open bar) during CML course.
Schematic representation of the different molecular events during progression of CML, supporting the co-occurrence of mutated RUNX1 alleles on one chromosome 21q22 and on the derived chromosome 1p36 harboring the RUNX1-PRDM16 fusion (example of UPN 11). (A) Sequencing data of the L29S mutation. Left: DNA-sequencing electrophoregram revealing the L29S mutation at 2/3 level. Right: RNA-sequencing electrophoregram of the full RUNX1 revealing the L29S mutation at 1/2 level. (B) Scenario of genetic and chromosomal events during the disease progression. Red cross: the mutated RUNX1 allele. Green cross: the remaining 3′ region of truncated RUNX1 gene after t(1;21)(p36;q22). (C) Representation of the corresponding various alleles of RUNX1 (white open bar) and PRDM16 (gray open bar) during CML course.
Acquired trisomy 21 is associated with poor prognostic factor in BCR-ABL+ leukemias
The acquisition of an additional chromosome 21 is associated with poor prognosis since among the 18 patients only 4 were alive at the latest follow-up (UPN 9, 12, 17, and 18). The median survival since diagnosis of trisomy 21 was 3 months (range: 1 to 144 months). It is of note that among the 4 patients remaining alive, UPN 9 and UPN 18 have received allografts and the 2 remaining (UPN 12 and UPN 17) are Ph+ ALL cases treated with dasatinib. Acquired trisomy 21 could also be related to IM resistance since all IM-treated patients reached only CHR or relapsed after CCyR. Here, the response loss in CML cases of secondary IM resistance is associated with emergence of BCR-ABL mutations with/without RUNX1 alterations (ie, L384M for UPN 12; T315I for UPN 14). RUNX1 alterations could not directly be related to the patients' outcome in our study. Among 7 of the patients investigated for both RUNX1 mutation and RUNX1/PRDM16 fusion, wild-type RUNX1 is observed in one de novo AML (UPN 16) and one AP-CML (UPN 13).
Discussion
For the first time, we report here a high frequency (7 of 18 patients, 38%) of RUNX1 disruptions among BCR-ABL+ leukemias, either by mutation within the Runt domain or by cryptic t(1;21)(p36;q22), or both. The observation that RUNX1-mutated allele is frequently duplicated by acquired trisomy of the altered chromosome 219,10 led us to focus our analyses on a selected cohort of patients presenting acquired trisomy 21: this strategy probably resulted in the high frequency of RUNX1 abnormalities observed in this study. However, this selection deprives us of the real incidence of RUNX1 alterations in Ph+ leukemias because a high association of RUNX1 mutations with acquired trisomy 13 in myeloid malignancies (especially AML-M0) has been recently reported. But acquired trisomy 13 in CML is a very rare event.23,25
To date, mutations within the COOH-terminal region have exclusively been described as strongly correlated to sporadic MDS/AML.26 In our study, we did not show any mutation in the COOH-terminal RUNX1 region but 33% (6/18) of patients were found to have RDB missense or frameshift mutations. Five of 6 mutations were localized within exon 3 of RUNX1, and 2 of them were R80C missense mutations. Surprisingly, R80C has already been reported in only one case, also a BC-CML.27 Missense RDB mutations trigger the loss of RUNX1-DNA–binding capacity but should keep the ability to bind CBFβ. Thereby, such mutated proteins lose their RUNX1 transactivation function but may efficiently sequester CBFβ from wild-type (wt) RUNX1, conferring a dominant negative inhibitor effect.10 However, this concept has recently been revised by Cammenga et al28 who reported that the disruption of the interface that binds CBFβ does not impair RDB mutant replating activity in a murine model. Indeed, these data argue against a dominant-negative effect and more accurately support an additional DNA-binding independent oncogenic function of RUNX1 in early progenitors. Frameshift mutations within the runt domain lead to a truncated RUNX1 transcription factor without transactivation potential, and lead to the ectopic expression of the truncated protein into the cytoplasm (when the arginine residues, localized on the C-terminal part of the runt domain, are lost).27
For all RUNX1 mutations described here, the wt allele was also detected. Song et al29 suggested that the RUNX1 monoallelic mutation was sufficient enough to induce predisposition to develop acute leukemia in familial thrombocytopenia, and monoallelic RUNX1 alteration was found in 26 of 51 reported cases of RDB mutations in AML/MDS and in AML-M0.10 Interestingly, in these latter cases, and when karyotype data were available, a high frequency of acquired trisomy 21 was found. Combined with the observation of the propensity of the first mutated allele to promote alteration of the second allele by mitotic recombination in AML-M0,9 a semidominant effect could be facilitated by the duplication of the mutated allele through the acquired trisomy 21 observed in this study. Likely, electrophoregram analysis of our sequencing data on DNA revealed a higher 2/3 level of the mutated sequence compared with the wt for 5 of the 6 mutated patients in our study, supporting a 2-fold dosage of the mutated allele by duplication of the altered chromosome 21. Furthermore, we described here 2 patients harboring both RUNX1 RDB mutations and RUNX1-PRDM16 fusion, resulting in a biallelic alteration of RUNX 1 with the co-occurrence of the mutated RUNX1 fused and not fused to PRDM16. As illustrated in Figure 3, this result could suggest that the RUNX1-PRDM16 fusion could be a secondary event to the acquired RUNX1 mutation in those cases. It is of note that this scenario further confirms our FISH data revealing the t(1;21)(p36;q22) as a rare subclone. Conversely, the co-occurrence of RUNX1/AML1 mutation and RUNX1/AML1-ETO fusion has recently been reported by Auewarakul et al30 in one AML patient, revealing a new mechanism for biallelic RUNX1 alterations.
Chromosomal translocations are infrequent secondary abnormalities in CML, however there is consistent evidence of their role in the Ph+ disease progression.12,31 In all reported cases thus far, the disruption of the RUNX1 gene by translocation triggers the loss of COOH-terminal region but the Runt domain is actually conserved. When the fusion is in frame, RUNX1-associated partners associate with a corepressor complex and act as dominant-negative inhibitors of the wt allele, as it has been described in detail in t(8,21)(q22;q22) and t(3;21)(q26;q22).12,16 In this study, we report for the first time at high frequencies the t(1;21)(p36;q22) in 21.4% (3/14) of investigated Ph+ leukemias associated with trisomy 21. Because only 7 of the 14 investigated patients for RUNX1-PRDM16 could be evaluated by RT-PCR, this frequency may be underestimated since FISH analysis could not detect the t(1;21)(p36;q22) in rare subclones. And the fusion transcript subsequently produced is probably transcribed at significant high levels since only few bone marrow metaphases with trisomy 21 were indeed t(1;21)(p36;q22) positive by FISH. To date, this event has been reported in only one case of advanced disease CML19 and has never been described in Ph+ ALL. PRDM16 is an EVI1-like gene encoding a zinc finger transcription factor recently identified as potentially involved in the immortalization of bone marrow progenitors.32 Its fusion to the RUNX1 runt domain may lead to the ectopic expression of the chimeric protein and/or mediate the recruitment of transcriptional corepressors resulting in a transcriptional repression, likely to target EVI1. Among the different RUNX1-PRDM16 reported cases,17-19 various fusion transcripts produced were described, leading to expression of full or truncated chimeric proteins. In our study, the presence of the RUNX1-PRDM16 fusion was investigated using a restricted primers design corresponding to the fusion domain only (ie, RUNX1 exon 5 and PRDM16 exon 2). Therefore, we cannot exclude the presence of various RUNX1-PRDM16 transcripts in our cohort that were not investigated here, or the coexistence of full and truncated RUNX1/PRDM16 proteins. It is of note that the stop mutation observed for UPN 9 impedes the translation of the full RUNX1 and, if the mutation arises in the fused transcript, of the PRDM16 remaining sequence. It is therefore likely that for this latter case the truncation of RUNX1 arising from the translation, the mutated sequence, and, possibly, also from the fused gene is the major event, giving to PRDM16 a poor or null effect for the disease progression.
Although acquired trisomy 21 is associated with other chromosomal abnormalities for most patients in our study and suggests a progression of the disease to a more advanced phase, it is associated with poor outcomes in our study: 14 of 18 patients died, with a median survival of 3 months after the diagnosis of the acquired trisomy 21. Furthermore, none of the patients in our study achieved durable responses to IM. It is of note that a recently prepublished study revealed a protective effect of Runx1 from oncogenic insult in mouse hematopoietic stem cells.33 Combined with the role of RUNX1 alterations observed in AML and its possible involvement in disease persistence in IM-resistant CML patients,6 we can hypothesize that RUNX1 insufficiency in advanced Ph+ leukemias could support the maintenance of leukemic subclones harboring differentiation arrest associated with a higher risk of additional genetic and/or chromosomal abnormalities. We therefore could speculate that a cooperative effect between RUNX1 disruption and BCR-ABL can induce differentiation arrest of leukemic CML cells, and initiate transformation into acute leukemia. Such RUNX1 alterations may occur either during the early phases of the disease (ie, at CML diagnosis,11 UPN 18) or later, during the progression of CML (ie, before the acquisition of trisomy 21, UPN 14). These observations strongly suggest the molecular heterogeneity of CML, reflecting the genomic instability inherent to these leukemic cells. In Ph+ ALL, we report RUNX1 disruption in one therapy-related disease through RUNX1-PRDM16 fusion (UPN 17), and the de novo cases of Ph+ ALLs or Ph+ AML (UPNs 4, 7, and 16) did not show the RUNX1 alterations investigated here. However, the RUNX1-PRDM16 fusion has been reported in diseases other than therapy-related: the 2 CML patients of our study (UPNs 9 and 11) harboring RUNX1-PRDM16 never received any chemotherapy, suggesting that this fusion could represent an intrinsic additional event for leukemogenesis.
Our results strengthen the hypothesis that there are 2 main genetic alterations that could be responsible for leukemogenesis: first, an alteration of signal transduction cascades associated with cell proliferation (mainly through molecular TK aberrations) and second, deregulation of hematopoietic differentiation (through the alteration of transcriptional regulation).34 Likewise, the results presented here support the idea that BCR-ABL collaborates with other genetic alterations, such as RUNX1 abnormalities, to induce transformation and IM resistance in BCR-ABL+ leukemias. Our findings also support that a heterogeneous genetic status at diagnosis of CML may exist, underlying the need for further investigations able to more precisely define the molecular disease characteristics for better therapeutic decision adjustments.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are indebted to the Fondation pour la Recherche Médicale for its grant. This work was supported by the Laurette Fugain association and by the Canceropôle Nord-Ouest.
Authorship
Contribution: C.R.-L. and C.P. designed the study, collected and analyzed the data, interpreted data, and wrote the paper; F.-E.N., S.C., I.T., and J.-L.L. acquired and collected data; L.D., N.P., S.G., and S.J. performed research; F.-E.N. and S.C. contributed to the final version of the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Catherine Roche-Lestienne, Laboratoire de Génétique Médicale, Hôpital Jeanne de Flandre, Avenue Eugène Avinée, 59037 Lille cedex – France; e-mail: c-roche-lestienne@chru-lille.fr.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal