Abstract
CD40 and its ligand, CD154, are major costimulatory molecules whose interactions are important in humoral and cellular immunity. We hypothesized that single nucleotide polymorphisms (SNPs) in TNFRSF5 and TNFSF5 encoding the CD40 and CD154 proteins, respectively, influence lymphoma risk, particularly a functional TNFRSF5 SNP (−1C>T, rs1883832) associated with reduced B-cell CD40 expression. TNFRSF5 and TNFSF5 SNPs were examined in a population-based case-control study of non-Hodgkin lymphoma (376 cases/801 controls with DNA), and compelling findings were followed up in 2 independent populations. Pooled analyses of all 3 case-control studies (total N = 1776 non-Hodgkin lymphoma cases, N = 2482 controls) revealed an increased risk of follicular lymphoma (FL) associated with the TNFRSF5 −1TT genotype (odds ratio = 1.6; 95% confidence interval, 1.1-2.4). In addition, among women, an inverse association was found between the variant A allele for a TNFSF5 6809G>A SNP and FL risk (OR = .61; 95% CI, 0.36-0.98). In genotype-phenotype studies, significantly reduced circulating soluble CD40 was observed in TNFRSF5 −1TT compared with −1CC carriers. Further, dendritic cells from those with −1TT versus −1CC genotypes exhibited lower CD40 cell surface expression. These results suggest that the TNFRSF5 −1C>T polymorphism may increase FL susceptibility through mechanisms that hinder cellular immune responses. Further studies are needed to explore these findings.
Introduction
Lymphomas are a heterogeneous group of malignancies derived from different stages of B-cell maturation.1 The germinal center reaction plays a pivotal role in the generation of the majority of B-cell lymphomas, such as follicular (FL), and some diffuse large B-cell (DLBCL)2,3 and Hodgkin lymphomas (HL).4 Thus, genetic and environmental factors that disrupt normal germinal center reactions may increase the risk of these lymphomas. Accumulating evidence underscores the essential roles of the costimulatory molecules CD40 receptor and its ligand CD154 (CD40L) in the generation and maintenance of humoral and cellular immune responses.5,6 CD40 is a member of the tumor necrosis factor (TNF) receptor family that is expressed constitutively on antigen-presenting cells (APCs).6 CD154 is primarily expressed on activated CD4+ T cells but also on a variety of cells, including CD8+ T cells, mast cells, monocytes, macrophages, natural killer cells, and dendritic cells.7 Soluble forms of CD40 (sCD40) and sCD154 also are present in the serum and plasma8,9 through transmembrane cleavage of bound CD40 and CD154 by ADAM family member proteases.10,11 sCD40 and sCD154 may regulate distinct signaling pathways or complement the biologic activity of their membrane-bound counterparts. CD40-CD154 ligation is crucial for the maturation and activation of B cells, dendritic cells, and other APCs.6 Activated B cells that receive low CD40-CD154 signals in the germinal center have limited lifespans and display impaired immunoglobulin (Ig) production, isotype switching, and B-cell memory.12 Moreover, dendritic cells and other APCs that express low CD40 have reduced tumor-specific immune responses involving cytotoxic T-lymphocyte activation.7
Mutations in the TNFSF5 and TNFRSF5 genes that encode CD154 and CD40, respectively, are associated with a severe form of immunodeficiency called hyper-immunoglobulin M (HIGM) syndrome.13 The HIGM disorders are characterized by reduced CD154 or CD40 protein expression, elevated IgM levels, defective class switched antibody formation, absence of germinal center reactions, and weakened humoral immune responses.6 Because CD40-CD154 interactions regulate multiple phases of the immune response, even slight imbalances in their expression may result in immune dysfunction. Enhanced expression may foster hyperimmune responses and autoimmune disease, whereas reduced levels may deregulate germinal center reactions and impair immune responses to invading pathogens and cancerous cells. The C allele of a −1C>T promoter single nucleotide polymorphism (SNP) located in the Kozak sequence of the TNFRSF5 gene (rs1883832)14 has been associated with enhanced CD40 translational efficiency15 and protein expression on B cells16 and increased susceptibility to Graves' disease.15,17 SNPs in the Kozak sequence, which is the 6 to 8 nucleotides before and after the ATG start codon,18 can influence initiation of translation of the CD40 protein.19 In light of the important roles of CD40 and CD154 in immune regulation, we hypothesized that SNPs in TNFRSF5 and TNFSF5 may influence susceptibility to non-Hodgkin lymphoma (NHL). To test this hypothesis, the −1C>T TNFRSF5 SNP and 8 others across the TNFRSF5 and TNFSF5 genes were investigated in a population-based case-control study of NHL. Associated SNPs were further explored in 2 independent case-control studies of lymphoma where we found that the TNFRSF5 −1TT SNP and an intronic SNP in the TNFSF5 gene influenced FL risk in pooled analyses. Given their potential importance in humoral and cellular immunity, we next investigated whether sCD40 and sCD154 levels were influenced by the functional TNFRSF5 −1C>T SNP. Here we found an inverse association with sCD40 and the −1T allele. We further explored genotype-phenotype associations using an ex vivo dendritic cell system and we relate a model delineating how this SNP could influence lymphoma risk.
Methods
Study populations
San Francisco Bay Area NHL1 study.
Detailed methods have been published previously.20-23 Briefly, incident NHL patients diagnosed between 1988 and 1993 were identified by the Northern California Cancer Center's rapid case ascertainment. Eligible patients were between 21 and 74 years of age, were residents of one of the 6 Bay Area counties at the time of diagnosis, and could complete an interview in English. A total of 1591 eligible patients (72% response rate) completed in-person interviews. A total of 2515 control participants (78% response rate) were identified by random-digit dial and by random sampling of the Health Care Financing Administration lists to supplement recruitment of participants aged 65 years of age and older. Controls were frequency-matched to patients by age within 5 years, sex, and county of residence. Eligibility criteria for controls were the same as for cases with the exception of NHL diagnosis. Eligible cases and controls were asked to participate in the laboratory portion of the study that consisted of providing a blood specimen for viral testing (63% and 66% participation rates, respectively). DNA was available for 376 case and 801 control white non-Hispanics for these analyses. Demographic characteristics of the participants for whom DNA was available for genotyping were similar to those of the total study population including case-control status (Fisher exact test, P = .09), age at interview/diagnosis (χ2, P = .74), sex (Fisher exact test, P = .36), race (χ2, P = .19), and median age at diagnosis among cases (Wilcoxon 2-sample test, P = .26; Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
San Francisco Bay Area NHL2 study.
A large population-based case-control study of NHL that included incident cases diagnosed from October 2001 through May 2006 was conducted in the San Francisco Bay Area. Eligible patients were identified by the Northern California Cancer Center's rapid case ascertainment, age 20 to 84 years, residents of one of the 6 Bay Area counties at the time of diagnosis and could complete an interview in English. Controls were frequency-matched to patients by age in 5-year groups, sex, and county of residence. Blood and/or buccal specimens were collected from eligible cases and controls who participated in the laboratory portion of the study (provisional participation rates, 87% and 89%, respectively). Participants were eligible for venipuncture if they had no medical contraindications, including current use of blood thinners, a portacath in place, or had a bleeding disorder. This report is based on interim analyses of the first 2468 study participants for whom biospecimens were available for analysis (1290 cases, 1358 controls).
NHL subtype classification for the San Francisco Bay Area NHL1 and NHL2 studies.
Diagnostic pathology materials for each study were rereviewed by the study's expert pathologist to confirm NHL diagnoses and to classify NHL subtypes consistently. The working formulation classification was used in the NHL1 study, whereas the Revised European-American Lymphoma/World Health Organization classification was used to classify subtypes in the NHL2 study.
German study.
Detailed information regarding the design of this population-based case-control study of lymphoma has been published previously.24 Briefly, the study was conducted from 1999 to 2002 in 6 regions of Germany among 18- to 80-year-old men and women. The study included 710 cases of DLBCL, FL, chronic lymphocytic leukemia, multiple myeloma, mucosa-associated lymphoid tissue lymphoma, marginal zone lymphoma, T-cell lymphoma, and HL and 710 controls individually matched by sex, age (±1 year of birth), and study region. Analyses for all NHL did not include multiple myeloma or HL. For subtype-specific analyses, only FL, DLBCL, and HL were considered. Patients were recruited from hospitals and office-based physicians involved in diagnosis and treatment of lymphoma within each study region. Controls were randomly selected from population registers within the study regions. The participation rate was 87% for cases and 44% for controls. DNA was available for 682 cases (96%) and 667 controls (94%).
Study protocols were approved by each institution's ethics review committee. All study participants provided informed consent before interview and biospecimen collection.
SNP selection
TNFRSF5 and TNFSF5 SNPs that were selected for genotyping in the San Francisco Bay Area NHL1 study are listed in Table S2 and were identified using dbSNP25 and SNPper.26 For TNFRSF5, 4 tags SNPs were chosen for investigation from a total of 10 validated SNPs with a 5% or greater minor allele frequency in the Centre d' Etude du Polymorphisme Humain (CEPH) population [Utah residents with ancestry from northern and western Europe from the HapMap database (Build 34, www.hapmap.org/)]. For TNFSF5, 5 of 7 validated SNPs with a more than or equal to 5% minor allele frequency in CEPH trios were chosen for investigation.
Genotyping
DNA was isolated from peripheral blood mononuclear cells using the QIAamp DNA Blood Maxi Kit protocol (Qiagen, Valencia, CA; Hilden, Germany) and quantified using PicoGreen dsDNA Quantitation kits (Invitrogen, Carlsbad, CA) according to the manufacturers' specifications. In the U.S. studies, genotyping was performed using TaqMan SNP Genotyping Assays (Applied Biosystems, Foster City, CA) on the ABI Prism 7700 Sequence Detection System or GeneAmp PCR System 9700 as previously described.27 In the German study, genotyping was performed using Pyrosequencing technology as previously described.28 Probe and primer sequences for all genotyping assays are listed in Table S2a/b. Replicate, blinded quality control samples were included to assess reproducibility of the above genotyping procedures by both groups. Ambiguous genotypes were regenotyped as necessary. All plates contained positive (all 3 genotypes) and negative (water) controls.
Enzyme-linked immunosorbent assays
sCD40 in plasma and serum and sCD154, CD80, IgM, IgG, and IgE levels in plasma were determined using ELISA (enzyme-linked immunosorbent assay) kits from Bender Medsystems (sCD40, sCD154; Vienna, Austria), R&D Systems (CD80; Minneapolis, MN), and Bethyl Laboratories (IgM, IgG and IgE; Montgomery, TX) according to the manufacturers' protocols.
Determination of CD40 cell surface levels on monocyte-derived activated dendritic cells
Using blood from healthy laboratory controls of the same age (± 5 years), negatively selected peripheral monocytes were obtained using a Monocyte Isolation Kit II according to manufacturer's recommendations (Miltenyi Biotec, Auburn, CA). Briefly, all nonmonocytes were stained with antibodies conjugated to biotin. A secondary antibiotin antibody conjugated to iron microbeads captured the stained cells on a magnetized capture column. Unstained monocytes were recovered in the column flow-through. Monocytes were stained with CD14-fluorescein isothiocyanate or RPE-Cy5.5 (phycoerythrin-cyanine dye conjugate), whereas nonmonocytes were stained with an antibiotin antibody conjugated with RPE. The monocyte-enriched fraction was cultured in RPMI 1640 (10% fetal bovine serum, penicillin and streptomycin, sodium pyruvate) with human interleukin-4 (50 ng/mL) and granulocyte-macrophage colony-stimulating factor (50 ng/mL; BD Biosciences, San Jose, CA) for 6 days at 37°C with 5% CO2. The CD11c surface marker was used to identify cultured myeloid dendritic cells. On day 6, lipopolysaccharide (LPS) was added to stimulate monocyte-derived dendritic cells. After 2 days of LPS stimulation, activated dendritic cells were identified with mouse antihuman CD80 conjugated to RPE-Cy5, along with the CD11c-PE and CD40-fluorescein isothiocyanate stains. CD40 levels were determined on activated dendritic cells (CD11c+ CD80+) by flow cytometry. Cell media were collected for cytokine determination by ELISA.
Statistical analyses
Genetic analyses.
For all 3 studies, analyses were restricted to HIV-negative white, non-Hispanic individuals (NHL1: 308 cases, 684 controls; NHL2: 980 cases, 1136 controls; Germany: 488 cases, 662 controls) to minimize potential population stratification due to differences in SNP frequencies across self-identified race and Hispanic ethnicity categories. Patients in each study were analyzed as all NHL and by the 2 major subtypes, DLBCL and FL. For the German study, an additional 111 HL patients also were analyzed. Unconditional logistic regression models were used to compute odds ratios (OR), expressed in the text as “risk” for NHL, and corresponding 95% confidence intervals (CI) adjusted for age and sex. For the German study, risk estimates from conditional logistic regression (for individually matched case-controls study designs) were similar to those from unconditional logistic regression based on the risk estimates and 95% CIs. Therefore, unconditional methods were used for the analyses of pooled data, which included a total of 1776 cases and 2482 controls. This maximized efficiency by allowing use of all available data for individuals. For the pooled analyses, ORs and 95% CIs adjusted for age, sex, and study geographic location were generated using unconditional logistic regression in analyses of white non-Hispanic participants. Analyses were conducted using Stata, version 8 (College Station, TX) or SAS, version 9.1 (SAS Institute, Cary, NC). For each SNP, Hardy-Weinberg equilibrium was determined among controls using standard chi-square methods.
Haplotype analysis.
Haplotype frequencies and the ORs for common haplotypes were estimated using methods for unphased data. The posterior probability of each haplotype for each participant was determined using the E-M algorithm as implemented in the gap package (gc.em function in the statistical package, R) with the referent group defined as the most common haplotype.29 The association of haplotype and cancer was estimated by multiple imputation of haplotypes from these posterior probabilities and then associating the presence and absence of each haplotype (relative to the referent haplotype) with case-control status using logistic regression. Bootstrapping (randomly resampling participants) was used to compute the variability estimates of these estimated ORs. Linkage disequilibrium between adjacent SNPs was measured using D′ where 1.0 indicated complete linkage.
Statistical analyses of ELISAs.
The overall associations between mean levels of CD40 serum/plasma markers and TNFRSF5 −1C>T genotypes were evaluated using one-way ANOVA. Tukey pairwise comparison procedure was used to evaluate the pair-wise differences between genotypes and serum and plasma marker levels in the case and control groups when the overall ANOVA results showed that mean levels differed across groups. Results were considered statistically significant for 2-sided P at .05 or less. Summary statistics for continuous variables are presented as mean plus or minus SEM.
Results
Distribution of the TNFRSF5 and TNFSF5 genotypes and their association with lymphoma
Control genotype frequencies for the 9 SNPs under study were in Hardy-Weinberg equilibrium (P > .05). Because the TNFSF5 gene is located on the X chromosome, genetic analyses for TNFSF5 polymorphisms were conducted by sex.
For TNFRSF5, no significant genotype or haplotype associations were found, although carriers of the low function TNFRSF5 −1C>T (rs1883832) homozygous variant genotype exhibited an elevated OR for FL (OR = 1.6; 95% CI, 0.80-3.2; Table S3). Hence, this SNP was further explored in 2 independent NHL case-control study populations (San Francisco Bay Area NHL2 and German studies). Here we observed elevated ORs for FL associated with the TNFRSF5 −1TT genotype in both studies (OR = 1.6; 95% CI, 0.93-2.9; and OR = 1.9; 95% CI, 0.78-4.6, respectively), although 95% CIs included 1.0 (Table S4). Moreover, in the German study, the −1TT genotype was associated with increased risks for DLBCL (OR = 2.0; 95% CI, 1.0-3.8) and HL (OR = 2.4; 95% CI, 0.98-5.6). Pooled analyses of all 3 studies revealed that the −1CT and −1TT genotypes were associated with significant 30% and 60% increased risks of FL, respectively (OR = 1.3; 95% CI, 1.1-1.7; OR = 1.6; 95% CI, 1.1-2.4; Table 1).
Pooled odds ratios (OR) and 95% confidence intervals (CI) by NHL subtype for TNFRSF5 −1C>T (rs1883832), TNFSF5 3005G>A (rs715762) and TNFSF5 6809 G>A (rs3092933) SNPs in the San Francisco Bay Area NHL1, NHL2, and German studies
| . | Controls, N (%) . | Cases, N (%) . | |||||
|---|---|---|---|---|---|---|---|
| All NHL . | OR (CI) . | DLBCL . | OR (CI) . | FL . | OR (CI) . | ||
| TNFRSF5 – 1C>T (rs1883832) | 2481 (100) | 1775 (100) | — | 520 (100) | — | 394 (100) | — |
| −1 CC | 1382 (56) | 933 (53) | 1.0 | 277 (53) | 1.0 | 188 (48) | 1.0 |
| −1 CT | 932 (38) | 695 (39) | 1.1 (0.97-1.3) | 199 (38) | 1.1 (0.87-1.3) | 168 (43) | 1.3 (1.1-1.7) |
| −1 TT | 166 (6.7) | 147 (8.2) | 1.3 (1.0-1.7) | 44 (8.5) | 1.3 (0.90-1.9) | 37 (9.4) | 1.6 (1.1-2.4) |
| CT/TT | 1098 (44) | 842 (47) | 1.1 (1.0-1.3) | 243 (47) | 1.1 (0.91-1.3) | 205 (52) | 1.4 (1.1-1.7) |
| P for trend* | — | — | .20 | — | 0.54 | — | .02 |
| TNFSF5 3005G>A (rs715762) | |||||||
| Men | 1402 | 1010 | — | 287 | — | 186 | — |
| 3005 G | 1262 (90) | 901 (89) | 1.0 | 256 (89) | 1.0 | 163 (88) | 1.0 |
| 3005 A | 140 (10) | 109 (11) | 1.1 (0.83-1.4) | 31 (11) | 1.1 (0.70-1.7) | 23 (12) | 1.3 (.76-2.1) |
| Women | 981 | 744 | — | 230 | — | 203 | — |
| 3005 GG | 790 (81) | 609 (82) | 1.0 | 186 (81) | 1.0 | 167 (82) | 1.0 |
| 3005 AG | 170 (12) | 119 (16) | 1.6 (1.2-2.1) | 40 (17) | 1.5 (1.0-2.4) | 31 (15) | 1.4 (.86-2.1) |
| 3005 AA | 21 (2.1) | 16 (2.1) | 0.99 (0.48-2.0) | 4 (1.7) | 0.81 (0.20-2.4) | 5 (2.5) | 1.1 (.33-3.1) |
| 3005 AG/AA | 191 (19) | 135 (18) | 0.92 (0.71-1.2) | 44 (19) | 0.77 (0.66-1.4) | 36 (18) | 0.89 (0.58-1.3) |
| TNFSF5 6809 G>A (rs3092933) | |||||||
| Men | 1399 | 1006 | — | 285 | — | 186 | — |
| 6809 G | 1276 (99) | 923 (92) | 1.0 | 265 (93) | 1.0 | 173 (93) | 1.0 |
| 6809 A | 123 (8.8) | 83 (8.4) | 0.93 (0.69-1.3) | 20 (7.0) | 0.78 (0.45-1.3) | 13 (6.7) | 0.78 (0.39-1.4) |
| Women | 984 | 744 | — | 226 | — | 205 | — |
| 6809 GG | 813 (83) | 640 (86) | 1.0 | 188 (83) | 1.0 | 182 (89) | 1.0 |
| 6809 AG | 159 (16) | 99 (13) | 0.78 (0.59-1.0) | 38 (17) | 1.0 (0.67-1.5) | 23 (11) | 0.65 (0.39-1.0) |
| 6809 AA | 10 (1.0) | 5 (.67) | 0.45 (0.13-1.3) | 0 | — | 0 | — |
| 6809 AG/AA | 169 (17) | 104 (14) | 0.75 (0.57-0.99) | 38 (17) | 0.94 (0.62-1.4) | 23 (11) | 0.61 (0.36-0.98) |
| . | Controls, N (%) . | Cases, N (%) . | |||||
|---|---|---|---|---|---|---|---|
| All NHL . | OR (CI) . | DLBCL . | OR (CI) . | FL . | OR (CI) . | ||
| TNFRSF5 – 1C>T (rs1883832) | 2481 (100) | 1775 (100) | — | 520 (100) | — | 394 (100) | — |
| −1 CC | 1382 (56) | 933 (53) | 1.0 | 277 (53) | 1.0 | 188 (48) | 1.0 |
| −1 CT | 932 (38) | 695 (39) | 1.1 (0.97-1.3) | 199 (38) | 1.1 (0.87-1.3) | 168 (43) | 1.3 (1.1-1.7) |
| −1 TT | 166 (6.7) | 147 (8.2) | 1.3 (1.0-1.7) | 44 (8.5) | 1.3 (0.90-1.9) | 37 (9.4) | 1.6 (1.1-2.4) |
| CT/TT | 1098 (44) | 842 (47) | 1.1 (1.0-1.3) | 243 (47) | 1.1 (0.91-1.3) | 205 (52) | 1.4 (1.1-1.7) |
| P for trend* | — | — | .20 | — | 0.54 | — | .02 |
| TNFSF5 3005G>A (rs715762) | |||||||
| Men | 1402 | 1010 | — | 287 | — | 186 | — |
| 3005 G | 1262 (90) | 901 (89) | 1.0 | 256 (89) | 1.0 | 163 (88) | 1.0 |
| 3005 A | 140 (10) | 109 (11) | 1.1 (0.83-1.4) | 31 (11) | 1.1 (0.70-1.7) | 23 (12) | 1.3 (.76-2.1) |
| Women | 981 | 744 | — | 230 | — | 203 | — |
| 3005 GG | 790 (81) | 609 (82) | 1.0 | 186 (81) | 1.0 | 167 (82) | 1.0 |
| 3005 AG | 170 (12) | 119 (16) | 1.6 (1.2-2.1) | 40 (17) | 1.5 (1.0-2.4) | 31 (15) | 1.4 (.86-2.1) |
| 3005 AA | 21 (2.1) | 16 (2.1) | 0.99 (0.48-2.0) | 4 (1.7) | 0.81 (0.20-2.4) | 5 (2.5) | 1.1 (.33-3.1) |
| 3005 AG/AA | 191 (19) | 135 (18) | 0.92 (0.71-1.2) | 44 (19) | 0.77 (0.66-1.4) | 36 (18) | 0.89 (0.58-1.3) |
| TNFSF5 6809 G>A (rs3092933) | |||||||
| Men | 1399 | 1006 | — | 285 | — | 186 | — |
| 6809 G | 1276 (99) | 923 (92) | 1.0 | 265 (93) | 1.0 | 173 (93) | 1.0 |
| 6809 A | 123 (8.8) | 83 (8.4) | 0.93 (0.69-1.3) | 20 (7.0) | 0.78 (0.45-1.3) | 13 (6.7) | 0.78 (0.39-1.4) |
| Women | 984 | 744 | — | 226 | — | 205 | — |
| 6809 GG | 813 (83) | 640 (86) | 1.0 | 188 (83) | 1.0 | 182 (89) | 1.0 |
| 6809 AG | 159 (16) | 99 (13) | 0.78 (0.59-1.0) | 38 (17) | 1.0 (0.67-1.5) | 23 (11) | 0.65 (0.39-1.0) |
| 6809 AA | 10 (1.0) | 5 (.67) | 0.45 (0.13-1.3) | 0 | — | 0 | — |
| 6809 AG/AA | 169 (17) | 104 (14) | 0.75 (0.57-0.99) | 38 (17) | 0.94 (0.62-1.4) | 23 (11) | 0.61 (0.36-0.98) |
Odds ratios (OR) and 95% confidence intervals (CI) were estimated using unconditional logistic regression adjusted for age and sex.
NHL indicates non-Hodgkin lymphoma; DLBCL, diffuse large B-cell lymphoma; FL, follicular lymphoma; and —, not applicable.
Linear trend estimated from age- and sex-adjusted logistic models where genotype was analyzed as an ordinal variable based on the number of variant alleles (0, 1, 2).
For TNFSF5, in the San Francisco Bay Area NHL1 study, the homozygous variant genotype for 3005G>A (rs715762) was associated with an increased risk of NHL (OR = 1.8; 95% CI, 1.1-2.9), DLBCL (OR = 2.1; 95% CI, 1.0-4.2), and FL (OR = 2.1; 95% CI, 1.0-4.4) in men, but not in women (Table S5). We also found that variant alleles for 2 completely linked (D′ = 1.0) SNPs, TNFSF5 6809G>A (rs3092933) and 10776T>C (rs3092923), conferred marginal reduced risks of FL in women (OR = 0.42; 95% CI, 0.14-1.2). No other TNFSF5 genotype or haplotype associations were observed in the NHL1 study.
In follow-up genetic analyses of TNFSF5 rs715762 and rs3092933 in the NHL2 and German studies, no significant associations were found with NHL (Table S6), although pooled analyses of all 3 studies revealed an inverse association between the rs3092933 variant A allele and FL risk in women (OR = 0.61; 95% CI, 0.36-0.98; Table 1).
TNFRSF5 −1C>T genotype influences sCD40 levels
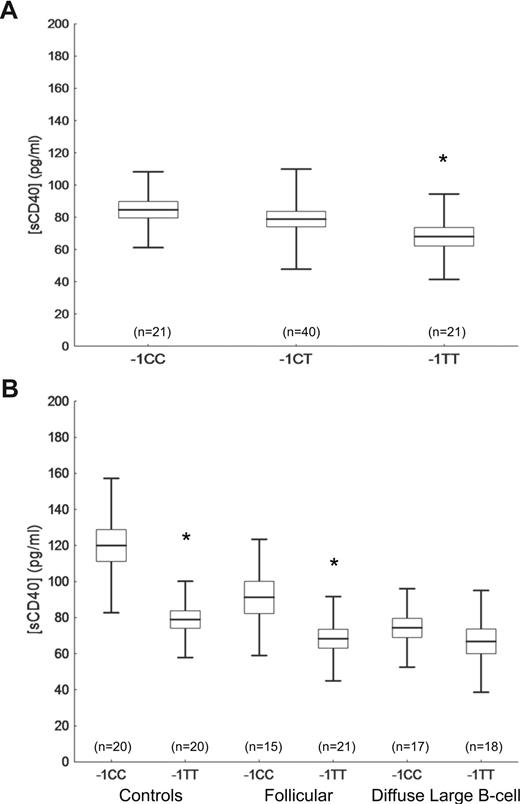
To investigate the effect of the TNFRSF5 −1C>T SNP on circulating sCD40, sCD154, CD80, IgG, IgE, and IgM levels, plasma concentrations of these markers were determined by ELISA in a subset of normal healthy NHL2 study controls who were randomly selected by genotype group. For sCD40, the data were nearly normally distributed. Nonparametric analyses provided essentially the same results (data not shown) as the analyses of non-transformed data. Lower sCD40 plasma levels were detected in controls who harbored the TNFRSF5 −1TT (67.9 ± 4.2 pg/mL) compared with the −1CC genotype (−1CC = 84.7 ± 6.2 pg/mL, P = .04; Figure 1A). Lower sCD40 serum levels also were detected in controls with the −1TT (79.03 ± 6.9 pg/mL) compared with −1CC carriers (119.97 ± 7.1 pg/mL, P < .001; Figure 1B). Significantly lower serum sCD40 levels also were observed for FL patients with the −1TT (68.36 ± 6.07 pg/mL) versus −1CC genotype (91.22 ± 7.53 pg/mL, P = .02), but no significant differences were observed for DLBCL patients (−1TT = 66.81 ± 6.11 pg/mL, −1CC = 74.30 ± 6.11 pg/mL; P = .39; Figure 1B). In overall analyses, the mean sCD40 levels between cases and controls were different across TNFRSF5 −1C>T genotype groups (P < .001). Regardless of genotype, sCD40 levels were lower among patients (77.36 ± 5.33 pg/mL for FL and 70.56 ± 5.26 pg/mL for DLBCL) than in controls (98.95 ± 5.04 pg/mL; P < .001). Adjustment for age and sex did not affect the results (data not shown). No other differences were found between sCD154, immunoglobulin, and CD80 plasma levels considering the TNFRSF5 −1C>T genotype or case-control status.
sCD40 blood concentrations by TNFRSF5 −1C>T genotypes. (A) sCD40 concentrations in plasma of healthy controls by TNFRSF5–1C>T genotypes. (B) sCD40 concentrations in sera of healthy controls and follicular and diffuse large B-cell lymphoma cases by TNFRSF5 −1CC and −1TT genotypes. Means (bars) with standard errors (box) and one SD (whiskers) are shown (*P < .05).
sCD40 blood concentrations by TNFRSF5 −1C>T genotypes. (A) sCD40 concentrations in plasma of healthy controls by TNFRSF5–1C>T genotypes. (B) sCD40 concentrations in sera of healthy controls and follicular and diffuse large B-cell lymphoma cases by TNFRSF5 −1CC and −1TT genotypes. Means (bars) with standard errors (box) and one SD (whiskers) are shown (*P < .05).
TNFRSF5 −1C>T genotype influences CD40 surface expression levels on LPS-activated dendritic cells
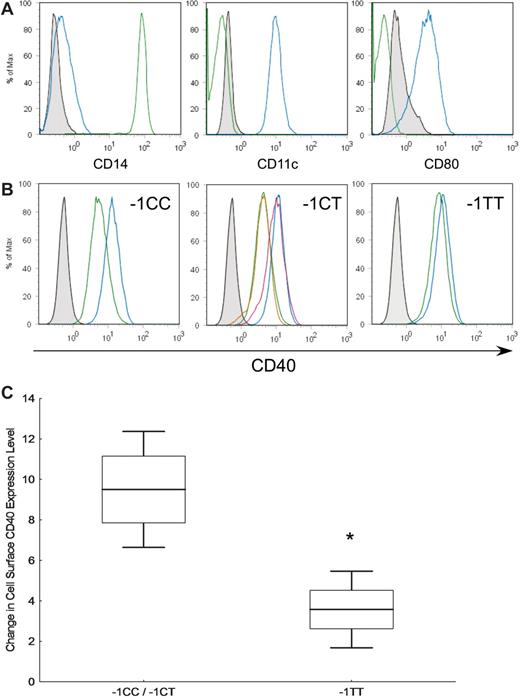
We assessed the functional consequences of the TNFRSF5 −1C>T polymorphism on CD40 surface expression on LPS-activated dendritic cells of healthy individuals. Figure 2A shows the differentiation of monocytes into dendritic cells indicated by the reduction in CD14 and increase in CD11c and CD80 after 6 days of culture. Results from pre- and post-LPS treatment of dendritic cells from participants with the 3 TNFRSF5 −1C>T genotypes revealed a downward shift in mean CD40 fluorescence intensity (FI) levels in −1CC/CT (9.6 FI) versus −1TT carriers (3.6 FI; Figure 2B). Collectively, CD40 cell surface expression levels on dendritic cells were 2.7 times lower in −1TT individuals (Figure 2C).
CD40 surface expression levels on lipopolysaccharide (LPS)–activated dendritic cells. (A) Monocytes fractionated from peripheral blood mononuclear cells were stained with CD14, CD11c, and CD80 (green) and after 6 days of IL-4 and GM-CSF stimulation, a dendritic cell phenotype was observed (blue). (B) Monocyte-derived dendritic cells treated with (blue) and without (green) LPS were stained with CD40 antibody. A replicate experiment for the TNFRSF5 −1CT individual is shown (red) overlapping the previous experiment. (C) Representative plots of differences between lipopolysaccharide (LPS) untreated and treated CD40 peak levels (TNFRSF5 −1CC/−1CT n = 7, −1TT n = 4). Means (bars) with standard errors (boxes) and one SD (whiskers) are plotted. Data from −1CC and −1CT carriers were pooled due to similarities in their CD40 shifts.
CD40 surface expression levels on lipopolysaccharide (LPS)–activated dendritic cells. (A) Monocytes fractionated from peripheral blood mononuclear cells were stained with CD14, CD11c, and CD80 (green) and after 6 days of IL-4 and GM-CSF stimulation, a dendritic cell phenotype was observed (blue). (B) Monocyte-derived dendritic cells treated with (blue) and without (green) LPS were stained with CD40 antibody. A replicate experiment for the TNFRSF5 −1CT individual is shown (red) overlapping the previous experiment. (C) Representative plots of differences between lipopolysaccharide (LPS) untreated and treated CD40 peak levels (TNFRSF5 −1CC/−1CT n = 7, −1TT n = 4). Means (bars) with standard errors (boxes) and one SD (whiskers) are plotted. Data from −1CC and −1CT carriers were pooled due to similarities in their CD40 shifts.
Discussion
In pooled analyses of 3 independent lymphoma studies comprising 1776 NHL cases and 2482 controls, we have demonstrated an important role for a −1C>T SNP located in the Kozak sequence of the TNFRSF5 gene as a susceptibility locus for FL. Here, we report that the TNFRSF5 −1TT genotype increased risk of FL by 30% in those harboring one copy of the variant −1T allele and up to 60% for those with 2 copies. Although an increased risk for DLBCL was observed for −1T heterozygotes in the NHL1 study and for homozygous variants in the German study, no association was found between the −1T allele and DLBCL in the NHL2 study or in the pooled analyses. Given the heterogeneity of DLBCL,30 this may be partially the result of different proportions of germinal center–derived DLBCL cases in the 3 study populations. Furthermore, in the German study, which is the only study with HL cases, the TNFRSF5 −1TT genotype also conferred a greater than 2-fold increased risk of HL. These studies suggest a potentially detrimental role for the TNFRSF5 −1C>T SNP in lymphomagenesis that may be particularly relevant to germinal center-derived lymphomas. In the pooled genetic analyses, we also observed that the variant A allele for an intronic SNP in TNFSF5, rs3092933, located on the X chromosome was inversely associated with FL risk in women. The relevance of this finding will require further study given the potential for inactivated alleles on the X chromosome and that this is a relatively rare SNP.
Previous functional studies revealed reduced CD40 surface expression on B cells in carriers of the TNFRSF5 −1T allele,16 suggesting that this SNP may affect humoral immune responses. As a follow-up to these studies, we determined whether the −1T allele influenced CD40 expression on dendritic cells because, among all APCs, CD40-activated dendritic cells are the most potent stimulators of antigen-specific CD4+ and CD8+ T-cell responses and antitumor immunity.31-33 In ex vivo studies of LPS-stimulated dendritic cells, we found that −1TT compared with −1CT/TT carriers exhibited a 2.7-fold reduction in CD40 dendritic cell surface expression, suggesting that the −1TT genotype may dampen cell-mediated immune responses. In vitro and animal studies in CD40 or CD154 knockout mice support the pivotal role of CD40 in germinal center reactions, particularly in the promotion of B-cell proliferation and differentiation, germinal center formation, and Ig production.34,35 Expression of CD40 on other APCs, such as dendritic cells, monocytes, and macrophages,6 also suggests the relevance of CD40-CD154 interaction in T cell–dependent cell-mediated immunity. The in vivo confirmation of these effects is evident in HIGM patients with CD154 or CD40 deficiency who exhibit defective Ig isotype switching and germinal center formation, high or normal IgM levels, low IgA, IgG, and IgE production, and increased susceptibility to opportunistic infections.13,36 In the present study, we hypothesized that the TNFRSF5 −1C>T SNP might modestly mimic CD40 deficiency and influence circulating CD40, IgM, IgG, or IgE levels. Although no differences were found in Ig levels when stratified by genotype, the inverse correlation between the TNFRSF5 −1T allele and sCD40 levels in healthy controls and FL cases may be biologically meaningful.
Soluble forms of membrane bound CD40 molecules can be released from cell surfaces by the actions of matrix metalloproteases.35 In vitro studies suggest that sCD40 acts as a natural agonist for CD40/CD154 ligation,38 although no comparable in vivo studies have been published. For T- and B-cell interaction, variations in sCD40 levels may influence germinal center reactions, somatic cell hypermutation, and class switch recombination as the B-cell attempts to mature its low affinity IgM to a higher affinity IgG antibody. Regarding T-cell and dendritic cell interactions, deviations in sCD40 levels may impact tumor-specific immune responses. In contrast to earlier studies that reported elevated circulating sCD40 levels in persons with arthritis, renal failure, lung cancer,10,38,39 and hematologic malignancies,40 our results showed that sCD40 levels were higher in healthy controls than in DLBCL and FL cases. The relevance of sCD40 in lymphomagenesis will need further investigation.
CD40 ligation facilitates the FAS-mediated growth inhibition of B-cell lymphomas through a caspase-dependent pathway of apoptosis.39 Indeed, activation of dendritic cell maturation through CD40 ligation is a current strategy in vaccine development to enhance primary and memory T-cell responses and antitumor immunity.42 Because CD40-CD154 interaction can elicit potent antitumor effects, clinical trials using recombinant CD154 currently are underway to treat human B-cell malignancies, such as chronic lymphocytic leukemia.43,44 Our studies provide additional evidence to support these clinical applications and highlight the important role of CD40-CD154 cross-linking to mediate a robust humoral immune response and tumor-specific immunity that may influence germinal center lymphomas. In future studies, we also plan to investigate the role of variation in genes that encode CD40 component proteins in NF-κB classic and alternative pathways, such as IKK (α, β, and γ), IκBα, NF-κB-p65, and others in lymphomagenesis and to determine how the TNFRSF5 −1C>T SNP may influence downstream NF-κB signaling pathways.
In conclusion, we provide novel evidence that the low function TNFRSF5 −1T allele is associated with increased risk of FL and possibly other lymphomas derived in the germinal center. Through subtle reductions in CD40 protein levels present on B cells, dendritic cells, and possibly other APCs, the TNFRSF5 −1C>T SNP may deregulate normal germinal center B- and T-cell or dendritic cell and T-cell interactions that can impede the immune system from mounting appropriate humoral and cell-mediated responses against infectious agents and precancerous cells. Our findings also suggest a role for the TNFSF5 6809G>A variant or a linked SNP in risk of FL in women. Further research is needed to evaluate the roles of the TNFRSF5 −1C>T and TNFSF5 6809G>A SNPs as risk factors in the pathogenesis of FL and in other germinal center lymphomas in larger populations. Studies also are warranted to focus on the functional effects of TNFRSF5 −1C>T on other CD40-expressing cells, its influence on dendritic cell subpopulations, and the prognostic significance of this SNP in lymphoma patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Institutes of Health grants CA122663 (C.F.S.), CA104862 (M.T.S.), and CA45614, CA89745, CA87014 (E.A.H.). Genotyping of the German study was supported by the German José Carreras Leukemia foundation (DJCLS_R04/08, A.N.).
National Institutes of Health
Authorship
Contribution: C.F.S. designed research and wrote the paper; L.A., A.N., J.D.C., and D.R.S. performed laboratory research; A.N. assisted in data analyses and writing of paper; A.H. and P.M.B. performed the epidemiologic data analyses; N.B. and E.A.H. headed the case-control epidemiology studies; M.T.S. contributed to research design and data interpretation.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Christine F. Skibola, School of Public Health, 237A Hildebrand Hall, University of California, Berkeley, CA 94720-7360; e-mail: chrisfs@berkeley.edu.