Abstract
Microarray gene expression profiling studies have demonstrated immune response gene signatures that appear predictive of outcome in follicular lymphoma (FL). However, measurement of these marker genes in routine practice remains difficult. We have therefore investigated the immune response in FL using real-time polymerase chain reaction (PCR) to measure expression levels of 35 candidate Indicator genes, selected from microarray studies, to polyA cDNAs prepared from 60 archived human frozen lymph nodes, in parallel with immunohistochemical analysis for CD3, CD4, CD7, CD8, CD10, CD20, CD21, and CD68. High levels of CCR1, a marker of monocyte activation, were associated with a shorter survival interval, and high levels of CD3 with better survival, while immunohistochemistry demonstrated association of high numbers of CD68+ macrophages with a shorter survival interval and of high numbers of CD7+ T cells with a longer survival interval. The results confirm the role of the host immune response in outcome in FL and identify CCR1 as a prognostic indicator and marker of an immune switch between macrophages and a T cell–dominant response. They demonstrate the utility of polyA DNA and real-time PCR for measurement of gene signatures and the applicability of using this type of “molecular block” in clinical practice.
Introduction
The non-Hodgkin lymphomas comprise a diverse group of malignancies that are currently subclassified according to the state of differentiation of the B cells, the characteristics of the morphology, and by the presence of specific cytogenetic abnormalities. Microarray gene expression profiling analyses have identified molecular heterogeneity and gene signatures, or “Indicator” genes, predictive of tumor behavior and patient outcome in many cancer types including diffuse large B-cell lymphoma (DLBCL) and follicular lymphoma (FL).1-4 Application of this approach to follicular lymphoma identified 2 prognostic signatures based upon nonmalignant tumor-infiltrating cells indicative of the host immune response rather than characteristics of the tumor cells. A predominant T-cell response was associated with good prognosis and a macrophage response was in contrast associated with a poor prognosis.2 Both of these immune signatures provided independent utility from established clinical prognostic markers. Given the extremely variable outcome in FL, there is an urgent need for validation of these immune signatures and for their translation to routine clinical use, which would allow better prognostication of outcome and would potentially direct and inform the application of more targeted molecular therapies in future.
Despite the promise of developing immune signatures in FL, relatively few studies have validated gene expression profiling using established immunohistochemical methods,5-10 and none has used real-time polymerase chain reaction (PCR) to measure the prognostic value of immune gene signatures using a clinically applicable methodology.
Measurement of gene expression using microarrays is difficult in routine practice,1 and there is a need for development of other, simpler, less expensive methods that are both sensitive and robust enough to translate into clinical practice. To test the use of Indicator genes as a diagnostic tool, in lymphoma, we have developed a simple, practical polyA PCR–based method for analysis of Indicator profiles.11 PolyA PCR enables global mRNA amplification from picogram amounts of RNA and has been routinely used to analyze expression in small samples including single cells.12 The polyA cDNA pool generated is also indefinitely renewable and as such represents a “molecular block.”11-13 The polyA cDNA can be then be assayed for the expression of particular genes, either by hybridization with cDNA microarrays14 or by real-time PCR. Real-time PCR measurement, however, enables more precise quantitation of the expression levels of specific Indicator genes. As such, it is better suited for measurement in clinical practice, while also focusing on diagnostically relevant Indicator genes. This approach thereby enables gene signatures to be detected within very small amounts of pathological material,15 and we have recently demonstrated its utility in diffuse large B-cell lymphoma and follicular lymphoma.11
In this study, we have used the same approach to measure immune gene signatures in follicular lymphoma. PolyA reverse-transcription (RT)–PCR was applied to RNA extracted from archived human frozen lymph nodes and the resultant cDNA analyzed by TaqMan real-time PCR for 35 Indicator genes that may potentially provide prognostic information chosen from candidates identified by microarray analysis (Dave et al2 and Glas et al16 ) together with genes associated with B-cell, T-cell, and macrophage differentiation. In addition, immunohistochemical markers of host immune cell tumor infiltration (B cell, T cell, and macrophage) were measured in parallel paraffin-embedded tissue sections. We have confirmed the applicability of this type of approach to clinical practice and the role of the host immune response in outcome in follicular lymphoma, as well as identifying CCR1 as a new potential prognostic indicator.
Methods
Clinical samples
Sixty frozen lymph nodes (with at least 5 years of follow-up) with a diagnosis of follicular lymphoma were obtained, with informed consent, from the archives of the Christie Hospital NHS Trust (Manchester, United Kingdom). Ethical permission for the study was granted by the North West Multicenter Research Ethics Committee and by the Central Manchester Research Ethics Committee. The samples were selected from the archive on a sequential chronologic basis, taking all samples for which consent was given, in the archive from 1995 backward to 1989. These were initially assessed for suitability on the basis of biopsy size, amount of necrosis, and confidence of diagnosis. All cases unlikely to have sufficient material for analysis, with more than 10% necrosis, or with uncertain or complex/mixed diagnoses were excluded. Paraffin-embedded sections from all cases were reviewed by 2 pathologists (R.J.B. and L.P.M.). A diagnosis of follicular lymphoma was confirmed in all cases, and biopsies in which there was morphologic evidence of transformation were excluded from the study. There were 33 males and 27 females with a median age of 66 years and an age range of 34 to 77 years. Forty-five were dead and 15 alive at the end of follow-up. None of the patients received rituximab as first-line treatment, as this treatment was not available at the time during which the cases were collected. All material used in the study was collected at initial diagnosis, prior to treatment.
Immunohistochemistry
Standard immunohistochemistry was used for the following antibodies: CD3, CD4, CD7, CD8, CD10, CD20, CD21, and CD68. All antibodies were diluted to working concentration in DAKO Real Antibody Diluent (DAKO, Glostrup, Denmark); primary antibody working concentrations are shown in Table 1. For each antibody, detection was carried out using a DAKO Envision kit (DAKO) with positivity disclosed by diaminobenzidine (DAB) staining. Details of retrieval method and antibody sources and working dilutions are given in Table 1; microwave pretreatment was performed for 4 minutes in 0.001 M EDTA (pH 8.0) using a pressure cooker and trypsin pretreatment at 37°C for 12 minutes in a 0.1% (vol/vol) solution of trypsin in Tris buffer (pH 7.8).
Details of antibodies used
| Antibody . | Clone . | Source . | Pretreatment . | Primary Ab dilution . |
|---|---|---|---|---|
| CD3 | PS1 | Vision Biosystems | Pressure cooker microwave | 1/100 |
| CD4 | 1F6 | Vector | Pressure cooker microwave | 1/20 |
| CD7 | CD7–272 | Vision Biosystems | Pressure cooker microwave | 1/50 |
| CD8 | CD8/144B | Cell Marque | Pressure cooker microwave | 1/25 |
| CD10 | 56C6 | Cell Marque | Pressure cooker microwave | 1/25 |
| CD20 | L26 | Vector | Pressure cooker microwave | 1/200 |
| CD21 | 2G9 | Vector | Pressure cooker microwave | 1/10 |
| CD68 | 514H12 | Vision Biosystems | Trypsin | 1/50 |
| Antibody . | Clone . | Source . | Pretreatment . | Primary Ab dilution . |
|---|---|---|---|---|
| CD3 | PS1 | Vision Biosystems | Pressure cooker microwave | 1/100 |
| CD4 | 1F6 | Vector | Pressure cooker microwave | 1/20 |
| CD7 | CD7–272 | Vision Biosystems | Pressure cooker microwave | 1/50 |
| CD8 | CD8/144B | Cell Marque | Pressure cooker microwave | 1/25 |
| CD10 | 56C6 | Cell Marque | Pressure cooker microwave | 1/25 |
| CD20 | L26 | Vector | Pressure cooker microwave | 1/200 |
| CD21 | 2G9 | Vector | Pressure cooker microwave | 1/10 |
| CD68 | 514H12 | Vision Biosystems | Trypsin | 1/50 |
All antibodies were detected with Envision detection kit supplied by DAKO. Locations of suppliers: Vision Biosystems, Newcastle, United Kingdom; Cell Marque, Rocklin, CA; Vector, Burlingame, CA.
The degree of positive immunohistochemical staining was measured by a semiautomated image analysis method using spectral unmixing to identify areas of DAB-positive staining, as validated in Das et al17 and Taylor and Levenson18 (detailed in Document S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Briefly, using a Leitz Diaplan fluorescence microscope and a CRI Nuance spectral analyzer (CRI, Woburn, MA), conventional bright-field image files were collected at 5-nm wavelength intervals from 450 nm (blue) to 700 nm (red), at 200× magnification. For each antibody, 5 high-power fields were collected for each case, at a constant magnification of 200×. Full sections were analyzed using exactly the same methodology as reported by Farinha et al.5 Specifically, 5 fields exhibiting strong and uniform staining were analyzed in each case. The Nuance system uses stacked liquid crystal filters to produce a solid-state tunable Lyot filter. The image files, each comprising the concatenated stack of images at each wavelength interval per pixel, were then used to reconstruct multiple spectral distributions via a maximum likelihood method, using CRI Nuance software. Specifically, the diaminobenzidine and hematoxylin spectral profiles were applied across all image cubes to determine the intensity of diaminobenzidine staining per pixel after which the staining intensities were converted to composite false color images, from which the percentage of positive pixels in the resultant images were measured using IPLab software (Scanalytics, Rockville, MD), according to the manufacturer's instructions. Since the area of field was constant over all cases and for all antibodies, this was taken as a measure of percentage cellular positive staining. The number of CD68+ macrophages was manually counted since the staining pattern was more diffuse than that for the other antibodies and the cell numbers were low, enabling this to be carried out practically in a manual manner. The method described by Farinha et al5 was used. Specifically, the number of positive cells per high-power field (400×) was counted using a mean of 3 high-power fields for each case.5
Extraction of RNA and global amplification of polyadenylated mRNAs
The lymph nodes were homogenized using a Mixer Mill MM 300 (Qiagen, Crawley, United Kingdom). Total RNA was then extracted an RNeasy mini kit (Qiagen), as recommended by the manufacturer; DNase was used to remove the contaminating genomic DNA, and polyA RT-PCR was carried out as previously described.13,19-21 Global amplification of cDNA corresponding to all expressed genes (polyA PCR) was carried out as previously reported.19-21
Specific RT-PCR
The 35 Indicator genes were chosen from previously published candidates (Dave et al2 and Glas et al16 ), together with genes associated with B-cell, T-cell, and macrophage differentiation, and are listed in Table 2. Taqman PCR primers and probes were designed for 35 Indicator genes and 4 housekeeping genes using Primer Express Software (Applied Biosystems, Foster City, CA) and are detailed in Document S1. All PCR primer pairs were designed for mRNA sequence within 300 bp of the 3′ end of each Indicator gene and were tested in PCR reactions carried out in 25 μL containing 1 ng polyA cDNA, 0.33 μM of each oligonucleotide, 0.5 units Ex-Taq polymerase (TaKaRa), and 0.25 μM dNTPs in the buffer supplied by the manufacturer. PCR was performed using the following thermal cycle: 5 minutes at 94°C, followed by 30 cycles of 30 seconds at 94°C, 30 seconds at 56°C, and one minute at 72°C.
List of genes measured by real-time PCR
| Genes . | Accession no. . | Markers . | Reference . |
|---|---|---|---|
| CD19 | NM 001770.3 | B-cell | Additional lineage marker |
| CDK2 | NM 001798.2 | Cell cycle control | Glas et al16 |
| CD47 | NM 001777.3 | Macrophage | Additional lineage marker |
| CD11D (ITGAD) | NM 005353.2 | Macrophage | Additional lineage marker |
| TNFSF13B | NM 006573.3 | Macrophage | Dave et al2 |
| ACTN1 | NM 001102.2 | Macrophage | Dave et al2 |
| CD68 | NM 001251 | Macrophage | Additional lineage marker |
| CCR1 | NM 001295.2 | Macrophage activation | Glas et al16 |
| FCGR1A (CD64) | NM 000566.2 | Macrophage and dendritic cells | Dave et al2 |
| C3AR1 | NM 004054 | Macrophage and dendritic cells | Dave et al2 |
| Sep-10 | NM 144710.2 | Macrophage and dendritic cells | Dave et al2 |
| LGMN | NM 005606.5 | Macrophage and dendritic cells | Dave et al2 |
| TLR5 | NM 003268.3 | Macrophage and dendritic cells | Dave et al2 |
| MAPK1 | NM 002745.4 | Mitogen-activated protein kinase 1 | Glas et al16 |
| CXCL12 | NM 000609.4 | Reactive infiltrate of T cells and macrophage | Glas et al16 |
| TSPAN7 | NM 004615.2 | Reactive infiltrate of T cells and macrophage | Glas et al16 |
| CD3D | NM 000732.3 | Reactive infiltrate of T cells and macrophage | Glas et al16 |
| NEK2 | NM 002497.2 | Signal transduction marker | Glas et al16 |
| CD2 | NM 001767.2 | T cell | Glas et al16 |
| CD4 | NM 000616.2 | T cell | Additional lineage marker |
| CD5 | NM 014207.2 | T cell | Additional lineage marker |
| CD25 (IL2RA) | NM 000417.1 | T cell | Additional lineage marker |
| ITK | NM 005546.3 | T cell | Dave et al2 |
| CD69 | NM 001781.1 | T cell | Glas et al16 |
| LEF1 | NM 016269.2 | T cell | Dave et al2 |
| STAT4 | NM 003151.2 | T cell | Dave et al2 |
| CD6 | NM 006725.2 | T cell | Additional lineage marker |
| CD7 | NM 006137.6 | T cell | Dave et al2 |
| CD8B1 | NM 172213.1 | T cell | Dave et al2 |
| CCL3 | NM 002983.1 | T-cell and macrophage activation | Glas et al16 |
| CCL5 | NM 002985.2 | T-cell and macrophage activation | Glas et al16 |
| CCL8 | NM 005623.2 | T-cell and macrophage activation | Glas et al16 |
| ILF3 | NM 012218.2 | T-cell and macrophage activation | Glas et al16 |
| GEM | NM 005261.2 | T-cell and macrophage activation | Glas et al16 |
| AKAP12 | NM 005100.2 | T-cell and macrophage activation | Glas et al16 |
| Beta actin | NM 001101 | Housekeeping gene | Housekeeping gene |
| Ribosomal S9 | U14971 | Housekeeping gene | Housekeeping gene |
| IF2b | U23028 | Housekeeping gene | Housekeeping gene |
| GAP | NM 002046 | Housekeeping gene | Housekeeping gene |
| Genes . | Accession no. . | Markers . | Reference . |
|---|---|---|---|
| CD19 | NM 001770.3 | B-cell | Additional lineage marker |
| CDK2 | NM 001798.2 | Cell cycle control | Glas et al16 |
| CD47 | NM 001777.3 | Macrophage | Additional lineage marker |
| CD11D (ITGAD) | NM 005353.2 | Macrophage | Additional lineage marker |
| TNFSF13B | NM 006573.3 | Macrophage | Dave et al2 |
| ACTN1 | NM 001102.2 | Macrophage | Dave et al2 |
| CD68 | NM 001251 | Macrophage | Additional lineage marker |
| CCR1 | NM 001295.2 | Macrophage activation | Glas et al16 |
| FCGR1A (CD64) | NM 000566.2 | Macrophage and dendritic cells | Dave et al2 |
| C3AR1 | NM 004054 | Macrophage and dendritic cells | Dave et al2 |
| Sep-10 | NM 144710.2 | Macrophage and dendritic cells | Dave et al2 |
| LGMN | NM 005606.5 | Macrophage and dendritic cells | Dave et al2 |
| TLR5 | NM 003268.3 | Macrophage and dendritic cells | Dave et al2 |
| MAPK1 | NM 002745.4 | Mitogen-activated protein kinase 1 | Glas et al16 |
| CXCL12 | NM 000609.4 | Reactive infiltrate of T cells and macrophage | Glas et al16 |
| TSPAN7 | NM 004615.2 | Reactive infiltrate of T cells and macrophage | Glas et al16 |
| CD3D | NM 000732.3 | Reactive infiltrate of T cells and macrophage | Glas et al16 |
| NEK2 | NM 002497.2 | Signal transduction marker | Glas et al16 |
| CD2 | NM 001767.2 | T cell | Glas et al16 |
| CD4 | NM 000616.2 | T cell | Additional lineage marker |
| CD5 | NM 014207.2 | T cell | Additional lineage marker |
| CD25 (IL2RA) | NM 000417.1 | T cell | Additional lineage marker |
| ITK | NM 005546.3 | T cell | Dave et al2 |
| CD69 | NM 001781.1 | T cell | Glas et al16 |
| LEF1 | NM 016269.2 | T cell | Dave et al2 |
| STAT4 | NM 003151.2 | T cell | Dave et al2 |
| CD6 | NM 006725.2 | T cell | Additional lineage marker |
| CD7 | NM 006137.6 | T cell | Dave et al2 |
| CD8B1 | NM 172213.1 | T cell | Dave et al2 |
| CCL3 | NM 002983.1 | T-cell and macrophage activation | Glas et al16 |
| CCL5 | NM 002985.2 | T-cell and macrophage activation | Glas et al16 |
| CCL8 | NM 005623.2 | T-cell and macrophage activation | Glas et al16 |
| ILF3 | NM 012218.2 | T-cell and macrophage activation | Glas et al16 |
| GEM | NM 005261.2 | T-cell and macrophage activation | Glas et al16 |
| AKAP12 | NM 005100.2 | T-cell and macrophage activation | Glas et al16 |
| Beta actin | NM 001101 | Housekeeping gene | Housekeeping gene |
| Ribosomal S9 | U14971 | Housekeeping gene | Housekeeping gene |
| IF2b | U23028 | Housekeeping gene | Housekeeping gene |
| GAP | NM 002046 | Housekeeping gene | Housekeeping gene |
TaqMan primers and probes for each gene are detailed in Table S1. For each gene, the reference from which it was selected is given (details are provided in references); additional lineage markers selected are indicated as such.
Taqman real-time quantitative PCR
For each gene, Taqman PCR was applied to 1 ng polyA cDNA from each sample and to 10 μL serially diluted human genomic standards, using a Taqman Gold kit. All samples were analyzed using an ABI Prism 7700 sequence detection system (Applied Biosystems). The copy number of each gene was determined by reference, after normalization to Mhouse (detailed in “Normalization”), of the real-time PCR expression level to human genomic DNA standards, previously described in Sakhinia et al.21
Normalization
The expression levels of 4 housekeeping genes (IF2-beta, GAP, human ribosomal protein S9 mRNA, and Beta actin) were measured by RT-PCR in each sample. Copy numbers obtained for the mean (Mhouse) of the 4 housekeeping genes (IF2-b, GAP, RbS9, and Beta actin) in each sample were divided by the highest Mhouse in all samples resulting in a normalization correction factor.21 Following real-time PCR amplification and quantification of the selected genes, this factor was then used for normalization of expression levels of each of the 35 genes measured. Specifically, the expression level of each gene (Ct value) was quantified at least twice against a standard curve obtained from a serial dilution of human sonicated DNA. For each gene, an equation, formulated from the best standard curve, was used to calculate copy number. The following specific example is given for GAP where copy number = 10^([A−38.325]/− 3.64) × B (A = mean of Ct values, B = dilution factor, slope = 3.64, Y-intercept = 38.325). Finally, copy numbers obtained for the mean of the IF2-b, GAP, RbS9, and Beta actin housekeeping genes (Mhouse) expression levels were divided by the highest mean value obtained in the experiment, resulting in a correction factor. All data were then normalized using this factor.
Statistical analysis
Kaplan-Meier survival analysis using a log-rank test was performed against the normalized real-time PCR expression levels of each of the genes and against the percentage immunohistochemical postivity for the antibodies CD3, CD4, CD7, CD8, CD10, CD20, and CD21. For CD68, Kaplan-Meier survival analysis was performed separately for cases with 15 or fewer and cases with more than 15 CD68+ cells per high-power field.
Results
Immunohistochemistry
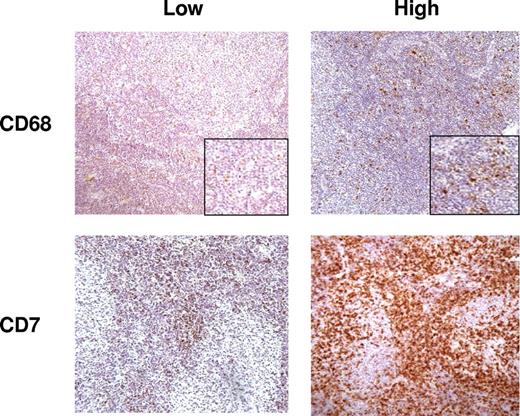
Measurement of the number of cells positive for CD3, CD4, CD7, CD8, CD10, CD20, and CD21 was performed by semiautomated image analysis using a spectral imager in view of the large number of positive cells in each case, while measurement of the number of CD68+ cells was performed manually using the same method detailed in Farinha et al.5 Figure 1 shows representative figures from cases with high and low numbers of positive cells for CD7 and CD68.
Representative sections from cases with low and high numbers of cells positive for CD7 and CD68; immunohistochemistry DAB staining (200×).
Representative sections from cases with low and high numbers of cells positive for CD7 and CD68; immunohistochemistry DAB staining (200×).
PolyA PCR
PolyA cDNA was generated from mRNA extracted from all 60 lymph nodes. The copy number for each gene in each sample was highly reproducible over at least duplicate tests, demonstrating reliability of the method. Specific PCR of these polyA cDNAs was positive for each of the selected Indicator genes, and bands of appropriate sizes were present in all samples, demonstrating presence of the target transcript in the polyA cDNA (data not shown). There was no statistically significant difference in the value of Mhouse for the different diagnostic groups (data not shown).
Survival analysis
Survival analysis was performed against real-time PCR expression levels and against immunopositivity for T-cell, B-cell, and macrophage reactive antibodies. The data for each gene and for each antibody were grouped into 4 quartiles for Kaplan-Meier survival analysis.
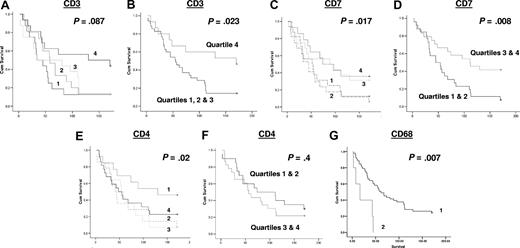
Of the T-cell antigens, the expression of CD7, a marker of T cells and specifically involved in T-cell activation, was positively associated with survival (P < .017). Other T-cell markers, namely CD3 (a pan T-cell marker), and CD8 (a marker of cytotoxic T cells), were not significantly associated with survival when analyzed in 4 quartiles, although CD3 was positively associated with survival when the highest quartile was compared with the lower 3 quartiles together (P = .023; Figure 2); the significance of association of CD7 with survival increased when the upper and lower quartiles were combined (P = .008; Figure 2). Curiously, when CD4 (a marker of T-helper cells) was analyzed as quartiles, it was negatively associated with survival, with higher numbers of CD4+ T cells being associated with worse survival (P = .02), although not when higher and lower quartiles were analyzed together (P = .4). CD68 expression was negatively associated with survival (P < .007) with cases with more than 15 macrophages per high-power field having a worse outcome than those with 15 or fewer per high-power field (Figure 2). The numbers of CD10+ (marker of follicle center cells), CD20+ (B-cell marker), and CD21+ (marker of follicular dendritic cells) cells were not statistically significantly associated with survival (data not shown).
Kaplan-Meier survival analysis based on numbers of cells positive by immunohistochemistry for each of the antibodies assayed. The data for each antibody were initially grouped into 4 quartiles for use in Kaplan-Meier survival analysis, shown for (A) CD3, (C) CD7, and (E) CD4: the survival curves as shown as 1 = 1st quartile, 2 = 2nd quartile, 3 = 3rd quartile & 4 = 4th quartile (1st quartile used for lower end of gene expression for each gene). Further Kaplan-Meier survival analysis was performed for CD3, CD7, and CD4, grouping the higher and lower quartiles, as shown for (B) CD3, (D) CD7, and (F) CD4. Quartile groupings used in these analyses are indicated in each panel. Kaplan-Meier survival analysis for CD68 was performed separately for cases with 15 or fewer (1) or greater than 15 macrophages (2) per high-power field, as shown in panel G.
Kaplan-Meier survival analysis based on numbers of cells positive by immunohistochemistry for each of the antibodies assayed. The data for each antibody were initially grouped into 4 quartiles for use in Kaplan-Meier survival analysis, shown for (A) CD3, (C) CD7, and (E) CD4: the survival curves as shown as 1 = 1st quartile, 2 = 2nd quartile, 3 = 3rd quartile & 4 = 4th quartile (1st quartile used for lower end of gene expression for each gene). Further Kaplan-Meier survival analysis was performed for CD3, CD7, and CD4, grouping the higher and lower quartiles, as shown for (B) CD3, (D) CD7, and (F) CD4. Quartile groupings used in these analyses are indicated in each panel. Kaplan-Meier survival analysis for CD68 was performed separately for cases with 15 or fewer (1) or greater than 15 macrophages (2) per high-power field, as shown in panel G.
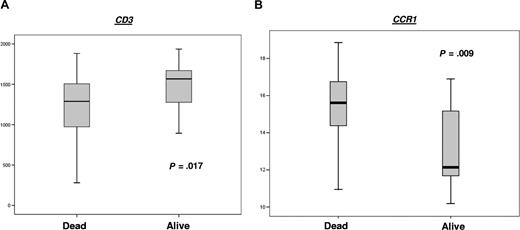
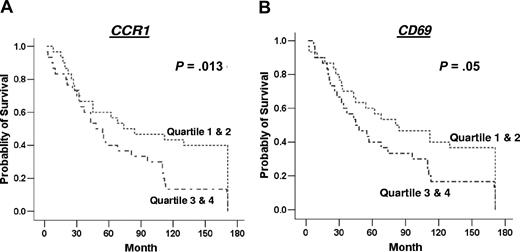
Of the 35 genes measured by real-time PCR, the expression of just 2, namely CCR1 and CD3, were expressed at different levels in those who had died from follicular lymphoma compared with those alive at the end of the follow-up period. CCR1 was significantly higher (P = .009) (Figure 3), while CD3 expression was lower (P = .017) in those who had died from follicular lymphoma (Figure 3). Of the genes measured by real-time PCR, only high expression of CCR1 (P = .013) was significantly associated with survival, with poorer outcome in those with higher expression of CCR1 (Figure 4). There was a trend of association of high expression levels of CD69 (P = .05), TNFSF13B (P = .07), CDK2 (P = .07), and CCL3 (P = .09) with poor survival (Figure 4; data not shown for nonsignificant genes).
Expression levels (displayed as natural logarithm) of genes with statically significant difference between patients either alive or dead from disease at the end of the study period. (A) CD3 expression levels; (B) CCR1 expression levels.
Expression levels (displayed as natural logarithm) of genes with statically significant difference between patients either alive or dead from disease at the end of the study period. (A) CD3 expression levels; (B) CCR1 expression levels.
Kaplan-Meier survival analysis based on gene expression level. Data are shown for genes significantly associated with survival or showing a trend toward association with survival, namely CCR1 (A) and CD69 (B). The data for each gene were grouped into 4 quartiles for use in Kaplan-Meier survival analysis, and these are indicated in the survival curves as 1 = 1st quartile, 2 = 2nd quartile, 3 = 3rd quartile, and 4 = 4th quartile; 1st quartile was used for lower end of gene expression for each gene.
Kaplan-Meier survival analysis based on gene expression level. Data are shown for genes significantly associated with survival or showing a trend toward association with survival, namely CCR1 (A) and CD69 (B). The data for each gene were grouped into 4 quartiles for use in Kaplan-Meier survival analysis, and these are indicated in the survival curves as 1 = 1st quartile, 2 = 2nd quartile, 3 = 3rd quartile, and 4 = 4th quartile; 1st quartile was used for lower end of gene expression for each gene.
Discussion
Predicting the biologic behavior and outcome of hematologic malignancies has traditionally been based on the pathological subclassification of tumor type and clinical staging of the extent of detectable disease. More recently, gene expression profiling has provided new insights into how the molecular heterogeneity may affect outcome, although the measurement of Indicator genes in routine practice remains difficult. We have previously demonstrated utility of real-time PCR measurement of Indicator genes in globally amplified polyA cDNA as a practical method for their clinical analysis11 and have demonstrated its utility in diffuse large B-cell lymphoma.21 PolyA PCR enables global mRNA amplification from picogram amounts of RNA, and the polyA cDNA pool generated is indefinitely renewable, representing a “molecular block.” Real-time PCR measurement of the expression levels of specific Indicator genes then allows gene signatures to be detected in the polyA cDNA.
In this project, we extended this method to measurement of the immune signature in follicular lymphoma, using real-time PCR to measure 35 Indicator genes reflective of T-cell and macrophage activation in polyA cDNAs prepared from 60 archived human frozen lymph nodes. In addition, immunohistochemistry for a range of T-cell and macrophage markers was performed on parallel formalin-fixed paraffin-embedded tissue blocks from the same cases.
Real-time PCR was used to measure the expression levels of a wide range of genes indicative of T-cell and macrophage activation, including several genes abstracted from previous microarray studies demonstrating the importance of the immune response in FL.2,5,16 Expression levels were measured for 35 genes, increased levels of just one of which, namely CCR1, were associated with poorer outcome (P = .013). In addition, comparison of the gene expression level in patients alive or dead at the end of the study interval confirmed the importance of CCR1 as a predictor of outcome (P = .009). Conversely, levels of CD3 were significantly higher in those alive compared with those who had died at the end of the study (P = .017).
Immunohistochemistical analyses demonstrated that high numbers of macrophages, identified by CD68 staining, were associated with a poor outcome, whereas high levels of CD7+ T cells were associated with a better outcome. High levels of CD3+ T cells were also associated with a better outcome, consistent with the finding of higher CD3 expression on those alive at the end of the period of study, while CD8 had no effect on survival. CD4 was negatively associated with survival in a quartile analysis but not when grouped together with upper and lower quartiles (Figure 2). The reason for one quartile being significant is unclear, although the quartiles do show a high degree of overlap. Whether this result is reproducible or biologically relevant remains unclear. Other markers, namely B-cell markers (CD20) and CD10 and CD21, appeared to have no effect on survival.
Our results are otherwise in keeping with previously published results demonstrating the importance of the immune signature in FL. CD7 was found to be present in the immune-response 1 signature associated with favorable outcome in Dave et al,2 while Farinha et al5 reported a poor outcome in those cases with high numbers of macrophages. However, in contrast to previous studies, measurement of gene expression has been achieved in this study using methods that can be routinely applied to clinical practice. Since the initial publication from Dave et al,2 there have been relatively few confirmatory studies,5-9 and these have supported the importance of the immune signature in terms of demonstrating either association of a poor outcome for macrophage activation5 or a good outcome for increased numbers of T cells,6-9 although no study has confirmed both a good outcome with increased numbers of T cells and a poor outcome for increased numbers of macrophages, as the results in the present study do. Furthermore, these associations were demonstrated at both the mRNA and protein levels in the present study, further strengthening the role of the immune response in outcome in follicular lymphoma.
CCR1, C-C chemokine receptor type 1, is a RANTES receptor22 and is expressed at high levels on human blood monocytes, while T cells express high levels of CCR5 and low levels of CCR1. This difference in expression has been hypothesized to indicate selective use of these 2 RANTES receptors during inflammatory recruitment. This hypothesis was tested by Weber et al,22 who demonstrated the importance of CCR1 for monocyte recruitment during inflammation. Up-regulation of CCR1 in the present study may therefore reflect increased monocyte recruitment, with subsequent increase in the number of macrophages, measured by the analyzing the number of CD68+ cells. The association of both with poor outcome in the present study is therefore concordant with the role of the immune signature identified by Dave et al2 and with the importance of CCR1 in immune modulation demonstrated by Weber et al.22 A role for CCR1 has also been identified in progression of myeloma, via osteoclast activation,23 and there is also interest in developing anatagonists to it for use in autoinflammatory diseases.24 This raises the possibility of macrophage-directed immunotherapy in FL, a field that has so far been concentrated on B-cell targeting.
The present study also highlighted the association of CD7+ T cells with good outcome. This supports the finding of CD7 in the favorable immune response 1 signature reported by Dave et al,2 while CD3 was also significant at P less than .023, reflecting the inclusion of CD7+ cells within this cellular fraction. In addition, levels of CD3 expression measured by real-time PCR were significantly raised in those alive compared with those dead at the end of the study. Conversely, CCR1 expression is low in T cells, and Weber et al22 proposed a central role for CCR1, in concert with CCR5, in modulating immune response between monocyte and T-cell recruitment, respectively, further supporting the finding of high CCR1 expression in those with poor survival. Dave et al found additive survival prediction when comparing the T-cell and macrophage signatures.2 We have tested for this using Cox multivariate analysis, which identified CD7 as the principal predictor of survival, followed by the macrophage marker CD68.
Numbers of CD3+ cells were significantly associated with survival when the upper and lower quartiles were grouped together. This concurs with the real-time PCR result for CD3 mRNA expression. Conversely, neither CD7 or CD68 mRNA expression levels were significantly associated with survival, although protein expression levels were for both. CCR1 was measured only by real-time PCR, and no comment as to its concordance with protein levels can be made. This indicates a discrepancy between mRNA and protein expression for CD7 and CD68, although there was good agreement for CD3. Discrepancies between mRNA and protein expression are commonly encountered,25 and this is one of the principal reasons for carrying out validation using multiple analyses, as has been done in this project. Notwithstanding this discrepancy, however, the results of both real-time PCR and immunohistochemistry demonstrated association of the immune response with survival, confirming the hypothesis the project set out to test. Furthermore, the set of genes and antibodies tested was deliberately large to maximize the possibility of identifying a prognostic association given the well-known discrepancy between mRNA expression and protein levels.25 A further point to consider is that the antibody levels were measured as numbers of positive cells rather than absolute quantity of protein. This represents a different metric to the absolute mRNA values measured by real-time PCR, and this may significantly contribute to differences between the 2 analyses. Quantitative measurement of protein level by immunohistochemistry was beyond the scope of this project and while technically feasible, using sophisticated imaging software or fluorescence, would not be practically useful in clinical practice.
This study represents extension of our previous application of this method to measure Indicator genes in DLBCL and FL to measurement of the immune signature in FL, confirming the broad utility of the technique. Furthermore, polyA cDNA generated for the initial study from archived samples of FL was used in this study to measure immune signature genes, underlining the value of the polyA cDNA central to this method as a “molecular block” that can be reanalyzed multiple times for different genes. The results confirm the role of the host immune response in outcome in FL and specifically demonstrate the degree of CD68+ macrophage and CD7+ T-cell infiltration as prognostically useful, together with identification of CCR1 as a putative novel prognostic indicator and marker of immune switch between macrophage and T cell–dominant response. The methods used are clinically applicable, particularly immunohistochemistry, while the clinical utility of polyA DNA and real-time PCR for measurement of gene signatures and the strength of this approach as a “molecular block” are confirmed.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The work was supported by a UK Department of Health, New and Emerging Applications of Technology (NEAT) Grant (C020) and by the National Translational Cancer Research Network (NTRAC), and was carried out in the Department of Histopathology and in the Manchester Molecular Diagnostic Center, Manchester Royal Infirmary, Central Manchester, and Manchester Children's University Hospitals NHS Trust. Statistical advice and analysis were provided by R. Swindell, Christie Hospital NHS Foundation Trust.
Authorship
Contribution: R.J.B. provided overall supervision, designed the experimental plan, analyzed data, and wrote the paper; E.S. performed real-time PCR technical work and data analysis; P.J. performed immunohistochemical analysis; C.G. performed immunohistochemical technical work and sample tracking; J.A.H. wrote the paper and provided scientific support; L.P.M. provided expert histopathology review; J.A.R. provided clinical support; T.I. provided experimental design, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Richard Byers, Department of Histopathology, Clinical Sciences Building One, Manchester Royal Infirmary, Oxford Road, Manchester, M13 9WL, United Kingdom; e-mail: r.byers@manchester.ac.uk; richard.byers@cmmc.nhs.uk.
References
Author notes
E.S. and P.J. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal