Abstract
Deletions on chromosome 9q are seen in a subset of acute myeloid leukemia (AML) cases and are specifically associated with t(8;21) AML. We previously defined the commonly deleted region in del(9q) AML and characterized the genes in this interval. To determine the critical lost gene(s) that might cooperate with the AML1-ETO fusion gene produced by t(8;21), we developed a set of shRNAs directed against each gene in this region. Within this library, shRNAs to TLE1 and TLE4 were the only shRNAs capable of rescuing AML1-ETO expressing U937T-A/E cells from AML1-ETO–induced cell-cycle arrest and apoptosis. Knockdown of TLE1 or TLE4 levels increased the rate of cell division of the AML1-ETO–expressing Kasumi-1 cell line, whereas forced expression of either TLE1 or TLE4 caused apoptosis and cell death. Knockdown of Gro3, a TLE homolog in zebrafish, cooperated with AML1-ETO to cause an accumulation of noncirculating hematopoietic blast cells. Our data are consistent with a model in which haploinsufficiency of these TLEs overcomes the negative survival and antiproliferative effects of AML1-ETO on myeloid progenitors, allowing preleukemic stem cells to expand into AML. This study is the first to implicate the TLEs as potential tumor suppressor genes in myeloid leukemia.
Introduction
One of the most common genetic aberrations in acute myeloid leukemia (AML) is the balanced chromosomal translocation t(8;21). This translocation, seen in 8% to 13% of de novo AML cases,1-3 creates the RUNX1-MTG8/AML1-ETO fusion gene. AML1-ETO is insufficient for leukemogenesis as evidenced by mouse models,4-7 the detection of fusion gene transcripts in AML patients in long-term remission,8,9 as well as the finding of transcripts in newborns who did not develop t(8;21) AML for more than 10 years.10 Although AML1-ETO expression promotes the maintenance of early hematopoietic precursors,11,12 it markedly inhibits short-term expansion of primary human bone marrow cells and the proliferation of committed progenitors and CD34+ cells.13 This suggests a model in which secondary mutations are required to further transform preleukemic stem cells and allow their progeny to expand.
Deletion of a portion of the long arm of chromosome 9, del(9q), is a recurring abnormality in malignant myeloid diseases reported in approximately 2% of AML cases and is nonrandomly associated with t(8;21). Approximately 36% to 50% of samples with del(9q) have t(8;21); conversely, 7% to 14% of pediatric AML samples with t(8;21) have del(9q).1,14-16 After numerical abnormalities, del(9q) is the single most common associated structural chromosomal abnormality seen with t(8;21) AML, indicating loss of function of a gene or genes on chromosome 9q may be one of the most important cooperating genes in t(8;21) AML.
In a search for this cooperating gene(s), we recently narrowed the commonly deleted region (CDR) in 43 del(9q) AML samples to less than 2.4 Mb at 9q21.32–9q21.33. There are 10 known genes within, or immediately adjacent to, this region -TLE (transducin like enhancer of split)-1, TLE-4, FRMD3, UBQLN1, GKAP42, KIF27, HNRPK, SLC28A3, RMI1 (Q9H9A7), and NTRK2, and 3 novel or potential genes, RASEF, C9orf103 (ENSG00000148057), and C9orf64 (Q8N2B1).17 Sequence analysis of the coding regions of these genes failed to identify clearly inactivating mutations in the remaining allele in del(9q) AML samples. However, the expression of several of these genes (TLE1, C9orf103, UBQLN1, KIF27, c9orf64, RMI1, and NTRK2) appeared specifically low in del(9q) AML samples, and we hypothesized that haploinsufficiency, or reduced expression of a critical gene(s) in this region resulting from promoter mutations or epigenetic changes, cooperated with AML1-ETO or other genetic events to cause leukemia.17 To identify this critical gene, we created shRNAs to knock down expression of genes in the del(9q) CDR and sought to determine whether any of these could rescue the cell death and apoptosis caused by inducible AML1-ETO expression in the U937T-A/E cell line.
Methods
Cell culture
The U937T-A/E cell line expresses AML1-ETO under a control of a “tetracycline-off” repressor. Withdrawal of tetracycline induces AML1-ETO expression, which causes a block in cell-cycle progression and apoptosis18 (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). The U937T-A/E cell line was cultured in RPMI with 10% fetal calf serum (FCS) (Cambrex, East Rutherford, NJ), 100 U/mL penicillin, and 100 μg/mL streptomycin with 1 mg/mL G418, 0.5 μg/mL puromycin, and 1 μg/mL tetracycline. To induce AML1-ETO expression, cells were washed 3 times and then cultured without tetracycline (using Tet System Approved Fetal Bovine Serum, Clontech, Mountain View, CA). The t(8;21) positive leukemia cell line Kasumi-1 was cultured in RPMI 1640 medium with 15% FCS. The Phoenix Ampho packaging cell line, the human embryonic kidney cell line 293T, and U2OS cells were cultured in Dulbecco modified Eagle medium supplemented with 10% FCS, 100 U/mL penicillin, and 100 μg/mL streptomycin. Kasumi-1, Phoenix Ampho, U2OS, and 293T cell lines were obtained from American Type Culture Collection (Manassas, VA).
Evaluation of gene expression
Total RNA extracted with Trizol Reagent (Invitrogen, Carlsbad, CA) was reverse transcribed into cDNA using the RETROscript kit (Ambion, Austin, TX). To evaluate AML1-ETO expression in U937T-A/E cells, we used the primers: 821-A1/821-E1 5′-AGCTTCACTCTGACCATCAC-3′/5′-TCAGCCTAGATTGCGTCTTC-3′, with reaction conditions as previously described.8 For beta-2-microglobulin, we used 95°C for 5 minutes; 35 cycles of 95°C for 30 seconds, 60°C for 30 seconds, 72°C for 30 seconds; followed by 72°C for 7 minutes. Expression of del(9q) CDR genes was evaluated by quantitative reverse-transcribed polymerase chain reaction (RT-PCR) (MyiQ, Bio-Rad, Hercules, CA) using the primers in Table S1 (iQ SYBR Green Supermix, Bio-Rad) and Assays on Demand, Applied Biosystems (Foster City, CA) TLE1 -Hs00270768_m1 for TLE1 (iQ Supermix, Bio-Rad) with normalization to β-2-microglobulin. HNRPK expression was evaluated by Western blot using a custom HNRPK polyclonal antibody from Invitrogen (gift of Emmett Schmidt). The specificities of the TLE primers were verified using cDNA containing plasmids for TLE1, TLE2, TLE3 (gifts of Stefano Stifani19 ), and TLE4 (KIAA1261 gift of Takahiro Nagase20 ). Representative products for each transcript were sequenced to verify specificity.
Del(9q) shRNA construction
We were able to demonstrate expression of all genes except UBQLN1, NTRK2, and RASEF in U937T-A/E cells, despite the strong expression of these latter 3 genes in U2OS cells (results not shown). ShRNA target sequences for the expressed genes TLE1, TLE4, FRMD3, GKAP1, SLC28A3, KIF27, C9orf103, C9orf64, RMI1, and HNRPK, as well as target sequences in common to both TLE1 and TLE4, were selected using the Whitehead siRNA selection program (http://jura.wi.mit.edu/bioc/siRNAext/home.php)21 and Ambion web-based resources (http://www.ambion.com/techlib/misc/siRNA_finder.html). Three to four shRNAs for each gene were individually cloned into the pSUPER_retro_GFP_neo retroviral vector (Oligoengine, Seattle, WA) as recommended by the manufacturer and transfected into 293T or U2OS cells. Those most effective in knocking down endogenous message levels (results not shown) were used for subsequent analyses. For the TLEs, an additional less effective shRNA was also used. The shRNAs and target sequences for these genes were TLE1si1 (GTTCACTATCCCGGAGTCC), TLE1si3 (AAGATAACCTCCTCAATGC), TLE4si2 (AGTGATGACAACT-TGGTGG), TLE1/4si2 (GGTCTGCTTCTCATGCTGC), TLE1/4si3 (TGA-TGGCACCAAGCTCTGG), FRMDsi4 (ATGGCCAGATGTCTGCAAA), GKAPsi2 (TACTGGAAAGTCTCAAACT), SLC28A3si3 (TGGCAGGACAGCTTTATGG), KIF27si3 (GGACCACAGCATGTTACAG), C9orf103si3 (AGTCAAGCTTAAATTGAAG), c9orf64si1 (TGCACCTGGTGGTTGAAAG), RMIsi3 (GGAATTGCAACCATTGACT), HNRPKsi8 (GATTTGGCTGGATCTATTA) along with a control scrambled shRNA T4si3SCR (CAGTCGCCATTAGTTCCAC).
Retrovirus and lentivirus production and cell infection
ShRNAs with the H1 promoter were transferred from pSUPER_retro_GFP_neo into the lentiviral vector FUGW (gift of Carlos Lois22 ). Full-length TLE1 and TLE4 cDNAs (gifts of Stefano Stifani19 and Takahiro Nagase20 ) were cloned into the MSCV-IRES-GFP retroviral vector. Retroviral and lentiviral supernatants were prepared as previously described (http://www.stanford.edu/group/nolan/protocols/pro_helper_dep.html).22 Retroviral supernatants were concentrated using the method described by Kanbe and Zhang.23 Two rounds of lentivirus or retrovirus infections 24 hours apart were performed with either U937T-A/E or Kasumi-1 cell lines. In each case, greater than 95% of cells were infected as judged by GFP expression (not shown). The efficacy of shRNA-mediated knockdown of the endogenous transcripts was assessed by RT-PCR, or Western blot for HNRPK, 4 days after infection.
Cell-cycle analysis and annexin V staining
For cell-cycle analysis, 1 × 106 cells were fixed in 80% ethanol for at least 1 hour at 4°C. Cells were then spun down for 5 minutes at 300g and incubated in 1 mL PI/RNase staining buffer (BD Pharmingen, San Diego, CA) for 15 minutes at RT. Cell-cycle distribution was analyzed using Modfit VS.2 software (Verity Software, Topsham, ME). For cell division assays, 5 × 105 cells were resuspended in 1 mL RPMI1640 and 5 μL of a 1 mM DiI stock solution (Invitrogen) was added. After incubation for 20 minutes at 37°C, cells were washed 3 times with RPMI 1640 and resuspended in RPMI 1640, 10% FCS. Cells were then incubated at 37°C, 5% CO2 until analysis. Intracellular staining with anti-Cyclin D1 (Cell Signaling Technology, Danvers, MA) and anti-Ki-67-PE (BD Pharmingen) was performed using Cytofix and Cytoperm (BD Pharmingen) following the manufacturer's protocol. In each assay, a gate was first set on GFP-positive cells, and these cells were further analyzed. To calculate the median fluorescence intensity, median fluorescence channel of isotype was subtracted from the specific antibody. Annexin-V Cy5-conjugated antibody and 7-AAD staining (BD Pharmingen) were used to determine cell death and apoptosis. All samples were analyzed on a FACS Calibur flow cytometer (BD Pharmingen) using the Cell Quest Pro software.
Patient samples
Cryopreserved diagnostic bone marrow (BM) specimens from de novo AML patients with t(8;21), including a subset with del (9q), were obtained from the Children's Oncology Group AML reference laboratory. The karyotypes were confirmed by central review. Twenty or more cells were analyzed to identify clonal abnormalities that were defined in accordance with International System for Human Cytogenetic Nomenclature guidelines.24 CD34+ cells from normal donors were also obtained. Informed consent and assent were obtained from parents and patients as appropriate using institutional review board (IRB)–approved protocols. The Massachusetts General Hospital IRB and the Children's Oncology Group Myeloid Disease Committee approved this study.
Immunoprecipitation
For immunoprecipitation, 293T cells were transfected with pFLAG-CMV2 (Sigma-Aldrich, St Louis, MO) or pFLAG-AML-ETO, along with HA-tagged TLE1 or TLE4 in a pcDNA3-zeo vector as indicated in the figure legends using Polyfect (Qiagen, Alameda, CA). After 48 hours, cells were washed once with phosphate-buffered saline (PBS) and lysed in radioimmunoprecipitation assay buffer (25 mM Tris, pH 7.4, 150 mM KCl, 5 mM EDTA, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS). Protein lysates were diluted in PBS and immunoprecipitated with anti-FLAG antibody (M2, Sigma-Aldrich). Immunocomplexes were recovered with Protein G-Sepharose (Amersham Biosciences) and washed 4 times with PBS containing 0.1% Nonidet P-40.
For Western blot analysis, total cell extracts or immunocomplexes recovered as described above were separated on gradient (4%-20%) polyacrylamide-SDS gels (Bio-Rad) and transferred to Immobilon-P (Millipore, Billerica, MA) or nitrocellulose membranes (GE Healthcare, Little Chalfont, United Kingdom). Membranes were probed with one of the following antibodies as indicated in the figure legends; anti-TLE1, anti-TLE4, antitubulin (all from Santa Cruz Biotechnology, Santa Cruz, CA), anti-HA (Covance Research Products, Princeton, NJ) or anti-FLAG (M2, Sigma-Aldrich), followed by appropriate HRP-conjugated secondary antibodies. The protein bands were detected by chemiluminescence.
Zebrafish
All zebrafish experiments were approved by the Massachusetts General Hospital Subcommittee on Research Animal Care and conform to national and institutional guidelines. The Tg(hsp:AML1-ETO) zebrafish line was generated using the pHSP/AML1-ETO construct containing zebrafish hsp70-4 promoter and human AML1-ETO cDNA.25
Morpholino oligonucleotides and microinjection
Examination of the annotated June 2004 zebrafish (Danio rerio) Zv4 sequence assembly (http://genome.ucsc.edu/) indicated the presence of 2 members of the Gro/TLE family, gro2 and gro3. We designed antisense morpholino oligonucleotides to each of these genes targeting the splice donor sites in the 5′ region of the gene. For microinjection, 300 μM of the morpholino oligonucleotides (obtained from Gene-Tools, LLC, Philomath, OR) in 0.3× Danieau's buffer (17 mM NaCl2, 2 mM KCl, 0.12 mM MgSO4, 1.8 mM Ca(NO3)2 and 1.5 mM HEPES, pH 7.6) were prepared and injected at the one to 4 cell stage as described.26 We subsequently determined only the gro3 morpholino oligonucleotides (sequences 5′-AGCGAGCAGAGATATTTACCTGTGG and 5′-ATAATTTATGATACTCACCGGATGC) were effective in knocking down the appropriate transcript and used these for our experiments.
Heat treatment and phenotyping of zebrafish embryos
Petri dishes containing zebrafish embryos were transferred from the growth temperature of 23°C to 28.5°C to a 37°C to 40°C incubator at 14 to 18 hours postfertilization (hpf) and incubated for 1 hour before returning them back to the growth temperature. The heat treatment was repeated once at 6 hours after the first heat treatment to maintain the induction. We have previously described a characteristic phenotype of AML1-ETO induction in zebrafish that included an expansion of primitive myeloblasts in the intermediate cell mass with associated lack of circulating cells.25 The percentages of embryos without circulation were scored by visual inspection between 30 and 40 hpf. The stages described in this report are based on the developmental stages of normal zebrafish embryos at 28.5°C.27 Fluorescence microangiography was done as described.28
In situ hybridization of zebrafish embryos
Wild-type and AML1-ETO transgenic fish embryos were injected with 300 μM Gro3 morpholino (MO). Noninjected and injected embryos were heat-shocked at 37°C for 1 hour at 18 hpf. The embryos were fixed at 32 hpf and were subjected to in situ hybridization of L-plastin (a macrophage marker) or mpo (a granulocyte marker). Digoxogenin-labeled antisense riboprobes for mpo and L-plastin were made as previously described.29 Whole-mount in situ hybridization was performed as described.30
Cytology
Blood cells collected from the zebrafish embryos were transferred onto glass slides by cytospin and stained with Protocol Wright-Giemsa stain (Fisher Diagnostics) following the manufacturer's instruction.
Statistics
Bar graphs indicate the means of the measured values plus or minus SEM. The Student t test or Mann-Whitney rank sum test was applied to determine significant differences (P < .05).
Results
TLE knockdown rescues AML1-ETO–induced death of U937T-A/E cells
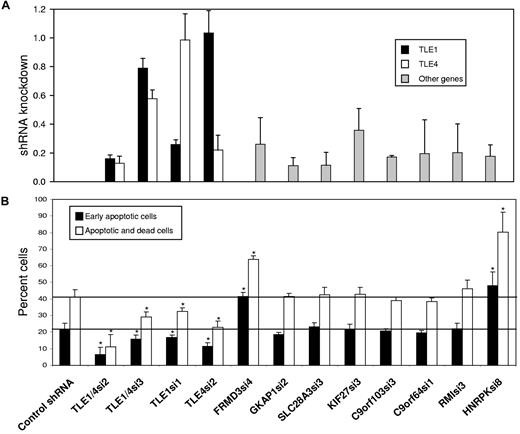
The effect of AML1-ETO in promoting the growth arrest and apoptosis of hematopoietic cell progenitors suggests that cooperating mutations must overcome this cell death in order for leukemia to develop.13,18,31 To identify such a cooperating gene within the del(9q) CDR, we attempted to rescue the U937T-A/E cell line from AML1-ETO induced cell death using shRNAs directed against all gene transcripts from the del(9q) CDR expressed in U937T-A/E cells. These shRNAs were introduced individually into U937T-A/E cells by lentiviral infection. After 48 hours, AML1-ETO expression was induced by tetracycline withdrawal. The shRNAs used for TLE1, TLE4, FRMD3, GKAP1, SLC28A3, c9orf103, C9orf64, RMI-1, and HNRPK knocked down their respective endogenous messages in U937T-A/E cells to 26% of control levels or lower when assayed by quantitative RT-PCR (or Western blot for HNRPK) 4 days after infection. The message levels for KIF27 was reduced to 36% (Figure 1A). The shRNAs directed against TLE1 and TLE4 were specific without knockdown of the other TLE. In this analysis, we also included 2 shRNAs of differing potency directed against both TLE1 and TLE4 (TLE1/4si2-16%/13% vs TLE1/4si3–79%/58%).
Knockdown of TLE levels inhibits the apoptosis and cell death induced by AML1-ETO expression in U937T-A/E cells. U937T-A/E cells were infected with lentivirus containing a control shRNA (TLE4scr3), an shRNA directed against both TLE1 and TLE4 (TLE1/4si2, TLE1/4si3), an shRNA specific for TLE1 (TLE1si1) or TLE4 (TLE4si2), or an shRNA against other del9q CDR genes: FRMD3, GKAP1, SLC28A3, KIF27, C9orf103, C9orf64, RMI1, and HNRPK. (A) Four days after infection, expression of del(9q) CDR genes was evaluated by quantitative RT-PCR or Western blot in the case of HNRPK and are shown relative to the level of expression of each gene after infection with a control shRNA. To evaluate the specificity of TLE knockdown with TLE shRNAs, the expression levels of both TLE1 (■) and TLE4 (□) were measured. (B). After 2 days of infection, tetracycline was withdrawn and AML1-ETO was inducted. Cells were cultured an additional 6 days and then stained with annexin-V and 7-AAD to detect cells undergoing early apoptosis (annexin-V positive, 7-AAD negative) or dead cells in late apoptosis (annexin-V positive, 7-AAD positive). Three independent experiments were performed, and results for percentage of cells undergoing early apoptosis (■) and total apoptotic and dead cells (□) are summarized and expressed as the mean plus or minus SEM.
Knockdown of TLE levels inhibits the apoptosis and cell death induced by AML1-ETO expression in U937T-A/E cells. U937T-A/E cells were infected with lentivirus containing a control shRNA (TLE4scr3), an shRNA directed against both TLE1 and TLE4 (TLE1/4si2, TLE1/4si3), an shRNA specific for TLE1 (TLE1si1) or TLE4 (TLE4si2), or an shRNA against other del9q CDR genes: FRMD3, GKAP1, SLC28A3, KIF27, C9orf103, C9orf64, RMI1, and HNRPK. (A) Four days after infection, expression of del(9q) CDR genes was evaluated by quantitative RT-PCR or Western blot in the case of HNRPK and are shown relative to the level of expression of each gene after infection with a control shRNA. To evaluate the specificity of TLE knockdown with TLE shRNAs, the expression levels of both TLE1 (■) and TLE4 (□) were measured. (B). After 2 days of infection, tetracycline was withdrawn and AML1-ETO was inducted. Cells were cultured an additional 6 days and then stained with annexin-V and 7-AAD to detect cells undergoing early apoptosis (annexin-V positive, 7-AAD negative) or dead cells in late apoptosis (annexin-V positive, 7-AAD positive). Three independent experiments were performed, and results for percentage of cells undergoing early apoptosis (■) and total apoptotic and dead cells (□) are summarized and expressed as the mean plus or minus SEM.
We could demonstrate expression of AML1-ETO 4 days after withdrawal of tetracycline. Most U937T-A/E cells were dead or dying in the subG1 population by 9 days (Figure S1). Six days after tetracycline withdrawal, 22% plus or minus 3% of cells infected with a control shRNA were in early apoptosis as quantified by FACS (annexin V positive, 7-AAD negative), with 41% plus or minus 4% either dead or undergoing apoptosis (annexin V positive, Figures 1B, S2). Simultaneous knockdown of TLE1 and TLE4 with TLE1/4si2 dramatically reduced the percentage of early apoptotic cells to 6% plus or minus 4% and the total percentage of dead and apoptotic cells to 11% plus or minus 7%. Reflecting the decreased effectiveness of TLE1/4si3 in knocking down TLE1 and TLE4, a significant but lesser effect was seen with 16% plus or minus 2% early apoptotic cells and 29% plus or minus 3% apoptotic or dead cells. Similar rescue was seen with individual knockdown of TLE1 (33% ± 2%) or TLE4 (23% ± 3%). In contrast, knockdown of all other genes either had no effect, or, in the case of FRMD3 or HNRPK, actually increased the rate of death and apoptosis to 64% plus or minus 2% or 80% plus or minus 12% (Figure 1B).
TLE1 and TLE4 levels are decreased in t(8;21) AML samples with del(9q)
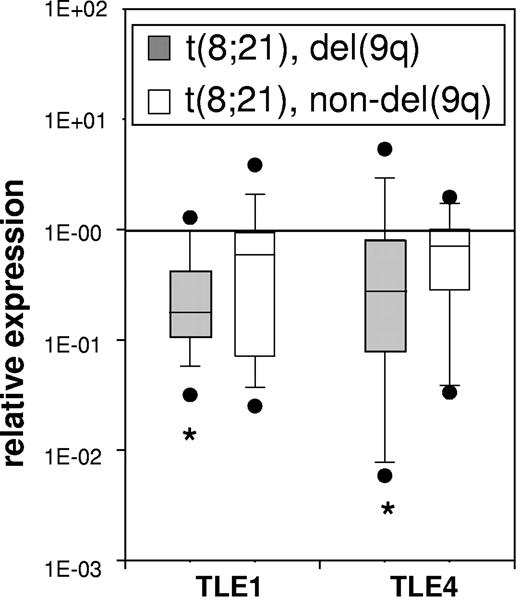
If low levels of TLE1 and/or TLE4 cooperate with AML1-ETO in leukemogenesis, we expected to see evidence of this in AML samples with t(8;21). We compared TLE1 and TLE4 expression in 20 t(8;21) AML samples with del(9q) and 19 t(8;21) AML samples without del(9q) to a control group of 11 normal control CD34+ samples. As shown in Figure 2, we found a significantly lower median level of expression of both TLE1 and TLE4 in t(8;21) samples with del(9q). The median level of TLE1 and TLE4 expression in t(8;21) samples without del(9q) was not significantly lower than control CD34 + samples.
TLE1 and TLE4 expression is specifically decreased in a subset of t(8;21) AMLS with del(9q). The median levels of TLE1 and TLE4 expression in 20 t(8;21) AML samples with del(9q) and 19 t(8;21) AML samples without del(9q) were compared with the expression of 11 normal CD34+ control samples. Boxes indicate measurements from 25th to 75th percentiles. A line within a box indicates median fold change in gene expression. The error bars represent the 10th and 90th percentiles; solid circles, outliers. (*P < .05, a significant change).
TLE1 and TLE4 expression is specifically decreased in a subset of t(8;21) AMLS with del(9q). The median levels of TLE1 and TLE4 expression in 20 t(8;21) AML samples with del(9q) and 19 t(8;21) AML samples without del(9q) were compared with the expression of 11 normal CD34+ control samples. Boxes indicate measurements from 25th to 75th percentiles. A line within a box indicates median fold change in gene expression. The error bars represent the 10th and 90th percentiles; solid circles, outliers. (*P < .05, a significant change).
Modulation of TLE levels affects cell proliferation and survival of the AML1-ETO–expressing Kasumi-1 cell line
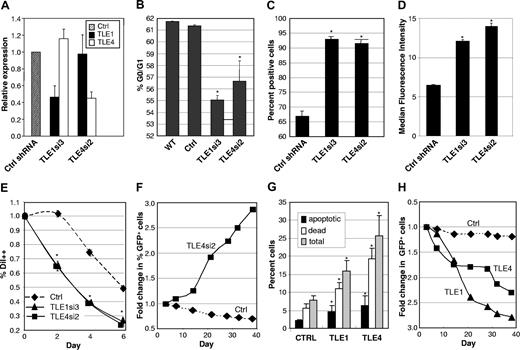
The Kasumi-1 myeloid cell line was established from a patient with t(8;21) AML. This cell line has a doubling time of 40 to 48 hours (per ATCC), significantly slower than most other myeloid cell lines such as HL-60 (34 hours,32 ) or THP-1 cells (26 hours, per ATCC). As shown in Figure 3A, Kasumi-1 cells transduced with shRNA against TLE1 or TLE4 demonstrated approximately 45% endogenous levels of expression relative to controls and the knockdown was specific to the directed gene. These transduced cells showed marked decrease in the resting G0/G1 fraction after PI staining (Figure 3B). This decrease was accompanied by a significant increase in cyclin D1-positive cells (Figure 3C), as evident by flow cytometric staining. We also observed a higher fraction of positive cells when looking at the proliferation marker Ki-67 in shRNA-transduced cells (Figure 3D). Furthermore, these cells had a much higher rate of cell division as monitored by a more rapid dilution of the lipophilic carbocyanine DiI (Figure 3E). These effects translated into an approximately doubled rate of cell division compared with control shRNA transduced cells. In accordance to these results, the proportion of GFP+ Kasumi-1 cells transduced with a TLE4 shRNA more than tripled in culture over a period of almost 40 days (Figure 3F), again indicating a proliferative advantage. This expansion was not seen with TLE1 shRNA or when TLE1 or TLE4 shRNAs were transfected into other myeloid cell lines not expressing AML1-ETO, including HL60, U937, or THP-1 (data not shown). In contrast, Kasumi-1 cells transduced with a control-scrambled shRNA failed to exhibit such a proliferative advantage (Figure 3F). Consistent with function as a tumor suppressor gene, we observed a significant increase in cell death and apoptosis with forced expression of either TLE1 or TLE4 (Figures 3G,S3) this combined with a slowing in cell-cycle progress (not shown) resulted in a progressive decrease in the percentage of cells coexpressing GFP and either TLE (Figure 3H).
Modulation of TLE levels affects the proliferation and survival of Kasumi-1 cells. (A) ShRNAs directed against TLE1 or TLE4 knockdown expression to approximately 45% control values and are specific for their respective target gene. (B) Knockdown of TLE1 or TLE4 increases cell-cycle progression. Flow cytometric determination of G0/G1 fraction in wild-type (WT) Kasumi-1 cells and cells infected with control (Ctrl) shRNA and shRNA against TLE1 or TLE4. (C) Knockdown of TLE1 or TLE4 increases cyclin D1 expression. Flow cytometric analysis of percentage of cyclin D1 positive Kasumi-1 cells infected with control (Ctrl) shRNA and shRNA against TLE1 or TLE4. (D) Knockdown of TLE1 or TLE4 increases expression of the cell proliferation marker Ki-67. Flow cytometric analysis of fluorescence intensity (FL) of Ki-67 in Kasumi-1 cells infected with control (Ctrl) shRNA and shRNA against TLE1 or TLE4. (E) Knockdown of TLE1 or TLE4 increases cell division. Flow cytometric cell proliferation assay with DiI in Kasumi-1 cells infected with control (Ctrl) shRNA and shRNA against TLE1 or TLE4. Shown is the percentage of bright DiI positive (DiI++) cells over time. (F) Knockdown of TLE4 leads to a proliferative advantage. Relative proportion over time of GFP positive Kasumi cells either infected with a control (Ctrl) or a specific shRNA against TLE4. (G) Expression of TLE1 or TLE4 leads to an increase in apoptosis and cell death. Percentage of apoptotic cells (annexin V+/7AAD−), dead and late apoptotic cells (annexin V+/7AAD+), and total apoptotic and dead cells in Kasumi cells infected with control empty MSCV-GFP retroviral vector (Ctrl) or retrovirus expressing TLE1 or TLE4 cDNAs. GFP positive cells were sorted before analysis. (H) Expression of TLE1 or TLE4 slows proliferation. Relative proportion over time of GFP positive Kasumi cells infected with either control empty MSCV-GFP retroviral vector (Ctrl) or retrovirus expressing TLE1 or TLE4 cDNAs. All diagrams denote results from 3 independent experiments. Results are expressed as the mean plus or minus SEM (*P < .05, significant difference vs controls).
Modulation of TLE levels affects the proliferation and survival of Kasumi-1 cells. (A) ShRNAs directed against TLE1 or TLE4 knockdown expression to approximately 45% control values and are specific for their respective target gene. (B) Knockdown of TLE1 or TLE4 increases cell-cycle progression. Flow cytometric determination of G0/G1 fraction in wild-type (WT) Kasumi-1 cells and cells infected with control (Ctrl) shRNA and shRNA against TLE1 or TLE4. (C) Knockdown of TLE1 or TLE4 increases cyclin D1 expression. Flow cytometric analysis of percentage of cyclin D1 positive Kasumi-1 cells infected with control (Ctrl) shRNA and shRNA against TLE1 or TLE4. (D) Knockdown of TLE1 or TLE4 increases expression of the cell proliferation marker Ki-67. Flow cytometric analysis of fluorescence intensity (FL) of Ki-67 in Kasumi-1 cells infected with control (Ctrl) shRNA and shRNA against TLE1 or TLE4. (E) Knockdown of TLE1 or TLE4 increases cell division. Flow cytometric cell proliferation assay with DiI in Kasumi-1 cells infected with control (Ctrl) shRNA and shRNA against TLE1 or TLE4. Shown is the percentage of bright DiI positive (DiI++) cells over time. (F) Knockdown of TLE4 leads to a proliferative advantage. Relative proportion over time of GFP positive Kasumi cells either infected with a control (Ctrl) or a specific shRNA against TLE4. (G) Expression of TLE1 or TLE4 leads to an increase in apoptosis and cell death. Percentage of apoptotic cells (annexin V+/7AAD−), dead and late apoptotic cells (annexin V+/7AAD+), and total apoptotic and dead cells in Kasumi cells infected with control empty MSCV-GFP retroviral vector (Ctrl) or retrovirus expressing TLE1 or TLE4 cDNAs. GFP positive cells were sorted before analysis. (H) Expression of TLE1 or TLE4 slows proliferation. Relative proportion over time of GFP positive Kasumi cells infected with either control empty MSCV-GFP retroviral vector (Ctrl) or retrovirus expressing TLE1 or TLE4 cDNAs. All diagrams denote results from 3 independent experiments. Results are expressed as the mean plus or minus SEM (*P < .05, significant difference vs controls).
Knockdown of Groucho3 in zebrafish potentiates the phenotype induced by AML1-ETO
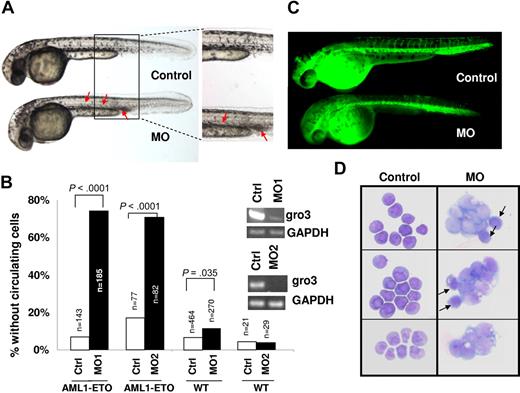
To further demonstrate cooperation between AML1-ETO and inhibition of Gro/TLE proteins, we used a zebrafish model with inducible AML1-ETO expression controlled by a heat shock promoter.25 In this transgenic zebrafish model, strong induction of AML1-ETO expression in zebrafish embryos results in highly penetrant accumulation of blast-like hematopoietic cells in the intermediate cell mass (ICM) and lack of circulating cells. These phenotypes are similar to the previous documented phenotypes caused by injecting AML1-ETO plasmid into zebrafish embryos.33 To detect synergistic effect of AML1-ETO expression and Gro/TLE knockdown, we induced AML1-ETO expression using a mild heat shock condition. Under this condition, more than 90% of the transgenic embryos exhibit circulating blood cells when scored at 30 to 40 hpf (Figure 4A,B). Injection of antisense morpholino oligonucleotides against the TLE homologue gro3 into the transgenic embryos results in absence of circulating cells and accumulation of hematopoietic cells in the ICM in more than 70% of the fish embryos (Figure 4A,B). Injection of gro3 morpholino oligonucleotides into wild-type (WT) embryos did not induce these phenotypes, indicating that the cooperating effects of mild AML1-ETO expression and Gro/TLE knockdown cause these phenotypes. Similar results were obtained from the second morpholino oligonucleotide against gro3 (Figure 4B). It has been shown that AML1-ETO expression results in abnormal patterning of the vasculature in zebrafish embryos.33 However, normal angiograms were obtained in embryos with accumulating hematopoietic cells without circulating cells (Figure 4C). This finding strongly suggests that the hematopoietic defects observed in these embryos are not secondary to any vascular defects that sometimes coexist. Using cytologic staining, we found that the noncirculating hematopoietic cells in the injected transgenic embryos contained abundant immature, blast-like cells, whereas the circulating blood in the uninjected transgenic embryos contained mostly red blood cells (Figure 4D). These data indicate that knockdown of gro3 in zebrafish enhances AML1-ETO-induced blast cell accumulation in vivo. To verify that these accumulated cells were indeed of the myeloid lineage, we performed in situ hybridization of these embryos with l-plastin and myeloperoxidase (mpo). L-plastin expression marks peripheral blood leukocytes with highest staining of monocytes and macrophages. As shown in Figure 5, l-plastin staining is seen along the body of the embryo with collection in the ICM. There is not a clear difference in l-plastin staining between WT embryos and embryos with Gro/TLE knockdown and/or AML1-ETO expression induced by mild heat shock. In contrast, myeloperoxidase, which is expressed in differentiated granulocytic and granulocyte progenitors, clearly demonstrated the accumulated immature appearing cells seen with AML1-ETO induction and knockdown of gro3 were of the granulocytic lineage.
Knockdown of gro3 in cooperation with AML1-ETO expression induces abnormal hematopoiesis and absence of circulating blood in zebrafish embryos.Gro3 MO (morpholino oligonucleotide)-injected or uninjected wild-type and Tg(hsp:AML1-ETO) zebrafish embryos were all subjected to the same heat induction. (A) Inspection of uninjected (Control) and gro3 MO injected Tg(hsp:AML1-ETO) zebrafish embryos showed the absence of circulating cells and accumulation of hematopoietic cells in the ICM and the ventral tail region (red arrows). These cells had a blast-like morphology with a large size and relatively little cytoplasm. (B) Percentages of embryos exhibiting loss-of-circulating cells were scored between 30 and 40 hpf. Control and MO indicate uninjected and morpholino oligonucleotide-injected embryos, respectively. AML1-ETO and WT indicate Tg(hsp:AML1-ETO) and wild-type embryos, respectively. n equals the number of embryos scored. P value was obtained using Student t test. The inset gels show the knockdown by RT-PCR of gro3 transcripts with 2 gro3 specific morpholino oligonucleotides. (C) Lack of circulating cells in AML1-ETO–expressing embryos is not caused by a vascular obstruction as evidenced by microangiography. The fluorescein-coupled latex beads injected into the inflow tract of the atrium was able to perfuse the whole vascular system of the Tg(hsp:AML1-ETO) embryos. It revealed a normal vascular pattern but reduced intersomitic vessels in gro3 morphant embryos. (D) Cytologic analysis of hematopoietic cells collected from the circulating blood in 3 uninjected Tg(hsp:AML1-ETO) embryos and the accumulating blood in 3 injected embryos. Whereas the uninjected embryos contain mostly red blood cells, the injected embryos contain abundant immature blast-like cells. Black arrows indicate red blood cells.
Knockdown of gro3 in cooperation with AML1-ETO expression induces abnormal hematopoiesis and absence of circulating blood in zebrafish embryos.Gro3 MO (morpholino oligonucleotide)-injected or uninjected wild-type and Tg(hsp:AML1-ETO) zebrafish embryos were all subjected to the same heat induction. (A) Inspection of uninjected (Control) and gro3 MO injected Tg(hsp:AML1-ETO) zebrafish embryos showed the absence of circulating cells and accumulation of hematopoietic cells in the ICM and the ventral tail region (red arrows). These cells had a blast-like morphology with a large size and relatively little cytoplasm. (B) Percentages of embryos exhibiting loss-of-circulating cells were scored between 30 and 40 hpf. Control and MO indicate uninjected and morpholino oligonucleotide-injected embryos, respectively. AML1-ETO and WT indicate Tg(hsp:AML1-ETO) and wild-type embryos, respectively. n equals the number of embryos scored. P value was obtained using Student t test. The inset gels show the knockdown by RT-PCR of gro3 transcripts with 2 gro3 specific morpholino oligonucleotides. (C) Lack of circulating cells in AML1-ETO–expressing embryos is not caused by a vascular obstruction as evidenced by microangiography. The fluorescein-coupled latex beads injected into the inflow tract of the atrium was able to perfuse the whole vascular system of the Tg(hsp:AML1-ETO) embryos. It revealed a normal vascular pattern but reduced intersomitic vessels in gro3 morphant embryos. (D) Cytologic analysis of hematopoietic cells collected from the circulating blood in 3 uninjected Tg(hsp:AML1-ETO) embryos and the accumulating blood in 3 injected embryos. Whereas the uninjected embryos contain mostly red blood cells, the injected embryos contain abundant immature blast-like cells. Black arrows indicate red blood cells.
Accumulated immature ICM cells in AML1-ETO transgenic zebrafish embryos with Gro3/TLE knockdown are myeloid cells of the granulocytic lineage. Wild-type and AML1-ETO transgenic fish embryos were injected with Gro3 morpholino (MO) and subjected to mild heat shock treatment to induce low levels of AML1-ETO expression. The embryos were fixed at 32 hpf and were subjected to in situ hybridization for (A) the macrophage marker, l-plastin or for (B) myeloperoxidase (mpo), a marker for cells of the granulocytic lineage. The data indicate that knockdown of groucho in the presence of AML1-ETO expression leads to accumulation of mpo+ cells in the ventral tail region (arrow).
Accumulated immature ICM cells in AML1-ETO transgenic zebrafish embryos with Gro3/TLE knockdown are myeloid cells of the granulocytic lineage. Wild-type and AML1-ETO transgenic fish embryos were injected with Gro3 morpholino (MO) and subjected to mild heat shock treatment to induce low levels of AML1-ETO expression. The embryos were fixed at 32 hpf and were subjected to in situ hybridization for (A) the macrophage marker, l-plastin or for (B) myeloperoxidase (mpo), a marker for cells of the granulocytic lineage. The data indicate that knockdown of groucho in the presence of AML1-ETO expression leads to accumulation of mpo+ cells in the ventral tail region (arrow).
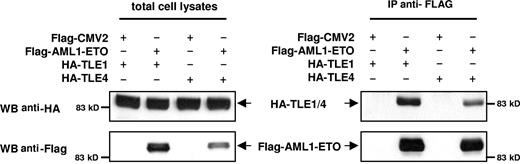
TLE1 and TLE4 complexes with AML1-ETO
Several models could explain the interactions of AML1-ETO and Gro/TLE, including effects on common downstream pathways. Alternatively, or in concert, the Gro/TLEs could physically interact with AML1-ETO to alter its activity. The Gro/TLE protein family is known to interact with RUNX family members, including RUNX1/AML1, through the c-terminal peptide sequence, WRPY, of AML1,34 which is missing from the AML1-ETO fusion gene. However, it has also been reported that TLE1 interacts with a peptide fragment containing the runt-binding domain of AML1.35 To confirm that AML1-ETO with the intact runt homology domain interacts with TLEs, we performed coimmunoprecipitation with AML1-ETO and either TLE1 or TLE4. These results showed that AML1-ETO can complex with either TLE1 or TLE4 (Figure 6). We recently reported truncated versions of AML1-ETO lacking C-terminal NHR3 and NHR4 domains involved in with NCoR/SMRT corepressors interaction no longer had a negative effect on the cell cycle and were potent inducers of leukemia.36,37 Thus, the loss of Gro/TLE corepressors, which mediate transcriptional silencing via the direct binding to HDAC138 and deacetylated histones,39 may similarly represent a cooperating event with AML1-ETO in promoting leukemogenesis.
TLE1 and TLE4 complex with AML1-ETO. 293T cells were transfected with HA-tagged TLE1 or TLE4 along with a FLAG empty vector (pFLAG-CMV2) or FLAG-tagged AML1-ETO (pFLAG-AML1-ETO). Total cell lysates immunoblotted with anti-HA and anti-FLAG antibodies show the expression of proteins. Lysates were immunoprecipitated with anti-FLAG antibodies, and the immunocomplexes were blotted with HA antibodies to demonstrate that both TLE1 and TLE 4 can interact with AML1-ETO.
TLE1 and TLE4 complex with AML1-ETO. 293T cells were transfected with HA-tagged TLE1 or TLE4 along with a FLAG empty vector (pFLAG-CMV2) or FLAG-tagged AML1-ETO (pFLAG-AML1-ETO). Total cell lysates immunoblotted with anti-HA and anti-FLAG antibodies show the expression of proteins. Lysates were immunoprecipitated with anti-FLAG antibodies, and the immunocomplexes were blotted with HA antibodies to demonstrate that both TLE1 and TLE 4 can interact with AML1-ETO.
Discussion
The identification of critical tumor suppressor genes within areas of recurrent deletions in AML has been particularly challenging. We had previously characterized the commonly deleted region in del(9q) AML and identified all the known genes and potential genes within and adjacent to this region.17 Given the close association of del(9q) and t(8;21), we designed a set of shRNAs to the del(9q) CDR genes and used them to attempt rescue of the cell death seen with induced expression of AML1-ETO in a myeloid cell line. This screen revealed that knockdown of TLE1 or TLE4, neighboring genes belonging to the Groucho/TLE family of corepressors, was capable of overcoming the apoptosis and cell death effects of AML1-ETO on myeloid progenitors. Although this effect was apparent with knockdown of expression of each TLE gene, the pronounced effects seen with simultaneous knockdown of TLE1 and TLE4 on the survival of AML1-ETO expressing cells suggest that the loss of both genes may be important in the pathogenesis of t(8;21) AML. A significant decrease in apoptosis was even demonstrated with a 20% to 40% reduction in TLE1 and TLE4 levels. Our results indicate that subtle modulation of the level of TLE expression can have quite significant effects. One implication of these findings is that haploinsufficiency of these proteins may cooperate with AML1-ETO and other genes in leukemogenesis. This is consistent with our previous study that did not demonstrate inactivating mutations in the remaining TLE alleles in del(9q) AMLs.17
Consistent with a cooperative effect of AML1-ETO and TLE reduction, we found a significantly lower median level of TLE expression in t(8;21) AML samples with del(9q) compared with CD34+ cells. This difference was not significant in t(8;21) without del(9q). We previously were unable to demonstrate occult deletions of TLE1 or TLE4 in t(8;21) samples without del(9q).17 These data suggest other mutations can cooperate with AML1-ETO in leukemogenesis. However, the large range of TLE expression levels seen in this group without del(9q), with several samples having quite low levels of TLE expression, is also consistent with a subset of t(8;21) samples without del(9q) having suppressed TLE expression by epigenetic mechanisms.
We cannot exclude the possibility that loss of expression of other genes within the CDR may cooperate with AML1-ETO in leukemogenesis. This experiment is subject to the limitations that genes with undetectable expression in this cell line, such as UBQLN1, NTRK2, and RASEF, could not be evaluated in this system, and that the shRNAs for some genes may have not been effective enough to reduce gene expression levels beyond a critical threshold.
In support of a role of the Gro/TLEs as tumor suppressor genes, we showed the AML1-ETO expressing Kasumi-1 cell line was very sensitive to the effects of modulating TLE levels. Knockdown of either TLE1 or TLE4 led to increased cell-cycle progression and cell division, with increased expression of the proliferation markers Ki-67 and of cyclinD1. With knockdown of TLE4 in Kasumi-1 cells, these effects translated into a proliferative advantage, which led to the expansion of these cells in culture. Furthermore, forced expression of TLE1 or TLE4 led to apoptosis and cell death of Kasumi-1 cells. The fact that TLE shRNAs did not confer a proliferative advantage to cell lines without AML1-ETO suggests that these 2 lesions may cooperate to overcome normal constraints on cell growth.
We also show that knockdown of the Gro/TLE family members cooperates with AML1-ETO in vivo using a zebrafish model. In this system, a high level of AML1-ETO expression at a defined period in zebrafish development results in an accumulation of immature-appearing granulocytic precursors and a lack of circulating blood cells. These effects were demonstrated at subthreshold levels of AML1-ETO expression in the presence of a specific morpholino oligonucleotide directed against the gro3 TLE homologue. Thus, gro3 knockdown appears to cooperate with AML1-ETO expression to arrest myeloid differentiation and prevent migration into the bloodstream. Although care must be taken in extending these conclusions to human leukemia, these experiments do provide important in vivo evidence for the cooperativity of Groucho/TLE knockdown and AML1-ETO.
These experiments demonstrate for the first time that the Gro/TLEs have tumor suppressor-type activities in regulating cell death and apoptosis and cellular proliferation in the context of myeloid cells expressing AML1-ETO, and implicate loss of the TLEs as potential second hits in the induction of AML1-ETO– associated AML.
The Groucho/TLE family of corepressors interacts with at least 5 families of transcription factors and plays critical roles in Drosophila and vertebrate development.40 During B-lymphocyte differentiation, the TLEs mediate the repressive effect of Pax5, via recruitment by Pu.1, in limiting alternative cell fate.41 However, the ability of the Gro/TLEs to interact with other signaling pathways suggests a potential broader role of the Gro/TLEs in both normal and malignant hematopoiesis. The Groucho/TLE family of corepressors bind to all known Tcf/LEF and act as inhibitors of Wnt/β-catenin signaling,42-45 a pathway implicated in expansion and self-renewal of the hematopoietic stem-cell compartment.46,47 Similarly, the Gro/TLEs inhibit NF-κB signaling,48 a pathway constitutively activated in AML and thought to play an important role in hematopoietic cell proliferation and survival and in chemoresistance.49-51 The Gro/TLE gene family is also a key effector of Notch signaling, a pathway implicated in HSC fate determination and self-renewal.52-54 In Drosophila, Groucho is one of the primary effectors maintaining silencing of downstream target genes in the absence of Notch signaling.55 Furthermore, the TLEs bind to and regulate RUNX1/AML1 and Pu.1, key myeloid transcription factors.34,56,57 It remains to be determined whether dysregulated pathways downstream of TLEs and AML1-ETO cooperate to promote leukemia or whether the direct interaction of TLEs modulates AML1-ETO activity. Although AML1-ETO was initially thought to behave primarily as a repressive transcription factor, it is now known to activate numerous genes potentially important in transformation. These include the macrophage colony stimulating factor receptor,58 the Notch ligand Jagged1,59 and the Wnt pathway mediator plakoglobin.60 Loss of TLE activity may be needed for AML1-ETO to exert its full effect in activating expression of such genes. One model for the interaction between TLE loss and AML1-ETO is suggested by our recent finding of AML1-ETO variants arising from alternative splicing or mutation. These variants delete C-terminal domains involved in the binding of NCoR/SMRT corepressors that recruit histone deacetylases and no longer had a negative effect on the cell cycle but were potent inducers of leukemia.36,37 The loss of Gro/TLE corepressors, which mediate transcriptional silencing via the direct binding to HDAC138 and deacetylated histones,39 may similarly represent cooperating events with AML1-ETO in promoting leukemogenesis.
The ability to interact with many different transcription factors suggests the function of the Gro/TLEs will probably vary, depending on the cell type and the timing of expression during development. Our studies demonstrating low levels of TLE1 and TLE4 expression in myeloid cell lines and subsets of AML samples, and potential tumor suppressor-like activities of the Gro/TLEs, stand in contrast to recent reports that overexpression of various Gro/TLE family members is seen in grade 1 astrocytomas,61 higher-grade meningiomas,62 pituitary adenomas,63,64 and synovial sarcomas.65,66 In addition, overexpression of murine Tle1/Grg1 in a transgenic model predisposed to the development of lung adenocarcinoma.67 Similar dual functions have been described for several other proteins, including WT1,68 E2F1,69 and K-RAS.70 This apparent dual Gro/TLE function mirrors the observation that RUNX1/AML1, which is inhibited by Gro/TLE binding, can act alternatively as an oncogene or a tumor suppressor gene,71-74 a finding that might reflect differential effects on stem cells and progenitors.75
In conclusion, we demonstrate that modulation of activity of 2 members of the Gro/TLE corepressor family, TLE1 and TLE4, cooperates with AML1-ETO to affect myeloid cell survival, apoptosis, and proliferation. These results provide a mechanistic rationale for the strong association of del(9q) and t(8;21) and implicate loss of TLE activity in the pathogenesis of AML1-ETO-associated AML. Further understanding of the mechanism of these interactions and of the downstream pathways affected could have therapeutic implications.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank David Scadden, Daniel Tenen, and Howard Weinstein for helpful discussions and support during the course of this work.
This work was supported by the Doris Duke Charitable Foundation Clinical Scientist Development Award (D.A.S.), American Society of Hematology Junior Faculty Scholar Award (D.A.S.), MassGeneral Hospital Marathon Fund (D.A.S.), Mattina Proctor Fund (D.A.S.), Lauri Strauss Leukemia Foundation (F.D.), Tosteson Fellowship from the Massachusetts Biomedical Research Corporation (J.-R.J.Y.), Lady Tata Memorial Trust (E.-Y.A.), American Cancer Society Clinical Research Professorship Award (I.D.B.), American Cancer Society PRD-95–124-12 (I.D.B.), Leukemia & Lymphoma Society SCOR 7-40-03 (I.D.B.), and National Institutes of Health grants 1R01CA115772 (D.A.S.), 5T32CA71345-09 (F.D.), CA118498 (R.T.P.), HL079267 (R.T.P.), CA96735 (D.E.Z.), CA98543 (I.D.B.), U10 CA98543 (I.D.B.), and U24 CA114766 (I.D.B.).
National Institutes of Health
Authorship
Contribution: F.D., J.W., J-R.J.Y., and E-Y.A. designed and performed research, analyzed data, and contributed to writing the paper; E.T. performed research; D.-E.Z., I.D.B., and R.T.P. designed research and contributed to writing the paper; D.A.S. designed research, supervised the study, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: David A. Sweetser, Massachusetts General Hospital, 55 Fruit Street, Jackson 904, Boston, MA 02114; e-mail: dsweetser@partners.org.
References
Author notes
*F.D. and J.W. contributed equally to this study.