Abstract
Activating mutations in tyrosine kinase (TK) genes (eg, FLT3 and KIT) are found in more than 30% of patients with de novo acute myeloid leukemia (AML); many groups have speculated that mutations in other TK genes may be present in the remaining 70%. We performed high-throughput resequencing of the kinase domains of 26 TK genes (11 receptor TK; 15 cytoplasmic TK) expressed in most AML patients using genomic DNA from the bone marrow (tumor) and matched skin biopsy samples (“germline”) from 94 patients with de novo AML; sequence variants were validated in an additional 94 AML tumor samples (14.3 million base pairs of sequence were obtained and analyzed). We identified known somatic mutations in FLT3, KIT, and JAK2 TK genes at the expected frequencies and found 4 novel somatic mutations, JAK1V623A, JAK1T478S, DDR1A803V, and NTRK1S677N, once each in 4 respective patients of 188 tested. We also identified novel germline sequence changes encoding amino acid substitutions (ie, nonsynonymous changes) in 14 TK genes, including TYK2, which had the largest number of nonsynonymous sequence variants (11 total detected). Additional studies will be required to define the roles that these somatic and germline TK gene variants play in AML pathogenesis.
Introduction
The importance of protein tyrosine kinases for the pathogenesis of malignancy has been recently demonstrated by the success of therapies targeted to erbB2 (herceptin) in a subset of breast cancers that overexpress this receptor,1 and to BCR/ABL (imatinib mesylate) in patients with chronic myelogenous leukemia (CML).2 Mutations that activate tyrosine kinases are found in numerous types of solid tumors and are the focus of intense study by many laboratories.
The genes encoding the receptor tyrosine kinases (RTK) are frequent targets for activating mutations in AML. RTKs can be activated by several mechanisms, including point mutations, internal tandem duplications (ITD), deletions, and insertions (eg, FLT3,3 CSF1R,4 KIT5 ), as well as by chromosomal translocations (eg, TEL-PDGFR,6 TEL-TRKC,7 ZNF198-FGFR1,8 BCR-ABL,9 TEL-JAK2,10 NPM-ALK,11 and TEL-ARG12 ) ITDs of FLT3 have been reported in approximately 30% of patients with AML13 and correlate with poor outcome. Mutations in KIT are present in approximately 7% of AML cases and frequently occur in association with t(8;21) or inv(16) (ie, core binding factor) cases.14 Activating mutations of KIT have been detected in 40% to 48% of patients with core binding factor leukemias.5,14
Based on the successful development of AML mouse models expressing combinations of oncogenes, a simple, 2-category system has been proposed to classify mutations in AML. “Type I mutations” result in constitutive activation of TK or Ras pathway genes (eg, FLT3-ITD, KITD816V, NRASV12D). “Type II mutations,” which result in altered hematopoietic transcription factors (eg, AML1, MLL, RARA), can arise via translocations or point mutations. Although expression of transcription factor fusion oncogenes, such as PML-RARA, AML1-ETO, and NUP98-HOXA9, can initiate leukemia in mice, they do so with a long latency; they can cooperate with type I mutations (eg, PML-RARA + FLT3-ITD15 ) to reduce the period of latency. Similarly, some activated tyrosine kinase oncogenes, such as BCR-ABL16 and TEL-PDGFRB17 can cooperate with type II mutations to cause AML in mice. Although mutations in tyrosine kinase genes occur commonly in AML, they are not found in all cases. For these reasons, many laboratories have suggested that there may be additional, currently uncharacterized tyrosine kinase gene mutations present in patients with AML.
In this report, we used targeted high-throughput resequencing of expression-prioritized tyrosine kinase genes to characterize the spectrum of sequence variants that occur in newly diagnosed cases of de novo AML without complex karyotypes. We assessed the frequencies not only of somatic mutations but also those of single nucleotide polymorphisms (SNPs), because some SNPs in cytokine signaling genes may contribute to AML development. For example, a rare nonsynonymous SNP in the granulocyte colony-stimulating factor receptor gene (GCSFR) encodes a hypomorphic receptor18 that appears to be associated with the development of high-risk myelodysplastic syndrome (MDS). Further, a SNP in FLT3 (FLT3D324N, rs35602083) occurs at increased frequency in AML patients versus controls.19
We resequenced 26 selected RTK and cellular tyrosine kinase (CTK) genes from 94 patients with de novo AML and further examined all sequence variants that were not known to be SNPs by resequencing their matched normal skin DNA samples. Sequence variants were also evaluated in a separate set of AML samples from an additional 94 patients. Finally, we identified nonsynonymous sequence variants in the normal tissue from our patients and determined the frequencies of these alleles in a set of ethnically matched normal controls to identify alleles that may contribute to AML predisposition. Several previously undescribed nonsynonymous sequence variants were found, but only 4 patients of the 188 examined had novel somatic mutations in TK genes. These data suggest that many additional TK genes may need to be sequenced to identify all relevant sequence variants; alternatively, the paucity of mutations beyond FLT3 might also suggest that many AML patients do not have TK mutations but rather have cooperating mutations in pathway genes or related genes that are currently unknown.
Methods
Patient characteristics
A total of 188 de novo AML samples were analyzed. The selection of these patients and their clinical characteristics have been described previously.20 This study was approved by Human Research Protection Office at the Washington University School of Medicine (WU) after patients provided informed consent in accordance with the Declaration of Helsinki. Briefly, a Discovery set of 94 de novo AML samples was obtained at WU, and both skin (“germline”) and leukemic cell genomic DNA were obtained. This allowed us to determine whether an observed nucleotide change in a leukemic sample was somatically acquired. Most of the de novo AML samples displayed normal or simple cytogenetic abnormalities. Sequence variants observed in our Discovery set were analyzed in a separate set of 94 genomic DNA samples obtained from the Cancer and Leukemia Group B (CALGB) cooperative group.
Sequencing strategy
The high-throughput sequencing pipeline at WU has been described previously.20,21 We used whole-genome amplified genomic DNA (Qiagen Repligene, Valencia, CA) isolated from unfractionated AML patient bone marrows and a semiautomated method to detect mutations. We assessed sequence quality and coverage as described.20 High-quality, double-stranded or single-stranded sequence was observed for nearly all of the samples. The primers used for amplification and resequencing are shown in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Sensitivity and specificity of sequencing pipeline
The sensitivity and specificity of our sequencing pipeline have been described previously.20 Briefly, we resequenced 12 genes with known mutation frequencies in AML (including FLT3, NPM, NRAS, and CEBPA) in the 188 de novo AML samples. Consistent with previous reports, we found the mutation frequencies of these genes in our Discovery set samples to be 28%, 24%, 9%, and 6%, respectively.20
Pyrosequencing
For selected SNPs, normal population frequencies were estimated by genotyping control genomic DNA samples. Ninety-five white samples (48 males and 47 females) were selected from the Human Variation Panel–Caucasian Panel of 100 (HD100CAU, Coriell Institute, Camden, NJ). Another 95 white samples (48 males and 47 females) were selected from the Cancer Free Control Samples collected by the Hereditary Cancer Core at the Siteman Cancer Center. This local resource consists of DNA derived from the peripheral blood of volunteers 64 years of age or older (mean 73.5 years; range, 64-94 years) with no personal history of cancer (with the possible exception of basal or squamous cell skin cancer). For rs3212723 (JAK3P132T), genotyping was also performed by the sequencing pipeline on an additional 95 samples (16 males and 79 females) selected from the Coriell Human Variation Panel–African American Panel of 100 (HD100AA).
PCR primers for pyrosequencing (sequences provided in Table S2) were designed using Pyrosequencing Assay Design Software, version 1.0.6 (Biotage, Uppsala, Sweden). Standard and 5′-biotinylated primers were synthesized by Sigma-Genosys (The Woodlands, TX). PCR reactions were carried out on a PTC-225 Programmable Thermal Cycler (MJ Research, Waltham, MA) using Hot StarTaq Master Mix (QIAGEN) and were run for 55 cycles. PCR reaction temperatures were selected from gradient temperature optimization experiments. Pyrosequencing and genotype analysis were performed using the Pyrosequencing HS 96A instrument and PyroMark MD software (Biotage) according to the vendor's recommended protocol.
RNA expression analysis using microarrays
Bone marrow aspirates were obtained from properly consented AML patients, and RNA was prepared from the unfractionated snap-frozen cell pellets. Total cellular RNA was purified using the Trizol reagent (Invitrogen, Carlsbad, CA), quantified using UV spectroscopy (Nanodrop Technologies, Wilmington, DE) and qualitatively assessed using a BioAnalyzer 2100 and RNANanoChip assay (Agilent Technologies, Palo Alto, CA). Samples were labeled and hybridized to Affymetrix Human Genome U133 Plus 2.0 Array GeneChip microarrays (Affymetrix, Santa Clara, CA) using standard protocols from the Siteman Cancer Center Multiplexed Gene Analysis Core Facility.22 To perform interarray comparisons, the raw scan data from each microarray were scaled to a target intensity of 1500 using the Affymetrix GCOS 1.2 (MAS 5) statistical algorithm. Scaled data for each array were exported to the Siteman Cancer Center Bioinformatics Server (http://bioinformatics.wustl.edu), merged with updated gene annotation data for each probe set on the array, and downloaded for further data visualization and analysis. The complete dataset has been analyzed in detail in a separate study (J.E.P., N. R. Grieselhuber, L. W. Chang, M. Murakami, W. Yuan, D.C.L., R.N., M. A. Watson, T.J.L., manuscript in preparation) and will be publicly deposited on publication.
Using the most recent annotations available from EntrezGene, UniGene, and Gene Ontology databases and manual curation, we identified probe sets representing all RTKs and CTKs on the U133 Plus 2.0 array. For genes with multiple probe sets, the one with the highest average intensity was retained for further analysis (Tables S3A,S3B). The most highly expressed RTKs and CTKs were selected for sequencing. Array data are available online at http://www.ncbi.nlm.nih.gov/geo/ as accession # GSE10358.
Statistical analysis
Statistical power for associations was estimated with AssocPow version 2.0.24 Statistical SNP-phenotype associations were performed using Prism 5 (GraphPad Software, San Diego, CA). Differences in allele frequency were evaluated using Fisher exact test. Genotype associations were further evaluated for significance according to genetic models of codominant, dominant, and overdominant expression by χ2 testing. Genetic modeling comparisons were corrected for multiple comparisons by the Bonferroni method.
TYK2 protein analysis
Parental 2fTGH cells and TYK2-deficient U1A cells were culture in DMEM supplemented with 10% fetal calf serum and penicillin/streptomycin. To test kinase activities of different TYK2 alleles, U1A cells were transduced with cDNAs encoding wild-type TYK2 and TYK2 alleles generated by site-directed mutagenesis. Transduced cells were isolated by flow cytometry based on equivalent GFP expression, then stimulated with 1000 units of human recombinant interferon alfa-2b (Shering, Kenilworth NJ) for 10 minutes. Total cell lysates were prepared and Western blots performed. Antibodies used were polyclonal anti-Tyk2 antibody (Cell Signaling, catalog no. 9312), anti phospho-Tyk2 (Tyr 1054/1055) antibody (Cell Signaling, catalog no. 9321), and monoclonal anti-β-actin antibody (Sigma-Aldrich, St Louis, MO). TYK2 V678F, a predicted homolog of JAK2V617F, was used as a positive control.
Results
Selecting genes for study by ranking tyrosine kinase gene expression
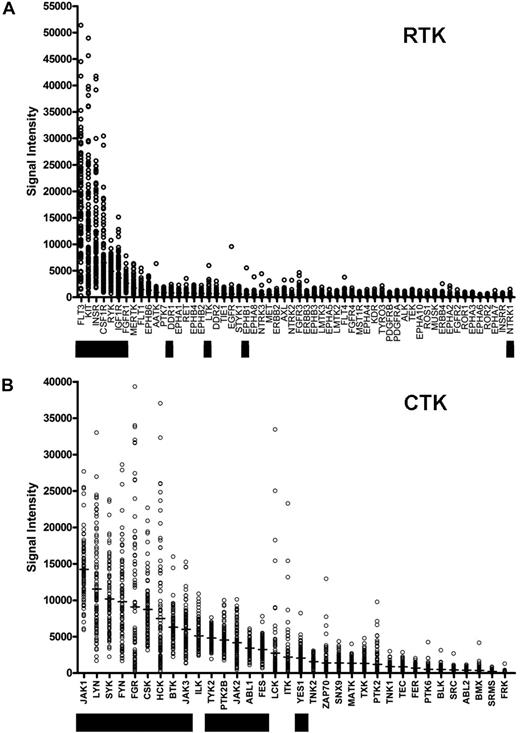
We sought to identify somatically acquired mutations relevant to AML disease biology, and we reasoned that the yield of biologically relevant nonsynonymous somatic mutations could potentially be increased by resequencing genes that are highly expressed in most AML samples. We therefore prioritized the expression of the annotated RTK and CTK genes on the Affymetrix U133 Plus 2 array platform using microarray data from AML patients banked at WU (Figure 1; Table S3). The genes that were chosen for resequencing are designated by the black boxes. The selection of genes to be sequenced was made based on the rank order of average expression values from the first 46 AML samples banked for the study, rather than the entire set of 92 that are presented here (2 of the 94 patients had inadequate or degraded RNA samples). As a consequence, some of the expressed genes (eg, ILK) were not sequenced in this study. The 2 most highly expressed RTK genes (in terms of absolute average expression values) were FLT3 and KIT, both known to contain somatic mutations in AML cells. We also included the minimally expressed gene NTRK1 (TrkA) because it has been reported to be mutated in AML.25
Ranking of tyrosine kinase (TK) genes based on expression in AML. Expression microarray data were obtained from 92 de novo AML patient bone marrow samples from the WU Discovery set samples. Normalized absolute expression values are shown for all annotated RTKs (A) and CTKs (B) on the Affymetrix U133 Plus 2 array platform. Genes that were sequenced in this study are indicated by the black bars.
Ranking of tyrosine kinase (TK) genes based on expression in AML. Expression microarray data were obtained from 92 de novo AML patient bone marrow samples from the WU Discovery set samples. Normalized absolute expression values are shown for all annotated RTKs (A) and CTKs (B) on the Affymetrix U133 Plus 2 array platform. Genes that were sequenced in this study are indicated by the black bars.
Sequence coverage
Genomic DNA was isolated from unfractionated bone marrow samples and matched skin biopsy samples from 94 patients with de novo AML, including all 92 that were included in the expression study. All samples were amplified approximately 1000× using the REPLI-g method (QIAGEN), and then subjected to high throughput automated exonic resequencing exactly as described.20 We sequenced the TK domains of 11 RTK genes and 15 CTK genes and all exons from the CTK genes TYK2 and JAK1 (Table 1). We sequenced all exons of these JAK family kinase genes because mutations are known to occur outside the kinase domain of JAK2 (V617F) and JAK3 (P132T). If nonsynonymous sequence changes were detected in the tumor sample, we determined whether the sequence variant had previously been reported as a SNP. The amplicons containing previously unreported variants were resequenced with the 94 matched skin samples from the same patients. All sequence variants were confirmed by direct sequencing from nonamplified sample templates. For sequence variants that were shown to be somatic or potentially relevant for AML susceptibility, we resequenced the tumor DNA from an additional 94 AML cases obtained from the CALGB. The characteristics of these cases have previously been reported.20
Cytoplasmic tyrosine kinase (CTK) and receptor tyrosine kinase (RTK) genes resequenced in primary AML patient samples
| Set . | Gene name . | Locus ID . | Total no. of exons . | No. of exons covered (discovery tumor) . | No. of amplicons sequenced (discovery tumor) . | No. of amplicons sequenced (discovery germline) . | No. of amplicons sequenced (CALGB) . |
|---|---|---|---|---|---|---|---|
| CTK | ABL1 | 25 | 10 | 6 | 6 | 2 | 1 |
| CTK | BTK | 695 | 19 | 6 | 6 | 0 | 0 |
| RTK | CSF1R | 1436 | 22 | 11 | 15 | 5 | 0 |
| CTK | CSK | 1445 | 12 | 8 | 19 | 0 | 0 |
| RTK | DDR1 | 780 | 19 | 6 | 8 | 5 | 1 |
| RTK | EPHB1 | 2047 | 16 | 7 | 8 | 2 | 0 |
| CTK | FES | 2242 | 19 | 7 | 6 | 1 | 1 |
| RTK | FGFR1 | 2260 | 18 | 8 | 7 | 2 | 0 |
| CTK | FGR | 2268 | 11 | 6 | 4 | 0 | 0 |
| RTK | FLT3 | 2322 | 24 | 2 | 5 | 2 | 2 |
| CTK | FYN | 2534 | 11 | 7 | 7 | 0 | 1 |
| CTK | HCK | 3055 | 13 | 6 | 6 | 2 | 2 |
| RTK | IGF1R | 3480 | 21 | 6 | 7 | 0 | 0 |
| RTK | INSR | 3643 | 22 | 6 | 10 | 3 | 0 |
| CTK | JAK1 | 3716 | 24 | 24 | 33 | 33 | 33 |
| CTK | JAK2 | 3717 | 25 | 14 | 20 | 10 | 1 |
| CTK | JAK3 | 3718 | 23 | 15 | 16 | 2 | 2 |
| RTK | KIT | 3815 | 21 | 2 | 2 | 2 | 1 |
| RTK | LTK | 4058 | 20 | 10 | 10 | 7 | 3 |
| CTK | LYN | 4067 | 13 | 6 | 22 | 1 | 1 |
| RTK | NTRK1 | 4914 | 17 | 5 | 7 | 4 | 3 |
| CTK | PTK2B | 2185 | 36 | 8 | 8 | 0 | 0 |
| RTK | RYK | 6259 | 14 | 8 | 8 | 0 | 0 |
| CTK | SYK | 6850 | 14 | 6 | 6 | 0 | 0 |
| CTK | TYK2 | 7297 | 25 | 23 | 33 | 7 | 9 |
| CTK | YES1 | 7525 | 11 | 6 | 7 | 0 | 0 |
| Total | 480 | 219 | 286 | 90 | 61 |
| Set . | Gene name . | Locus ID . | Total no. of exons . | No. of exons covered (discovery tumor) . | No. of amplicons sequenced (discovery tumor) . | No. of amplicons sequenced (discovery germline) . | No. of amplicons sequenced (CALGB) . |
|---|---|---|---|---|---|---|---|
| CTK | ABL1 | 25 | 10 | 6 | 6 | 2 | 1 |
| CTK | BTK | 695 | 19 | 6 | 6 | 0 | 0 |
| RTK | CSF1R | 1436 | 22 | 11 | 15 | 5 | 0 |
| CTK | CSK | 1445 | 12 | 8 | 19 | 0 | 0 |
| RTK | DDR1 | 780 | 19 | 6 | 8 | 5 | 1 |
| RTK | EPHB1 | 2047 | 16 | 7 | 8 | 2 | 0 |
| CTK | FES | 2242 | 19 | 7 | 6 | 1 | 1 |
| RTK | FGFR1 | 2260 | 18 | 8 | 7 | 2 | 0 |
| CTK | FGR | 2268 | 11 | 6 | 4 | 0 | 0 |
| RTK | FLT3 | 2322 | 24 | 2 | 5 | 2 | 2 |
| CTK | FYN | 2534 | 11 | 7 | 7 | 0 | 1 |
| CTK | HCK | 3055 | 13 | 6 | 6 | 2 | 2 |
| RTK | IGF1R | 3480 | 21 | 6 | 7 | 0 | 0 |
| RTK | INSR | 3643 | 22 | 6 | 10 | 3 | 0 |
| CTK | JAK1 | 3716 | 24 | 24 | 33 | 33 | 33 |
| CTK | JAK2 | 3717 | 25 | 14 | 20 | 10 | 1 |
| CTK | JAK3 | 3718 | 23 | 15 | 16 | 2 | 2 |
| RTK | KIT | 3815 | 21 | 2 | 2 | 2 | 1 |
| RTK | LTK | 4058 | 20 | 10 | 10 | 7 | 3 |
| CTK | LYN | 4067 | 13 | 6 | 22 | 1 | 1 |
| RTK | NTRK1 | 4914 | 17 | 5 | 7 | 4 | 3 |
| CTK | PTK2B | 2185 | 36 | 8 | 8 | 0 | 0 |
| RTK | RYK | 6259 | 14 | 8 | 8 | 0 | 0 |
| CTK | SYK | 6850 | 14 | 6 | 6 | 0 | 0 |
| CTK | TYK2 | 7297 | 25 | 23 | 33 | 7 | 9 |
| CTK | YES1 | 7525 | 11 | 6 | 7 | 0 | 0 |
| Total | 480 | 219 | 286 | 90 | 61 |
We evaluated the sequence coverage of all exons using previously described approaches.20 For “adequate” sequence coverage, an exon had to have high-quality single-stranded coverage with no gaps in the coding region of more than 10 bp. The coverage was extremely high for all genes analyzed, and nearly all sequences were obtained on both strands (Table S4). In our discovery set of 94 patients, we sequenced a total of 20 586 exons (219 exons × 94 patients) with a mean coverage of 96% (19 838 covered exons/20 586 sequenced exons). Including sequence data obtained from selected amplicons from our skin samples and from the 94 additional AML cases from the CALGB, a total of approximately 14.3 million base pairs of sequence data were obtained for analysis.
Identification of novel sequence variants
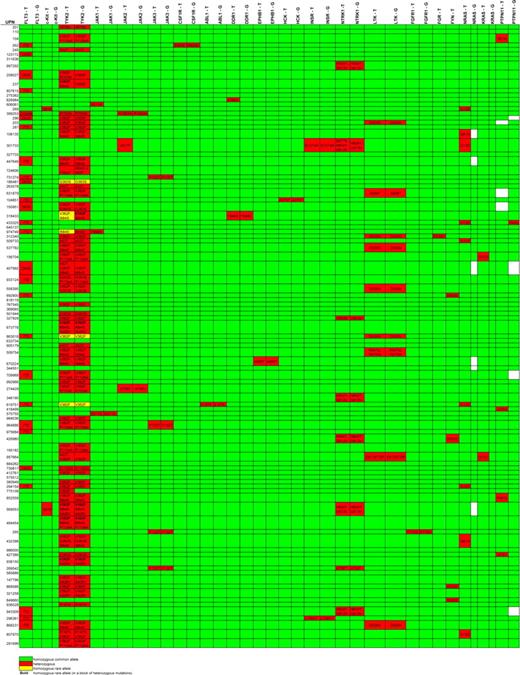
Sequence variants identified in our sequencing pipeline were validated by sequencing a second, nonamplified genomic DNA sample. Twenty-one potential mutations were assessed by second sample sequencing (“hand validation”). Seven sequence variants (33%) were found to be unreported germline polymorphisms, and 10 variants (48%) failed hand validation. By sequencing the tumor and germline DNA from candidate amplicons, we verified a total of 4 novel somatic mutations in 4 different patients: JAK1V623A, JAK1T478S, DDR1A803V, and NTRK1S677N (Figures 2). The functional validation of the JAK1 mutations is being reported elsewhere.25
Nonsynonymous base changes identified by exonic resequencing of DNAs from 94 AML patients. Unique patient numbers (UPNs) are shown in the rows, and the names of sequenced genes are shown in the columns. T indicates tumor (sequence variants in the AML tumor sample); G, germline (sequence variants in the “germline” (skin) sample from the same patient). Green indicates that no nonsynonymous sequence variants were identified in that sample; red identifies samples with nonsynonymous variants; the predicted consequences of all sequence variants detected are listed; yellow indicates the presence of a homozygous sequence variant; white boxes indicate that no sequence was obtained. All somatic mutations were confirmed by automated resequencing and/or hand validation. No nonsynonymous sequence variants were detected in FES, LYN, YES1, BTK, PTK2B, IGF1R, SYK, RYK, or CSK (data not shown). Mutations found in FLP3, KIT, N-RAS, KRAS, and PTPN11 are exactly the same as described previously.20 The nonsynonymous sequence variants shown for FGR (D230V) and FYN (D502E) are known to be SNPs and therefore were not sequenced in the skin samples.
Nonsynonymous base changes identified by exonic resequencing of DNAs from 94 AML patients. Unique patient numbers (UPNs) are shown in the rows, and the names of sequenced genes are shown in the columns. T indicates tumor (sequence variants in the AML tumor sample); G, germline (sequence variants in the “germline” (skin) sample from the same patient). Green indicates that no nonsynonymous sequence variants were identified in that sample; red identifies samples with nonsynonymous variants; the predicted consequences of all sequence variants detected are listed; yellow indicates the presence of a homozygous sequence variant; white boxes indicate that no sequence was obtained. All somatic mutations were confirmed by automated resequencing and/or hand validation. No nonsynonymous sequence variants were detected in FES, LYN, YES1, BTK, PTK2B, IGF1R, SYK, RYK, or CSK (data not shown). Mutations found in FLP3, KIT, N-RAS, KRAS, and PTPN11 are exactly the same as described previously.20 The nonsynonymous sequence variants shown for FGR (D230V) and FYN (D502E) are known to be SNPs and therefore were not sequenced in the skin samples.
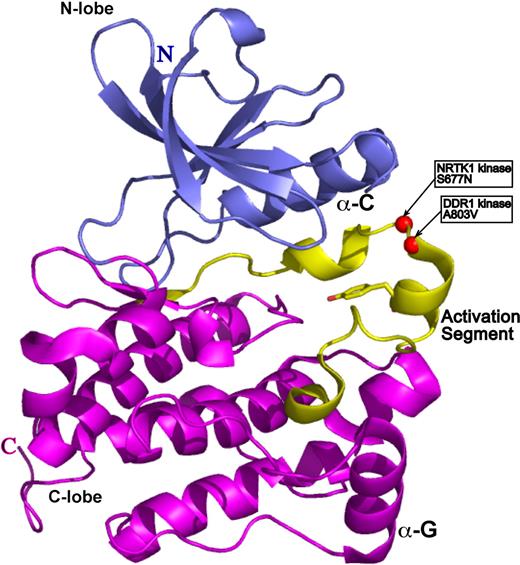
To gain insight into the functional significance of the DDR1 and NTRK1 mutants, we sought to map mutated residues onto the three-dimensional structure of a prototypical kinase domain. We used a structural homology analysis strategy (3D-PSSM26 ) that recognizes structural similarity among proteins with low sequence identity based on three-dimensional position-specific scoring algorithms. This search algorithm can also generate three-dimensional models based on threading of the submitted protein sequence onto existing structural data. The kinase model for both DDR1 and NTRK1 was based on the structure of the Hck kinase in complex with Src kinase inhibitor.27 The structure is in the auto-inhibited form with the electron density for activation loop being entirely resolved. A803V of DDR1 and S677N of NTRK1 map to the most critical structural element of kinase domain: its activation loop (Figure 3). The activation loop is a site of kinase transphosphorylation and subsequent activation. We therefore speculate that the DDR1 and NTRK1 mutants may disrupt kinase function by directly interfering with the site of ATP hydrolysis.
DDR1 and NTRK1 somatic mutations map onto activation loop of a prototypical kinase domain. The kinase model was based on the alignment of the DDR1 and NTRK1 sequences onto the structure of the Hck kinase in complex with Src kinase inhibitor (PDB ID 1qcf).26 The N-terminal domain (N-lobe) is in blue, the C-terminal domain (C-lobe) is in magenta, and the activation segment is in yellow. The catalytic Tyr is in ball-and-stick representation, whereas the mutations are represented by red spheres.
DDR1 and NTRK1 somatic mutations map onto activation loop of a prototypical kinase domain. The kinase model was based on the alignment of the DDR1 and NTRK1 sequences onto the structure of the Hck kinase in complex with Src kinase inhibitor (PDB ID 1qcf).26 The N-terminal domain (N-lobe) is in blue, the C-terminal domain (C-lobe) is in magenta, and the activation segment is in yellow. The catalytic Tyr is in ball-and-stick representation, whereas the mutations are represented by red spheres.
Frequency of germline TK alleles in normal and AML populations
The majority of nonsynonymous sequence variants found in RTK and CTK genes were found to be sequence variants that were present in the matched skin DNA samples. In addition to previously reported SNPs, we also discovered novel germline sequence changes not previously described in any SNP database (Figures 2, S2). No nonsynonymous sequence variants were detected in the TK domains of the FES, LYN, YES1, BTK, PTK2B, IGF1R, SYK, RYK, or CSK genes (data not shown). Analysis of paired tumor and skin samples identified 42 nonsynonymous germline sequence variants in TK genes (Figures 2, S2). We examined the clinical outcomes (event-free survival and overall survival) for all of the individual SNPs detected; none conferred a significant difference in any outcome parameter (data not shown). To address the possibility that some of these variants might be susceptibility alleles for de novo AML, we asked whether their frequencies differ in AML versus non-AML populations. Of the 42 identified SNPs, sufficient data were available in dbSNP to demonstrate that there was no significant difference in allele or genotype frequencies between the AML population and race-matched normal individuals for 4 previously identified SNPs (rs12720263, rs12720356, rs6336, and rs6339). Eight SNPs were chosen for further study based on a minor allele frequency more than 0.04 (rs35932273, rs34536443, rs2304256, rs2304255) or because of particular biologic interest (predicted functional consequences based on sequence alignments). Although the remaining 30 rare alleles are potentially relevant for AML pathogenesis, they were not considered further in this analysis.
Genotypes for the 8 selected SNPs were obtained by pyrosequencing (for controls) or resequencing (for cases). Because the majority of the AML subjects were white (WU 88%, CALGB 94%), a white control population was initially selected. Combining cases (WU AML and CALGB AML samples) and combining controls (WU Cancer Free Controls and Coriell Caucasian Controls) resulted in 2 populations of similar size. Comparison of all cases with control subjects demonstrated a positive association for rs2304255 (TYK2G363S; Table 2). In control subjects, the minor allele was significantly more common (0.092 vs 0.032, P = .0013). This variation was not observed in any of the black subjects, and the statistical association remained significant when only white cases and controls were compared (P < .002). In addition, the genotype distribution of rs3212723 (JAK3P132T) was significantly different (P = .024) in AML cases versus controls (Table 2). Further review of JAK3 genotype results revealed that, in every instance, the variant allele was detected only in blacks. Subsequent genotyping of DNA from 95 black control subjects revealed that the observed frequency of the variant allele in the 16 black AML subjects was not significantly different from the observed frequency in the race-matched control population (Table 3).
Frequency of LTK, FYN, JAK3, and TYK2 alleles and genotypes in AML and control populations
| Gene name and rsID . | Chr . | Position . | Variant . | Codon . | Controls . | AML . | P . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n . | AA . | AB . | BB . | n . | AA . | AB . | BB . | Allele . | Genotype . | |||||
| LTK | ||||||||||||||
| rs35932273 | 15 | 39585434 | G>A | D474N | 162 | 155 (0.96) | 7 (0.04) | 0 (0) | 188 | 179 (0.95) | 9 (0.05) | 0 (0) | 1 | 1 |
| novel | 15 | 39583644 | G>T | E813/752ter* | 188 | 185 (0.98) | 3 (0.02) | 0 (0) | 188 | 186 (0.99) | 2 (0.01) | 0 (0) | 1 | 1 |
| FYN | ||||||||||||||
| rs28763975 | 6 | 112089731 | C>G | D502/506E* | 138 | 133 (0.96) | 5 (0.04) | 0 (0) | 188 | 180 (0.96) | 8 (0.04) | 0 (0) | 1 | 1 |
| JAK3 | ||||||||||||||
| rs3212723 | 19 | 17815215 | C>A | P132T | 188 | 188 (1.0) | 0 (0) | 0 (0) | 188 | 183 (0.97) | 5 (0.03) | 0 (0) | .062 | .024 |
| TYK2 | ||||||||||||||
| rs34536443 | 19 | 10324118 | C>G | P1104A | 94 | 87 (0.93) | 7 (0.07) | 0 (0) | 188 | 175 (0.93) | 13 (0.07) | 0 (0) | 1 | 1 |
| rs35018800 | 19 | 10325843 | C>T | A928V | 176 | 172 (0.98) | 4 (0.02) | 0 (0) | 188 | 183 (0.97) | 5 (0.03) | 0 (0) | 1 | 1 |
| rs2304256 | 19 | 10336652 | G>T | V362F | 146 | 82 (0.56) | 53 (0.36) | 11(0.08) | 184 | 99 (0.54) | 71 (0.39) | 14 (0.08) | .79 | .91 |
| rs2304255 | 19 | 10336649 | G>A | G363S | 147 | 120 (0.82) | 27 (0.18) | 0 (0) | 188 | 177 (0.94) | 10 (0.05) | 1 (0.01) | .0013 | .006† |
| Gene name and rsID . | Chr . | Position . | Variant . | Codon . | Controls . | AML . | P . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n . | AA . | AB . | BB . | n . | AA . | AB . | BB . | Allele . | Genotype . | |||||
| LTK | ||||||||||||||
| rs35932273 | 15 | 39585434 | G>A | D474N | 162 | 155 (0.96) | 7 (0.04) | 0 (0) | 188 | 179 (0.95) | 9 (0.05) | 0 (0) | 1 | 1 |
| novel | 15 | 39583644 | G>T | E813/752ter* | 188 | 185 (0.98) | 3 (0.02) | 0 (0) | 188 | 186 (0.99) | 2 (0.01) | 0 (0) | 1 | 1 |
| FYN | ||||||||||||||
| rs28763975 | 6 | 112089731 | C>G | D502/506E* | 138 | 133 (0.96) | 5 (0.04) | 0 (0) | 188 | 180 (0.96) | 8 (0.04) | 0 (0) | 1 | 1 |
| JAK3 | ||||||||||||||
| rs3212723 | 19 | 17815215 | C>A | P132T | 188 | 188 (1.0) | 0 (0) | 0 (0) | 188 | 183 (0.97) | 5 (0.03) | 0 (0) | .062 | .024 |
| TYK2 | ||||||||||||||
| rs34536443 | 19 | 10324118 | C>G | P1104A | 94 | 87 (0.93) | 7 (0.07) | 0 (0) | 188 | 175 (0.93) | 13 (0.07) | 0 (0) | 1 | 1 |
| rs35018800 | 19 | 10325843 | C>T | A928V | 176 | 172 (0.98) | 4 (0.02) | 0 (0) | 188 | 183 (0.97) | 5 (0.03) | 0 (0) | 1 | 1 |
| rs2304256 | 19 | 10336652 | G>T | V362F | 146 | 82 (0.56) | 53 (0.36) | 11(0.08) | 184 | 99 (0.54) | 71 (0.39) | 14 (0.08) | .79 | .91 |
| rs2304255 | 19 | 10336649 | G>A | G363S | 147 | 120 (0.82) | 27 (0.18) | 0 (0) | 188 | 177 (0.94) | 10 (0.05) | 1 (0.01) | .0013 | .006† |
Alternative transcripts.
P < .002 (white), P < .0004 (dominant), P < .0002 (overdominant).
Numbers in parentheses are percentage values.
Frequency of JAK3P132T allele and genotypes in white and black populations
| . | Controls . | AML . | P . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N . | AA . | AB . | BB . | N . | AA . | AB . | BB . | Allele . | Genotype . | |
| Pooled | 283 | 265 (0.94) | 17 (0.06) | 1 (0.004) | 188 | 183 (0.97) | 5 (0.03) | 0 (0) | .0587 | .1706 |
| White | 188 | 188 (1.0) | 0 (0) | 0 (0) | 171 | 171 (1.0) | 0 (0) | 0 (0) | 1.00 | 1.00 |
| Black | 95 | 77 (0.81) | 17 (0.18) | 1 (0.01) | 16 | 11 (0.69) | 5 (0.3) | 0 (0) | .3566 | .4356 |
| . | Controls . | AML . | P . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N . | AA . | AB . | BB . | N . | AA . | AB . | BB . | Allele . | Genotype . | |
| Pooled | 283 | 265 (0.94) | 17 (0.06) | 1 (0.004) | 188 | 183 (0.97) | 5 (0.03) | 0 (0) | .0587 | .1706 |
| White | 188 | 188 (1.0) | 0 (0) | 0 (0) | 171 | 171 (1.0) | 0 (0) | 0 (0) | 1.00 | 1.00 |
| Black | 95 | 77 (0.81) | 17 (0.18) | 1 (0.01) | 16 | 11 (0.69) | 5 (0.3) | 0 (0) | .3566 | .4356 |
Data are numbers (proportion of total) for JAK3 (rs3212723), chromosome 19, position 17815215, variant C>A, codon P132T.
Numbers in parentheses are percentage values.
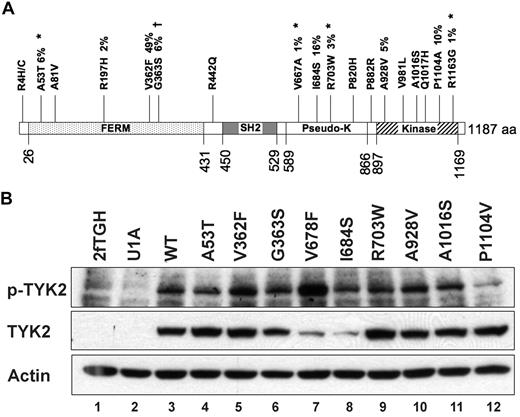
Of the 42 nonsynonymous germline sequence variants we identified, 18 (43%) occurred in a single gene, TYK2 (Figure 4A). To determine whether any of these alleles possesses altered function that might contribute to AML biology, we assessed TYK2 expression and phosphorylation in response to interferon-alpha in TYK2-deficient cells engineered to express 10 patient-derived TYK2 alleles (A53T, A81V, R197H, V362F, G363S, I684S, R703W, A928V, A1016S, and P1104V), and also an artificial variant, V678F, which is a predicted homolog of the JAK2V617F allele. Protein abundance and phosphorylation in response to ligand were indistinguishable from wild-type for 8 of these 10 TYK2 alleles; the V678F allele is an activated kinase, as predicted. In contrast, the TYK2 I684S and P1104V variants appeared different from wild-type in this assay. The steady-state level of TYK2 I684 protein was consistently reduced, and TYK2 P1104V autophosphorylation in response in IFN was consistently reduced compared with wild-type TYK2 (Figure 4B).
Nonsynonymous germline sequence variants in the TYK2 gene. (A) Diagram of the TYK2 protein showing nineteen (19) nonsynonymous changes. Four novel germline SNPs (*) were detected in the 94 Discovery set samples. Previously identified SNPs are also shown. Percentages indicate the frequency of the indicated sequence variant in the 94 Discovery set samples. †The TYK2G363S sequence change is found at a significantly different frequency between AML patients and normal controls. (B) Autophosphorylation of variant TYK2 alleles in response to stimulation by interferon-α (IFN). The human TYK2-deficient cell line U1A was transduced with cDNAs encoding patient-derived variant TYK2 alleles. An artificial allele, V678F, containing an amino acid substitution homologous to the V617F activating mutation in JAK2, was used as a positive control. Most patient-derived alleles were indistinguishable from wild-type. In contrast, TYK2I684S consistently demonstrated reduced total TYK2 protein levels, whereas the absolute level of phosphorylated TYK2I684S appeared no different from wild-type (lane 8). Autophosphorylation of the TYK2P1104V was reduced after IFN stimulation, suggesting a decreased level of kinase activity (lane 12). TYK2V362F phosphorylation appears slightly increased (lane 5), but this finding was not reproduced in replicate experiments. This experiment was performed 3 times, and a representative blot is shown.
Nonsynonymous germline sequence variants in the TYK2 gene. (A) Diagram of the TYK2 protein showing nineteen (19) nonsynonymous changes. Four novel germline SNPs (*) were detected in the 94 Discovery set samples. Previously identified SNPs are also shown. Percentages indicate the frequency of the indicated sequence variant in the 94 Discovery set samples. †The TYK2G363S sequence change is found at a significantly different frequency between AML patients and normal controls. (B) Autophosphorylation of variant TYK2 alleles in response to stimulation by interferon-α (IFN). The human TYK2-deficient cell line U1A was transduced with cDNAs encoding patient-derived variant TYK2 alleles. An artificial allele, V678F, containing an amino acid substitution homologous to the V617F activating mutation in JAK2, was used as a positive control. Most patient-derived alleles were indistinguishable from wild-type. In contrast, TYK2I684S consistently demonstrated reduced total TYK2 protein levels, whereas the absolute level of phosphorylated TYK2I684S appeared no different from wild-type (lane 8). Autophosphorylation of the TYK2P1104V was reduced after IFN stimulation, suggesting a decreased level of kinase activity (lane 12). TYK2V362F phosphorylation appears slightly increased (lane 5), but this finding was not reproduced in replicate experiments. This experiment was performed 3 times, and a representative blot is shown.
Discussion
In this report, we resequenced the TK domains of 26 highly expressed RTKs and nonreceptor (cytoplasmic) tyrosine kinases (CTK) in a discovery set of 94 genomic DNA tumor and skin samples from patients with de novo AML (Figures 5, 6). We identified 4 novel somatic mutations in the JAK1, DDR1, and NTRK1 genes. These mutations were confirmed as somatic changes, and each occurred in conserved residues within functional domains. The generally low number of somatic mutations found is consistent with high-throughput resequencing studies performed in other cancer types.28-31 A number of nonsynonymous sequence variants were found in the skin samples of many of these patients as well, and several are clearly polymorphisms that are not related to disease susceptibility because they were also identified in the samples of normal, ethnically matched control samples.
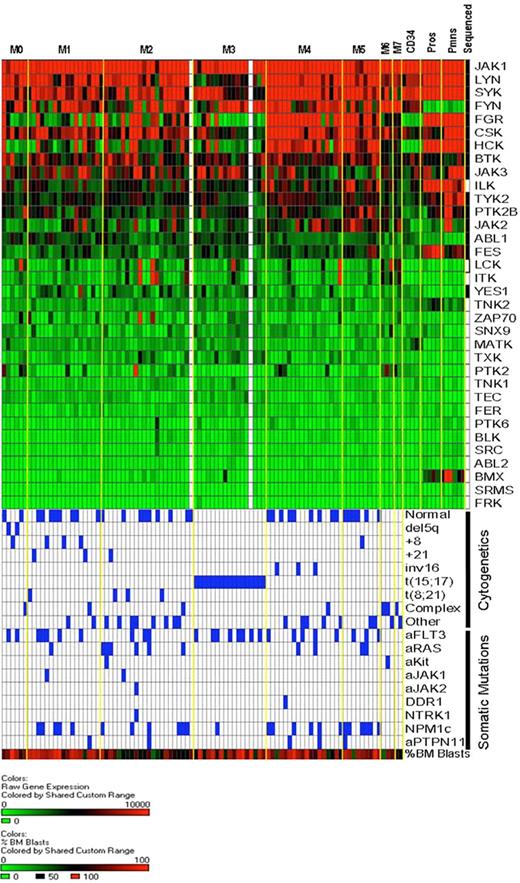
Cytoplasmic tyrosine kinase (CTK) gene expression in AML and normal myeloid cells. RNA isolated from unfractionated AML bone marrow samples or from normal human bone marrow CD34+ cells, flow-sorted promyelocytes, and flow-sorted polymorphonuclear leukocytes was hybridized to Affymetrix U133 Plus 2 microarrays. Expression of CTK genes in our discovery set AML samples is rank-ordered from the highest to lowest levels of mean expression (92 of 94 samples had successful array studies performed; the 2 samples without array data are shown as white columns). AML samples are arranged by FAB classification. Expression levels are based on scaled signal intensity values for each probe set (mean value for all probe sets on each array was scaled to a value of 1500 using the MAS 5.0 algorithm), with a value of 0 represented in green and 10 000 or greater as red. Black squares in the right-most column indicate the CTK genes that were resequenced in this study. Below the expression data, the cytogenetic findings and somatic mutation analysis for each sample are shown. Blue boxes indicate the presence of the indicated cytogenetic abnormalities and/or somatic mutations. The bottom-most row indicates the percentage of bone marrow blasts in the AML samples, scaled from 0% (green) to 100% (red); the average blast count for the 94 samples analyzed was 70.5% (range, 30%-100%).
Cytoplasmic tyrosine kinase (CTK) gene expression in AML and normal myeloid cells. RNA isolated from unfractionated AML bone marrow samples or from normal human bone marrow CD34+ cells, flow-sorted promyelocytes, and flow-sorted polymorphonuclear leukocytes was hybridized to Affymetrix U133 Plus 2 microarrays. Expression of CTK genes in our discovery set AML samples is rank-ordered from the highest to lowest levels of mean expression (92 of 94 samples had successful array studies performed; the 2 samples without array data are shown as white columns). AML samples are arranged by FAB classification. Expression levels are based on scaled signal intensity values for each probe set (mean value for all probe sets on each array was scaled to a value of 1500 using the MAS 5.0 algorithm), with a value of 0 represented in green and 10 000 or greater as red. Black squares in the right-most column indicate the CTK genes that were resequenced in this study. Below the expression data, the cytogenetic findings and somatic mutation analysis for each sample are shown. Blue boxes indicate the presence of the indicated cytogenetic abnormalities and/or somatic mutations. The bottom-most row indicates the percentage of bone marrow blasts in the AML samples, scaled from 0% (green) to 100% (red); the average blast count for the 94 samples analyzed was 70.5% (range, 30%-100%).
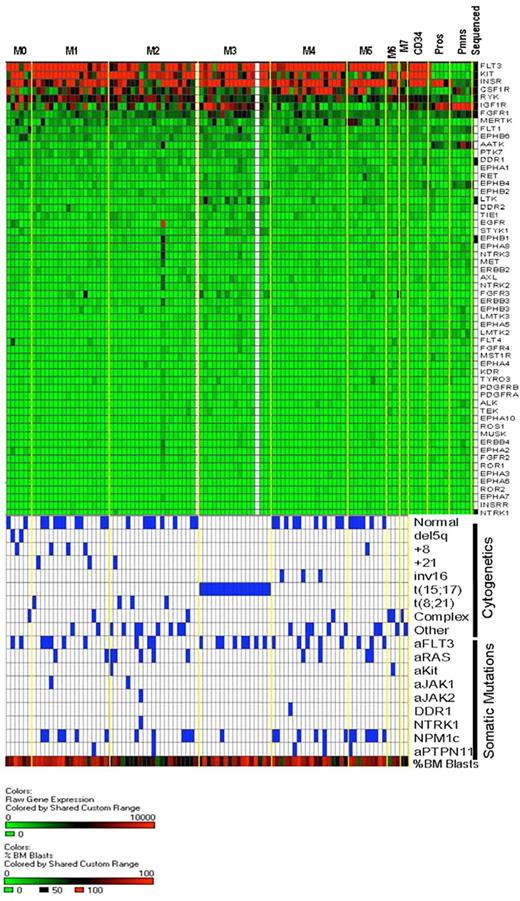
Heat map of receptor tyrosine kinase gene expression in AML and normal myeloid development. The same expression arrays were used to plot data for the RTK genes, using the same approach outlined in Figure 5. The bottom half of the figure is identical to that of Figure 5 and is shown to allow for direct comparisons of the data within samples.
Heat map of receptor tyrosine kinase gene expression in AML and normal myeloid development. The same expression arrays were used to plot data for the RTK genes, using the same approach outlined in Figure 5. The bottom half of the figure is identical to that of Figure 5 and is shown to allow for direct comparisons of the data within samples.
Functional validation will be required to prove that the somatic mutations identified in this study contribute to AML pathogenesis. The JAK1 mutations appear to contribute to the activation of JAK1 kinase and downstream signaling pathways, yet do not “score” in typical transformation assays.25 NTRK1 (TRK-A) has previously been implicated in AML pathogenesis. A small deletion of the juxtamembrane region of the gene encoding the TRKA RTK has been found in a patient with AML,25 and TRKA MRNA is up-regulated by AML1-ETO.32 DDR1 mutations have not previously been described in AML, but this gene was found to be highly expressed in B-ALL cases without other molecular abnormalities.32 DDR1 gene locus amplification has been identified by fluorescence in situ hybridization in one patient with AML (Olivier Bernard, personal oral communication). Regardless, functional characterization is required to definitively determine whether the DDR1 and NTRK1 mutations also change protein function and how they contribute to disease development and progression. Our preliminary experiments have yet to demonstrate unique biologic properties of DDR1A803V or NTRK1S677N; however, additional studies will be required to determine whether either of these somatic mutations is relevant for AML pathogenesis.28
In contrast to the rarity of somatic mutations in TK genes other than FLT3, nonsynonymous germline TK gene sequence variants were common in many of the genes that we sequenced. We therefore sought to determine whether any of the nonsynonymous germline sequence variants might mediate a predisposition for the development of AML. We genotyped 8 sequence variants using genomic DNA samples from 94 to 188 normal controls, and we compared the allele and genotype frequencies with our 188 AML samples. Our initial analysis indicated that 2 alleles (JAK3P132T and TYK2G363S) displayed significantly different frequencies between AML and cancer-free controls (Table 2). However, the JAK3P132T allele was found in nearly 20% of black control samples, and no statistical difference between AML and cancer-free controls was apparent when race was taken into account. Expression of the JAK3P132T allele in Ba/F3 cells results in factor independent growth and the ligand-independent activation of downstream signaling pathways.34 Although we did not see an association between the JAK3P132T allele and the development of AML, it is important to note that this statistical comparison must be viewed in light of its small sample size. Future studies with larger numbers of black AML cases will required to determine whether there is a significant association of the JAK3P132T with AML development within this population.
The difference in frequencies between AML and cancer-free controls for the TYK2G363S allele was not related to race and was statistically significant (Table 2). Models of gene effect were examined, and dominant and overdominant models demonstrated the greatest significance, suggesting that the presence of the minor allele may exert a protective effect in the healthy population. Germline polymorphisms of TYK2 are also seen in association with rheumatoid arthritis,35,36 but the effect of these changes on gene function is unknown. We expressed a variety of TYK2 alleles in TYK2-deficient cells and found no difference between TYK2WT and TYK2G363S activity. However, 2 other variant TYK2 alleles appeared to have altered function. TYK2I684S consistently demonstrated reduced steady-state levels of total TYK2 protein, suggesting a possible effect on protein stability. We found TYK2P1104V to be consistently hypo-phosphorylated in response to IFN, suggesting that this allele is hypofunctional. The TYK2P1104A variant was recently predicted to have altered kinase function using computer algorithms to distinguish functional cancer-associated missense mutations from common polymorphisms.37 Our protein data confirm this prediction and suggest that some TYK2 SNPs may influence AML susceptibility or biology.
Inherited mutations of RUNX1 and CEBPA confer a strong predisposition to AML.38,39 Although these high-penetrance alleles cause rare cases of familial AML, they explain little of the AML risk in the general population. Combinatorial effects of more common, low-penetrance polymorphisms are likely to be more relevant for susceptibility to nonsyndromic AML. To date, most gene association studies in AML have focused on variants involved in drug metabolism and DNA repair. We reasoned that polymorphisms in genes with established connections to AML biology were strong candidate susceptibility factors for both de novo and therapy-related AML.40 Support for this hypothesis is provided by the association between variants of CSF3RE785K and FLT3D324N previously reported in MDS and AML,18,19 and the association between TYK2G363S and AML in this report. Further validation of the importance of these findings will require replication by other groups and results from other ongoing gene association studies in AML.
Our data demonstrate that very few somatic changes occur in the tumor DNA of AML samples. We found 45 somatic mutations (41 previously characterized and 4 novel mutations) in 14.3 million base pairs of sequence data from AML patients. The observation of frequent p53 mutations in colon cancer samples had previously suggested that hypermutability was a general feature of human cancers.41 Indeed, cancers that arise in the setting of familial cancer predisposition syndromes (eg hereditary nonpolyposis colorectal carcinoma) are frequently associated with mutations of specific DNA repair machinery components, and they display microsatellite instability and are associated with a hypermutator phenotype. Most cancer types do not have a hypermutator phenotype but rather display rates of spontaneous mutation that are approximately equal to that of normal cells.21,42 The large amounts of normal sequence data generated in our studies20,21 strongly suggest that de novo AML cells with few cytogenetic changes generally contain intact DNA repair pathways and that bona fide somatic point mutations in AML genomes occur rarely.
Several groups have suggested that activating mutations in TK genes may be necessary for AML development and that mutations in one or more of these genes would be found in virtually all cases. Our resequencing data might suggest a different model: TK mutations, although sometimes contributing to disease progression, may not be specifically required. Data on clinical outcomes have demonstrated that FLT3-ITD and KITD816V mutations confer a poor prognosis in AML,43-48 which logically infers that equivalent TK mutations may not occur in all AML cases. It is possible that further resequencing studies of additional expressed TK genes, and sequencing of all exons of these genes, not just the TK domains, may yet identify common mutations. TK genes can also be activated independently of ligand when overexpressed in the absence of activating mutations.49,50 We suggest that mechanisms other than activating mutations (such as altered miRNA or transcription factor networks) may also dysregulate TK gene expression in some AML cases.
The activating mutations of TK genes are dominant and exhibit gain-of-function properties. This finding has suggested that mutations in one of these genes may preclude the need for mutations in additional family members (ie, they may be mutually exclusive). However, the finding that activating mutations in FLT3 and JAK2 are sometimes homozygous suggests that multiple “hits” in this pathway may be additive.51-55 In the 188 cases in our study, 7 of 40 patients with FLT3 ITD were found to have additional somatic mutations in the TK signaling pathway; 5 had concurrent activating mutations in Ras genes, one had the JAK1T478S mutation, and one had FLT3 D835Y. One patient with JAK2V617F had additional somatic mutations in NRAS and NTRK1. Because such a small number of patients displayed multiple mutations, our study was underpowered to detect differences in outcomes; however, as additional sequencing studies are performed, this question should be continuously revisited.56,57
We focused here on somatic, nonsynonymous mutations in the coding regions of known genes. Germline sequence variants, noncoding base changes, and even synonymous mutations will probably be found to contribute to cancer development because synonymous mutations have recently been shown to affect protein function.58 Future studies will be required to define the role of these changes for AML pathogenesis.
In conclusion, we have found that, outside of the known somatic mutations in FLT3 and KIT, acquired mutations in expressed RTK and CTK genes occur infrequently in AML, suggesting that TK mutations may not be a prerequisite for AML development; they may represent later, disease-modifying events. We also found nonsynonymous germline sequence changes in several TK genes. Notably, the TYK2G363S allele occurred significantly less frequently in patients with AML in this study. We determined that the germline TYK2P1104V allele encodes a hypofunctional kinase. Large-scale gene association studies are warranted to explore the role of germline TK gene variants in AML pathogenesis.
An Inside Blood analysis of this article appears at the front of this issue
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the CALGB tumor bank for providing the CALGB AML tumor samples and also our patients for their participation in this study.
This work was supported by National Institutes of Health grant CA101937 and the Barnes-Jewish Hospital Foundation.
National Institutes of Health
Authorship
Contribution: M.H.T., T.A.G., D.C.L., J.F.D., M.J.W., R.K.W., and T.J.L. designed research and wrote the paper; Z.X., Y.Z., T.M., Y.K., R.E.R., E.R.M., and J.E.P. performed research; R.W. performed research and wrote the paper; P.W., M.W., and C.D.B. provided vital reagents; M.D.M., J.B., S.H., W.D.S., O.L., R.N., and D.F. analyzed data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michael H. Tomasson, Division of Oncology, Department of Medicine, 660 South Euclid Avenue, Campus Box 8007, St Louis, MO 63110; e-mail: tomasson@wustl.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal