Abstract
Warfarin is an effective, commonly prescribed anticoagulant used to treat and prevent thrombotic events. Because of historically high rates of drug-associated adverse events, warfarin remains underprescribed. Further, interindividual variability in therapeutic dose mandates frequent monitoring until target anticoagulation is achieved. Genetic polymorphisms involved in warfarin metabolism and sensitivity have been implicated in variability of dose. Here, we describe a novel variant that influences warfarin requirements. To identify additional genetic variants that contribute to warfarin requirements, screening of DNA variants in additional genes that code for drug-metabolizing enzymes and drug transport proteins was undertaken using the Affymetrix drug-metabolizing enzymes and transporters panel. A DNA variant (rs2108622; V433M) in cytochrome P450 4F2 (CYP4F2) was associated with warfarin dose in 3 independent white cohorts of patients stabilized on warfarin representing diverse geographic regions in the United States and accounted for a difference in warfarin dose of approximately 1 mg/day between CC and TT subjects. Genetic variation of CYP4F2 was associated with a clinically relevant effect on warfarin requirement.
Introduction
Warfarin is the only oral anticoagulant approved by the US Food and Drug Administration. Although it has been used for more than 50 years, initiation of therapy remains problematic because of interindividual variability in degree of anticoagulation achieved in response to the same warfarin dose. Thus, an appropriate warfarin dose in one patient may induce a hemorrhagic event in another
Warfarin initiation is associated with one of the highest adverse event rates for any single drug.1-4 Because of concerns regarding warfarin-induced bleeding, particularly in the elderly, physicians are often reluctant to initiate warfarin therapy in patients where it is warranted. Up to one-half of patients with atrial fibrillation and no contraindication to warfarin therapy, who are also at high risk of stroke (annual risk > 4%), are currently not receiving anticoagulation therapy because of the risk perceived to be associated with such therapy by both patients and healthcare providers.5 Thus, more reliable dosing strategies for warfarin initiation that approximate optimal maintenance dosing could improve the risk-benefit ratio, allowing a broader range of patients to safely benefit from warfarin treatment.
Modeling stable dose requirements based on clinical, physiologic, environmental, and genetic factors has shown promise as a strategic approach to predict individualized stable warfarin dose requirements.6-12 Patients who have variant alleles of cytochrome P450 (CYP) 2C9, the primary enzyme that metabolizes S-warfarin, require reduced maintenance doses compared with those having wild-type alleles.13-15 Warfarin dosing variability is also attributable to genetic polymorphisms in vitamin K 2,3 epoxide reductase complex 1 (VKORC1),6,16-19 the rate-limiting enzyme in the warfarin sensitive, vitamin K-dependent gamma carboxylation system. By incorporating these factors, it may be possible to decrease the time to achieve stable dose and risk of serious or life-threatening hemorrhagic events in patients with variant alleles compared with patients with the wild-type genotype. To date, however, most pharmacogenetic models explain just more than one-half of the variation in warfarin dose,6-9 suggesting that additional genetic, environmental, medical, or personal factors are important.
Genetic screening using the Affymetrix drug-metabolizing enzymes and transporters (DMET) chip was undertaken to explore whether additional polymorphisms of drug-metabolizing enzymes might contribute to warfarin dosing variability in a clinically important way. Here we provide evidence that a polymorphism in CYP4F2 affects warfarin dose requirements, and we speculate on its mechanism of action in the vitamin K pathway.
Methods
Participating subjects
Screening/discovery cohort.
The initial cohort was recruited at Marshfield Clinic, a multispecialty group practice in Wisconsin as described.6,15 All patients recruited at Marshfield Clinic were established on a stable warfarin dose and had stable control of anticoagulation. Patients were excluded from the study if they were known to have cancer, renal or hepatic insufficiency, or congestive heart failure.
Validation cohorts.
A second cohort of patients stabilized on warfarin therapy was recruited at the University of Florida as described.7 Patients were excluded from this study if they had liver cirrhosis, advanced malignancy, hospitalization within the previous 4 weeks of the index visit, or febrile/diarrheal illness within 2 weeks of the index visit.
A third cohort of patients was recruited at Washington University in St Louis as described.20 Patients were excluded from this study if they had liver cirrhosis, advanced malignancy, or febrile/diarrheal illness within 2 weeks of the index visit.
Protocols at all 3 participating institutions were approved by the respective Institutional Review Board, and all subjects provided informed written consent in accordance with the Declaration of Helsinki for participation, and use of their genetic sample in studies aimed at understanding warfarin dose variability. To limit population stratification, all subjects in this study were white.
DMET panel testing
The Targeted Human DMET 1.0 assay (Affymetrix, Santa Clara, CA) uses molecular inversion probes and GeneChip universal tag arrays to target 1228 single nucleotide polymorphism (SNP) positions within 170 DMETs.21,22 Polymorphisms were selected for the panel as known biomarkers or likely candidates to affect drug response, including SNPs targeting CYP proteins, non-CYP enzymes, and transport proteins (Table 1). Each molecular inversion probe recognizes a specific locus but lacks the polymorphic nucleotide at the SNP site. Separate incubations with each of the 4 deoxyribonucleotide triphosphates (dNTPs) led to polymerization and ligation only if the added dNTP is complementary to the base at the target site. If this gap-filling step is successful, the probe will subsequently be polymerase chain reaction (PCR)-amplified and labeled with a different fluorescent dye for each dNTP incubation. The specificity of the allele signal is therefore determined at the gap-filling step rather than by chip hybridization, because the latter is accomplished through generic tags on the probe constructs. In addition to the biallelic assays, there are also 27 insertion/deletion probes and one triallelic probe. The DMET assay has been shown to be highly accurate, at approximately 99.8% correctly called genotypes.23
Genes containing SNPs with more than 1% minor allele frequency in the Marshfield population
| DMET metabolism . | DMET metabolism . | DMET metabolism . | DMET transport and other genes . | ||||
|---|---|---|---|---|---|---|---|
| Gene . | SNP count . | Gene . | SNP count . | Gene . | SNP count . | Gene . | SNP count . |
| ADH1B | 3 | CYP2C18 | 4 | GSTM4 | 2 | ABCB1 | 14 |
| ADH4 | 4 | CYP2C19 | 1 | GSTO1 | 1 | ABCB4 | 9 |
| ADH7 | 2 | CYP2D6 | 7 | GSTP1 | 3 | ABCB11 | 15 |
| ALDH1A1 | 1 | CYP2E1 | 7 | HNMT | 1 | ABCC1 | 10 |
| ALDH2 | 1 | CYP2F1 | 2 | MAOB | 1 | ABCC2 | 8 |
| ALDH3A1 | 2 | CYP2J2 | 1 | NAT1 | 7 | ABCC3 | 3 |
| AOX1 | 1 | CYP2S1 | 1 | NAT2 | 6 | ABCC4 | 12 |
| CDA | 2 | CYP39A1 | 2 | NQO1 | 4 | ABCC5 | 3 |
| CHST1 | 1 | CYP3A4 | 2 | POR | 1 | ABCC6 | 4 |
| CHST2 | 1 | CYP3A43 | 3 | PTGIS | 1 | ABCG2 | 1 |
| CHST3 | 13 | CYP3A5 | 1 | SULT1A1 | 1 | AHR | 2 |
| CHST5 | 4 | CYP3A7 | 1 | SULT1A2 | 1 | ATP7A | 1 |
| CHST7 | 1 | CYP4A11 | 1 | SULT1A3 | 1 | ATP7B | 10 |
| CHST10 | 5 | CYP4B1 | 6 | SULT1C1 | 1 | NR3C1 | 3 |
| CHST11 | 7 | CYP4F2 | 7 | SULT1C2 | 1 | PPARD | 38 |
| CHST13 | 2 | CYP4F8 | 4 | SULT1E1 | 2 | PPARG | 1 |
| CHST8 | 1 | CYP4F11 | 5 | SULT2A1 | 1 | RALBP1 | 3 |
| COMT | 2 | CYP4F12 | 4 | SULT2B1 | 2 | SLC10A1 | 1 |
| CYP11A1 | 1 | CYP4Z1 | 1 | SULT4A1 | 2 | SLC10A2 | 7 |
| CYP11B1 | 10 | CYP51A1 | 3 | TBXAS1 | 2 | SLC13A1 | 2 |
| CYP11B2 | 4 | CYP7A1 | 1 | TPMT | 3 | SLC15A1 | 5 |
| CYP17A1 | 3 | CYP8B1 | 1 | UGT1A1 | 5 | SLC15A2 | 5 |
| CYP19A1 | 5 | DPYD | 2 | UGT1A3 | 3 | SLC16A1 | 2 |
| CYP1A1 | 2 | EPHX1 | 7 | UGT1A4 | 1 | SLC19A1 | 2 |
| CYP1A2 | 6 | FMO1 | 4 | UGT1A6 | 2 | SLC22A1 | 7 |
| CYP1B1 | 1 | FMO2 | 10 | UGT1A7 | 1 | SLC22A2 | 2 |
| CYP20A1 | 2 | FMO3 | 6 | UGT2A1 | 2 | SLC22A3 | 2 |
| CYP24A1 | 4 | FMO5 | 1 | UGT2B4 | 2 | SLC22A4 | 3 |
| CYP2A13 | 3 | FMO6 | 4 | UGT2B11 | 1 | SLC22A5 | 3 |
| CYP2A6 | 13 | GSTA1 | 1 | UGT2B15 | 1 | SLC22A6 | 1 |
| CYP2A7 | 3 | GSTA2 | 4 | UGT2B28 | 1 | SLC22A8 | 2 |
| CYP2B6 | 8 | GSTA4 | 4 | UGT8 | 1 | SLC28A1 | 8 |
| CYP2C8 | 2 | GSTA5 | 1 | XDH | 4 | SLC28A2 | 1 |
| CYP2C9 | 2 | GSTM3 | 2 | SLC28A3 | 2 | ||
| SLC29A1 | 1 | ||||||
| SLC5A6 | 2 | ||||||
| SLC7A5 | 1 | ||||||
| SLC7A7 | 5 | ||||||
| SLCO1A2 | 3 | ||||||
| SLCO1B1 | 5 | ||||||
| SLCO1B3 | 4 | ||||||
| SLCO2B1 | 1 | ||||||
| SPG7 | 2 | ||||||
| DMET metabolism . | DMET metabolism . | DMET metabolism . | DMET transport and other genes . | ||||
|---|---|---|---|---|---|---|---|
| Gene . | SNP count . | Gene . | SNP count . | Gene . | SNP count . | Gene . | SNP count . |
| ADH1B | 3 | CYP2C18 | 4 | GSTM4 | 2 | ABCB1 | 14 |
| ADH4 | 4 | CYP2C19 | 1 | GSTO1 | 1 | ABCB4 | 9 |
| ADH7 | 2 | CYP2D6 | 7 | GSTP1 | 3 | ABCB11 | 15 |
| ALDH1A1 | 1 | CYP2E1 | 7 | HNMT | 1 | ABCC1 | 10 |
| ALDH2 | 1 | CYP2F1 | 2 | MAOB | 1 | ABCC2 | 8 |
| ALDH3A1 | 2 | CYP2J2 | 1 | NAT1 | 7 | ABCC3 | 3 |
| AOX1 | 1 | CYP2S1 | 1 | NAT2 | 6 | ABCC4 | 12 |
| CDA | 2 | CYP39A1 | 2 | NQO1 | 4 | ABCC5 | 3 |
| CHST1 | 1 | CYP3A4 | 2 | POR | 1 | ABCC6 | 4 |
| CHST2 | 1 | CYP3A43 | 3 | PTGIS | 1 | ABCG2 | 1 |
| CHST3 | 13 | CYP3A5 | 1 | SULT1A1 | 1 | AHR | 2 |
| CHST5 | 4 | CYP3A7 | 1 | SULT1A2 | 1 | ATP7A | 1 |
| CHST7 | 1 | CYP4A11 | 1 | SULT1A3 | 1 | ATP7B | 10 |
| CHST10 | 5 | CYP4B1 | 6 | SULT1C1 | 1 | NR3C1 | 3 |
| CHST11 | 7 | CYP4F2 | 7 | SULT1C2 | 1 | PPARD | 38 |
| CHST13 | 2 | CYP4F8 | 4 | SULT1E1 | 2 | PPARG | 1 |
| CHST8 | 1 | CYP4F11 | 5 | SULT2A1 | 1 | RALBP1 | 3 |
| COMT | 2 | CYP4F12 | 4 | SULT2B1 | 2 | SLC10A1 | 1 |
| CYP11A1 | 1 | CYP4Z1 | 1 | SULT4A1 | 2 | SLC10A2 | 7 |
| CYP11B1 | 10 | CYP51A1 | 3 | TBXAS1 | 2 | SLC13A1 | 2 |
| CYP11B2 | 4 | CYP7A1 | 1 | TPMT | 3 | SLC15A1 | 5 |
| CYP17A1 | 3 | CYP8B1 | 1 | UGT1A1 | 5 | SLC15A2 | 5 |
| CYP19A1 | 5 | DPYD | 2 | UGT1A3 | 3 | SLC16A1 | 2 |
| CYP1A1 | 2 | EPHX1 | 7 | UGT1A4 | 1 | SLC19A1 | 2 |
| CYP1A2 | 6 | FMO1 | 4 | UGT1A6 | 2 | SLC22A1 | 7 |
| CYP1B1 | 1 | FMO2 | 10 | UGT1A7 | 1 | SLC22A2 | 2 |
| CYP20A1 | 2 | FMO3 | 6 | UGT2A1 | 2 | SLC22A3 | 2 |
| CYP24A1 | 4 | FMO5 | 1 | UGT2B4 | 2 | SLC22A4 | 3 |
| CYP2A13 | 3 | FMO6 | 4 | UGT2B11 | 1 | SLC22A5 | 3 |
| CYP2A6 | 13 | GSTA1 | 1 | UGT2B15 | 1 | SLC22A6 | 1 |
| CYP2A7 | 3 | GSTA2 | 4 | UGT2B28 | 1 | SLC22A8 | 2 |
| CYP2B6 | 8 | GSTA4 | 4 | UGT8 | 1 | SLC28A1 | 8 |
| CYP2C8 | 2 | GSTA5 | 1 | XDH | 4 | SLC28A2 | 1 |
| CYP2C9 | 2 | GSTM3 | 2 | SLC28A3 | 2 | ||
| SLC29A1 | 1 | ||||||
| SLC5A6 | 2 | ||||||
| SLC7A5 | 1 | ||||||
| SLC7A7 | 5 | ||||||
| SLCO1A2 | 3 | ||||||
| SLCO1B1 | 5 | ||||||
| SLCO1B3 | 4 | ||||||
| SLCO2B1 | 1 | ||||||
| SPG7 | 2 | ||||||
DMET indicates drug-metabolizing enzymes and transporters; and SNP, single nucleotide polymorphism.
The laboratory protocol followed the standard process described in the Targeted Genotyping System User Guide,24 except for a preliminary PCR amplification step aimed at resolving genotypes from 29 SNPs in 14 regions containing pseudogenes and close homologs.25 The region containing CYP4F2 was not included in this class of preamplified genes.
GeneChip Targeted Genotyping Analysis software was used to make genotype calls and generate quality control (QC) metrics, including call rates, average signal, cluster quality, Hardy-Weinberg equilibrium, and reproducibility. Of the 1228 SNP assays, 11 failed because of low call rate (< 70%) and were removed from the analysis. The average call rate of the remaining 1217 SNP assays was more than 98%. Genotypes represented by the passed SNP assays were called according to either predefined boundaries (derived from historical training sets) or dynamic clustering using the current dataset. From an initial sample population of 497 patients, 491 passed all QC metrics and produced useable genotypes. In the Marshfield population, 517 SNPs across 144 genes showed a minor allele frequency more than 1% and were used in the association study. The remaining polymorphisms target rare mutations or SNPs prevalent in other racial groups. Linkage disequilibrium estimates were generated with Haploview software (Broad Institute, Cambridge, MA).26
Genotyping methods
VKORC1 genotypes were determined on 436 Marshfield Clinic samples as described6 and on the remaining 61 samples using Invader chemistry in a laboratory test developed with analyte specific reagents manufactured by Third Wave Technologies (TWT, Madison, WI). The TWT assay used analyte-specific reagents that detect CYP2C9*2, CYP2C9*3, and VKORC1 (position −1639).27
Genotyping QC procedures
To confirm results from the DMET panel, CYP4F2 genotypes were subsequently validated with 100% concordance for 9 samples using Big Dye Terminator, version 3.1 cycle sequencing with results read on an Applied Biosystems Prism 3100 Genetic Analyzer. Twenty-one additional samples were tested using Invader CYP4F2 research use only reagents provided by TWT with 100% concordance of genotype. This TWT assay was previously used in the Japanese Millennium project28,29 and the International Haplotype Map project. Analytic performance of the TWT research-use-only assay was validated using synthetic targets for both alleles and 4 sets of trios from the HapMap project obtained from Coriell Institute for Medical Research (Camden, NJ).
QC procedures for CYP4F2 at the University of Florida and Washington University in St Louis included genotyping blinded to therapeutic dose. Approximately 5% of samples were duplicate genotyped at Washington University in St Louis with concordance. Genotyping of 15 samples of known genotype was also performed at the University of Florida.
Statistical analysis
Candidate SNPs were screened for association with the residuals from the Marshfield pharmacogenetic model6 for 429 Marshfield subjects. The Kruskal-Wallis test was used to compare the distribution of residuals among observed SNP categories for each SNP. Because a large number of SNPs were to be screened, a Bonferroni correction to the usual 5% nominal level (P < .05/1228 = .00004) was set as a stringent initial threshold for statistical significance.
To validate the single SNP (rs2108622) that stood out strongly and was biologically plausible, the result was initially validated in 61 additional Marshfield subjects. The observed association was next validated in 2 independent cohorts from the University of Florida (295 white subjects, 3 missing VKORC1) and Washington University in St Louis (269 white subjects). These data were classified according to SNP genotype and evaluated relative to raw therapeutic dose and residuals from the Marshfield model. Table 2 shows the allele frequencies at the 3 sites. Genetic data were tested for deviation from Hardy-Weinberg proportions using weighted least squares estimates of allele frequencies and χ2 goodness-of-fit tests. The rs2108622 SNP did not deviate from Hardy-Weinberg equilibrium (P = .37).
Genotype by site
| . | Marshfield, % . | University of Florida, % . | Washington University in St Louis, % . |
|---|---|---|---|
| CYP2C9 | |||
| *1/*1 | 64.69 | 71.19 | 64.31 |
| *1/*2 | 18.98 | 15.93 | 20.07 |
| *1/*3 | 12.45 | 8.81 | 11.52 |
| *2/*2 | 1.43 | 3.39 | 1.49 |
| *2/*3 | 2.04 | 0.68 | 1.86 |
| *3/*3 | 0.41 | 0.00 | 0.74 |
| VKORC1 | |||
| GG | 38.78 | 36.99 | 37.92 |
| GC | 48.37 | 48.63 | 47.21 |
| CC | 12.86 | 14.38 | 14.87 |
| rs2108622 | |||
| CC | 49.59 | 47.80 | 52.42 |
| CT | 42.04 | 44.75 | 40.15 |
| TT | 8.37 | 7.46 | 7.43 |
| . | Marshfield, % . | University of Florida, % . | Washington University in St Louis, % . |
|---|---|---|---|
| CYP2C9 | |||
| *1/*1 | 64.69 | 71.19 | 64.31 |
| *1/*2 | 18.98 | 15.93 | 20.07 |
| *1/*3 | 12.45 | 8.81 | 11.52 |
| *2/*2 | 1.43 | 3.39 | 1.49 |
| *2/*3 | 2.04 | 0.68 | 1.86 |
| *3/*3 | 0.41 | 0.00 | 0.74 |
| VKORC1 | |||
| GG | 38.78 | 36.99 | 37.92 |
| GC | 48.37 | 48.63 | 47.21 |
| CC | 12.86 | 14.38 | 14.87 |
| rs2108622 | |||
| CC | 49.59 | 47.80 | 52.42 |
| CT | 42.04 | 44.75 | 40.15 |
| TT | 8.37 | 7.46 | 7.43 |
In a final set of analyses, data from all 3 sites were pooled (n = 1051 total subjects) and used to fit a series of new regression models to fully evaluate the potential contribution of rs2108622 to interindividual variability in therapeutic warfarin dose and to investigate the consistency of the effect across study sites. All models were fit with the log of stable warfarin dose as the response. The association of CYP2C9 and therapeutic warfarin dose has been well established with substantially reduced stable dose requirements in individuals with no wild-type CYP2C9 alleles (ie, no *1 alleles). However, given the uncertain effect and relatively low frequency of multiple variant CYP2C9 alleles, we did not try to quantify their effect. Instead, we conducted multiple regression analyses only on patients with at least one wild-type (CYP2C9*1) allele (n = 1009) and used the observed mean dose (log scale) as the predicted value for those with 2 CYP2C9 variants (ie, *2/*2, *2/*3, and *3/*3, n = 42). To assess the relative contribution of clinical and genetic factors, 3 models were developed using different sets of potential predictor variables: clinical factors only (sex, age, body surface area, and target international normalized ratio), clinical factors plus CYP2C9 and VKORC1, and clinical factors CYP2C9, VKORC1, and rs2108622. A modified step-down approach30 was used, starting with a “full” model, which included all potential predictors pertinent to that model, effects to allow the 3 study sites to differ, 2-factor interactions (including those by study site), and terms allowing deviations from linearity for the genetic factors age, body surface area, and target international normalized ratio. Terms were then cautiously sequentially excluded from this full model in a hierarchical fashion (no interactions retained without main effects) if they showed very weak associations with dose (P < .5 in the initial steps, P < .1 for the final model). The regression models were finalized after evaluating residuals and excluding cases with large residuals as outliers (Studentized residual > 3). At most, 8 cases (< 1% of the cohort) were excluded as outliers during model fitting (these outliers were included for model assessment, as described in the next paragraph).
The explanatory power for the 3 final models was assessed graphically and by using the adjusted r-squared (R2adj). The R2adj statistic measures the proportion of the total variability explained by the model with adjustment for the number of parameters in the model. The statistic was directly calculated to use all cases under analysis by pooling across both parts of the composite models and by applying the final regression model to any outliers dropped when estimating the parameters in the regression models.
Results
Discovery of novel SNPs affecting warfarin dose requirements
We used the DMET genotyping platform to assay 1228 SNPs. Of these, 517 were more than 1% polymorphic in the Marshfield (white) population. The 517 polymorphisms are distributed within 144 genes thought to be medically relevant (Table 1).
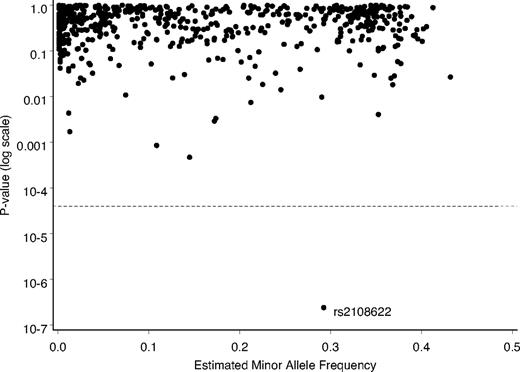
In our initial screening analysis, we tested each SNP for association with the residual variability from the Marshfield prediction model, evaluating each SNP for the proportion of residual variability explained by it after adjusting for known clinical factors and CYP2C9 and VKORC1. One SNP, rs2108622, which represents a polymorphism in CYP4F2, stood out dramatically in the analysis (Figure 1). The estimated P value of 2.4 × 10−7 was well below our stringent initial threshold for statistical significance of 4.0 × 10−5.
Depiction of relative statistical relationships of SNPs with predicted warfarin therapeutic dose, Marshfield model. The P value (log scale) for each polymorphism, comparing residuals for warfarin dose, was plotted. Dashed line shows the adjusted threshold for significance.
Depiction of relative statistical relationships of SNPs with predicted warfarin therapeutic dose, Marshfield model. The P value (log scale) for each polymorphism, comparing residuals for warfarin dose, was plotted. Dashed line shows the adjusted threshold for significance.
Signals from SNP rs2108622 on the DMET panel produced 3 well-formed and distinct genotype clusters with a Hardy-Weinberg equilibrium P value of .83 and a 100% call rate. The minor allele frequency for rs2108622 in the Marshfield population was 29.1%. Minor allele frequencies for this SNP in the Centre d'Etude du Polymorphisme Humain (from various locations in the state of Utah), Han Chinese (Beijing, China), and Japanese (Tokyo, Japan) HapMap populations31 were 23.3%, 26.7%, and 23.3%, respectively, but only 5.8% in the Yoruban (Ibadan, Nigeria) population.
Three additional SNPs on the DMET panel, rs3093114, rs3093106, and rs3093105, were in partial linkage disequilibrium with rs2108622 with r2 values of 0.42, 0.49, and 0.48, respectively, in the Marshfield population. Each showed strong association with warfarin therapeutic dose, although none of these SNPs attained statistical significance under our stringent threshold. Several other SNPs in CYP4F2 (rs3093216, rs3093211, rs3093209, rs3093207, rs3093199, rs3093193) from the PGA European Panel (http://pga.gs.washington.edu) were found in linkage disequilibrium, with rs2108622 with r2 values ranging from 0.70 to 0.75. Of these listed SNPs in linkage disequilibrium with rs2108622, only rs3093105 produces a nonsynonymous change in the coding sequence.
Validation of SNP rs2108622
Results for 61 new Marshfield cases were obtained for initial confirmation of the association between rs2108622 and warfarin dose. In these new cases, the residuals showed association with rs2108622 (P = .023). Based on validation in this small Marshfield sample, the association was further tested in 2 independent cohorts from geographically distinct regions of the country.
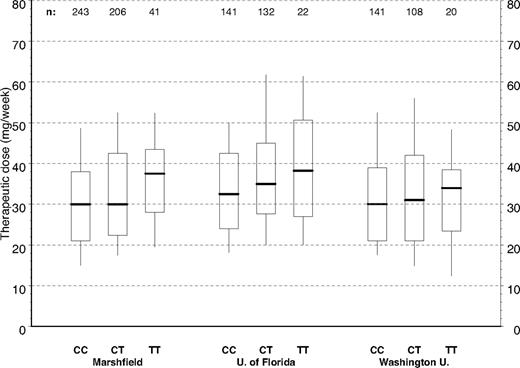
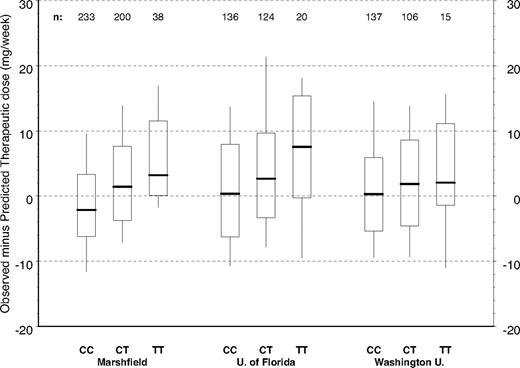
Figure 2 depicts the distribution of warfarin therapeutic dose by CYP4F2 genotype for the full Marshfield cohort and the 2 validation cohorts. The raw data in Figure 2 have not been adjusted for other clinical or genetic factors; but by inspection of the relationships between dose and CYP4F2, genetic variants are consistent across the 3 study groups with CC homozygotes requiring less warfarin. TT homozygotes required more warfarin and heterozygotes (CT) required intermediate warfarin doses. This pattern is repeated in Figure 3 where we have controlled for clinical factors, as well as CYP2C9 and VKORC1 genotypes, by evaluating the distributions of residuals from the Marshfield model. Study site–specific analyses of these relationships yielded statistically significant results for the Marshfield (P < .001) and University of Florida (P = .027) study cohorts; results for the Washington University in St Louis cohort (P = .382) were not statistically significant, but the trend was consistent across sites.
Warfarin therapeutic dose by CYP4F2 genotype and study site. Box plots of dose by study site and CYP4F2 (all cases). Boxes extend from the 25th to the 75th percentiles, with a horizontal line at the median and vertical lines extending to the 10th and 90th percentiles.
Warfarin therapeutic dose by CYP4F2 genotype and study site. Box plots of dose by study site and CYP4F2 (all cases). Boxes extend from the 25th to the 75th percentiles, with a horizontal line at the median and vertical lines extending to the 10th and 90th percentiles.
Marshfield model residuals by CYP4F2 genotype and study site (*1 genotypes only). Model adjusts for age, gender, body surface area, indication for warfarin, VKORC1, and CYP2C9. Boxes extend from the 25th to the 75th percentiles, with a horizontal line at the median and vertical lines extending to the 10th and 90th percentiles.
Marshfield model residuals by CYP4F2 genotype and study site (*1 genotypes only). Model adjusts for age, gender, body surface area, indication for warfarin, VKORC1, and CYP2C9. Boxes extend from the 25th to the 75th percentiles, with a horizontal line at the median and vertical lines extending to the 10th and 90th percentiles.
Multiple regression models were fit to the combined data from all 3 study sites to better assess the overall contribution of CYP4F2 to variability in dose. Results from the models are summarized by predictor contribution to variation in therapeutic warfarin dose in Table 3. Clinical factors alone explained approximately 17% of the overall variability among patients in therapeutic warfarin dose. Clinical factors together with CYP2C9 and VKORC1 genotype explained approximately 54% of the variability, whereas the addition of CYP4F2 genotype increased this R2adj to 56%.
Explained variation in therapeutic warfarin dose pooled models
| Predictor/predictor group . | Adjusted R2 . |
|---|---|
| Clinical only | 0.17 |
| Clinical plus CYP2C9 and VKORC1 | 0.54 |
| Clinical plus CYP2C9 and VKORC1 and CYP4F2 | 0.56 |
| Predictor/predictor group . | Adjusted R2 . |
|---|---|
| Clinical only | 0.17 |
| Clinical plus CYP2C9 and VKORC1 | 0.54 |
| Clinical plus CYP2C9 and VKORC1 and CYP4F2 | 0.56 |
Clinical variables were sex, age, body surface area, and target international normalized ratio.
Study site–specific effects were evaluated in the multiple regression models to examine the consistency of dosing and CYP4F2 effects. Some difference in the general level of dosing was observed by study site (Figure 2), with somewhat higher doses at the University of Florida (P = .006) and slightly higher doses at Washington University in St Louis (P = .15) relative to Marshfield. With respect to CYP4F2, study site effects were significant at Washington University in St Louis (P = .029) but not the University of Florida (P = .19) relative to Marshfield. Study site–specific estimates of the CYP4F2 effect in the pooled model showed a 12% increase with each T allele at Marshfield compared with 7% at the University of Florida and 4% at Washington University in St Louis.
Discussion
We have identified a DNA variant in CYP4F2 that has a clinically important impact on stable warfarin dose. In our pooled analyses, patients with 2 TT alleles require approximately 1 mg/day more warfarin than patients with 2 CC alleles.
Because it is frequently difficult to generalize the original association between a gene variant and phenotype,32 we replicated our original observation in 2 additional white cohorts of patients stabilized on warfarin. Importantly, all 3 cohorts demonstrated a consistent effect of CYP4F2 rs2108622 on warfarin dose. There was a 4% to 12% increase in the warfarin dose per T allele. We postulate that the observed study site–related variation in effect may be the result of differences among cohorts with respect to other genetic and clinical factors not included in our current model. However, because no other studies have pooled data from multiple, independent study sites to estimate the effects of polymorphisms on warfarin stable dose, we do not know whether the effects we have observed are typical of pooled study site models compared with single study site models. This would more likely be the result if single study site models were developed on populations that were more genetically homogeneous as is the case with the Marshfield model.
Because of differences in the frequency of the underlying genetic variants among major racial groups, the potential clinical benefit from prospective CYP4F2 genotyping varies by race. In whites and Asians, the minor allele frequency for CYP4F2 is approximately 30% compared with approximately 7% in blacks (http://www.ncbi.nlm.nih.gov/SNP/index.html). Accordingly, from a population perspective, the expected contribution of this polymorphism to stable warfarin dose in blacks is likely to be less than in whites and Asians.
The physiologic role of CYP4F2 in the vitamin K/warfarin pathway is unknown. It is known, however, that CYP4F2 hydroxylates the tocopherol phytyl side chain as the first step in the inactivation pathway of vitamin E.33 Given the similarity of the vitamin E and vitamin K side chains, CYP4F2 may hydroxylate the vitamin K phytyl side chain. Because CYP4F2 is the major cytochrome responsible for synthesis of 20-hydroxyeicosatetraenoic acid (20-HETE) in human kidney,34 the effect of CYP4F2 on warfarin dose could also be mediated through 20-HETE production.
The rs2108622 polymorphism in CYP4F2 affects enzyme activity. It decreased 20-HETE production in a reconstituted recombinant protein system to approximately 60% of the wild-type enzyme.35 We postulate that individuals with T alleles have decreased function of the enzyme, thereby increasing the individual's requirement for warfarin.
Although recent studies11,12,35,36 demonstrate that using dosing models that include genetic testing may improve warfarin dosing, we have not presented a clinical model for use in prospectively dosing patients initiating warfarin therapy. However, we expect that such models will be forthcoming and that these models will continue to evolve as data from more patients at additional study sites become available. We also expect that, as those models improve with the discovery of additional genes or other factors, they will be important in clinical warfarin dosing.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Marshfield Clinic Research Foundation for its support through the assistance of Linda Weis and Alice Stargardt in the preparation of this manuscript. The authors also thank WaiWai Chang, Leewin Chern, Queeny Dong, Sai Htun, Richard Lao, Carlos Montesinos, Karen Tran, and Hanna Viernes of the Affymetrix Services Laboratory, and Duncan Foster of TWT for his assistance teaching the Third Wave assays.
This work was supported by Affymetrix Inc, Marshfield Clinic Research Foundation, and National Heart, Lung, and Blood Institute grant R01 HL074724.
National Institutes of Health
Authorship
Contribution: M.D.C., T.A., P.G., R.L.B., J.K.B. designed the research; J.K.B. wrote the first draft of the manuscript; M.D.C., T.A., M.F., P.G., J.H., Y.T., J.A.J., T.Y.L., B.F.G., C.E., C.K., A.B., J.R.S., I.G., H.J.V., S.H.Y., K.Q.Z., and R.L.B. provided critical review and revisions to the manuscript; K.Q.Z. provided technical support.
Conflict-of-interest disclosure: T.A., M.F., P.G., J.H., and Y.T. are employees of Affymetrix Inc, the manufacturer of the DMET GeneChip. A.B. is an employee of TWT, the company that supplied reagents. All other authors declare no competing financial interests.
A provisional patent application for the use of CYP4F2 genetic testing has been filed.
Correspondence: James K. Burmester, Center for Human Genetics, Marshfield Clinic Research Foundation, 1000 North Oak Avenue, Marshfield, WI 54449; e-mail: burmester.jim@mcrf.mfldclin.edu.