Abstract
CD8+ T-cell responses to persistent viral infections are characterized by the accumulation of an oligoclonal T-cell repertoire and a reduction in the naive T-cell pool. However, the precise mechanism for this phenomenon remains elusive. Here we show that human cytomegalovirus (HCMV)–specific CD8+ T cells recognizing distinct epitopes from the pp65 protein and restricted through an identical HLA class I allele (HLA B*3508) exhibited either a highly conserved public T-cell repertoire or a private, diverse T-cell response, which was uniquely altered in each donor following in vitro antigen exposure. Selection of a public T-cell receptor (TCR) was coincident with an atypical major histocompatibility complex (MHC)–peptide structure, in that the epitope adopted a helical conformation that bulged from the peptide-binding groove, while a diverse TCR profile was observed in response to the epitope that formed a flatter, more “featureless” landscape. Clonotypes with biased TCR usage demonstrated more efficient recognition of virus-infected cells, a greater CD8 dependency, and were more terminally differentiated in their phenotype when compared with the T cells expressing diverse TCR. These findings provide new insights into our understanding on how the biology of antigen presentation in addition to the structural features of the pMHC-I might shape the T-cell repertoire and its phenotype.
Introduction
The αβ T-cell receptor (TCR) recognizes immunogenic peptides in association with the major histocompatibility complex (pMHC) on the surface of virus-infected cells.1,2 Upon recognition of foreign antigen, the naive T-cell repertoire responds to produce either polyclonal, oligoclonal, or clonal immune responses. The diversity of the TCR repertoire is created primarily due to differences in the sequence of the hypervariable complementarity determining region 3 (CDR3) of each chain of the TCR.3,4 The CDR3 loops are formed at the junction of the V(D)J gene segments as a result of random nucleotide addition and removal during the gene rearrangement process in the thymus.5,6 Numerous factors shape the T-cell repertoire, including thymic selection of the naive T-cell repertoire, TCR avidity for the pMHC complex, antigen load, and duration of the pMHC-TCR interaction.7-10 However, the relative contribution of these in shaping the TCR repertoire remains unclear
TCR repertoire selection is also complicated by MHC class I–restricted T cells interacting with peptides of noncanonical and canonical length. Typically, noncanonical peptides of more than 10 amino acids in length bulge from the peptide-binding cleft,11-17 and this unusual feature can result in biased selection of the TCR repertoire, suggesting the structure of the pMHC complex plays an important role in shaping the TCR repertoire. Conversely, featureless canonical 8- to 9- amino acid long epitopes from both persistent and nonpersistent viruses can also induce a highly biased T-cell repertoire.18-21 Collectively, these studies highlight that a variety of factors shape the TCR repertoire.
Here we have examined how endogenous antigen presentation and the structural landscape of the pMHC complex might together determine the character of CD8+ T-cell responses during a persistent viral (human cytomegalovirus; HCMV) infection in humans. As such, we have developed a model system based on 2 immunodominant HCMV epitopes derived from the structural protein, pp65. These epitopes, the noncanonical 103CPSQEPMSIYVY111 (CPS) and the canonical 188FPTKDVAL195 (FPT), are restricted through HLA B*3508.22 Given that these 2 epitopes are highly immunogenic, derived from the same antigenic source, and restricted through the identical MHC alleles, this model provides an ideal opportunity to investigate the role of pMHC in shaping the TCR repertoire. Here, we present evidence that competing levels of endogenous epitope presentation is pivotal in shaping the T-cell repertoire during a persistent viral infection.
Methods
These studies were approved by the Human Research Ethics Committee of the Queensland Institute of Medical Research. Informed consent from all blood donors was obtained in accordance with the Declaration of Helsinki.
Cell lines
The human fibroblast cell line, MRC-5 was used as host for HCMV infection and was maintained in growth medium (10% fetal calf serum [FCS] in RPMI 1640 medium). Phytohemagglutinin blasts were generated as described previously.23 Peripheral blood mononuclear cells (PBMCs), isolated from healthy donors after informed consent, were used to expand HCMV-specific CTL clones and polyclonal T-cell lines.23 CTL cultures were generated by culturing PBMCs (2 × 106 per 2 mL well) in 10% FCS/RPMI 1640 with autologous PBMCs that had been precoated with an HCMV peptide epitope (0.1 μM for 1 hour, responder:stimulator ratio of 2:1). Cultures were supplemented with rIL2 (20 U/mL) on day 3, split on day 7, and analyzed on day 10. CTL cultures were tested in for cytotoxicity in the standard 5-hour chromium release assay. HLA-peptide multimers (ProImmune, Oxford, United Kingdom) were also used to confirm the specificity of some CTL cultures.
MHC-peptide pentamer and TRBV staining
PBMC or HCMV-specific T cells from healthy virus carriers were incubated for 30 minutes at 4°C with an APC-labeled HLA-B*3508-CPS pentamer or with an APC-labeled HLA-B*3508-FPT pentamer (Pro-Immune). Cells were then washed twice and labeled for 30 minutes at 4°C with Tricolor-labeled antibody to human CD8 (Invitrogen, Carlsbad, CA). Cells were again washed and analyzed on a FACSCanto with FACSDiva software (BD Biosciences, Palo Alto, CA). In some experiments these T cells were also co-stained with anti-Vβ 1 (TRBV9), 2 (TRBV20), 3 (TRBV28), 5.1 (TRBV5), 5.2 (TRBV5), 5.3 (TRBV5), 6.7 (TRBV7), 7 (TRBV4), 8 (TRBV12), 11 (TRBV25), 12 (TRBV10), 13.1 (TRBV6), 13.6 (TRBV6), 14 (TRBV27), 16 (TRBV14), 17 (TRBV19), 20 (TRBV30), 21.3 (TRBV11), 22 (TRBV2) and 23 (TRBV13)-FITC (Serotec, Raleigh, NC); anti-Vβ4 (TRBV29), 9 (TRBV3), and 18 (TRBV18)-PE (Immunotech, Marseille, France). Briefly, these cells were initially stained with anti–CD3 PE-Cy7 and anti–CD8 PE-Cy5 for 30 minutes at 4°C and then stained with the relevant pentamer. After extensive washing with phosphate-buffered saline (PBS) (supplemented with 1% FCS) these cells were stained with a panel of monoclonal Vβ antibodies for 30 minutes at 4°C. Cells were washed and fixed in 1% paraformaldehyde and analyzed on a FACS Canto using FACS Diva software (BD Biosciences).
IFN-γ ELISPOT assay
The Enzyme-linked Immunospot assay used in this study has been described previously.23 Briefly, 96-well multiscreen plates were coated overnight at 4°C with 1 μg/mL anti–human interferon (IFN)–γ mAb 1-D1K (Mabtech, Mosman, NSW, Australia). The plates were washed with PBS and blocked with culture medium. Between 2.5 × 104 and 2 × 105 cells/well were incubated for 24 hours at 37°C with 1 μg/mL of the relevant peptide (Mimotopes, Clayton, Australia). The plates were washed to remove cells, and cytokine bound to the nitrocellulose was detected using biotinylated anti–human IFN-γ, 7-B6–1 (Mabtech), followed by streptavidin alkaline phosphatase (Sigma-Aldrich, Castle Hill, Australia). ELISPOTS were developed using the substrate 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium (Sigma-Aldrich) and counted automatically using image analysis software.
Cytotoxicity assay
MRC-5 were infected with AD169 at an MOI of 5, 2.5, 0.5, 0.25, or 0.05 for 2 or 16 hours and then labeled with 51Cr (Amersham Biosciences, Pittsburgh, PA) for 1 hour at 37°C. After incubation, these cells were washed in growth medium and used as targets in a standard cytotoxicity assay.24
Intracellular cytokine staining
2.5 × 104 MRC-5 cells were seeded into a 48-well plate containing 500 μL growth medium, and 24 hours later were infected with the Ad5F35 HLA B*3508 construct at an MOI of 5:1. Twelve hours later these cells were infected with AD169 at an MOI of 0.5 for 1, 2, 16, or 24 hours. HCMV-specific CD8+ T cells (5 × 105) were added to the HCMV-infected and HLA B*3508+ MRC-5 cells, along with GolgiPlug. After 4 hours' incubation at 37°C, the HCMV-specific CD8+ T cells were then analyzed for intracellular IFNγ production and CD107a mobilization using the Cytofix/Cytoperm Plus (with GolgiPlug) kit (BD Pharmingen, San Diego, CA) according to the manufacturer's instructions. Cells were analyzed using the FACS Canto and FACS Diva software (BD Biosciences) and expressed as a percentage of the positive control (peptide-pulsed target cells).
Protein expression, purification, and crystallization
The HLA B*3508-CPS and HLA B*3508-FPT complexes were expressed and purified as described.25 Crystals of the HLA B*3508-CPS and HLA B*3508-FPT complexes were formed at 4°C with the hanging-drop vapor-diffusion technique, in which an equal volume of HLA B*3508-CPS or HLA B*3508-FPT (10 mg/mL) was mixed with the reservoir (200 mM ammonium acetate, 16% PEG 3350, and 100 mM cacodylate, pH 7.6).
Data collection, structure determination, and refinement
Crystals were soaked in reservoir solution containing increasing increments of glycerol as a cryoprotectant (5%, 10%, and 15%) and then flash frozen before data collection. Data were collected at Bio-CARS (Advanced Photon Source, Argonne, IL) and was processed and scaled using the HKL suite.26 The HLA-B35 complex structures were refined from an HLA B*3508 structure that was previously determined in our laboratory.14 The model was built/refined using ARP/wARP and Refmac/COOT. The progress of refinement was monitored by Rfactor and Rfree values. Rigid-body refinement and simulated annealing were used in the first instance, but in later rounds energy minimization and B-individual refinement were used to improve the quality of the model. See Table S1 for the final refinement and model statistics (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Generation of recombinant adenovirus encoding HLA B*3508
The assembly and production of the recombinant Ad5F35-based adenovirus was completed in 3 stages using a highly efficient, ligation-based protocol of the Adeno-X System (Clontech, Palo Alto, CA). Firstly, the wild-type or mutated (with CD8-binding site modified) HLA B*3508 cDNA sequence was cloned into the pShuttle expression vector. Following amplification in Escherichia coli, the expression cassette from pShuttle was cloned into an Ad5F35 expression vector. The recombinant Ad5F35 vector was transfected into human embryonic kidney (HEK) 293 cells, and recombinant adenovirus (referred to as HLA B*3508 wt or mut) was harvested from the transfected cells by successive freeze-thawing cycles.
Molecular analysis of TRAV and TRBV usage
RNA was extracted from whole PBMCs or antigen-specific CD8+ T cells using TRIzol reagent (Invitrogen Life Technologies, Mount Waverley, Australia) as per the manufacturer's instructions. RNA was subjected to SuperScript III Reverse Transcriptase (Invitrogen Life Technologies) using antisense TCRα and TCRβ chain primers (5′-GTT GCT CCA GGC CAC AGC ACT G-3′ and 5′-TAT CTG GAG TCA TTG AGG GCG GGC T-3′, respectively). PCR was done in a 25-μL reaction containing a final concentration of 0.2 mM dNTP (Promega, Madison, WI), 2 mM MgCl2 (Applied Biosystems, Foster City, CA), 10× buffer (Applied Biosystems), and 1.25 U AmpliTaq Gold (Applied Biosytems) with 0.2 μM TCR Cβ constant primer and one of the region-specific primers (0.2 μM). Thirty-five cycles were completed on a GeneAmp PCR System 9700 (Applied Biosystems) under the following conditions: 95°C 20 seconds, 58°C 40 seconds, and 72°C 1 minute. A 30-minute final extension was included. Samples were run on a 2% agarose gel at 90 V to confirm correct band size.
TCR immunoscope analysis
PCR product from the TRBV PCR was used in an elongation reaction using Platinum GenoTYPE Tsp DNA Polymerase (Invitrogen Life Technologies). A 5′-6-FAM-labeled CβP primer was used (5′-TTC TGA TGG CTC AAA CAC-3′). After a 95°C 10-minute denaturation stage, 7 cycles of 95°C 20 seconds, 55°C 40 seconds, and 72°C 1 minute were completed. Products were diluted 1:10 in DEPC H2O and added to GeneScan-500 ROX Standards (Applied Biosystems) and formamide. Samples were heat denaturated by boiling for 2 minutes before being run on an Applied Biosystems 3700 Sequencer. Results were analyzed using ABI PRISM GeneScan software (Applied Biosystems).
CDR3 sequencing
TRBV PCR product was purified using the Qiagen MinElute PCR Purification Kit (Qiagen, Doncaster, Australia) and was used to transform DH5α competent cells (Subcloning Efficiency DH5α Cells; Invitrogen Life Technologies) using the pGEM-T Vector System I kit (Promega) according to manufacturer's instructions. Cells were plated out onto agarose plates containing X-galactose and isopropyl-beta-D-thiogalactopyranoside (IPGT; Sigma-Aldrich) and left to grow overnight at 37°C. Colonies were picked and a PCR screen done using vector-specific primers to confirm correct product size. The DNA was purified as above and a sequencing reaction done using the ABI PRISM BigDye Termination Reaction kit (Applied Biosystems). Sequencing products were purified using the Qiagen DyeEx 2.0 Spin Kit according to the manufacturer's instructions. Sequences were analyzed on the Applied Biosystems 3700 Sequencer (Applied Biosystems) and using the IMGTV-Quest software.
Results
CD8+ T cells specific for HCMV epitopes restricted through identical HLA class I allele display contrasting TCR architecture
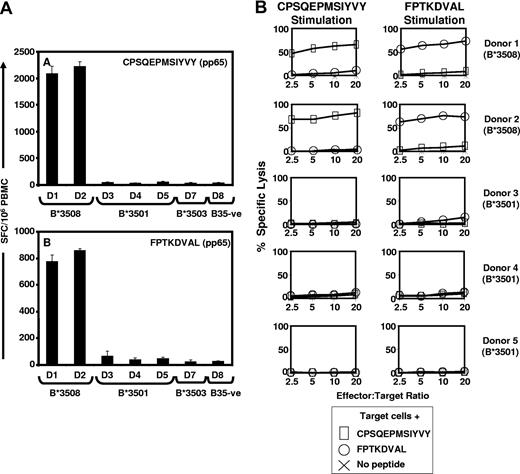
Recent studies have shown that HLA micropolymorphisms can significantly impact on the selection of CD8+ T-cell epitopes from HCMV antigens.22 This is illustrated by HLA B35-restricted CD8+ T-cell responses to the pp65 epitopes, CPSQEPMSIYVY and FPTKDVAL (herein referred to as CPS and FPT, respectively), which are exclusively recognized by HLA B*3508+ individuals, while HLA B*3501+, HLA B*3503+, and HLA B35− donors showed no significant ex vivo response to these epitopes (Figure 1A). These observations were also confirmed by in vitro stimulation of T cells, which showed that these epitopes selectively expanded T cells from HLA B*3508+ individuals (Figure 1B).
The HCMV-encoded CD8+ T-cell epitopes CPS and FPT are exclusively recognized by HLA B*3508 individuals. (A) Healthy HCMV seropositive donors (shown as D1-D8) who expressed different HLA B35 subtypes were assessed for CPS- and FPT-specific T-cell responses using ELISPOT assays as described in “Methods.” The results are expressed as spot forming cells (SFC) per 106 PBMC. (B) In vitro stimulation of T cells with CPS and FPT epitopes confirmed effector-memory responses exclusively in HLA B*3508 individuals. Polyclonal CTL cultures were generated from 2 HLA-B*3508+ and 3 HLA-B3501+ healthy HCMV-exposed individuals by stimulation with CPS or FPT peptide epitopes. CTLs were tested as effectors at a range of E:T ratios for recognition of autologous phytohemagglutinin (PHA) blast target cells pretreated with the peptide used to stimulate the CTLs or left untreated. The HLA-A/B types of the donors were as follows: donor 1: HLA A2, A2, B*3508, B57; donor 2: HLA A23, A32, B*3508, B49; donor 3: HLA A1, A3, B8, B*3501; donor 4: HLA A11, A32, B*3501, B44, and donor 5: HLA A11, A24, B*3501, B60.
The HCMV-encoded CD8+ T-cell epitopes CPS and FPT are exclusively recognized by HLA B*3508 individuals. (A) Healthy HCMV seropositive donors (shown as D1-D8) who expressed different HLA B35 subtypes were assessed for CPS- and FPT-specific T-cell responses using ELISPOT assays as described in “Methods.” The results are expressed as spot forming cells (SFC) per 106 PBMC. (B) In vitro stimulation of T cells with CPS and FPT epitopes confirmed effector-memory responses exclusively in HLA B*3508 individuals. Polyclonal CTL cultures were generated from 2 HLA-B*3508+ and 3 HLA-B3501+ healthy HCMV-exposed individuals by stimulation with CPS or FPT peptide epitopes. CTLs were tested as effectors at a range of E:T ratios for recognition of autologous phytohemagglutinin (PHA) blast target cells pretreated with the peptide used to stimulate the CTLs or left untreated. The HLA-A/B types of the donors were as follows: donor 1: HLA A2, A2, B*3508, B57; donor 2: HLA A23, A32, B*3508, B49; donor 3: HLA A1, A3, B8, B*3501; donor 4: HLA A11, A32, B*3501, B44, and donor 5: HLA A11, A24, B*3501, B60.
Next we analyzed fresh PBMCs from HLA B*3508+ healthy virus carriers (donor 1: HLA A2, A2, B*3508, B57; donor 2: HLA A23, A32, B*3508, B49, and donor 6: HLA A30, A32, B*3508, B42) using pMHC pentamers in combination with a panel of monoclonal TRBV antibodies. Ex vivo staining of fresh PBMCs from donors 1 and 2 revealed that 5.9% and 3.71% of CD8+ T cells stained with CPS-pentamers, respectively, while 2.32% and 2.34% of CD8+ T cells stained with FPT pentamers. Co-staining of CPS-specific T cells with TCR-specific antibodies revealed a highly biased TCR repertoire with more than 80% of the T cells expressing TRBV28 (Figure 2A,B). Conversely, we could not detect a predominating TRBV usage for FPT-specific T cells (Figure 2C,D), indicative of an unbiased T-cell response. Next we expanded CPS- and FPT-specific T cells following stimulation with synthetic peptides and reanalyzed TCR usage by using pMHC pentamers in combination with the panel of monoclonal TRBV antibodies (Figure 2E-H). Although in vitro–expanded CPS-specific T cells showed no change in the TCR usage (Figure 2E,F); a donor-specific focusing of FPT-specific T cells was observed with unique TRBV expansions in each donor (Figure 2G,H).
Ex vivo analysis of TRBV usage by CPS- and FPT-specific T cells. PBMCs from healthy virus carriers were co-stained with pMHC pentamers (CPS or FPT) and a panel of TRBV monoclonal antibodies. Data based on fresh PBMCs is presented in panels A and B (for CPS-specific T cells) and panels C and D (for FPT-specific T cells). After in vitro culture these T cells were re-analyzed using pMHC-pentamers and a panel of TRBV monoclonal antibodies. Data for CPS-specific T-cell cultures is presented in panels E and F, while data for FPT-specific T cells is displayed in panels G and H. The results are expressed as percent pMHC-pentamer-specific cells. Data presented in panels A, C, E, and G are from donor 1; data in panel B and D are from donor 2; while data in panels F and H are from donor 6 (HLA A30, A32, B*3508, B42).
Ex vivo analysis of TRBV usage by CPS- and FPT-specific T cells. PBMCs from healthy virus carriers were co-stained with pMHC pentamers (CPS or FPT) and a panel of TRBV monoclonal antibodies. Data based on fresh PBMCs is presented in panels A and B (for CPS-specific T cells) and panels C and D (for FPT-specific T cells). After in vitro culture these T cells were re-analyzed using pMHC-pentamers and a panel of TRBV monoclonal antibodies. Data for CPS-specific T-cell cultures is presented in panels E and F, while data for FPT-specific T cells is displayed in panels G and H. The results are expressed as percent pMHC-pentamer-specific cells. Data presented in panels A, C, E, and G are from donor 1; data in panel B and D are from donor 2; while data in panels F and H are from donor 6 (HLA A30, A32, B*3508, B42).
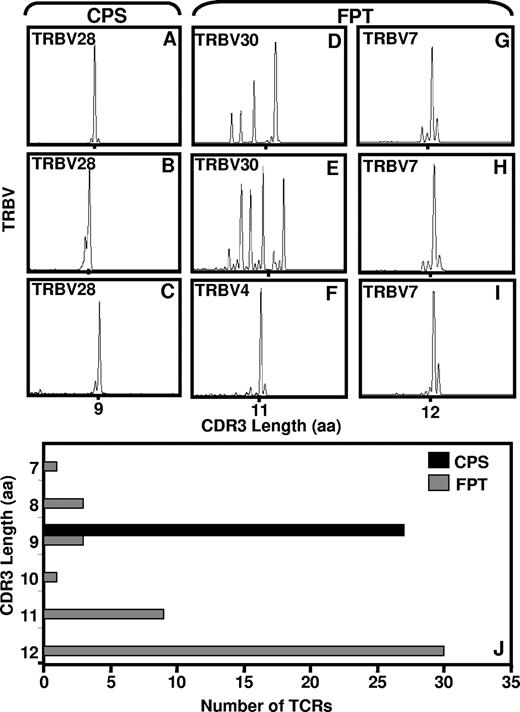
To examine the repertoire diversity of the CPS- and FPT-specific CD8+ T cells in finer detail, we further analyzed the CDR3 region of the TCR using immunoscope technology. Figure 3A-C shows the immunoscope profile of CPS-specific CD8+ T cells from fresh blood or in vitro–expanded T-cell lines. In addition to strongly biased TRBV28 usage, CPS-specific T cells further showed a highly conserved CDR3 region, with all CDR3 regions being 9 amino acids in length (Figure 3J). By comparison, ex vivo analysis of the CDR3 region of the FPT-specific T cells revealed an oligoclonal profile of CDR3 lengths (Figure 3D), which is consistent with the data presented in Figure 2C,D. However, when these cells were cultured in vitro, the CDR3 profile remained either oligoclonal (Figure 3E) or showed a highly biased clonal CDR3 profile (Figure 3F-I) with an average length of 12 amino acids (Figure 3J).
Immunoscope analysis of the CDR3 region of TCR specific for CPS- and FPT-specific T cells. cDNA samples from PBMCs or in vitro–expanded T cells cultures were used for PCR reaction with Vβ specific primers as outlined in “Methods,” and results were analyzed using ABI PRISM GeneScan software. Panel A shows the representative immunoscope profile of CPS-specific CD8+ T cells sorted from fresh PBMCs using pMHC-peptide pentamer; panels B and C show data based on in vitro–expanded T-cell lines. Data in panel A was obtained from donor 2, while data in panels B and C was acquired from donors 1 and 2, respectively. Panel D shows representative data for FPT-specific T cells sorted from fresh PBMCs using pMHC-peptide pentamer; panels E-I show data based on in vitro–cultured T-cell lines. Data presented in panels D and E are from donor 1; data in panels F-H were from donor 2; while data in panel I was from donor 6, respectively. Panel J shows a comprehensive summary of multiple CDR3 profiles for CPS- and FPT-specific T cells. Data presented in panel J is based on the analysis carried out in 3 different HLA B*3508 + individuals.
Immunoscope analysis of the CDR3 region of TCR specific for CPS- and FPT-specific T cells. cDNA samples from PBMCs or in vitro–expanded T cells cultures were used for PCR reaction with Vβ specific primers as outlined in “Methods,” and results were analyzed using ABI PRISM GeneScan software. Panel A shows the representative immunoscope profile of CPS-specific CD8+ T cells sorted from fresh PBMCs using pMHC-peptide pentamer; panels B and C show data based on in vitro–expanded T-cell lines. Data in panel A was obtained from donor 2, while data in panels B and C was acquired from donors 1 and 2, respectively. Panel D shows representative data for FPT-specific T cells sorted from fresh PBMCs using pMHC-peptide pentamer; panels E-I show data based on in vitro–cultured T-cell lines. Data presented in panels D and E are from donor 1; data in panels F-H were from donor 2; while data in panel I was from donor 6, respectively. Panel J shows a comprehensive summary of multiple CDR3 profiles for CPS- and FPT-specific T cells. Data presented in panel J is based on the analysis carried out in 3 different HLA B*3508 + individuals.
To complement the immunoscope analysis, we next sequenced the CDR3 regions of the TCR. Analysis of the CPS-specific T cells from the fresh blood revealed a high level of conservation at the nucleotide level, which has been described as a “type 3 bias,” whereby both the TRBV usage and CDR3 sequence are identical or highly homologous10 (Figure 4A top). Furthermore, when the CDR3 region from the TRAV chain was sequenced, it too showed high levels of bias (Figure 4A bottom). Conversely, analysis of the FPT-specific T cells from fresh blood showed highly diverse CDR3 regions (Figure 4B). In vitro expanded clonal FPT-specific T cells showed donor-specific CDR3 sequences, while polyclonal cultures included a mixture of CDR3 sequences with varying length (Figure 4B). Taken together, these analyses clearly indicate that the noncanonical HLA B*3508-CPS complex drives a biased TCR selection, while canonical HLA B*3508-FPT complex favors oligoclonal TCR usage.
Nucleotide and amino acid sequences of the CDR3 regions of the TRBV and TRAV chains expressed by CPS- (A) and FPT- (B) specific T cells. Data presented in this Figure is based on either clonal or pMHC-pentamer sorted CPS- and FPT-specific T cells from fresh PBMCs or polyclonal T-cell lines (as indicated in left-hand column). Sequences from pMHC-pentamer sorted cells were carried out using pooled cells from an individual donor. *Donor 2 PBMCs were used for TRBV sequencing only.
Nucleotide and amino acid sequences of the CDR3 regions of the TRBV and TRAV chains expressed by CPS- (A) and FPT- (B) specific T cells. Data presented in this Figure is based on either clonal or pMHC-pentamer sorted CPS- and FPT-specific T cells from fresh PBMCs or polyclonal T-cell lines (as indicated in left-hand column). Sequences from pMHC-pentamer sorted cells were carried out using pooled cells from an individual donor. *Donor 2 PBMCs were used for TRBV sequencing only.
HLA B*3508CPS and HLA B*3508FPT display distinct structural landscape
Considering the emerging evidence on the impact of pMHC-1 structural constraints on the binding of the TCR and its selection10 within an immune repertoire, we next determined whether the structural landscape of the HLA B*3508CPS and HLA B*3508FPT complexes correlated with the pattern of TCR repertoire. Crystal structures of HLA B*3508CPS (PDB accession code 3BW9) and HLA B*3508FPT (PDB accession code 3BWA) complexes were resolved to 1.75 and 1.30 Å, respectively (Figure 5 and Table S1). The HLA B*3508CPS and HLA B*3508FPT complexes were crystallized under the same conditions and thus, any structural differences observed were attributable to the peptide antigen, although the overall structure of HLA-B*3508 from both complexes was very similar (r.m.s.d. = 0.165 Å for all Cα atoms within the Ag-binding cleft).
Crystal structures of HLA B*3508CPS and HLA B*3508FPT. Panels A and C showing the electron density 2mfo-Dfc at 1σ of the FPT (orange stick) or CPS (purple stick) epitope bound to HLA B*3508. Panel B represents the structures of the featureless FPT peptide (orange) superposed with the one of the bludged CPS epitope (purple), which made one helix turn in its c-terminus. Panels D and E show surface view of the CPS and FPT epitopes bound to the HLA B*3508 molecule. The solvent-exposed surface of the peptide, which is accessible to the TCR, is represented by the green surface. The FPT epitope (panel E) has a smaller solvent-exposed surface than the CPS epitope (panel D).
Crystal structures of HLA B*3508CPS and HLA B*3508FPT. Panels A and C showing the electron density 2mfo-Dfc at 1σ of the FPT (orange stick) or CPS (purple stick) epitope bound to HLA B*3508. Panel B represents the structures of the featureless FPT peptide (orange) superposed with the one of the bludged CPS epitope (purple), which made one helix turn in its c-terminus. Panels D and E show surface view of the CPS and FPT epitopes bound to the HLA B*3508 molecule. The solvent-exposed surface of the peptide, which is accessible to the TCR, is represented by the green surface. The FPT epitope (panel E) has a smaller solvent-exposed surface than the CPS epitope (panel D).
The CPS peptide adopted an unusual conformation, whereby the epitope displayed a helical conformation that bulged from the Ag-binding cleft. The single helical turn was observed at the C-terminus of the peptide between the Met7 and the Tyr10 (Figure 5A-B). In addition to the Pro2 and Tyr12 main anchor positions, Glu5 acted as a secondary anchor, salt bridging with Arg157 and Arg97 (Table S2). The CPS peptide bulged approximately 6 Å out of the HLA-B*3508 cleft such that the side chains at positions Gln4, Pro6, Met7, Ser8, and Ile9 of the peptide were surface exposed and available for interaction with the TCR (Figure 5D). By comparison, the FPT peptide bound in an extended conformation, adopting a flatter “featureless” conformation in the Ag-binding cleft (Figure 5C), with all positions of the peptide interacting with HLA B*3508 (Table S2). In addition to the main anchor positions at P2-Pro and Leu8 the Asp5 acted as a secondary anchor, forming a salt bridge with the Arg97 of HLA B*3508 (Table S2) while the side chains at positions Phe1, Lys4, and Val6 of the peptide were surface exposed and available for interaction with the TCR (Figure 5E). These observations suggested that the structural features of the pMHC-I complexes may have minimal impact on the architecture of the antigen-specific T-cell repertoire during a persistent viral infection.
TCR selection correlates with functional properties of CPS- and FPT-specific CD8+ T cells
To further investigate whether this contrasting TCR selection was influenced by the presentation differences between the CPS and FPT epitopes, we developed ex vivo T-cell assays that used virus-infected cells as APCs. These assays allowed an assessment of clonal competition for the respective epitopes on virus-infected cells. Firstly, MRC-5 cells were co-infected with HCMV strain AD169 (MOI 0.5:1) and a recombinant adenoviral construct encoding HLA B*3508 and then exposed to CPS- or FPT-specific T cells and analyzed at different time intervals. The levels of INF-γ expression by each of these T cells were measured using intracellular cytokine assays. Data presented in Figure 6A showed that CPS-specific T cells were consistently stimulated more efficiently when compared with the FPT-specific T cells. An 8- to 10-fold difference in the IFN-γ expressing antigen-specific T cells numbers was observed over a period of 2 to 24 hours. To determine whether the decreased functional response of FPT-specific T cells was primarily due to limited availability of FPT epitopes in the context of natural HCMV infection or to low TCR affinity/avidity of FPT-specific T cells, we assessed functional avidity of CPS and FPT-specific T cells through peptide titration studies. MRC-5 cells expressing HLA B*3508 were sensitized with serially diluted synthetic CPS and FPT peptide epitopes and then exposed to antigen-specific T cells. Data presented in Figure 6B showed that the FPT-specific T cell recognized peptide-sensitized target cells more efficiently when compared with the CPS-specific T cells. These observations were also confirmed by intracellular cytokine assays, which showed almost 1.75-fold higher levels of IFN-γ expression by FPT-specific cells when CPS- and FPT-specific T cells were exposed to peptide-sensitized MRC-5 cells (Figure 6C). These results clearly show that the decreased functional response of FPT-specific T cells was primarily due to limited availability of FPT epitopes in the context of natural HCMV infection rather than low TCR affinity/avidity.
CD8+ T-cell recognition of target cells sensitized with CPS or FPT epitopes using either viral infection or synthetic peptides. (A) MRC-5 cells expressing HLA B*3508 were infected with HCMV at an MOI of 5:1 and then exposed to CPS- or FPT-specific T cells (responder to stimulator ratio of 2:1) at different time intervals (indicated on x-axis). At each time point, T cells were assessed for IFN-γ expression using intracellular cytokine assay. Data presented here is summary of 3 different experiments based on 3 different HLA B*3508+ donors. (B) CTL recognition of HLA B*3508 expressing target cells sensitized with synthetic CPS and FPT peptide epitopes. MRC-5 cells expressing HLA B*3508 were sensitized with serial dilutions of the peptides and then exposed to CPS- or FPT-specific CTL lines. These clonal T cells were derived from donor 1. (C) MRC-5 cells expressing HLA B*3508 were sensitized with synthetic CPS- or FPT-peptide epitopes (1 μg/mL) and then exposed to CPS- or FPT-specific T cells (responder to stimulator ratio of 2:1) for 16 to 18 hours. T cells were assessed for IFN-γ expression using intracellular cytokine assay. Data presented here is a summary of 3 different experiments. (D) HCMV-infected MRC-5 cells expressing either wild-type or mutated (to disallow CD8 co-receptor binding) HLA*B3508 were exposed to CPS- or FPT-specific T cells at 2 different responder-to-stimulator ratios, and IFN-γ expression measured using an intracellular cytokine assay. These data are representative of 3 different experiments and shows mean fluorescence intensity (MFI) of IFN-γ expression in CPS- or FPT-specific T cells. (E) MRC-5 cells expressing HLA B*3508 were pre-sensitized with synthetic peptide epitopes (CPS or FPT) and then exposed to CPS- or FPT-specific T cells (responder to stimulator ratio of 2:1). These T cells were assessed for IFN-γ expression as described above. MFI indicates mean fluorescence intensity.
CD8+ T-cell recognition of target cells sensitized with CPS or FPT epitopes using either viral infection or synthetic peptides. (A) MRC-5 cells expressing HLA B*3508 were infected with HCMV at an MOI of 5:1 and then exposed to CPS- or FPT-specific T cells (responder to stimulator ratio of 2:1) at different time intervals (indicated on x-axis). At each time point, T cells were assessed for IFN-γ expression using intracellular cytokine assay. Data presented here is summary of 3 different experiments based on 3 different HLA B*3508+ donors. (B) CTL recognition of HLA B*3508 expressing target cells sensitized with synthetic CPS and FPT peptide epitopes. MRC-5 cells expressing HLA B*3508 were sensitized with serial dilutions of the peptides and then exposed to CPS- or FPT-specific CTL lines. These clonal T cells were derived from donor 1. (C) MRC-5 cells expressing HLA B*3508 were sensitized with synthetic CPS- or FPT-peptide epitopes (1 μg/mL) and then exposed to CPS- or FPT-specific T cells (responder to stimulator ratio of 2:1) for 16 to 18 hours. T cells were assessed for IFN-γ expression using intracellular cytokine assay. Data presented here is a summary of 3 different experiments. (D) HCMV-infected MRC-5 cells expressing either wild-type or mutated (to disallow CD8 co-receptor binding) HLA*B3508 were exposed to CPS- or FPT-specific T cells at 2 different responder-to-stimulator ratios, and IFN-γ expression measured using an intracellular cytokine assay. These data are representative of 3 different experiments and shows mean fluorescence intensity (MFI) of IFN-γ expression in CPS- or FPT-specific T cells. (E) MRC-5 cells expressing HLA B*3508 were pre-sensitized with synthetic peptide epitopes (CPS or FPT) and then exposed to CPS- or FPT-specific T cells (responder to stimulator ratio of 2:1). These T cells were assessed for IFN-γ expression as described above. MFI indicates mean fluorescence intensity.
To examine the relative dependence on the CD8 co-receptor, we examined the recognition of virus-infected cells expressing wild-type or CD8 mutant HLA B*3508 molecules. HCMV-infected MRC-5 cells were co-infected with adenoviral constructs encoding either wild-type HLA B*3508 or a mutant form of HLA B*3508 (with D227K/T228A mutations on the CD8-binding site on the α3 domain) for 4 hours and then exposed to CPS- or FPT-specific T cells at a responder to stimulator ratio of 20:1 or 80:1. The levels of INF-γ expression by each of these T cells were measured using intracellular cytokine assays. Representative data presented in Figure 6D showed that CPS-specific T cells produced 1.5- to 2.5-fold higher IFN-γ levels when compared with FPT-specific T cells following exposure to HCMV-infected cells. More importantly, the T-cell recognition of HLA B*3508-CPS complex was significantly affected when the CD8-binding site was mutated, while the loss of the CD8-binding site on the HLA B*3508 molecule had minimal effect on the T-cell recognition of the FPT epitope. Notably, following presensitization with synthetic peptide, both the wild-type and mutant HLA B*3508 resulted in comparable levels of IFN-γ production by CPS- and FPT-specific T cells (Figure 6E).
To determine whether these differences in the endogenous presentation were related to the binding efficiency of the CPS and FPT epitopes to HLA B*3508 molecules, we conducted MHC stabilization assays based on T2.B*3508 cells and thermal stability assays, which demonstrated that these epitopes bound with similar affinity to HLA B*3508 (data not shown). Although these experiments indicated that both the FPT and CPS epitopes bind to HLA B*3508 with equal efficiency, our recent studies have shown that the dissociation half-life for B*3508CPS and B*3508FPT complexes was 37.1 hours and 21.1 hours, respectively.22 Taken together, these observations suggested the differences in the disassociation kinetics of the CPS and FPT epitopes for HLA B*3508 directly impacts on their antigen presentation efficiency in HCMV-infected cells, which was consistent with our previously documented evidence on the impact of pMHC binding on determinant selection.27
CPS- and FPT-specific T cells display contrasting phenotype
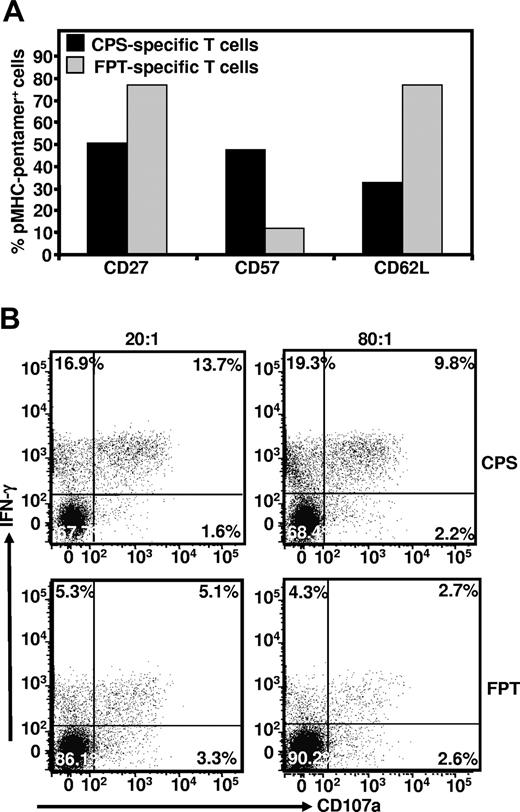
Previous studies have proposed that competition for cognate epitopes can impact on the clonal differences in the maturational phenotype of antigen-specific T cells.28 To explore this possibility, we conducted ex vivo phenotypic analysis of CPS- and FPT-specific T cells from fresh PBMCs. For these experiments cells were co-stained with HLA B*3508CPS or HLA B*3508FPT pentamers and mAbs specific for CD27, CD57, and CD62L. CPS- and FPT-specific T cells showed dramatic differences in the expression of these markers (Figure 7A). The majority of the CPS-specific T cells displayed a terminally differentiated (CD27−, CD57+, and CD62L−) phenotype, while FPT-specific T cells were predominantly CD27+ CD57−, and CD62L+, a phenotype typical of central memory T cells (Figure 7A).
Ex vivo phenotypic analysis of CPS- and FPT-specific T cells. Panel A: PBMC from HCMV-infected healthy individual were co-stained with pMHC-pentamers and antibodies specific for CD27, CD57 and CD62L. Data presented here is a summary of 3 different experiments. Panel B: To determine any functional differences between CPS and FPT-specific T cells, these effector cells were exposed to MRC-5 cells expressing HLA B*3508 and co-infected with HCMV at an MOI of 0.5:1. 2 different responder to stimulator ratios (80:1 and 20:1) were used in these assays. Following incubation, these cells were co-stained with pMHC pentamer, anti-IFN-γ and anti-CD107a as outlined in the Material and Methods section. This data are representative of 3 different experiments.
Ex vivo phenotypic analysis of CPS- and FPT-specific T cells. Panel A: PBMC from HCMV-infected healthy individual were co-stained with pMHC-pentamers and antibodies specific for CD27, CD57 and CD62L. Data presented here is a summary of 3 different experiments. Panel B: To determine any functional differences between CPS and FPT-specific T cells, these effector cells were exposed to MRC-5 cells expressing HLA B*3508 and co-infected with HCMV at an MOI of 0.5:1. 2 different responder to stimulator ratios (80:1 and 20:1) were used in these assays. Following incubation, these cells were co-stained with pMHC pentamer, anti-IFN-γ and anti-CD107a as outlined in the Material and Methods section. This data are representative of 3 different experiments.
To determine whether clonal antigen competition was also associated with functional heterogeneity of CPS- and FPT-specific T cells in response to cognate antigen, we examined the IFN-γ expression and perforin degranulation reflected by CD107a mobilization by in vitro–expanded T cells following stimulation with HCMV-infected cells. Representative data presented in Figure 7B shows that co-expression of IFN-γ and CD107a mobilization in CPS-specific T cells was approximately 2.6- to 3.6-fold higher when compared with the FPT-specific T cells. This difference in the levels of CD107a mobilization following antigen stimulation was observed over 2 different responder-to-stimulator ratios (Figure 7B). Taken together, the contrasting phenotypic profile of CPS- and FPT-specific T cells and increased mobilization of CD107a and/or IFN-γ by CPS-specific T cells correlated well with the data presented in Figure 6 and suggests that enhanced antigen stimulation of CPS-specific T cells may be driving selective expansion of terminally differentiated effector cells and a public TCR response.
Discussion
We have attempted to dissect the relative roles of antigen presentation kinetics and the structural landscape of pMHC-I in dictating the T-cell repertoire selection using stoichiometrically equivalent peptide determinants presented by same MHC allotypes. Our model, based on 2 immunodominant HCMV epitopes derived from the structural protein, pp65 (the noncanonical CPS and the canonical FPT) antigen, has the advantage of being independent of any virus or host-associated variables. A number of conclusions were drawn from these studies.
First, structural analysis revealed that the CPS epitope bound to the HLA B*3508 molecule adopted an unusual helical conformation that bulged from the Ag-binding cleft. Thus, the CPS-HLA B*3508 structural data were consistent with the previous finding that the bulged peptide epitopes select highly restricted public TCR.10,11,16,29 On the other hand, the FPT epitope, which stimulated a highly diverse TCR repertoire, adopted a flatter “featureless” conformation. By comparison, the FPT peptide was deeply embedded in the MHC groove with fewer exposed residues for TCR interaction. This was an unexpected finding, as previous studies with influenza virus, which indicated that viral epitopes that have been termed “featureless” and presumably too similar to self, select a biased TCR repertoire.18,30 Similarly, noncanonical epitopes that adopt bulged conformation when bound to the MHC class I molecules also exhibited highly biased TCR usage.13,17 These studies highlight that it is likely that in addition to the pMHC-I landscape, other factors also contribute toward the selection of TCR repertoire in persistent viral infection.
Second, dominant CPS-specific clonotypes with a “type 3” biased TCR10 usage consistently showed much higher IFN-γ expression following exposure to virus-infected cells when compared with FPT-specific T cells expressing diverse TCR. This difference in their ability to produce IFN-γ was not due an inherent disparity, as these T cells produced comparable levels of this cytokine when stimulated with synthetic peptides. Although the CPS and FPT epitopes showed similar efficiency in stabilizing HLA B*3508, our recent studies have shown that the B*3508CPS complexes display a much longer dissociation half-life when compared with B*3508FPT complexes.22 These observations suggested that the more efficient endogenous loading of the CPS epitope on HLA B*3508 molecules in virus-infected cells may drive increased expansion of CPS-specific T cells and thus clonal selection of a public TCR. Indeed, previous studies have shown that chronic stimulation of antigen-specific T cells results in continuous selection and accrual of high avidity clonotypes during the latent phase of infection.31,32 Interestingly, CPS-specific T cells showed higher dependency on pMHC-CD8 interaction when compared with FPT-specific T cells, which is consistent with ongoing competition for antigen in vivo. Indeed, these observations are consistent with an earlier hypothesis that high levels of viremia during viral infection might lead to an expansion of clonotypes, which are highly dependent on the pMHC-CD8 interaction.28 Although previous studies have argued that higher dependency on pMHC-CD8 interaction indicates lower avidity and may reflect a function of virus biology,28,33,34 we propose that CD8 dependency is an intrinsic property of the epitopes and this interaction may provide increased stability for the binding of TCR for noncanonical peptide-MHC conformations.
Third, ex vivo phenotypic analysis of CPS- and FPT-specific T cells revealed that the CPS-specific T cells displayed a terminally differentiated effector phenotype (CD27+/−CD57+ and CD62L−), while FPT-specific T cells were predominantly CD27+ CD57− and CD62L+, a phenotype typical of central memory T cells.35,36 This phenotypic difference was also reflected in their functional ability. The CPS-specific polyclonal T cells showed 2.6- to 3.6-fold higher IFN-γ expression and CD107a mobilization when compared with the FPT-specific T cells. These 2 different sets of experiments clearly showed that enhanced in vivo stimulation of CPS-specific T cells is driving selective expansion of terminally differentiated effector cells and a public TCR response. These studies clearly demonstrate that in vivo clonal competition for pMHC-1 complexes also plays a crucial role in the selection of TCR repertoire.
Taken together, data presented here further emphasize that TCR selection is a highly complex process and additional selection forces other than structural constraints would also have significant impact on this process.7,9,28,37-39 Indeed, after a decade of intense interest into what dictates TCR selection, there is an increasing realization that many different parameters, all operating to varying degrees, seem to be at play. No single or even set of several parameters seem to universally apply for every pMHC combination. In fact, quite the opposite appears to be true. In latent or otherwise benign viral infections there is an apparent flexibility/plasticity inherent within the sum total T-cell response. This is particularly exemplified here with the selection of equally opposed repertoire responses despite the co-immunodominance and shared HLA restriction. The data provide further evidence that selection is a highly ordered yet incredibly faceted process that ultimately operates to ensure a hugely diverse collective response to a particular pathogen. To get such a model operational, one could imagine that clonal competition for endogenously processed epitopes could provide one facet of such a dynamic environment. Similarly, the pMHC-1 structural landscape, avidity, and precursor frequency would also interplay. More importantly, it would be incorrect to view TCR selection as a 2-dimensional model and that rather a model of integrating and competing variables need to be explored. It will be important to expand these observations to other epitopes (within individual viral infections) to provide further support for this proposition.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors wish to thank Dr Judy Tellam for assistance in preparation of the adenovirus constructs.
This work was supported by the National Health and Medical Research Council (NH&MRC), Queensland Cancer Fund (QCF), and the Australian Research Council (ARC). R.K. and S.R.B. are supported by NH&MRC Principal Research Fellowship and Senior Research Fellowship, respectively. J.R. is supported by an ARC Federation Fellowship. K.K.W. is supported by a scholarship from the QCF.
Authorship
Contribution: K.K.W., Z.F., L.C., S.G., J.K.A., F.E.T., and J.J.M. performed the research; S.L.S., S.R.B., J.M., and J.R. analyzed the data and contributed toward the design of some of the experiments. R.K. designed the study and wrote the manuscript with J.R. and K.K.W.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Rajiv Khanna, QIMR, 300 Herston Rd, Herston (Qld) Australia 4006; e-mail: rajiv.khanna@qimr.edu.au; or Jamie Rossjohn, Department of Biochemistry, Monash University, Clayton, Australia 3800; e-mail: jamie.rossjohn@med.monash.edu.au.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal