Abstract
Rhesus macaque rhadinovirus (RRV) is closely related to Kaposi sarcoma-associated herpesvirus (KSHV) and is associated with the development of B-cell hyperplasia and persistent lymphadenopathy resembling multicentric Castleman disease in rhesus macaques (RMs) coinfected with simian immunodeficiency virus (SIV). Here we investigated whether RMs experimentally infected with SIV and RRV can develop other disease manifestations observed in HIV- and KSHV-infected patients. As reported earlier, inoculation of SIV-infected RMs with RRV results in persistent RRV infection, whereas immunocompetent animals infected with RRV exhibit viremia 2 weeks after infection, followed by a period of no virus detection until they are subsequently made immunodeficient by SIV infection. A subset of animals developed abnormal cellular proliferations characterized as extranodal lymphoma and a proliferative mesenchymal lesion. In situ hybridization and immunohistochemistry analysis indicate RRV is present in both malignancies, and DNA microarray analysis detected viral interleukin-6 (vIL-6) and viral FLICE-like inhibitory protein (vFLIP) transcripts. Reverse-transcriptase polymerase chain reaction analysis confirmed vIL-6 and vFLIP expression, and that of RRV open reading frames 72 and 73, homologs of KSHV open reading frames shown to be expressed in primary effusion lymphoma. These data support the utility of the RRV-/SIV-infected RM as an excellent animal model to investigate KSHV-like pathogenesis.
Introduction
Kaposi sarcoma-associated herpesvirus (KSHV) is widely considered to be the etiologic agent of Kaposi sarcoma (KS),1 primary effusion lymphoma (PEL),2 multicentric Castleman disease (MCD),3 and some non-Hodgkin lymphomas (NHLs)4 in patients infected with the human immunodeficiency virus (HIV). The ability of KSHV to induce these different disease manifestations suggests that KSHV possesses a complex pathogenic potential. Indeed, DNA sequence analysis of the virus genome reveals that the virus encodes several open reading frames (ORFs) considered to be associated with these disease manifestations.5,6 Deciphering the role of each ORF either alone or in concert with the whole virus is difficult in the absence of a reliable animal model that closely recapitulates KSHV-associated syndromes. Unfortunately, an effort to develop an experimental animal model via inoculation with KSHV has not resulted in success, despite evidence of infection.7 Several laboratories have developed small animal models, some of which can evaluate the potential contribution specific viral ORFs have in abnormal cellular processes. For example, transgenic mice models encoding KSHV ORF74, the viral G-protein coupled receptor or KSHV ORF73, the latency-associated nuclear antigen (LANA), have developed abnormal cellular proliferations that resemble those observed in humans.8-10 Although these rodent models are valuable and provide considerable information, they do not take into account the dynamic host/pathogen interactions that are invoked during a natural infection and development of disease
An alternative is to develop an animal model that naturally harbors a viral pathogen that is phylogenetically related to KSHV. Rhesus macaques (RMs), which are evolutionarily related to humans, harbor a virus referred to as rhesus macaque rhadinovirus (RRV) that is closely related to KSHV.11 RRV, like KSHV, is a gamma-2 herpesvirus, and DNA sequence analysis of RRV reveals that the genome is essentially colinear to KSHV, encoding several of the unique viral ORFs probably associated with disease progression.12 More importantly, an earlier study has shown that experimental infection of rhesus macaque (RM) with simian immunodeficiency virus (SIV) and RRV17577 leads to B-cell lymphoproliferative disease (LPD), characterized as persistent angiofollicular lymphadenopathy, which closely resembles MCD, and hypergammaglobulinemia, both clinical manifestations frequently observed in AIDS patients coinfected with KSHV.13
To determine whether RRV17577 is associated with other malignancies, SIV-infected RMs were experimentally inoculated with RRV17577 or plaque-purified isolates and monitored for disease development. As reported previously, most, if not all, animals infected by this approach develop B-cell hyperplasia; however, a percentage of these animals go on to develop abnormal cellular proliferations presenting as B-cell lymphoma and a proliferative mesenchymal lesion referred to as retroperitoneal fibromatosis (RF).14 Here we characterize the malignancies and demonstrate the involvement of RRV in NHL. Because RRV is closely related to KSHV, this experimental animal infection model represents an ideal model to study KSHV-like disease in an animal model of AIDS pathogenesis.
Methods
Experimental animal infections
All aspects of the animal studies were conducted in accordance with institutional guidelines for animal care and use at the Oregon National Primate Research Center and were performed as described.13 For viral load determination, peripheral blood mononuclear cells (PBMCs) or tissues were isolated at scheduled blood draws, biopsies, or at necropsy and DNA purified and analyzed by real-time polymerase chain reaction (PCR) or cocultured on primary rhesus fibroblasts.15 To screen for the presence of other infectious agents in the lymphomas and retroperitoneal fibromatosis, we used PCR primers specific for rhesus lymphocryptovirus latent membrane protein-113,16 and retroperitoneal fibromatosis-associated herpesvirus DNA polymerase,17 respectively.
Immunohistochemistry and in situ hybridization for viral DNA in tissue sections
Sections of formalin-fixed, paraffin-embedded tissues were deparaffinized, hydrated, and prepared for antigen recovery. Slides were blocked with 10% bovine serum albumin and endogenous biotin blocked with Avidin D (Vector Laboratories, Burlingame, CA). Endogenous peroxidase was quenched by standard techniques. Slides were treated with primary antibodies rabbit polyclonal α-CD3 (Dako Denmark, Glostrup, Denmark) or mouse monoclonal α-CD20 (clone L26; Dako Denmark), and mouse IgG1 isotype (R&D Systems, Minneapolis, MN) was used as an isotype control. Detection of biotinylated secondary antibodies was performed with peroxidase ABC (Vector Laboratories) and visualization with DAB (Dako Denmark) for CD20 and Vector SG for CD3 (Vector Laboratories). For detection of RRV by in situ hybridization, slides previously stained for CD3 and CD20 were treated with Proteinase K (100 μg/mL; Promega, Madison, WI). To detect the presence of RRV DNA in tissue, negative control SuperCos DNA and RRV cosmid clones 28b and 4412 were biotinylated and used for probes. After hybridization, probes were identified within tissue using peroxidase ABC and Vector VIP (Vector Laboratories). Stained slides were covered with DPX imaging medium (Invitrogen, Carlsbad, CA). Sections were examined using a Zeiss Axioscope 2 plus microscope (Carl Zeiss, Thornwood, NY) using Plan NeoFluar objective lenses (20×/0.5 NA, 40×/0.75 NA and 63×/1.4 NA). Optical images were obtained with standard conditions of illumination and exposure on Zeiss Axiocam camera (Carl Zeiss). Images were acquired using Metamorph software (Molecular Devices, Sunnyvale, CA) and were processed with Adobe Illustrator C52 version 12.0.1 software (Adobe Systems, San Jose, CA).
RNA isolation and hybridization
RNA was isolated from mammary lymphoma tissue from animal 19185 using a commercial RNA isolation kit (Ambion, Austin, TX). Two separate preparations of lymphoma-derived RNA were subsequently quantified and amplified by the Oregon Health & Science University Shared Gene Microarray facility, using linear T7 amplification (MessageAmp; Ambion). Target is generated by reverse transcription to synthesize a cDNA containing aminoallyl-modified dUTP (CyScribe Post-labeling; GE Healthcare, Little Chalfont, United Kingdom). Aminoallyl-modified cDNA is incubated with Cy-dye esters for a nonenzymatic, covalent attachment of either Cy5 or Cy3 to the cDNA, and applied to each of 2 identical slides containing 252 70mers (QIAGEN, Valencia, CA), representing 84 RRV ORFs. Each viral ORF was represented by 3 70mers to detect each transcript or a transcript going in the predicted antisense direction. RRV-specific slides were printed on aminosilane-coated slides (Corning, Corning, NY) using the Cartesian PixSys 5500 XL microarray printer. The oligos were chosen with a 3′ bias and compared against the National Center for Biotechnology Information database for alignment to the genome sequence of the RRV17577 and for possible cross-hybridization to cellular sequences. DNA sequences for each of the oligos are included in the Supplemental data (available on the Blood website; see the Supplemental Materials link at the top of the online article). Arrays are hybridized using M-series LifterSlips (Erie Scientific, Portsmouth, NH), and hybridized arrays are scanned on a ScanArray 4000 XL (PerkinElmer Life and Analytical Sciences, Waltham, MA), using ScanArray Express software. Images are stored as TIFF files and analyzed using ImaGene (BioDiscovery, El Segundo, CA). Data derived from each replicate sample are provided in the Supplemental data. Microarray data have been deposited at Gene Expression Omibus under accession number GSE12608.
Confirmation of microarray results was performed using lymphoma-derived RNA. RNA was pretreated with DNase (Ambion) before reverse transcriptase (RT)–PCR using primers specific to ORF-R2 (viral interleukin-6 [vIL-6]): 5′-CCAACGCACGCACGGTTTTTTC-3′ and 5′-CGCTGGTTCGTTTACCTGTGCC-3′), ORF-71 (viral FLICE-like inhibitory protein [vFLIP]: 5′-ATCGAGAGGCTCCGGTCCAG-3′ and 5′-TTGGC-GGCCACGACCTT-3′), ORF-72 (v-cyclin: 5′-CCAGGTGGTGGAGTCTGTTCACAAA-3′ and 5′-GAACCGCGGGCGTTACTTAAAAT-3′), ORF-73 (LANA: 5′-AGTCATCTTTTGTAACTCAGACC C-3′ and 5′-GAATTGGCAGTCCTCTGTCCAT-3′) and glyceraldehyde-3-phosphate dehydrogenase as a positive control for RNA. RT-PCR was performed with SuperScript III One-Step RT-PCR System with Platinum Taq DNA Polymerase (Invitrogen). As control for DNA contamination, the RNA sample was subjected to PCR amplification without reverse transcriptase.
Immunofluorescence staining for viral IL-6
vIL-6-positive cells were detected with a mouse monoclonal α-vIL-6 (clone #17D5 developed in Vaccine and Gene Therapy Institute Antibody Core facility) overnight at room temperature. Mouse IgG1 isotype antibody (R&D Systems) was used as a background staining control. Sections were washed and subsequently incubated with monovalent goat antimouse Fab fragment antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA) for 1 hour at room temperature. The slides were washed and incubated with biotin-conjugated rabbit antigoat secondary antibodies (Vector Laboratories) for 30 minutes at room temperature. Sections were once again washed and then incubated for 30 minutes with peroxidase ABC (Vector Laboratories), washed again, and incubated with streptavidin Alexa Fluor 488 conjugate (Invitrogen) for 30 minutes at room temperature.
vIL-6-stained tissues were blocked with solution of 5% bovine serum albumin + 1% normal horse serum in phosphate-buffered saline for 1 hour at room temperature, and potential biotin binding sites after first round of staining were blocked again with Avidin/Biotin blocking kit (Vector Laboratories). Next, tissue samples were incubated with either mouse monoclonal α-vimentin (clone 3B4; Dako Denmark) or mouse monoclonal α-CD20 or biotinylated goat antihuman IgM (SouthernBiotech, Birmingham, AL) for 1 hour at room temperature. Mouse antibody was followed by biotinylated horse antimouse antibody (Vector Laboratories) for 30 minutes at room temperature. Sections were once again washed and then incubated for 30 minutes with peroxidase ABC (Vector Laboratories), washed again, and incubated with streptavidin Alexa Fluor 594 conjugate (Invitrogen) for 30 minutes at room temperature. Nuclei were visualized with Hoechst dye (Sigma-Aldrich, St Louis, MO). Stained slides were covered with Slow Fade imaging medium (Invitrogen). Pictures were analyzed at room temperature on a Zeiss Axioskop 2 plus epifluorescence microscope equipped with filter cubes documenting specific fluorescein isothiocyanate, Alexa 488, Alexa 594, and Hoechst fluorescence (Carl Zeiss). Images were taken with digital camera (AxioCam; Carl Zeiss), using Plan NeoFluar oil objective lens (63×/1.4 NA), and were edited by OpenLab 4.0.3 (Improvision, Waltham, MA). Images were processed with Adobe Illustrator C52 version 12.0 software (Adobe Systems).
Results
Persistent RRV infection in immunodeficient macaques
We previously reported that experimental RRV infection of immunodeficient RM leads to B-cell hyperplasia and persistent angiofollicular lymphadenopathy resembling MCD.13 To evaluate whether RRV is associated with additional manifestations, 13 animals were assigned to the experimental animal infection study. The animals were separated into 3 cohorts for different experimental infection schemes to evaluate whether order of infection and/or different RRV isolates contribute to diverse clinical outcomes. Three of 5 animals in cohort 2 (cohort 2A) were inoculated intravenously with SIVmac239 to induce an immunodeficient condition in the animals and at 8 weeks after SIV infection or as defined otherwise, the animals were intravenously inoculated with wild-type RRV17577. RRV17577 represents a potential mixture of RRV genotypes isolated from animal 17577 that developed SIV-associated LPD.13 The 2 remaining animals in cohort 2 (cohort 2B) were intravenously inoculated with RRV17577 and 8 weeks later with SIVmac239, to determine whether the order of virus infection is important for clinical syndromes and RRV-associated disease outcomes. Cohort 3 animals were first experimentally inoculated with SIVmac239; and at 8 weeks after SIV infection, the animals were intravenously inoculated with a plaque-purified isolate of RRV17577 that exhibited a small plaque size phenotype on primary rhesus fibroblasts. Cohort 4 animals were infected similarly but with a plaque-purified isolated of RRV17577 that exhibited a large plaque size phenotype.
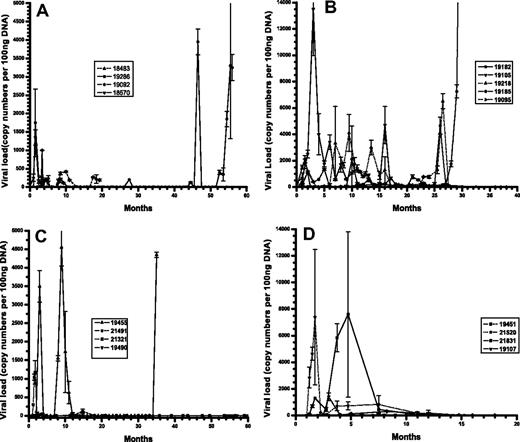
To assess the potential differences in viral kinetics in immunodeficient vs immunocompetent animals, and the potential difference in replicative ability in vivo between different viruses based on plaque size, viral loads were assayed at different times after infection. Total cellular DNA was purified from PBMCs from each animal and samples quantitated for RRV DNA by real-time PCR (Figure 1). Cohort 1 animals, which were described previously,13 showed marked differences in RRV levels throughout the infection period depending primarily on their SIV status. Animals 18483 and 18570, which were SIV-infected, exhibited persistent RRV infection, with RRV levels reaching a high of 400 copies or 1000 copies per 100 ng PBMC-derived DNA. Immunocompetent animals 19092 and 19286 both yielded detectable viral loads shortly after infection and very little thereafter for several years, until 19286 exhibited high levels (18 000 copies) at 54 months after infection, which coincided with detection of lymphoma. Cohort 2A and 2B animals, which were infected with SIV and RRV in different order, exhibited distinct viral loads between each cohort and from animals within cohorts. Specifically, animals 19105 and 19185 exhibited different levels of viral load at 2 months after RRV infection, 14 000 copies per 100 ng DNA versus 1200 copies, respectively. Animal 19185 had high viral loads late during infection, which corresponded to the discovery of widespread B-cell lymphomas. In addition, animals initially infected with RRV displayed differences in persistent infection after SIV infection, with animal 19218 demonstrating higher viral loads throughout the study period, whereas 19182 remained low overall.
RRV load in peripheral blood compartment of experimentally inoculated RMs. Real-time PCR analysis of viral DNA loads using DNA isolated from PBMCs and RRV-specific primers from each cohort of animals. (A) Cohort1. (B) Cohort 2A and 2B. (C) Cohort 3. (D) Cohort 4.
RRV load in peripheral blood compartment of experimentally inoculated RMs. Real-time PCR analysis of viral DNA loads using DNA isolated from PBMCs and RRV-specific primers from each cohort of animals. (A) Cohort1. (B) Cohort 2A and 2B. (C) Cohort 3. (D) Cohort 4.
Animals in cohorts 3 and 4, which were experimentally infected with the plaque-purified isolates, also displayed significant differences between the cohorts and within each cohort. Two of 4 animals in cohort 3 had viral loads of greater than 1000 copies, whereas 3 of 4 animals in cohort 4 had viral loads of at least 1000 copies, with highs of nearly 8000 copies per 100 ng DNA. Cohort 3 animal 19455 displayed high viral load, which was coincident with widespread LPD and MCD detected when the animal became moribund and was killed.
Lymphoma and retroperitoneal fibromatosis in SIV/RRV-infected macaques
To monitor for clinical syndromes associated with RRV and SIV infection, all animals were routinely evaluated by physical examinations and for alterations in lymphocyte subsets (CD4, CD8, and CD20) by fluorescence-activated cell sorter analysis and by real-time PCR or coculture for viral loads. Consistent with our first cohort of animals experimentally inoculated with SIV and/or RRV (animals 18483, 18570, 19092, and 19286), 9 of the 13 animals developed B-cell hyperplasia in the peripheral blood defined as a 2-fold or greater increase in CD20+ lymphocytes (Table 1). The range of B-cell hyperplasia differed among the animals, with animals 19218 and 19451 each exhibiting nearly 6-fold increase in B cells, whereas animals 19185 and 21831 exhibited a 1.3-fold and 1.47-fold increase, respectively. Animals 19107 and 19490 achieved slightly higher increases of 1.91-fold and 1.97-fold, respectively. These data are consistent with our previous cohort of animals (cohort 1, Table 1), which are included for comparison.
Summary of experimental RRV infection studies
| Animal ID . | Virus infection . | CD4+ T-cell preinfection cells/μL . | CD4+ T-cell nadir, cells/μL . | CD20+ B-cell preinfection cells/μL . | CD20+ B-cell at peak, cells/μL . | Survival (days) after RRV infection . | Disease . |
|---|---|---|---|---|---|---|---|
| Cohort 1 | |||||||
| 18483 | SIVmac239/RRV17577 | 1697 | 422 | 899 | 2782 | 531 | RF, MCD |
| 18570 | SIVmac239/RRV17577 | 903 | 270 | 630 | 5543 | 286 | MCD, autoimmune hemolytic anemia |
| 19092 | RRV17577 | 1142 | 693 | 717 | 1246 | Alive | |
| 19286 | RRV17577 | 793 | 190 | 447 | 1686 | 1552 | B-cell lymphoma (liver) |
| Cohort 2 | |||||||
| 19095 | 2A:SIVmac239/RRV17577 | 902 | 1472 | 330 | 960 | 93 | Lymphadenopathy/cryptosporidium |
| 19105 | 2A:SIVmac239/RRV17577 | 788 | 287 | 251 | 1367 | 352 | Staphyloccocal infection in heart, splenomegaly/lymphadenopathy |
| 19185 | 2A:SIVmac239/RRV17577 | 1537 | 162 | 1103 | 1488 | 676 | Widespread B-cell lymphoma/MCD |
| 19182 | 2B:RRV17577/SIVmac239 | 1502 | 5 | 1174 | 2724 | 1135 | Chronic diarrhea, splenomegaly/lymphadenopathy |
| 19218 | 2B:RRV17577/SIVmac239 | 1531 | 93 | 495 | 2964 | 1119 | Splenomegaly/lymphadenopathy |
| Cohort 3 | |||||||
| 19455 | SIVmac239/RRV17577 #3 | 1792 | 188 | 2039 | 5859 | 1331 | MCD |
| 19490 | SIVmac239/RRV17577 #3 | 910 | 502 | 735 | 1449 | Alive | Lymphadenopathy |
| 21321 | SIVmac239/RRV17577 #3 | 2543 | 623 | 906 | 3092 | Alive | |
| 21491 | SIVmac239/RRV17577 #3 | 2196 | 1010 | 1760 | 4090 | 713 | Splenomegaly/lymphadenopathy |
| Cohort 4 | |||||||
| 19107 | SIVmac239/RRV17577 #7 | 1480 | 359 | 937 | 1793 | 680 | Lymphadenopathy |
| 19451 | SIVmac239/RRV17577 #7 | 592 | 99 | 308 | 1796 | 296 | Lymphadenopathy |
| 21520 | SIVmac239/RRV17577 #7 | 2525 | 598 | 1167 | 3870 | 887 | Splenomegaly |
| 21831 | SIVmac239/RRV17577 #7 | 2297 | 588 | 1665 | 2455 | 475 |
| Animal ID . | Virus infection . | CD4+ T-cell preinfection cells/μL . | CD4+ T-cell nadir, cells/μL . | CD20+ B-cell preinfection cells/μL . | CD20+ B-cell at peak, cells/μL . | Survival (days) after RRV infection . | Disease . |
|---|---|---|---|---|---|---|---|
| Cohort 1 | |||||||
| 18483 | SIVmac239/RRV17577 | 1697 | 422 | 899 | 2782 | 531 | RF, MCD |
| 18570 | SIVmac239/RRV17577 | 903 | 270 | 630 | 5543 | 286 | MCD, autoimmune hemolytic anemia |
| 19092 | RRV17577 | 1142 | 693 | 717 | 1246 | Alive | |
| 19286 | RRV17577 | 793 | 190 | 447 | 1686 | 1552 | B-cell lymphoma (liver) |
| Cohort 2 | |||||||
| 19095 | 2A:SIVmac239/RRV17577 | 902 | 1472 | 330 | 960 | 93 | Lymphadenopathy/cryptosporidium |
| 19105 | 2A:SIVmac239/RRV17577 | 788 | 287 | 251 | 1367 | 352 | Staphyloccocal infection in heart, splenomegaly/lymphadenopathy |
| 19185 | 2A:SIVmac239/RRV17577 | 1537 | 162 | 1103 | 1488 | 676 | Widespread B-cell lymphoma/MCD |
| 19182 | 2B:RRV17577/SIVmac239 | 1502 | 5 | 1174 | 2724 | 1135 | Chronic diarrhea, splenomegaly/lymphadenopathy |
| 19218 | 2B:RRV17577/SIVmac239 | 1531 | 93 | 495 | 2964 | 1119 | Splenomegaly/lymphadenopathy |
| Cohort 3 | |||||||
| 19455 | SIVmac239/RRV17577 #3 | 1792 | 188 | 2039 | 5859 | 1331 | MCD |
| 19490 | SIVmac239/RRV17577 #3 | 910 | 502 | 735 | 1449 | Alive | Lymphadenopathy |
| 21321 | SIVmac239/RRV17577 #3 | 2543 | 623 | 906 | 3092 | Alive | |
| 21491 | SIVmac239/RRV17577 #3 | 2196 | 1010 | 1760 | 4090 | 713 | Splenomegaly/lymphadenopathy |
| Cohort 4 | |||||||
| 19107 | SIVmac239/RRV17577 #7 | 1480 | 359 | 937 | 1793 | 680 | Lymphadenopathy |
| 19451 | SIVmac239/RRV17577 #7 | 592 | 99 | 308 | 1796 | 296 | Lymphadenopathy |
| 21520 | SIVmac239/RRV17577 #7 | 2525 | 598 | 1167 | 3870 | 887 | Splenomegaly |
| 21831 | SIVmac239/RRV17577 #7 | 2297 | 588 | 1665 | 2455 | 475 |
RF indicates retroperitoneal fibromatosis; and MCD, multicentric Castleman disease.
Of the 17 experimentally infected animals represented in Table 1, 8 developed persistent splenomegaly and lymphadenopathy resembling MCD (animals 18483, 18570, 19105, 19182, 19218, 19455, 21491, and 21520), and 4 developed persistent lymphadenopathy alone (animals 19095, 19490, 19107, and 19451). More interesting is the finding that 5 animals (29.4%) developed abnormal cellular proliferations, characterized as lymphoma (animals 19185 and 19286), RF (animal 18483; Figure 2A,D,G), and systemic MCD (18483, 18570, 19185, and 19455; Table 1). Of the 3 animals presenting with lymphoma or RF, animal 19286 was infected with RRV alone.
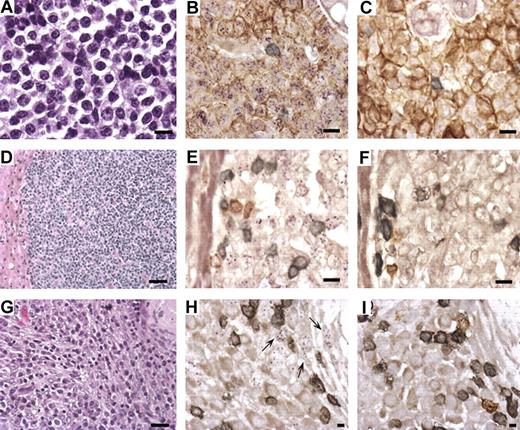
Lymphoma and retroperitoneal fibromatosis (RF) in RRV-infected RMs. (A) Hematoxylin and eosin stain of lymphoma isolated at biopsy from the breast of animal 19185 (original magnification ×630). (B) Combined in situ hybridization of lymphoma with RRV cosmid probe (purple) and immunohistochemistry with mouse anti-CD20 (brown) and anti-CD3+ (gray) demonstrates that RRV is present in CD20+ cells comprising the lymphoma (original magnification ×630). (C) Same as panel B, except cosmid vector control replaces the RRV cosmid probe. (D) Hematoxylin and eosin stain of lymphoma isolated from liver of animal 19286 during necropsy (original magnification ×200). (E) Same as panel B, except RRV cosmid probe detected by immunofluorescence (purple) with no evidence of CD20 staining. (F) Similar to panel C; this is the control for panel E. (G) Hematoxylin and eosin stain of RF attached to the stomach of animal 18483 (original magnification ×400). (H) Combined in situ hybridization with RRV cosmid probe (purple) and immunohistochemistry with anti-CD20 (brown) and anti-CD3 (gray) shows that RRV is not present in the CD20+ or CD3+ cells (original magnification ×1000). Arrows point to RRV-positive cells that have spindle-like morphology. (I) Identical to panel H, except vector control replaces the RRV cosmid (original magnification ×630).
Lymphoma and retroperitoneal fibromatosis (RF) in RRV-infected RMs. (A) Hematoxylin and eosin stain of lymphoma isolated at biopsy from the breast of animal 19185 (original magnification ×630). (B) Combined in situ hybridization of lymphoma with RRV cosmid probe (purple) and immunohistochemistry with mouse anti-CD20 (brown) and anti-CD3+ (gray) demonstrates that RRV is present in CD20+ cells comprising the lymphoma (original magnification ×630). (C) Same as panel B, except cosmid vector control replaces the RRV cosmid probe. (D) Hematoxylin and eosin stain of lymphoma isolated from liver of animal 19286 during necropsy (original magnification ×200). (E) Same as panel B, except RRV cosmid probe detected by immunofluorescence (purple) with no evidence of CD20 staining. (F) Similar to panel C; this is the control for panel E. (G) Hematoxylin and eosin stain of RF attached to the stomach of animal 18483 (original magnification ×400). (H) Combined in situ hybridization with RRV cosmid probe (purple) and immunohistochemistry with anti-CD20 (brown) and anti-CD3 (gray) shows that RRV is not present in the CD20+ or CD3+ cells (original magnification ×1000). Arrows point to RRV-positive cells that have spindle-like morphology. (I) Identical to panel H, except vector control replaces the RRV cosmid (original magnification ×630).
In situ hybridization for RRV
To assess the presence of RRV in the lymphomas, DNA was isolated from the tissue samples and analyzed by PCR. RRV was readily detected in each tissue sample but were negative for rhesus lymphocryptovirus, a simian gamma-herpesvirus that is related to Epstein-Barr virus and shown to be associated with lymphoma in some SIV-infected RMs (data not shown).18 To further evaluate the extent of RRV infection and to determine the cell type(s) harboring RRV genomes, combined immunohistochemistry and in situ hybridization were performed. Detection of the RRV genome was accomplished with 2 cosmid clones representing approximately 50% of the RRV genome, whereas the cosmid vector lacking RRV-specific sequences was used as negative control. By this analysis, RRV genomes were detected in all of the CD20+ B cells in the lymphoma collected from the mammary tissue of animal 19185 (Figure 2B). RRV was also present in cells comprising the lymphoma in animal 19286 (Figure 2E). Interestingly, the RRV-containing cells from the liver lymphoma were negative for CD20 but were later shown to be positive for IgM, confirming these cells to be of B-cell origin. Thus, these molecular analyses demonstrate that RRV17577 is present in extranodal lymphoma in these experimentally infected RMs.
The single RF case was similarly analyzed. RF is a rare disease in captive macaques, initially observed in macaques that have simian acquired immunodeficiency syndrome caused by a simian type D retrovirus referred to as SRV. More recent studies suggest an agent considered to be closely related to KSHV, referred to as retroperitoneal fibromatosis-associated herpesvirus (RFHVmmu) is associated with RF.17 To determine whether RRV or RFHVmmu is present in RF tissue of the SIV-infected animal, DNA from RF tissue and other tissue from animal 18483 were isolated and assayed by PCR for the presence of RRV and RFHVmmu. Here RRV was found to be present in higher quantities in tissue derived from the RF lesion than in other tissues, including those presenting with LPD, and no evidence of RFHVmmu was detected (data not shown).
To further define the cell types harboring RRV genomes in the RF lesion, in situ hybridization and immunohistochemistry were performed and positive staining for RRV was observed; however, cells found to be positive for RRV did not stain positively for CD3 and CD20 antigen (Figure 2H). The positive staining is similar to the in situ hybridization of KS lesions for KSHV genomes, which required PCR in situ for detection.19
RRV ORF expression in lymphoma
To further evaluate whether RRV is associated with the development of the lymphomas and not just a passenger within the malignancies, we isolated RNA from the mammary lymphoma of animal 19185 and amplified the RNA for analysis with an RRV-specific DNA microarray. Labeled cDNA target, derived from amplified RNA, was applied to slides printed with RRV-specific probes. Analysis of the array for presence and absence of viral gene expression detected specific transcript expression corresponding to RRV ORF-R2 and ORF-71, which encode the vIL-620 and the vFLIP homolog, respectively. Interestingly, the analysis did not detect expression of RRV ORF-73 and ORF-72, which encode the RRV homologs of KSHV LANA and v-cyclin, respectively. Transcripts encoding KSHV LANA are readily detected in body cavity-based lymphoma cells and are derived from a cluster of transcripts that also encode KSHV ORF-72 and ORF-71.21 RRV LANA is thought to be expressed from 3.0- and 4.0-kb transcripts that are polycistronic for RRV ORF-72 and ORF-71.22 Thus, the lack of detection of RRV LANA transcripts could be because the cDNA was amplified using oligo-dT primers, which impose a 3′ bias to the probes, which could explain detection of ORF-71 transcripts, but not that of ORF-72 and ORF-73. Alternatively, RRV LANA may function differently from KSHV LANA and not be transcribed in these tissues. However, given the conservation of function among the γ2-herpesviruses, we think that RRV LANA is most probably transcribed and expressed similarly to KSHV LANA, despite the difference in amino acid sequence identity.
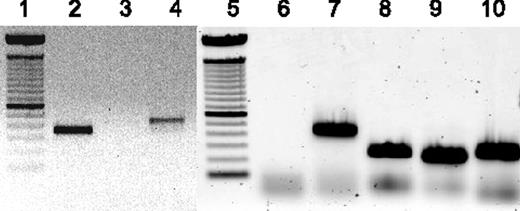
To verify the presence or absence of ORF-R2, ORF-71, ORF-72, and ORF-73 expression, we performed RT-PCR on the lymphoma-derived RNA with primers specific for RRV ORF-R2 (vIL-6), RRV ORF-71 (vFLIP), RRV ORF-72 (v-cyclin), and RRV ORF-73 (LANA). By this method, expression of vIL-6 and ORF 71 was confirmed, as were RNA transcripts encoding ORFs 72 and 73 (Figure 3). Expression of these RRV ORFs in the lymphoma of animal 19185 parallel what is found in body cavity-based lymphoma cells obtained from KSHV/HIV-infected patients.
Analysis of RRV gene expression in lymphoma of animal 19185. RNA isolated from lymphoma of animal 19185 was analyzed by reverse transcriptase (RT)-PCR to confirm expression of vIL-6 and vFLIP and to analyze expression of ORFs 72 and 73. Lanes 1 and 5 indicate 100 base pair ladder; lane 2, RNA from lymphoma amplified with vIL-6-specific primers; lane 3, same as lane 2, minus RT; lanes 4 and 7, GADPH-specific primers as control for RNA; lane 6, no template control; lane 8, ORF-73–specific primers; lane 9, ORF-72–specific primers; and lane 10, ORF-71–specific primers.
Analysis of RRV gene expression in lymphoma of animal 19185. RNA isolated from lymphoma of animal 19185 was analyzed by reverse transcriptase (RT)-PCR to confirm expression of vIL-6 and vFLIP and to analyze expression of ORFs 72 and 73. Lanes 1 and 5 indicate 100 base pair ladder; lane 2, RNA from lymphoma amplified with vIL-6-specific primers; lane 3, same as lane 2, minus RT; lanes 4 and 7, GADPH-specific primers as control for RNA; lane 6, no template control; lane 8, ORF-73–specific primers; lane 9, ORF-72–specific primers; and lane 10, ORF-71–specific primers.
RRV vIL-6 is present in lymphoid and mesenchymal proliferative lesions
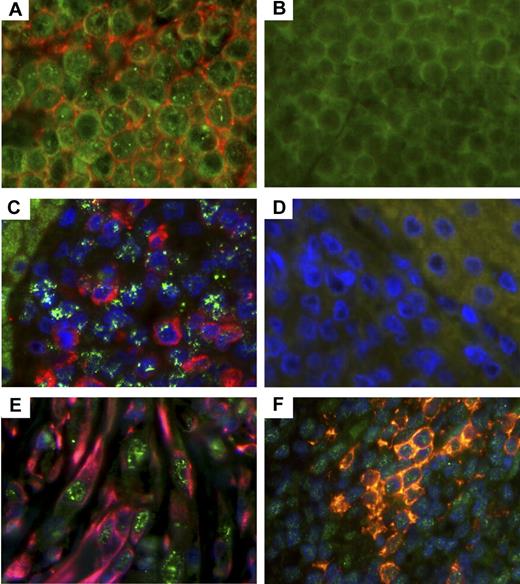
Further validation of RRV involvement in lymphoid lesions was demonstrated via immunohistochemistry to detect RRV vIL-6 within these tissue sections. RRV vIL-6 possesses IL-6-like activity and is capable of stimulating IL-6-dependent B cells to proliferate in cell culture by interacting with the IL-6 receptor or directly through gp130.20 More importantly, body cavity-based lymphoma cells have been shown to express KSHV vIL-6. Tissue sections from lymphomas in animals 19185 and 19286 were stained with a monoclonal antibody specific for RRV vIL-6 and for CD20 along with appropriate isotype controls. By this analysis, both tumors stained positively for vIL-6 (Figure 4A,C), and the majority of cells from animal 19185's lymphoma stained positive for CD20 (Figure 4A), whereas cells from animal 19286's lymphoma were positive for IgM (Figure 4C).
Detection of vIL-6 in RRV-associated lymphomas, RF, and MCD. (A) Lymphoma from RM 19185 was stained with murine monoclonal antibodies specific for RRV vIL-6 and CD20. (C) Lymphoma from animal 19286 was stained with murine monoclonal antibodies specific for RRV vIL-6 and goat antihuman IgM, and nuclei. (B,D) Isotype controls (original magnification ×630). (A) vIL-6 (green) and CD20 (red). (B) Isotype controls for vIL-6 and CD20. (C) vIL-6 (green) and IgM (red), and nuclei (blue). (D) Isotype controls for vIL-6 and IgM. (E) RF from animal 18483 was stained for vIL-6 (green), vimentin (red), and nuclei (blue). Original magnification ×630. (F) MCD lesion from animal 19455 was stained for vIL-6 (green), CD20 (red), and nuclei (blue). Original magnification ×630.
Detection of vIL-6 in RRV-associated lymphomas, RF, and MCD. (A) Lymphoma from RM 19185 was stained with murine monoclonal antibodies specific for RRV vIL-6 and CD20. (C) Lymphoma from animal 19286 was stained with murine monoclonal antibodies specific for RRV vIL-6 and goat antihuman IgM, and nuclei. (B,D) Isotype controls (original magnification ×630). (A) vIL-6 (green) and CD20 (red). (B) Isotype controls for vIL-6 and CD20. (C) vIL-6 (green) and IgM (red), and nuclei (blue). (D) Isotype controls for vIL-6 and IgM. (E) RF from animal 18483 was stained for vIL-6 (green), vimentin (red), and nuclei (blue). Original magnification ×630. (F) MCD lesion from animal 19455 was stained for vIL-6 (green), CD20 (red), and nuclei (blue). Original magnification ×630.
The RF lesion from animal 18483 was also analyzed for RRV vIL-6 expression, as earlier reports have demonstrated KSHV vIL-6 expression in KS lesions, although differing results have been reported.23,24 In addition, previous publications have reported that RF requires IL-6 for growth, similar to what has been reported for KS cells.25,26 Thus, using the RRV vIL-6–specific monoclonal antibody, numerous cell types stained positive for vIL-6 (Figure 4E). Some, but not all, of the vIL-6-positive cells also stained positive for vimentin, an intermediate filament protein found in mesenchymal cells, which has been used in the differential diagnosis of undifferentiated neoplasms of mesenchymal origin. By contrast, these same cells were negative for desmin (data not shown), implying that the RF lesion found in this animal is a proliferative mesenchymal lesion.
Because the lesions shown in Figure 4A, 4C, and 4E were from animals experimentally infected with the parental wild-type RRV, we analyzed lesions from the kidney of animal 19455. This animal was experimentally inoculated with the plaque-purified isolate of RRV17577 having small plaque phenotype. Histologic examination of the kidney indicated that the organ contained numerous Castleman-like lymphoproliferative lesions, and these lesions also stained positively for vIL-6 (Figure 4F). Thus, this plaque isolate is pathogenic and represents an isogenic clone of RRV17577 for which we can begin to identify viral determinants of pathogenesis in an experimental animal model.15
Discussion
We earlier reported that experimental RRV infection of SIV-infected RMs leads to B-cell hyperplasia and LPD that resembles MCD observed in some HIV- and KSHV-infected humans. In this report, we present results from experimental animal infection studies that further support the utility of the RRV/SIV-infected RM as an animal model to study aspects of KSHV-like pathogenesis. Specifically, this study establishes that experimental rhadinovirus infection with this particular strain of RRV can yield virus-positive lymphoma and RF. The most common outcome of the experimental infections is B-cell hyperplasia, lymphadenopathy, and persistent virus infection, which can eventually lead to more complicated clinical manifestations. Overall, 20% to 30% of the coinfected animals developed lymphoma, RF, or systemic MCD, which is in agreement with the percentage of humans coinfected with HIV and KSHV that develop KS, MCD, PEL, or NHL.4,27,28
The experimental animal infection studies also extend our current understanding of acute RRV infection in immunocompetent and immunodeficient animals. We found that immunocompetent animals 19092, 19286, 19182, and 19218 exhibited detectable levels of DNA viremia in PBMCs between weeks 2 and 4 after RRV infection and became essentially undetectable until the animals were subsequently infected with SIV (animals 19182 and 19218) or when the animal developed a lymphoma (animal 19286). By contrast, immunodeficient animals exhibited detectable levels of DNA viremia throughout the infection period, either sporadically or persistently, depending on the individual animal. Differences in viral load between the 2 immune statuses suggest that SIV-infected animals lack functional immune components required to maintain RRV in a latent state. Thus, identifying and/or elucidating the immune components associated with maintaining a latent RRV infection could lead to an approach to prevent RRV reactivation and disease, which could translate into an effective treatment for KSHV-associated disease in HIV patients.
Primary characterization of the disease outcomes in these animals strongly supports the utility of the RRV/SIV RM model as a model of KSHV pathogenesis in an immunocompromised host. Specifically, we detect the expression of vIL-6 by DNA microarray and immunohistochemistry in the lymphoma tissue of animal 19185 and subsequently in the lymphoma of animal 19286 and within RF tissue from animal 18483. These results are essentially identical to what has been reported in KSHV-associated PEL, MCD, and KS.23,24,29,30 Moreover, by RT-PCR analysis, we confirmed the RNA transcripts specific for RRV ORF-71 (vFLIP) and detected RNA transcripts for ORF-72 (v-cyclin) and ORF-73 (LANA) in the lymphoma of animal 19185, which is consistent with KSHV gene expression in PEL cells.21 Thus, the RRV-associated lesions display viral signatures that parallel those observed in KSHV-associated disease.
What is interesting to note is the single incidence of RF in the RRV/SIV-infected animals, in which we show RRV is present and expressing vIL-6. As mentioned earlier, RF is a rare disease in nonhuman primates, and another γ2 herpesvirus (RFHVmmu) is considered to be responsible for this disease. Unfortunately, we could not detect the presence of this potential infectious agent using specific PCR primers. Although this does not prove RRV is responsible for RF, it suggests that RRV may play a role, which needs to be further explored. Host factors most probably play a role in RF and the lymphomas observed, and we are starting to evaluate this by characterizing the animals with disease.
The advantage of this nonhuman primate model is that a genetic system exists to investigate the role specific viral ORFs play in different aspects of infection, ie, latency, persistence, and disease progression. In addition, with the creation of an infectious RRV bacterial artificial chromosome,15 it is now possible to generate novel chimeric KSHV and RRV viruses to investigate specific contributions KSHV ORFs have in disease. This should lead to new insights into KSHV pathogenesis and provide an animal model to test novel vaccines or treatments to prevent KSHV-mediated disease.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Ryan Estep and Ilhem Messaoudi, and John Jones and Bridget Robinson for helpful comments; Alfred Legasse and Shannon Planar for assistance with the animals; and Lori Boshears for editorial assistance.
This work was supported by the Public Health Service (grants RR00163 and CA 75922; S.W.W.).
National Institutes of Health
Authorship
Contribution: B.U.O. and M.F.P. designed and performed experiments, analyzed data, and wrote portions of the manuscript; J.S., H.L., and B.Y. designed and performed experiments; R.P.S. designed the primers, performed the DNA microarray experiment, and analyzed the data; M.K.A. assisted in the design of the experimental animal infection studies and performed pathologic examinations; and S.W.W. designed the experiments and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Scott W. Wong, Vaccine and Gene Therapy Institute, Oregon Health & Science University, West Campus, 505 NW 185th Avenue, Beaverton, OR 97006; e-mail: wongs@ohsu.edu.
References
Author notes
*B.U.O. and M.F.P. contributed equally to the manuscript.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal