Abstract
TP53 is a tumor suppressor gene that functions as transcriptional regulator influencing cellular responses to DNA damage. Here we explored the clinical and transcriptional effects of TP53 expression in multiple myeloma (MM). We found that low expression of TP53, seen in approximately 10% of newly diagnosed patients, is highly correlated with TP53 deletion, an inferior clinical outcome, and represents an independent risk factor. Analysis of the expression of 122 known TP53 target genes in TP53-high vs -low MM cells from 351 newly diagnosed cases, revealed that only a few were highly correlated with TP53 expression. To elucidate TP53 regulatory networks in MM, we overexpressed TP53 in 4 MM cell lines. Gene expression profiling of these cell lines detected 85 significantly differentially expressed genes, with 50 up-regulated and 35 down-regulated. Unsupervised hierarchical clustering of myeloma samples from 351 newly diagnosed and 90 relapsed patients using the 85 putative TP53 target genes revealed 2 major subgroups showing a strong correlation with TP53 expression and survival. These data suggest that loss of TP53 expression in MM confers high risk and probably results in the deregulation of a novel set of MM-specific TP53-target genes. TP53 target gene specificity may be unique to different cell lineages.
Introduction
The genetic lesions important in the pathogenesis and prognosis of multiple myeloma (MM) continue to be elucidated. We recently showed that gene expression profiles could be used to identify high-risk disease.1 One of the surprising findings of this former study was that variation in TP53 gene expression was not indicative of high-risk disease
Using high-resolution array comparative genomic hybridization, we recently identified 87 discrete minimal common regions of recurrent copy number alterations in 65 newly diagnosed MM patients. Fourteen minimal common regions, including a deletion at chromosome 17p13.1-17p12, where the TP53 gene resides, were found to be associated with poor survival.2
In MM, TP53 mutations are rare and may represent late events in disease progression.3-7 The frequency of TP53 deletions detected by fluorescence in situ hybridization (FISH) ranges from 9% to 34% in newly diagnosed cases of MM and is related to survival.8-13 However, the role of TP53 loss in the pathogenesis of MM, its relationship to gene expression, and its relevance as an independent prognostic variable remain to be elucidated. In this study, we determined TP53 mRNA expression, DNA sequence integrity, and copy number loss in newly diagnosed and relapsed disease. We also correlated these features with disease progression and outcome. Low TP53 gene expression, which is highly correlated with loss of heterozygosity of the TP53 locus, was associated with shorter event-free survival (EFS) and overall survival (OS), even in the context of other high-risk disease features.
As a transcription factor, TP53 regulates the expression of genes involved in a variety of cellular functions, including cell-cycle arrest, DNA repair, and apoptosis,14-19 but the function of TP53 and the signaling pathways regulated by it in MM are still not clear. The TP53-dependent expression of 122 target genes identified by coupled chromatin immunoprecipitation (CHIP) with the paried-end ditag (PET) sequencing analysis was recently demonstrated20 ; however, expression of only a few of these 122 previously identified TP53 target genes was correlated with TP53 expression in tumor cells from our cohort of 351 MM patients. This suggested that TP53 may regulate a distinct set of genes in MM. In the current study, we identified novel TP53-associated genes and demonstrated the clinical relevance of these alterations to disease progression.
Methods
Study subjects
Purified plasma cells were obtained from newly diagnosed MM patients who were treated on the National Institutes of Health-sponsored clinical trials UARK 98-026 (Total Therapy 2 [TT2], n = 351) and UARK 03-033 (Total Therapy 3 [TT3], n = 214).1,21-26 Both protocols used induction regimens followed by melphalan-based tandem autotransplantation, consolidation chemotherapy, and maintenance treatment. Studies are approved by the Institutional Review Board of the University of Arkansas for Medical Sciences. Informed consent was obtained in accordance with the Declaration of Helsinki.
Human MM cell lines ARP-1, JJN3, OCI-MY5, and Delta 47 were cultured in RPMI 1640 containing 10% heat-inactivated fetal calf serum, 2 mM l-glutamine (Invitrogen, Carlsbad, CA), penicillin (100 U/mL), and streptomycin (100 μg/mL) at 37°C in humidified 95% air and 5% CO2. In JJN3, OCI-MY5, and ARP-1, TP53 mRNA is absent, and Delta 47 cell line has middle level of TP53 expression but has increased MDM2, which can trigger the degradation of TP53.14
Fluorescence in situ hybridization
To detect TP53 deletions, a SpectrumRed-labeled DNA probe (LSI p53; Vysis, Downers Grove, IL) was combined with a SpectrumGreen-labeled probe (CEP17, Vysis) for the chromosome 17 α-satellite-DNA centromere. The interphase-FISH procedure used to analyze the samples has previously been described.27,28 On the basis of FISH studies of normal bone marrow mononuclear cells, the upper limit of normal plus 3 SD was less than 10% for deletions of TP538 ; therefore, we used 10% as the background cutoff level for the probe sets.
Gene expression profiling
Bone marrow plasma cells from 565 newly diagnosed (351 TT2 and 214 TT3) patients and from 90 patients with relapsed disease were purified by CD138+ selection.29,30 Gene expression levels in purified plasma cells and MM cell lines were profiled with the U133Plus2.0 array (Affymetrix, Santa Clara, CA), and the signal of probe set 201746_at representing TP53 was used in this current analysis. Signal intensities were preprocessed and normalized by GCOS1.1 software (Affymetrix).29-31 Gene expression data on this patient cohort can be found at the National Institutes of Health GEO omnibus under accession number GSE2658.1,21,22,25,27
Overexpression of the TP53 gene in MM cell lines
The amplified TP53 cDNA sequence was cloned into the pWPI lentiviral vector, which was a generous gift from Dr Didier Trono (National Center for Competence in Research, Lausanne, Switzerland).32 Recombinant lentivirus was produced by transient transfection of 293T cells according to a standard protocol.33,34 Crude virus was concentrated by ultracentrifugation at 26 000g for 90 minutes. Viral titers were determined by measuring the amount of HIV-1 p24 antigen by enzyme-linked immunosorbent assay (PerkinElmer Life and Analytical Sciences, Waltham, MA). A 99% transduction efficiency of MM cell lines was achieved with 3000 ng of lentiviral p24 particles per 106 cells.
Western blotting
To test TP53 protein levels in MM cell lines and for TP53 overexpression studies, nuclear protein was isolated with the Nuclear/Cytosol Fractionation Kit (BioVision, Mountain View, CA). Nuclear protein (30 μg) was separated by electrophoresis on 4% to 12% sodium dodecyl sulfate-polyacrylamide gels, and Western blotting was performed with the WesternBreeze Chemiluminescent Immunodetection protocol (Invitrogen). Antibodies to antipoly (ADP-ribose) polymerase (PARP), anti–β-tubulin, and anti-histone 4 were purchased from Upstate Biotechnology (Charlottesville, VA); anti-p53 was purchased from Chemicon International (Temecula, CA).
Cell cycle/DNA content analysis
Cells (106) from each sample were fixed in 75% ethanol at −20°C overnight. The next day, the cells were washed with cold phosphate-buffered saline, treated with 100 μg RNase A (QIAGEN, Valencia, CA), and stained with 50 μg of propidium iodide (Roche Applied Science, Indianapolis, IN). Flow cytometric acquisition was performed with a 3-color FACScan flow cytometer and CellQuest software (BD Biosciences, San Jose, CA). For each sample, 10 000 events were gated. Data were analyzed with Modfit LT software (Verity Software House, Topsham, ME).
TP53 mutations detected by sequencing analysis
Mononuclear cells were obtained from bone marrow specimens and enriched by a Ficoll-gradient centrifugation method. Genomic DNA was used as a template (100 ng/reaction) for polymerase chain reaction (PCR) analysis with intronic primer pairs (TP53 Ex2-4-F and TP53 Ex7-9-R) covering exons 2 to 9 of the TP53 gene, where most TP53 mutations were detected.35 Sequencing primers were nested within the PCR products (TP53-Ex2-4-F: 5′-CAGCCATTCTTTTCCTGCTC-3′; TP53-Ex2-4-R: 5′-AGGGTGTGATGG GATGGATA-3′; TP53-Ex5-6-F: 5′-GTTTCTTTGCTGCCGTCTTC-3′; TP53-Ex5-6-R: 5′-TTG CACATCTCATGGGGTTA-3′; TP53-Ex7-9-F: 5′-GGAGGCTGAGGAAGGAGAAT-3′; and TP53-Ex7-9-R: 5′-TTGAAAGCTG GTCTGGTCCT-3′).
Statistical analysis
The Kaplan-Meier method was used to estimate OS, with group comparisons made with the log-rank test. OS was defined as the time from date of registration until death from any cause; survivors were censored at the time of last contact. Significance analysis of microarray36 was used to determine statistically significant expression changes of genes in high- and low-TP53–expressing MM plasma cells. Univariate and multivariate analyses of prognostic factors were performed with Cox regression.
Results
Low TP53 expression, highly correlated with deletion, is a significant and independent adverse prognostic factor in newly diagnosed MM
FISH analyses for TP53 deletion were available for 194 TT2 cohort patients with newly diagnosed disease. TP53 deletion was observed in 40 (20.6%) samples, 4 of which had biallelic deletion. Patients with TP53 deletion were associated with shorter EFS and OS (P = .005 and P < .001, respectively; Figure 1A). However, we did not find increased incidence of deletion in 28 cases for which a sample was tested at diagnosis and at disease relapse paired patients with relapsed disease (data not shown).
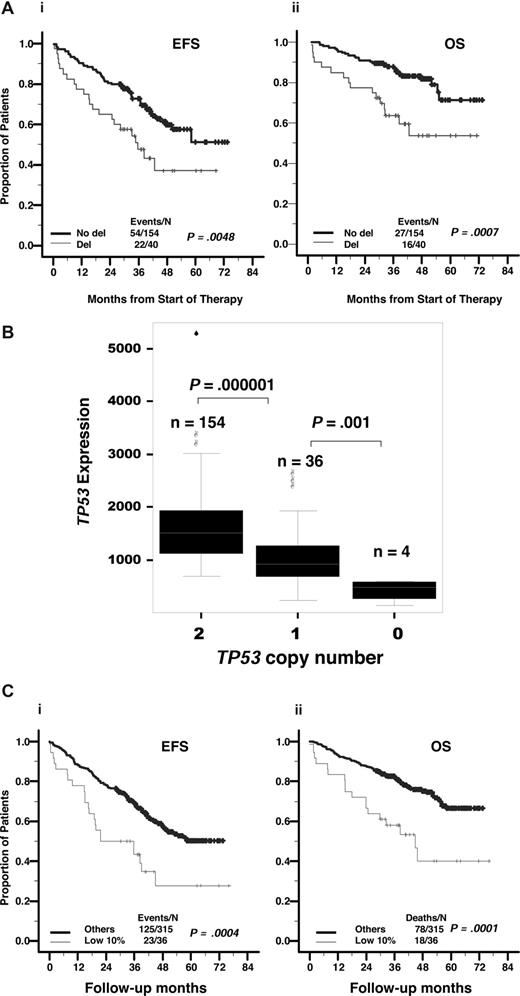
Low expression levels of TP53, highly correlated with deletion and adversely affects outcome. (A) The 194 patients with newly diagnosed MM on TT2 were divided into 2 groups based on TP53 deletion. Kaplan-Meier estimates show 5-year actuarial probabilities of 36% being event-free and 52% alive in cases with TP53 deletion and 51% event-free and 70% alive in those without TP53 deletion (P < .005). (B) TP53 deletion is highly correlated with low TP53 expression. TP53 expression levels, as measured by Affymetrix microarray signal, relative to TP53 copy number by FISH is displayed. Expression levels in 154 cases with no evidence of deletion (minimum, 680; maximum, 5241; median, 1599; mean, 1487) are significantly higher than in 36 cases with monoallelic deletion (minimum, 226; maximum, 2600; median, 889; mean, 1044), which are higher than in 4 cases with biallelic deletion (minimum, 138; maximum, 599; median, 470; mean, 419). (C) Low TP53 gene expression is related to outcome. Samples from 351 patients with newly diagnosed MM on TT2 were divided into 2 groups based on TP53 Affymetrix signal being greater than or less than 733 (lowest 10%). Kaplan-Meier estimates showed 5-year actuarial probabilities of 28% being event-free and 41% alive in cases with TP53 less than 733 and 50% event-free and 68% alive in those with TP53 more than 733 (P < .001).
Low expression levels of TP53, highly correlated with deletion and adversely affects outcome. (A) The 194 patients with newly diagnosed MM on TT2 were divided into 2 groups based on TP53 deletion. Kaplan-Meier estimates show 5-year actuarial probabilities of 36% being event-free and 52% alive in cases with TP53 deletion and 51% event-free and 70% alive in those without TP53 deletion (P < .005). (B) TP53 deletion is highly correlated with low TP53 expression. TP53 expression levels, as measured by Affymetrix microarray signal, relative to TP53 copy number by FISH is displayed. Expression levels in 154 cases with no evidence of deletion (minimum, 680; maximum, 5241; median, 1599; mean, 1487) are significantly higher than in 36 cases with monoallelic deletion (minimum, 226; maximum, 2600; median, 889; mean, 1044), which are higher than in 4 cases with biallelic deletion (minimum, 138; maximum, 599; median, 470; mean, 419). (C) Low TP53 gene expression is related to outcome. Samples from 351 patients with newly diagnosed MM on TT2 were divided into 2 groups based on TP53 Affymetrix signal being greater than or less than 733 (lowest 10%). Kaplan-Meier estimates showed 5-year actuarial probabilities of 28% being event-free and 41% alive in cases with TP53 less than 733 and 50% event-free and 68% alive in those with TP53 more than 733 (P < .001).
TP53 deletion was highly correlated with low TP53 expression. Comparison of TP53 expression level on the basis of deletion status revealed that TP53 expression was lower in 36 monoallelic deletion cases (P < .001) and even lower in 4 biallelic deletion cases (P = .001) than in 154 cases lacking deletion (Figure 1B).
TP53 expression in the 351 newly diagnosed cases varied from an Affymterix signal output (a quantitative measure of the level of activity of a given gene) from a low of 10 to a high of 5241. Using a running log-rank test, we defined a 10% cutoff as those cases with a TP53 expression level lower than 733 on the basis of the Affymetrix microarray signal represented by 36 of 351 patients with newly diagnosed disease. Genes with an expression level less than 500 typically have an absent-detection call and are not detectable by sensitive quantitative RT-PCR. The cases with low TP53 expression were associated with a shorter EFS and OS (P < .001 and P < .001, respectively; Figure 1C). Myeloma samples from 16 of 18 (88.9%) patients with low TP53 expression (TP53 Affymetrix signal < 733) were detected with TP53 deletion, and all of 4 myeloma cell samples with TP53 bi-allelic deletion had low TP53 expression (TP53 Affymetrix signal < 733), whereas cell samples from only 24 of 176 (13.6%) myeloma patients with TP53 high expression (TP53 Affymetrix signal > 733) were detected with TP53 deletion, and none of them had TP53 bi-allelic deletion (P < .001).
With the same cutoff point, a low TP53 gene expression level predicted short postrelapse survival (P = .03; Figure S1A, available on the Blood website; see the Supplemental Materials link at the top of the online article) in 90 TT2 patients with relapsed disease and short EFS and OS (P = .017 and P = .022, respectively; Figure S1B) in a separate cohort of 214 patients treated on the successor protocol TT3.
With regard to clinical and biologic features, patients with a low TP53 expression level had high levels of lactate dehydrogenase (P = .036), increased numbers of bone lesions on magnetic resonance imaging (P = .012), and an increased incidence of deletion of chromosome 13 (P < .001) and amplification of chromosome 1q21 (P = .002; Table 1).
Baseline TT2 patient characteristics according to TP53 expression level
| Characteristic . | Low TP53, % (n = 36) . | High TP53, % (n = 315) . | P† . |
|---|---|---|---|
| Age at least 65 y | 29 | 22 | NS |
| Female sex | 51 | 42 | NS |
| White race | 94 | 88 | NS |
| IgA isotype | 20 | 26 | NS |
| Creatinine at least 2.0 mg/dL (221 μmol/L) | 20 | 10 | NS |
| MRI focal bone lesions, at least 3 | 77 | 55 | NS |
| CRP at least 8.0 mg/L | 49 | 34 | NS |
| LDH at least 190 IU/L | 49 | 31 | .036 |
| β2-microglobulin | |||
| Less than 3.5 mg/L | 60 | 59 | |
| At least 3.5 and less than 5.5 mg/L | 14 | 20 | |
| 5.5 mg/L or more | 26 | 21 | NS |
| Albumin less than 3.5 g/dl | 17 | 14 | NS |
| Hemoglobin less than 10 g/dL | 23 | 26 | NS |
| Bone marrow plasma cells (by aspiration) 33% or greater | 61 | 66 | NS |
| Chromosomal abnormalities (defined by G-banding) | 40 | 34 | NS |
| Deletion of chromosome 13 | 84 | 46 | < .001 |
| Amplification of 1q21 | 75 | 42 | .002 |
| High-risk model (17-gene)* | 19 | 14 | NS |
| Subgroups with poor prognosis* (Proliferation/MMSET/FGFR3/MAF/MAFB) | 59 | 35 | .023 |
| Characteristic . | Low TP53, % (n = 36) . | High TP53, % (n = 315) . | P† . |
|---|---|---|---|
| Age at least 65 y | 29 | 22 | NS |
| Female sex | 51 | 42 | NS |
| White race | 94 | 88 | NS |
| IgA isotype | 20 | 26 | NS |
| Creatinine at least 2.0 mg/dL (221 μmol/L) | 20 | 10 | NS |
| MRI focal bone lesions, at least 3 | 77 | 55 | NS |
| CRP at least 8.0 mg/L | 49 | 34 | NS |
| LDH at least 190 IU/L | 49 | 31 | .036 |
| β2-microglobulin | |||
| Less than 3.5 mg/L | 60 | 59 | |
| At least 3.5 and less than 5.5 mg/L | 14 | 20 | |
| 5.5 mg/L or more | 26 | 21 | NS |
| Albumin less than 3.5 g/dl | 17 | 14 | NS |
| Hemoglobin less than 10 g/dL | 23 | 26 | NS |
| Bone marrow plasma cells (by aspiration) 33% or greater | 61 | 66 | NS |
| Chromosomal abnormalities (defined by G-banding) | 40 | 34 | NS |
| Deletion of chromosome 13 | 84 | 46 | < .001 |
| Amplification of 1q21 | 75 | 42 | .002 |
| High-risk model (17-gene)* | 19 | 14 | NS |
| Subgroups with poor prognosis* (Proliferation/MMSET/FGFR3/MAF/MAFB) | 59 | 35 | .023 |
MRI indicates magnetic resonance imaging; CRP, C-reactive protein; LDH, lactate dehydrogenase; and NS, not significant.
χ2 was used to compare the clinical and biologic parameters between cases with the lowest 10% of TP53 expression and the other 90% of cases with higher expression levels.
In the context of a recently defined molecular subgroup classification,25 the proportion of cases with low levels of TP53 expression was greater in the high-risk molecular subgroups than in the low-risk subgroups. The high-risk molecular subgroups included the MMSET subtype with a t(4;14) translocation, the MAF/MAFB subtype with a t(14;16) or a t(14;20) translocation, and the proliferation subtype; the low-risk groups consisted of subtypes designated hyperdiploid or low bone disease or marked by CCND1/CCND3 spike signatures (CD-1 or CD-2) (59% vs 35%; P = .023). In the context of molecular risk stratification based on 17 genes,1 TP53 Affymetrix signal < 733 was seen in 30 (9.8%) of 305 low-risk and in 6 (13%) of 46 high-risk-disease cases. Low TP53 expression adversely affected both EFS and OS in low-risk disease. Because of the small sample size, it is not clear whether low TP53 expression influences survival in high-risk disease (Figure S1C). Given the strong correlation between low TP53 expression and high-risk MM subtypes, we investigated whether low TP53 expression levels simply reflected the poor prognostic features of high-risk MM1 or whether it held independent prognostic significance. In a multivariate analysis, low TP53 gene expression was an independent poor-prognostic factor with respect to both EFS and OS (Table 2). Thus, although associated with a number of high-risk-MM features, reduced TP53 gene expression independently confers a poor clinical outcome.
Multivariate analysis of clinical characteristics affecting OS and EFS
| Variable . | n/N (%) . | Overall survival . | Event-free survival . | ||
|---|---|---|---|---|---|
| HR (95% CI) . | P† . | HR (95% CI) . | P† . | ||
| TT2 | |||||
| Creatinine ≥ 2.0 mg/dL | 37/334 (11) | 1.75 (1.07, 2.84) | .024 | 1.70 (1.19, 2.43) | .004 |
| LDH ≥ 190 U/L | 114/334 (34) | 1.81 (1.24, 2.66) | .002 | 1.44 (1.13, 1.82) | .003 |
| Cytogenetic abnormalities | 108/334 (32) | 1.77 (1.20, 2.62) | .004 | NS | NS |
| Randomization to thalidomide | 166/334 (50) | NS | NS | 0.75 (0.60, 0.93) | .009 |
| t(4;14) | 47/334 (14) | 1.81 (1.15, 2.85) | .010 | 1.87 (1.35, 2.57) | < .001 |
| 17 gene-defined GEP high-risk* | 50/334 (15) | 2.47 (1.58, 3.85) | < .001 | 2.15 (1.56, 2.96) | < .001 |
| TP53 high-risk | 35/334 (10) | 2.01 (1.22, 3.30) | .006 | 1.44 (1.00, 2.07) | .049 |
| TT3 | |||||
| Age 65 y or older | 50/176 (28) | 2.32 (1.04, 5.22) | .041 | NS | NS |
| β2-microglobulin > 5.5 mg/L | 38/176 (22) | NS | NS | 3.33 (1.68, 6.62) | < .001 |
| Creatinine ≥ 2.0 mg/dL | 58/176 (33) | 3.54 (1.54, 8.16) | .003 | NS | NS |
| Cytogenetic abnormalities | 30/176 (17) | 2.42 (1.08, 5.39) | .031 | NS | NS |
| 17-gene high-risk | 20/176 (11) | 3.19 (1.32, 7.68) | .010 | 3.97 (1.98, 7.97) | < .001 |
| TP53 high-risk | 50/176 (28) | 2.32 (1.04, 5.22) | .041 | 2.51 (1.13, 5.57) | .024 |
| Variable . | n/N (%) . | Overall survival . | Event-free survival . | ||
|---|---|---|---|---|---|
| HR (95% CI) . | P† . | HR (95% CI) . | P† . | ||
| TT2 | |||||
| Creatinine ≥ 2.0 mg/dL | 37/334 (11) | 1.75 (1.07, 2.84) | .024 | 1.70 (1.19, 2.43) | .004 |
| LDH ≥ 190 U/L | 114/334 (34) | 1.81 (1.24, 2.66) | .002 | 1.44 (1.13, 1.82) | .003 |
| Cytogenetic abnormalities | 108/334 (32) | 1.77 (1.20, 2.62) | .004 | NS | NS |
| Randomization to thalidomide | 166/334 (50) | NS | NS | 0.75 (0.60, 0.93) | .009 |
| t(4;14) | 47/334 (14) | 1.81 (1.15, 2.85) | .010 | 1.87 (1.35, 2.57) | < .001 |
| 17 gene-defined GEP high-risk* | 50/334 (15) | 2.47 (1.58, 3.85) | < .001 | 2.15 (1.56, 2.96) | < .001 |
| TP53 high-risk | 35/334 (10) | 2.01 (1.22, 3.30) | .006 | 1.44 (1.00, 2.07) | .049 |
| TT3 | |||||
| Age 65 y or older | 50/176 (28) | 2.32 (1.04, 5.22) | .041 | NS | NS |
| β2-microglobulin > 5.5 mg/L | 38/176 (22) | NS | NS | 3.33 (1.68, 6.62) | < .001 |
| Creatinine ≥ 2.0 mg/dL | 58/176 (33) | 3.54 (1.54, 8.16) | .003 | NS | NS |
| Cytogenetic abnormalities | 30/176 (17) | 2.42 (1.08, 5.39) | .031 | NS | NS |
| 17-gene high-risk | 20/176 (11) | 3.19 (1.32, 7.68) | .010 | 3.97 (1.98, 7.97) | < .001 |
| TP53 high-risk | 50/176 (28) | 2.32 (1.04, 5.22) | .041 | 2.51 (1.13, 5.57) | .024 |
The multivariate model uses stepwise selection with entry level 0.1 and variable remains if meets the 0.05 level. A multivariate P > .05 indicates a variable forced into the model, with significant variables chosen by stepwise selection.
HR indicates hazard ratio; CI, 95% confidence interval; P, probability value from Wald χ2 test in Cox regression; NS, not statistically significant at the .05 level on multivariate analysis; LDH, lactate dehydrogenase; GEP, gene expression profile; and PI, proliferation index.25
17 gene-defined GEP high-risk has been described elsewhere.1
Variables for which P > .05: age, race, sex, isotype, hemoglobin, C-reactive protein, MRI lesions, and albumin.
Effects of overexpressing TP53 on MM cell growth and survival
Recently, PET analysis in a colorectal cancer cell line identified 122 TP53 target genes, whose TP53-dependent expression was verified in breast tumors.20 However, expression of only a few of the previously identified TP53 target genes was correlated with TP53 expression in MM cells (Figure S2A). This suggested that TP53 might regulate a distinct set of genes in MM. To elucidate the TP53 regulatory networks in MM, we used lentiviral transduction to overexpress TP53 in 4 MM cell lines: OCI-MY5, JJN3, ARP-1, and Delta 47.
Stable expression of TP53 in OCI-MY5 cells was confirmed by Western blot 24 hours after lentiviral infection (Figure 2A,B). To verify that TP53 was capable of activating target genes, TP53 DNA binding activity was examined. These studies confirmed that TP53 overexpression was correlated with increased DNA binding activity at 24 hours after lentiviral infection (data not shown).
Effect of TP53 overexpression on MM cell survival. (A) TP53 (in both nuclear and cytosolic extracts) and cleaved PARP (in nuclear extracts) were evaluated by Western blot performed in OCI-MY5 cells after lentiviral infection of TP53 and empty vector (EV) at 4, 8, 12, and 24 hours. Histone H4 and β-tubulin were used as loading controls. (B) TP53 and cleaved PARP were evaluated by Western blot performed in OCI-MY5, JJN3, ARP-1, and Delta 47 cells after lentiviral infection of TP53 and empty vector (EV) at 24 hours. (C) Effect of overexpression of TP53 on cell viability in JJN3, OCI-MY5, ARP-1, and Delta 47 cells. Cell viability was evaluated by trypan blue exclusion every 12 hours after lentiviral infection of TP53 compared with the EV. (D) Overexpression of TP53 induces apoptosis. Cell cycle distribution and apoptosis were evaluated by flow cytometry performed 24 hours after lentiviral infection in JJN3, OCI-MY5, ARP-1, and Delta 47 cells infected with EV or TP53 cDNA. Note that overexpression of TP53 induced a dramatic increase in the percentage of cells with sub-G0-phase DNA content (indicative of apoptosis).
Effect of TP53 overexpression on MM cell survival. (A) TP53 (in both nuclear and cytosolic extracts) and cleaved PARP (in nuclear extracts) were evaluated by Western blot performed in OCI-MY5 cells after lentiviral infection of TP53 and empty vector (EV) at 4, 8, 12, and 24 hours. Histone H4 and β-tubulin were used as loading controls. (B) TP53 and cleaved PARP were evaluated by Western blot performed in OCI-MY5, JJN3, ARP-1, and Delta 47 cells after lentiviral infection of TP53 and empty vector (EV) at 24 hours. (C) Effect of overexpression of TP53 on cell viability in JJN3, OCI-MY5, ARP-1, and Delta 47 cells. Cell viability was evaluated by trypan blue exclusion every 12 hours after lentiviral infection of TP53 compared with the EV. (D) Overexpression of TP53 induces apoptosis. Cell cycle distribution and apoptosis were evaluated by flow cytometry performed 24 hours after lentiviral infection in JJN3, OCI-MY5, ARP-1, and Delta 47 cells infected with EV or TP53 cDNA. Note that overexpression of TP53 induced a dramatic increase in the percentage of cells with sub-G0-phase DNA content (indicative of apoptosis).
We then examined the effect of TP53 overexpression on MM cell proliferation and viability. TP53 overexpression decreased cell viability in the 4 MM cell lines within 24 hours (viability, 60%-67%, measured by trypan blue exclusion), and massive cell death occurred within 36 hours (viability, 15%-26%), compared with 90% viability of control MM cells infected with empty vector. At 48 hours after lentiviral transduction, virtually all cells expressing TP53 had died, whereas the control cells continued to proliferate (Figure 2C).
Cell proliferation and apoptosis were quantitatively assessed by flow cytometry. The results showed that TP53 expression induced strong apoptosis at 24 hours after infection (Figure 2D). Analysis of apoptotic mechanisms revealed that TP53 overexpression in MM cells was also associated with cleavage of PARP, an apoptotic marker (Figure 2A,B).
On the basis of our analysis of protein expression and DNA binding activity, we chose to identify TP53-regulated genes 24 hours after lentiviral infection. Therefore, gene expression levels were profiled in the 4 MM cell lines (JJN3, OCI-MY5, ARP-1, and Delta 47) at 24 hours after lentiviral infection to identify potential TP53-regulated genes.
Identification and classification of genes associated with TP53 expression in MM
Gene expression profiling revealed that a total of 85 genes were affected by TP53 overexpression (50 being up-regulated and 35 down-regulated) of 1.5-fold or greater in at least 3 of the 4 MM cell lines. Consistent with TP53 cellular functions, we found that 69 of the 85 genes in MM were involved in apoptosis, cell cycle regulation, cell growth and differentiation, DNA repair and chromatin modification, or transcription regulation (Table 3; Figure 3A). The 85 genes associated with TP53 overexpression also exhibited differential expression in primary MM, when the lowest relative to the highest TP53 expressers were compared (Figure 3B; Table S1). These data suggest that TP53 may directly or indirectly regulate the expression of these genes. Consistent with the independent prognostic role of TP53 loss in MM, none of the differentially expressed genes was identified in our recently described 70-gene high-risk model.1 To identify the most relevant biologic mechanisms, pathways, and functional categories of the 85 genes affected by TP53 expression, we used Ingenuity Pathways Analysis software (Ingenuity Systems, Redwood City, CA). Three networks were identified, representing proteins involved in cancer and the cell cycle (Figure 4); cell cycle and cellular movement, assembly, and organization (Figure S3A); and cell morphology and DNA replication, recombination, and repair (Figure S3B).
Categories of TP53-associated genes in MM cell lines and newly diagnosis primary tumors*
| Apoptosis . | Cell cycle . | DNA repair/chromatin modifier . | Cell growth differentiation . | Signal transduction . | Biosynthesis metabolism . | Posttranslational modification . | Transcription regulation . | Transport and ion channel . | Unknown . |
|---|---|---|---|---|---|---|---|---|---|
| TP53 | APP | IFIT5 | FXYD5 | ARHGAP25 | STT3B | SAT2 | ZNF600 | LAPTM4B | CYB5D2 |
| TRIM13 | ABCB9 | ANKRA2 | MCC | RTN2 | ECHDC2 | SLAMF7 | L3MBTL4* | OSBPL10 | UNC93B1 |
| NADSYN1 | GAA | PHLDB1 | MKNK2 | WNT16 | ENPP4* | MAN1C1 | KCNS3 | SIDT1 | |
| TRIM22† | CEP55* | TUBA1A* | KLHL24 | DEPDC1* | INTS7* | THEX1* | TMEM57 | ||
| AGRN | BRCA1* | CDCA7* | DLC1 | HIGD2A | |||||
| CENTD2 | ANLN* | CDCA2* | OPN3* | FKSG44 | |||||
| SESN1 | PYGL* | HFE* | B3GALNT1* | C14orf28 | |||||
| TM7SF2 | CCNE2* | RIF1* | SPRED1* | LOC387763 | |||||
| NCKAP1 | ASPM* | NEIL3* | TncRNA | ||||||
| COPG | SUV39H2* | SLC4A7* | C18orf1 | ||||||
| STAT3 | CDC25A* | DCUN1D4* | |||||||
| ALOX5 | SEPP1* | FANCI* | |||||||
| ZMAT3† | RIT1* | CD302* | |||||||
| NOTCH1† | KIF2C* | C5orf34* | |||||||
| BTG2 | S100A4* | FAM111B* | |||||||
| RAB1A | MEIS1* | C18orf54* | |||||||
| TNFRSF10B† | SGOL2* | ||||||||
| HDLBP |
| Apoptosis . | Cell cycle . | DNA repair/chromatin modifier . | Cell growth differentiation . | Signal transduction . | Biosynthesis metabolism . | Posttranslational modification . | Transcription regulation . | Transport and ion channel . | Unknown . |
|---|---|---|---|---|---|---|---|---|---|
| TP53 | APP | IFIT5 | FXYD5 | ARHGAP25 | STT3B | SAT2 | ZNF600 | LAPTM4B | CYB5D2 |
| TRIM13 | ABCB9 | ANKRA2 | MCC | RTN2 | ECHDC2 | SLAMF7 | L3MBTL4* | OSBPL10 | UNC93B1 |
| NADSYN1 | GAA | PHLDB1 | MKNK2 | WNT16 | ENPP4* | MAN1C1 | KCNS3 | SIDT1 | |
| TRIM22† | CEP55* | TUBA1A* | KLHL24 | DEPDC1* | INTS7* | THEX1* | TMEM57 | ||
| AGRN | BRCA1* | CDCA7* | DLC1 | HIGD2A | |||||
| CENTD2 | ANLN* | CDCA2* | OPN3* | FKSG44 | |||||
| SESN1 | PYGL* | HFE* | B3GALNT1* | C14orf28 | |||||
| TM7SF2 | CCNE2* | RIF1* | SPRED1* | LOC387763 | |||||
| NCKAP1 | ASPM* | NEIL3* | TncRNA | ||||||
| COPG | SUV39H2* | SLC4A7* | C18orf1 | ||||||
| STAT3 | CDC25A* | DCUN1D4* | |||||||
| ALOX5 | SEPP1* | FANCI* | |||||||
| ZMAT3† | RIT1* | CD302* | |||||||
| NOTCH1† | KIF2C* | C5orf34* | |||||||
| BTG2 | S100A4* | FAM111B* | |||||||
| RAB1A | MEIS1* | C18orf54* | |||||||
| TNFRSF10B† | SGOL2* | ||||||||
| HDLBP |
Down-regulated gene.
Known TP53 target.
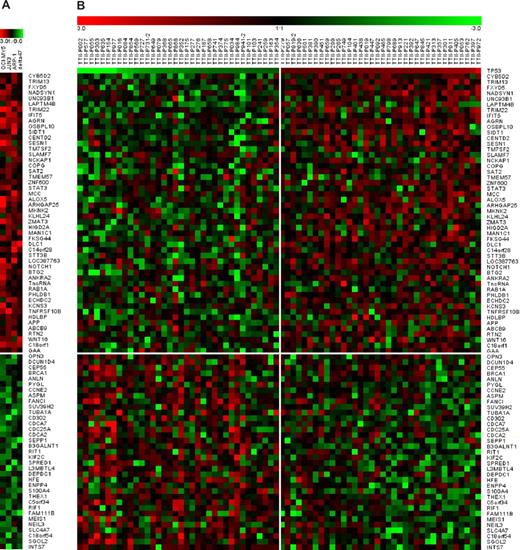
Gene expression profile of 85 TP53-regulated genes in MM cell lines and patients. A total of 85 genes were up-regulated (n = 50) or down-regulated (n = 35) at least 1.5-fold in at least 3 of 4 MM cell lines and also exhibited differential expression in a comparison of primary MM between the lowest relative to the highest TP53 expressers. (A) In the normalized log ratio (TP53 overexpression vs empty vector) of the 4 MM cell lines (JJN3, OCI-MY5, ARP-1, and Delta 47), red represents induction and green represents repression. (B) Differences in gene expression between 36 patients on TT2 with lowest TP53 expression and 36 patients with the highest TP53 expression.
Gene expression profile of 85 TP53-regulated genes in MM cell lines and patients. A total of 85 genes were up-regulated (n = 50) or down-regulated (n = 35) at least 1.5-fold in at least 3 of 4 MM cell lines and also exhibited differential expression in a comparison of primary MM between the lowest relative to the highest TP53 expressers. (A) In the normalized log ratio (TP53 overexpression vs empty vector) of the 4 MM cell lines (JJN3, OCI-MY5, ARP-1, and Delta 47), red represents induction and green represents repression. (B) Differences in gene expression between 36 patients on TT2 with lowest TP53 expression and 36 patients with the highest TP53 expression.
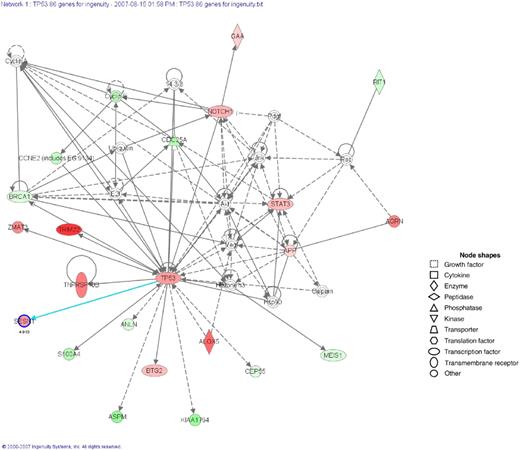
Networks of TP53-regulated genes in MM cell lines and patients. Ingenuity Pathways Analysis software (Ingenuity Systems) was used to analyze the identified genes (n = 85). Three networks were identified. The network representing proteins involved in the biologic functions of cancer and the cell cycle is shown. The genes written in bold letters with a shaded node were identified by microarray analysis, and the other genes were those related to the regulated genes based on the network analysis. The intensity of a node color indicates the degree of up-regulation (red). The meanings of node shapes are indicated in the figure.
Networks of TP53-regulated genes in MM cell lines and patients. Ingenuity Pathways Analysis software (Ingenuity Systems) was used to analyze the identified genes (n = 85). Three networks were identified. The network representing proteins involved in the biologic functions of cancer and the cell cycle is shown. The genes written in bold letters with a shaded node were identified by microarray analysis, and the other genes were those related to the regulated genes based on the network analysis. The intensity of a node color indicates the degree of up-regulation (red). The meanings of node shapes are indicated in the figure.
From the group of 122 TP53 target genes identified by PET analysis,20 only 11 up-regulated genes were consistently expressed in all 4 MM cell lines (≥ 1.5-fold in at least 3 of the 4 MM cell lines; Figure S2B), and only 4 of these 11 genes (ZMAT3, TNFRSF10B, TRIM22, and NOTCH1) were correlated with TP53 expression in primary MM cells.
Clinical relevance of genes associated with TP53 expression
Using the 85 genes identified as associated with TP53 overexpression and the expression data derived from the 351 TT2 patients with newly diagnosed MM, we performed unsupervised hierarchical clustering. This resulted in 2 primary tumor clusters that were significantly associated with TP53 expression, with TP53 Affymetrix signal from 10 to 2898 in lower TP53 expression cluster versus 226 to 5241 in the higher TP53 expression cluster (P < .001, Figure 5A). The subtype associated with lower TP53 expression had a significantly shorter EFS (P = .001; Figure 5B) and OS (P = .001; Figure 5C).
Gene expression profiles of TP53-regulated genes and their clinical relevance. (A) Two-dimensional unsupervised hierarchical cluster analysis of 85 (rows) TP53-regulated genes in CD138-enriched plasma cells of newly diagnosed MM patients on TT2 (n = 351). The right branch consists of MM samples that have a gene expression profile associated with a high TP53 expression level (horizontal green bar), and the left branch contains MM samples that have a gene expression profile associated with low TP53 expression level (horizontal red bar). Kaplan-Meier estimates of (B) EFS and (C) OS in newly diagnosed MM patients on TT2 show superior 5-year actuarial probabilities of EFS (51% vs 39%; P = .001) and OS (69% vs 46%; P = .001) in the right-branch patients whose 85-gene expression profile was associated with a high TP53 expression level.
Gene expression profiles of TP53-regulated genes and their clinical relevance. (A) Two-dimensional unsupervised hierarchical cluster analysis of 85 (rows) TP53-regulated genes in CD138-enriched plasma cells of newly diagnosed MM patients on TT2 (n = 351). The right branch consists of MM samples that have a gene expression profile associated with a high TP53 expression level (horizontal green bar), and the left branch contains MM samples that have a gene expression profile associated with low TP53 expression level (horizontal red bar). Kaplan-Meier estimates of (B) EFS and (C) OS in newly diagnosed MM patients on TT2 show superior 5-year actuarial probabilities of EFS (51% vs 39%; P = .001) and OS (69% vs 46%; P = .001) in the right-branch patients whose 85-gene expression profile was associated with a high TP53 expression level.
With regard to clinical and biologic features, the subtype of patients associated with low TP53 expression had high levels of creatinine (P = .011) and lactate dehydrogenase (P = .017), low levels of albumin (P = .041), an increased number of bone lesions on magnetic resonance imaging (P = .001), and an increased incidence of chromosomal abnormalities defined by G-banding (P = .002), deletion of chromosome 13 (P < .001), and amplification of chromosome 1q21 (P = .002) (Table 4).
Baseline TT2 patient characteristics according to TP53-regulated gene cluster
| Characteristic . | Cluster 1, % (n = 98) . | Cluster 2, % (n = 253) . | P† . |
|---|---|---|---|
| Age at least 65 y | 21 | 22 | NS |
| Female sex | 42 | 44 | NS |
| White race | 91 | 88 | NS |
| IgA isotype | 30 | 24 | NS |
| Creatinine at least 2.0 mg/dL | 19 | 9 | .011 |
| MRI focal bone lesions, at least 3 | 72 | 53 | .001 |
| C-reactive protein at least 4.0 mg/L | 45 | 32 | .023 |
| Lactate dehydrogenase at least 190 IU/L | 44 | 30 | .017 |
| β2-microglobulin at least 4.0 mg/L | 38 | 33 | NS |
| Albumin less than 3.5 g/dL | 45 | 33 | .041 |
| Hemoglobin less than 10 g/dL | 31 | 23 | NS |
| Bone marrow plasma cells (by aspiration) at least 33% | 47 | 56 | NS |
| Chromosomal abnormalities (defined by G-banding) | 48 | 30 | .002 |
| Deletion of chromosome 13 | 66 | 43 | < .001 |
| Amplification of chromosome 1q21 | 65 | 36 | .002 |
| High-risk model (17-gene)* | 36 | 7 | < .001 |
| Subgroups with poor prognosis* (Proliferation/MMSET/FGFR3/MAF/MAFB) | 50 | 51 | NS |
| Characteristic . | Cluster 1, % (n = 98) . | Cluster 2, % (n = 253) . | P† . |
|---|---|---|---|
| Age at least 65 y | 21 | 22 | NS |
| Female sex | 42 | 44 | NS |
| White race | 91 | 88 | NS |
| IgA isotype | 30 | 24 | NS |
| Creatinine at least 2.0 mg/dL | 19 | 9 | .011 |
| MRI focal bone lesions, at least 3 | 72 | 53 | .001 |
| C-reactive protein at least 4.0 mg/L | 45 | 32 | .023 |
| Lactate dehydrogenase at least 190 IU/L | 44 | 30 | .017 |
| β2-microglobulin at least 4.0 mg/L | 38 | 33 | NS |
| Albumin less than 3.5 g/dL | 45 | 33 | .041 |
| Hemoglobin less than 10 g/dL | 31 | 23 | NS |
| Bone marrow plasma cells (by aspiration) at least 33% | 47 | 56 | NS |
| Chromosomal abnormalities (defined by G-banding) | 48 | 30 | .002 |
| Deletion of chromosome 13 | 66 | 43 | < .001 |
| Amplification of chromosome 1q21 | 65 | 36 | .002 |
| High-risk model (17-gene)* | 36 | 7 | < .001 |
| Subgroups with poor prognosis* (Proliferation/MMSET/FGFR3/MAF/MAFB) | 50 | 51 | NS |
NS indicates not significant.
χ2 was used to compare the clinical and biologic parameters between cases in hierarchical cluster 1 (correlated with low TP53 expression) and cluster 2 (correlated with high TP53 expression).
By the same unsupervised hierarchical clustering, the subcluster of MM associated with lower TP53 expression levels had a significantly shorter postrelapse survival (P < .001; Figure S4A) in 90 TT2 cases with relapsed disease and short EFS (P = .001) and OS (P = .053) in a separate cohort of 214 patients treated on the successor protocol TT3 (Figure S4B).
Taken together, these results strongly suggest that the 85 putative TP53-target genes may play an important role in myeloma progression associated with low TP53 expression.
No significant increase in deletion of TP53 or decrease in expression of TP53 in relapsed disease
TP53 deletion did not increase in frequency or extent in a paired comparison of 28 baseline and relapse samples (data not shown). Fifty-one patients had TP53 gene expression data available at both diagnosis and relapse. Consistent with paired FISH results, in these 51 patients, there were also no significant differences in TP53 gene expression at baseline compared with expression at relapse, only 8 patients had at least a 2-fold change in TP53 expression level, 5 had a decreased level, and 3 had an increased level at relapse (Figure 6A).
TP53 expression and its associated gene expression profile in 51 paired MM patients on TT2 at baseline and disease relapse. (A) A total of 51 paired TT2 patients had similar TP53 expression levels at baseline and relapse (P = .455); only 8 patients had at least a 2-fold TP53 signal change (5 decreased and 3 increased) at relapse. (B) Two-dimensional unsupervised hierarchical cluster analysis of 85 TP53-regulated genes in 51 MM patients with paired gene expression data at baseline and relapse shows that 36 of 51 patients have very similar gene expression patterns of the 85 TP53-regulated genes, and these genes cluster closely together (marked by red bracket). BL indicates baseline; and RL, relapse.
TP53 expression and its associated gene expression profile in 51 paired MM patients on TT2 at baseline and disease relapse. (A) A total of 51 paired TT2 patients had similar TP53 expression levels at baseline and relapse (P = .455); only 8 patients had at least a 2-fold TP53 signal change (5 decreased and 3 increased) at relapse. (B) Two-dimensional unsupervised hierarchical cluster analysis of 85 TP53-regulated genes in 51 MM patients with paired gene expression data at baseline and relapse shows that 36 of 51 patients have very similar gene expression patterns of the 85 TP53-regulated genes, and these genes cluster closely together (marked by red bracket). BL indicates baseline; and RL, relapse.
Interestingly, 36 of 51 patients had very similar gene expression patterns of the 85 TP53-associated genes at baseline and relapse. When the gene expression data on the 51 paired baseline and relapse cases were combined and unsupervised hierarchical clustering was performed, the data clustered closely together (Figure 6B).
TP53 mutation is a rare and late event in MM
TP53 mutations were detected by sequencing exons 2 to 9 in 44 patients, 24 of whom had newly diagnosed disease and 27 had relapsed disease; in 7 of the 44 cases, there were paired baseline and relapse samples. No mutations were detected in the 24 newly diagnosed cases or the 7 paired baseline-relapse samples, whereas mutations were seen in exons 7, 8, and 9 in 5 of 20 unpaired relapsed-disease samples (P = .014).
Discussion
We recently described a powerful prognostic model in MM based on the expression of 17 genes.1 This risk-stratification model for newly diagnosed MM treated with high-dose chemotherapy was also predictive of outcome in relapsed disease with the single agents bortezomib or high-dose dexamethasone37 or high-dose therapy.38 Interestingly, TP53 gene expression was not one of the genes in this model perhaps because of the method of model development using expression quartiles. In the current study, we found that using a 10% cutoff (rather than the 25% and 75% cutoff points used to identify the genes high risk model), tumors with TP53 expression levels in the lowest 10th percentile were associated with a significantly shorter EFS and OS than those in the 90th percentile. We went on to show that low expression levels of TP53 were correlated with monoallelic or biallelic deletion of the TP53 locus. Multivariate regression analyses revealed that low TP53 expression was an independent adverse prognostic factor in 2 independent trials, TT2 and TT3. It is noteworthy that in the multivariate analysis we found that, although the t(4:14) translocation was a significant variable in TT2, it was not retained as an independent factor in TT3 (Table 2), and may imply that the use of bortezomib, the only fundamental difference between TT2 and TT3, overcomes negative impact of t(4:14) but cannot overcome the negative impact of low TP53 expression or our high-risk 17-gene model.24 Low TP53 expression was able to further dissect the survival of low-risk patients defined by the 17-gene model (Figure S1A). These data add to the continued refinement of molecular prognostics in MM.
In addition to identifying TP53 as a poor prognostic factor, this study also provides, for the first time, a comprehensive list of genes that are differentially expressed in association with TP53 expression in MM. The TP53 tumor suppressor gene plays a key role in prevention of tumor formation through transcriptional-dependent and -independent mechanisms. Transcriptional-dependent mechanisms are mainly mediated by TP53 regulation of downstream targets, leading to growth arrest and apoptosis.39 Recently, a global map of TP53 transcription factor binding sites in the human genome was identified in a colorectal cancer cell line by PET analysis, and 122 TP53 target genes were characterized.20 Their TP53-dependent expression was verified in breast cancer patients20 ; however, expression of only a few of these genes was correlated with TP53 expression in MM cells. This suggested that TP53 might regulate a distinct set of genes in MM and for that matter may regulate a unique set of tissue specific genes. By overexpressing TP53 in TP53-negative MM cell lines, we identified 85 differentially expressed genes associated with TP53 expression. Importantly, the expression of these same genes was correlated with TP53 expression and with clinical outcome in primary disease. None of the 85 TP53-associated genes was included in our previous high-risk 70-gene model.1 This is consistent with the finding that TP53 expression is an independent prognostic variable and may complement our recently described high-risk model.
It is noteworthy that 69 of the 85 TP53-regulated genes have a defined function in apoptosis and the cell cycle, DNA repair, chromatin modification, cell growth, cell differentiation, and transcriptional regulation. Identification and characterization of these genes and their pathways may lead to a better understanding of the critical role of TP53 loss in MM. TP53-induced growth arrest is achieved mainly by transactivation of p21 (for G1-phase arrest), of 14-3-3σ (for G2-phase arrest), or of placenta transforming growth factor-β. Under a transcriptional-dependent mechanism, TP53 induces apoptosis by transactivating the genes in both mitochondrial and death receptor pathways, as well as transrepressing cellular survival genes.39 The results of our analysis show that TP53 up-regulates death receptor pathway apoptotic genes (eg, TNFRSF10B) and down-regulates cell cycle genes (eg, BRCA1, cyclin E, S100A4, and CDCs) in MM.
Of the 85 genes associated with expression of TP53 in MM, only 4 genes, TNFRSF10B, NOTCH1, ZMAT3, and TRIM22, were previously identified as TP53 target genes. Both TNFRSF10B and NOTCH1 gene products are cell membrane proteins. TNFRSF10B, also named KILLER/DR5, is a member of the tumor necrosis factor-receptor superfamily and plays a key role in the death receptor pathway. It is located in a minimal region of loss at 8p21.3-p12 in MM.2 TNFRSF10B is a TP53-inducible receptor for the cytotoxic ligand TNFSF10/TRAIL and induces caspase-dependent apoptotis.40 A recombinant form of the death ligand TRAIL is not cytotoxic for normal human cells and is a good candidate for the treatment of MM.41,42 NOTCH1 functions as a receptor for membrane-bound ligands Jagged1, Jagged2, and Delta1 to regulate cell-fate determination, and affects the implementation of differentiation, proliferation, and apoptotic programs.43 Recent results show that NOTCH1 signaling is involved in bone marrow stroma-mediated de novo drug resistance in MM.44 ZMAT3, also named WIG1, is a TP53-regulated gene that encodes a growth inhibitory zinc finger protein.45 Wig-1 can bind short-interfering/micro RNAs in vitro, which raises the possibility that it is involved in miRNA-mediated regulation of cell growth and survival, acting to promote TP53-induced cell growth arrest and/or apoptosis.46 TRIM22, and another TRIM/RBCC family member, TRIM13, were identified as being associated with TP53 expression in the current study. The interferon-inducible protein TRIM22 has been identified as a TP53 target gene, with possible involvement in hematopoietic proliferation and differentiation.47 TRIM13 is one of the most likely candidates for tumor suppressor gene for B-cell chronic lymphocytic leukemia.48 TRIM13 has also been found to exhibit copy number-sensitive expression in MM.2
We found no significant differences in TP53 deletion and expression at baseline and in relapsed disease in 51 paired samples, and it is noteworthy that most (36 of 51) paired samples had a gene expression pattern similar to that observed when TP53 is expressed. This result may imply that current strategies for MM have no ability to alter expression of TP53 and its associated genes. We also provide evidence that 90% of TP53 deletions in MM are monoallelic deletions. Furthermore, TP53 mutation is not a frequent event in MM.3-7 Consistent with previous studies, TP53 mutation was not detected in our 24 newly diagnosed patients. We have been able to show that overexpression of TP53 can induce strong apoptosis in vitro. Taken together, our results may inform discovery of an ideal strategy for induction of apoptosis in apoptosis-resistant cancer cells through the modulation of TP53 or MM-specific TP53 signaling pathways.
In conclusion, we have demonstrated that low TP53 gene expression is strongly correlated with 17p13 deletion and is an independent adverse prognostic marker in newly diagnosed MM treated with autotransplantation. In addition, we have identified by expression profiling, TP53-regulated genes in both cultured MM cells and primary tumors that are correlated with clinical outcome. Our data suggest that low levels of expression of TP53 and its regulated genes are associated with a malignant phenotype in MM, and these findings may provide important new insights into the role of TP53 loss in MM pathogenesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Hongwei Xu, Yan Xiao, David R. Williams, and Christopher Randolph for assistance, and the Office of Grants and Scientific Publications at the University of Arkansas for Medical Sciences for editorial assistance during the preparation of this manuscript.
This work was supported by National Institutes of Health grants CA55819 (J.D.S., F.Z., B.B.) and CA97513 (J.D.S.), and by the Fund to Cure Myeloma and the Peninsula Community Foundation.
National Institutes of Health
Authorship
Contribution: J.D.S. and F.Z. designed the research, analyzed and interpreted the data, and drafted the manuscript; W.X. designed and performed the research, analyzed and interpreted the data, and drafted the manuscript; X.W., S.S., S.K.J., J.H., S.W., and L.C. performed the research; and B.B. performed the research, contributed clinical samples, analyzed and interpreted the data, and drafted the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Fenghuang Zhan, Department of Hematology, School of Medicine, University of Utah, Salt Lake City, UT 84132; e-mail: fenghuang.zhan@hsc.utah.edu.