Abstract
The inflammasomes are macromolecular cytosolic complexes involved in the production of interleukin-1β (IL-1β) and IL-18 in response to several pathogen-derived stimuli. Such interleukins have been implicated in the origin of severe allogeneic stem cell transplant (allo-SCT) complications. We analyzed the relationship between the interindividual variability in inflammasome protein-encoding genes in donors and patients and clinical outcome after allo-SCT. Fourteen common genetic variants in 5 genes of the inflammasome, namely, NLRP1, NLRP2, NLRP3, CARD8, and CASP5, were genotyped in 133 human leukocyte antigen-identical sibling pairs undergoing allo-SCT. In the multivariate analysis, donor variants in NLRP2 and NLRP3 were the most important prognostic factors for the clinical outcome after allo-SCT. Thus, donor TT genotype at rs10925027 in NLRP3 was associated with disease relapse (odds ratio (OR) = 6.3, P = 1 × 10−7), and donor GG genotype at rs1043684 in NLRP2 was associated with nonrelapse mortality (OR = 4.4, P = 6 × 10−4) and overall survival (OR = 3.1, P = .001). In addition, patient AA genotype at rs5862 in NLRP1 was associated with nonrelapse mortality (OR = 2.8, P = .005) and overall survival (OR = 2.0, P = .009). These results suggest that inflammasome genetic variants are important prognostic factors for the outcome of allo-SCT. If validated in larger studies, including unrelated allo-SCT, NLRPs genotype would become an important factor in donor selection.
Introduction
The inflammasomes are macromolecular cytosolic complexes that stimulate pro-caspase-1 autocatalysis and promote the processing of different pro-inflammatory cytokines, such as pro-interleukin (IL)-1β and pro-IL-18, into its active forms. They are composed by the assembly of several proteins, some of which are constitutive (pro-caspase-1, ASC), whereas others are present depending on the inflammasome type (Cardinal, pro-caspase-5, FIIND). These complexes are involved in the innate immune response by the recognition of both endogenous signals (adenosine triphosphate, urate, and calcium pyrophosphate crystals) as well as external pathogen-derived products (bacterial RNA, bacterial toxins).1 To date, 4 different types of inflammasomes have been described, which are, respectively, termed Nalp1, Nalp2, Nalp3, and Ipaf-inflammasome.2,3
Nalp proteins play a central role in the inflammasomes. They belong to the NACHT-leucine-rich repeat (NLR) family that includes different intracellular receptors that share a common structural organization, with an N-terminal effector domain, a central NACHT domain involved in self-oligomerization, and multiple C-terminal leucine-rich repeats, which act as sensors.4 Those NLR proteins with a CARD domain at N-terminus are distributed into the NOD, IPAF, and CIITA subfamilies, whereas those with a pyrin domain at N-terminus belong to the NALP subfamily and are the key initiators of the inflammasome assembly.5 Interestingly, dominantly inherited “gain-of-function” mutations in the NLRP3 gene have been associated with the cryopyrinopathies familial cold-induced autoinflammatory syndrome, Muckle-Wells syndrome, and chronic infantile neurologic cutaneous and articular syndrome/neonatal-onset multisystem inflammatory disease syndrome.6-8 All these diseases are characterized by recurrent episodes of fever and serosal inflammation resulting from an increased production of IL-1β. The recombinant human IL-1 receptor antagonist anakinra improves clinical manifestations of such patients.9,10 In addition, common genetic variants in a different member of the NALP subfamily, the NLRP1 gene, have been recently associated with the risk of several vitiligo-associated autoimmune diseases.11
In the allogeneic stem cell transplantation (allo-SCT) setting, IL-1β is a key inflammatory cytokine involved in the development of graft-vs-host disease (GVHD).12 As regulators of the activation of IL-1β, it has been proposed that inflammasomes could play a role in the pathogenesis of such complications and in the outcome of allo-SCT.13 Indeed, we and others have demonstrated an association of common variants in NOD2 gene, another member of NLR family, with severe acute GVHD (aGVHD),14 bronquiolitis obliterans,15 relapse,16 and nonrelapse mortality (NRM)17 after allo-SCT.
In this study, we have investigated the association of 14 common variants in genes encoding for inflammasome proteins to the outcome of 133 patients undergoing unmanipulated human leukocyte antigen (HLA)-identical sibling allo-SCT, focusing on the incidence of aGVHD, relapse, NRM, and overall survival (OS).
Methods
Patients
A total of 133 patients who underwent unmanipulated HLA-identical sibling allo-SCT, and of whom stored donor and patient DNA was available, were included in the study. All transplants were performed in the Hematology Department of the Hospital Clínic of Barcelona between February 1997 and February 2007 according to standard protocols. All donor and recipient pairs were white. Main patient and procedure characteristics are given in Table 1. The local Ethics Committee, Hospital Clinic of Barcelona provided institutional review board–approval for this study, and informed consent was obtained from all patients in accordance with the Declaration of Helsinki.
Patient characteristics (N = 133)
| Characteristic . | Value . |
|---|---|
| Age at transplantation, y [median (range)] | 41 (17-64) |
| Sex, no. (%) female | 52 (39) |
| Underlying disease | |
| Acute leukemia, no. (%) | 51 (38) |
| Lymphoma/myeloma, no. (%) | 43 (32) |
| Myeloproliferative disorder, no. (%) | 25 (19) |
| Myelodysplasia/aplasia, no. (%) | 10 (8) |
| Chronic lymphocytic leukemia, no. (%) | 4 (3) |
| Advanced phase, no. (%) | 95 (71) |
| Source, peripheral blood, no. (%) | 108 (81) |
| Conditioning | |
| Myeloablative, no. (%) | 84 (63) |
| Cy/TBI,* no. (%) | 57 (43) |
| Bu/Cy,† no. (%) | 13 (10) |
| Other myeloablative,‡ no. (%) | 14 (10) |
| Reduced intensity conditioning, no. (%) | 49 (37) |
| Flu/Mel,§ no. (%) | 28 (21) |
| Flu/Bu,‖ no. (%) | 13 (10) |
| Other reduced intensity conditioning,¶ no. (%) | 8 (6) |
| GVHD prophylaxis | |
| Cyclosporin A/methotrexate, no. (%) | 102 (77) |
| Cyclosporin A/mycophenolate mophetil, no. (%) | 24 (18) |
| Cyclosporin A/prednisone, no. (%) | 7 (5) |
| CD34+ cells infused [median (range)] | 4.3 (0.9-11.0) |
| Characteristic . | Value . |
|---|---|
| Age at transplantation, y [median (range)] | 41 (17-64) |
| Sex, no. (%) female | 52 (39) |
| Underlying disease | |
| Acute leukemia, no. (%) | 51 (38) |
| Lymphoma/myeloma, no. (%) | 43 (32) |
| Myeloproliferative disorder, no. (%) | 25 (19) |
| Myelodysplasia/aplasia, no. (%) | 10 (8) |
| Chronic lymphocytic leukemia, no. (%) | 4 (3) |
| Advanced phase, no. (%) | 95 (71) |
| Source, peripheral blood, no. (%) | 108 (81) |
| Conditioning | |
| Myeloablative, no. (%) | 84 (63) |
| Cy/TBI,* no. (%) | 57 (43) |
| Bu/Cy,† no. (%) | 13 (10) |
| Other myeloablative,‡ no. (%) | 14 (10) |
| Reduced intensity conditioning, no. (%) | 49 (37) |
| Flu/Mel,§ no. (%) | 28 (21) |
| Flu/Bu,‖ no. (%) | 13 (10) |
| Other reduced intensity conditioning,¶ no. (%) | 8 (6) |
| GVHD prophylaxis | |
| Cyclosporin A/methotrexate, no. (%) | 102 (77) |
| Cyclosporin A/mycophenolate mophetil, no. (%) | 24 (18) |
| Cyclosporin A/prednisone, no. (%) | 7 (5) |
| CD34+ cells infused [median (range)] | 4.3 (0.9-11.0) |
Cyclophosphamide 120 mg/kg and total body irradiation 12 to 13 Gy.
Busulfan 16 mg/kg and cyclophosphamide 200 mg/kg.
Cy/TBI/etoposide 8, Cy/total nodal irradiation 4, melphalan/TBI 2.
Fludarabine 150 mg/m2 and melphalan 140 mg/m2.
Fludarabine 150 mg/m2 and busulfan 10 mg/kg.
Carmustine/etoposide/citarabine/melphalan 4; IDAFLAG 3, fludarabine/citarabine 1.
Selection of genetic variants
NLRP1, NLRP2, NLRP3, CARD8, and CASP5 genes were screened at the National Center for Biotechnology Information database searching for variants that fulfilled the following criteria: located in promoter regions or in coding regions if associated with an amino acid exchange, and a frequency of the less frequent genotype higher than 10% in whites. Based on these criteria, 14 single nucleotide polymorphisms (SNPs) were selected for the study (Table 2).
Candidate genes and SNP
| Gene . | NCBI code . | Nucleotides . | Location . | Amino acid change . |
|---|---|---|---|---|
| NLRP1 | rs925598 | C/T | UTR 5′ | None |
| rs1156989 | C/T | UTR 5′ | None | |
| rs12150220 | A/T | Exon 3 | His-155-Leu | |
| rs2301582 | A/G | Exon 11 | Met-1029-Val | |
| rs5862 | A/G | UTR 3′ | None | |
| NLRP2 | rs1043673 | A/C | Exon 13 | Glu-1052-Ala |
| rs1043684 | A/G | UTR 3′ | None | |
| NLRP3 | rs10925027 | C/T | UTR 5′ | None |
| rs10754558 | C/G | UTR 3′ | None | |
| rs10802501 | A/T | UTR 3′ | None | |
| CARD8 | rs11665831 | C/T | UTR 5′ | None |
| rs4802445 | C/T | UTR 3′ | None | |
| CASP5 | rs477163 | C/T | UTR 5′ | None |
| rs523104 | C/G | Exon 6 | Leu-318-Val |
| Gene . | NCBI code . | Nucleotides . | Location . | Amino acid change . |
|---|---|---|---|---|
| NLRP1 | rs925598 | C/T | UTR 5′ | None |
| rs1156989 | C/T | UTR 5′ | None | |
| rs12150220 | A/T | Exon 3 | His-155-Leu | |
| rs2301582 | A/G | Exon 11 | Met-1029-Val | |
| rs5862 | A/G | UTR 3′ | None | |
| NLRP2 | rs1043673 | A/C | Exon 13 | Glu-1052-Ala |
| rs1043684 | A/G | UTR 3′ | None | |
| NLRP3 | rs10925027 | C/T | UTR 5′ | None |
| rs10754558 | C/G | UTR 3′ | None | |
| rs10802501 | A/T | UTR 3′ | None | |
| CARD8 | rs11665831 | C/T | UTR 5′ | None |
| rs4802445 | C/T | UTR 3′ | None | |
| CASP5 | rs477163 | C/T | UTR 5′ | None |
| rs523104 | C/G | Exon 6 | Leu-318-Val |
UTR indicates untranslated region.
Genotyping
Genomic DNA from whole blood samples from patients and donors was isolated before transplant using a QIAmp DNABlood Mini Kit (QIAGEN, Hilden, Germany) following the manufacturer's instructions and stored at −20°C. Allelic variants were genotyped using the TaqMan SNP Genotyping Assay (Applied Biosystems, Foster City, CA). The specific primers and FAM- and VIC- dye labeled probes used were designed by the Applied Biosystems assay-on-demand service. Reactions were performed as previously reported.18
Data analysis
Five endpoints were considered: aGVHD grades II to IV and III and IV, relapse, NRM, and OS. Acute GVHD, NRM, and relapse were analyzed using the Gray test.19 The cumulative incidence was computed with the cmprsk package for R 2.6.2 software.20
The competing events were defined as follows. In the case of acute GVHD and relapse, the competing event was death. For NRM, the competing event was relapse. OS was obtained by the Kaplan-Meier method and compared using the log-rank test. P values were 2-sided and represent raw values. Variables included in the univariate analysis were donor and patient genotype for each variant (3 groups: homozygous for the most frequent allele, heterozygous, and homozygous for the less frequent allele), patient age (younger or older than the median), relation donor/patient gender (donor female into recipient male vs others), cytomegalovirus serology of donors and patients, stem cell source (peripheral blood vs bone marrow), conditioning (myeloablative vs reduced intensity), GVHD prophylaxis (cyclosporin A and methotrexate vs cyclosporin A and mycophenolate), and phase of the disease (early vs advanced). Early phase was considered chronic myeloid leukemia in first chronic phase, acute leukemia in first complete remission, and myelodysplasia with low or low to intermediate international prognostic scoring system. All prognostic variables in the univariate analysis with a P value less than or equal to .1 as well as patient age and conditioning were included in the multivariate analysis. Multivariate analysis of OS was performed using Cox regression with backward method and Wald statistic, by SPSS 15.0 software (Chicago, IL). The proportional hazard assumption was tested for each variable by both analytical and graphical methods. Multivariate analysis of NRM and relapse was performed using the subdistribution regression model of Fine and Gray21 with the cmprsk package mentioned in the first paragraph of “Data analysis.” The proportional hazard assumption was tested for each variable by analyzing the Schoenfeld residuals. Analysis of linkage disequilibrium for homozygous cases was performed by Fisher exact test and Bonferroni procedure using DnaSP 4.10.9 software.22
Results
Genotype frequency
Fourteen SNPs were analyzed in 266 cases (133 donors and 133 patients), which meant 3724 samples. Successful genotyping was achieved in 3392 (91%) SNPs. Allele and genotype frequencies were similar to those observed in National Center for Biotechnology Information database and were in Hardy-Weinberg equilibrium (Tables S1, S2, available on the Blood website; see the Supplemental Materials link at the top of the online article).
aGVHD
Eighty-four of 133 (63%) patients developed aGVHD. Of them, 39 (29%) patients had aGVHD grade I, 35 (26%) grade II, 8 (6%) grade III, and 2 (1%) grade IV. There were neither clinical nor genetic characteristics associated with aGVHD grades II to IV. When the analysis was restricted to aGVHD grades III to IV, factors associated with this complication in the univariate analysis were patient genotype CC versus CA/AA at rs1043673 (22% vs 5%, P = .04), and for the genotype GG versus GA/AA at rs1043684 (14% vs 5%, P = .15), both located in the NLRP2 gene. Alleles rs1043673 and rs1043684 were in linkage disequilibrium (Table S2; D = 0.169, P < .001). Multivariate analysis for acute GVHD grades III to IV was not performed because of the relatively low number of events.
Relapse or progression
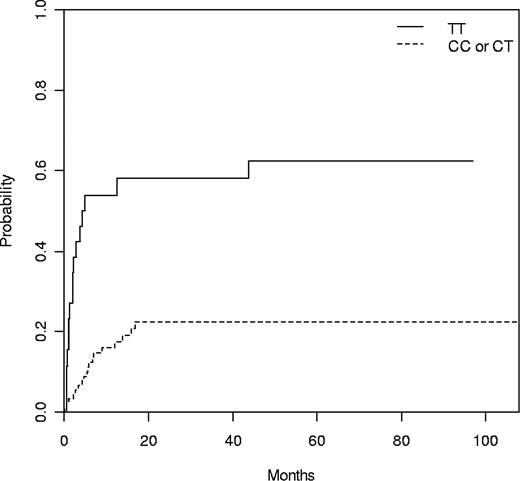
Four patients transplanted for aplastic anemia were removed for relapse analysis. Of the remaining 129 patients, 39 (30%) relapsed or progressed after transplantation, the median time to progression being 6 months (range, 1-44 months). The only clinical risk factor associated with relapse was to be transplanted in advanced phase (34% vs 8% at 1 year, P = .02). In addition, 3 genetic variants, 2 within the NLRP3 gene from the donor (rs10925027 and rs10754558) and 1 within the CARD8 gene from the patient (rs11665831), were associated with relapse (Table 3). Alleles rs10925027 and rs10754558 in the NLRP3 gene were in linkage disequilibrium (Table S3; D = 0.221, P < .001). Cumulative incidence for relapse of donor NLRP3 rs10925027 genotype at 1 year was: TT 53%, TC 18%, and CC 17% (P = .001; Table 3). TT genotype at rs10925027 in the NLRP3 gene was compared with the combination of TC and CC alleles (TT 54% vs TC/CC 18% at 1 year, P = 3.04 × 10−5; Figure 1). In the multivariate analysis, the following variables were included: donor rs10925027 genotype, patient rs11665831 genotype, phase of the disease, conditioning, and patient age at transplant. Of them, only donor rs10925027 TT genotype and patient rs11665831 CC genotype were associated with relapse (Table 4).
Univariate analysis
| Gene/rs code . | Relapse events/at risk (cumulative incidence*) . | P . | NRM events/at risk (cumulative incidence*) . | P . | OS events/at risk . | P . |
|---|---|---|---|---|---|---|
| NLRP1 | ||||||
| 5862 | ||||||
| Patient | ||||||
| GG | 8/41 (18) | NS | 8/41 (17) | .04 | 11/41 (72) | .02 |
| GA | 21/57 (34) | 12/57 (21) | 23/57 (58) | |||
| AA | 9/31 (20) | 14/31 (42) | 18/31 (42) | |||
| Donor | ||||||
| GG | 8/31 (19) | NS | 10/31 (32) | NS | 14/31 (54) | NS |
| GA | 24/68 (34) | 14/68 (19) | 25/68 (61) | |||
| AA | 3/22 (10) | 9/22 (39) | 10/22 (51) | |||
| NLRP2 | ||||||
| 1043673 | ||||||
| Patient | ||||||
| CC | 25/83 (27) | NS | 15/83 (17) | .007 | 25/83 (68) | .001 |
| CA | 2/14 (15) | 7/14 (45) | 8/14 (38) | |||
| AA | 4/9 (33) | 4/9 (44) | 7/9 (22) | |||
| Donor | ||||||
| CC | 24/73 (29) | NS | 17/73 (21) | .04 | 27/73 (61) | 0.1 |
| CA | 3/18 (18) | 4/18 (25) | 5/18 (64) | |||
| AA | 4/16 (21) | 8/16 (51) | 10/16 (35) | |||
| 1043684 | ||||||
| Patient | ||||||
| GG | 7/21 (30) | NS | 9/21 (43) | .11 | 14/21 (31) | .006 |
| GA | 19/64 (30) | 16/64 (22) | 23/64 (62) | |||
| AA | 11/41 (18) | 8/41 (20) | 13/41 (67) | |||
| Donor | ||||||
| GG | 5/23 (19) | NS | 13/23 (53) | .001 | 14/23 (38) | .003 |
| GA | 15/55 (23) | 10/55 (18) | 19/55 (64) | |||
| AA | 13/38 (33) | 8/38 (19) | 13/38 (63) | |||
| NLRP3 | ||||||
| 10925027 | ||||||
| Patient | ||||||
| TT | 9/18 (44) | .12 | 4/18 (23) | NS | 8/18 (52) | NS |
| TC | 17/61 (25) | 13/61 (20) | 20/61 (66) | |||
| CC | 1/49 (23) | 16/49 (31) | 24/49 (50) | |||
| Donor | ||||||
| TT | 16/26 (53) | .001 | 4/26 (12) | NS | 12/26 (52) | NS |
| TC | 10/48 (18) | 15/48 (30) | 17/48 (63) | |||
| CC | 8/43 (17) | 12/43 (28) | 17/43 (59) | |||
| 10754558 | ||||||
| Patient | ||||||
| GG | 6/24 (27) | NS | 7/24 (30) | NS | 12/24 (47) | NS |
| GC | 17/68 (22) | 17/68 (23) | 25/68 (62) | |||
| CC | 14/34 (35) | 8/34 (24) | 13/34 (59) | |||
| Donor | ||||||
| GG | 3/27 (12) | .002 | 10/27 (37) | NS | 13/27 (52) | NS |
| GC | 14/56 (22) | 17/56 (29) | 21/56 (61) | |||
| CC | 20/40 (42) | 7/40 (15) | 18/40 (53) | |||
| CARD8 | ||||||
| 11665831 | ||||||
| Patient | ||||||
| TT | 14/64 (19) | .05 | 16/64 (23) | NS | 23/64 (63) | NS |
| TC | 13/42 (25) | 13/42 (30) | 18/42 (55) | |||
| CC | 10/19 (53) | 4/19 (21) | 10/19 (43) | |||
| Donor | ||||||
| TT | 16/53 (25) | NS | 14/53 (26) | NS | 24/53 (54) | NS |
| TC | 12/47 (26) | 15/47 (31) | 19/47 (56) | |||
| CC | 6/17 (30) | 3/17 (12) | 4/17 (74) |
| Gene/rs code . | Relapse events/at risk (cumulative incidence*) . | P . | NRM events/at risk (cumulative incidence*) . | P . | OS events/at risk . | P . |
|---|---|---|---|---|---|---|
| NLRP1 | ||||||
| 5862 | ||||||
| Patient | ||||||
| GG | 8/41 (18) | NS | 8/41 (17) | .04 | 11/41 (72) | .02 |
| GA | 21/57 (34) | 12/57 (21) | 23/57 (58) | |||
| AA | 9/31 (20) | 14/31 (42) | 18/31 (42) | |||
| Donor | ||||||
| GG | 8/31 (19) | NS | 10/31 (32) | NS | 14/31 (54) | NS |
| GA | 24/68 (34) | 14/68 (19) | 25/68 (61) | |||
| AA | 3/22 (10) | 9/22 (39) | 10/22 (51) | |||
| NLRP2 | ||||||
| 1043673 | ||||||
| Patient | ||||||
| CC | 25/83 (27) | NS | 15/83 (17) | .007 | 25/83 (68) | .001 |
| CA | 2/14 (15) | 7/14 (45) | 8/14 (38) | |||
| AA | 4/9 (33) | 4/9 (44) | 7/9 (22) | |||
| Donor | ||||||
| CC | 24/73 (29) | NS | 17/73 (21) | .04 | 27/73 (61) | 0.1 |
| CA | 3/18 (18) | 4/18 (25) | 5/18 (64) | |||
| AA | 4/16 (21) | 8/16 (51) | 10/16 (35) | |||
| 1043684 | ||||||
| Patient | ||||||
| GG | 7/21 (30) | NS | 9/21 (43) | .11 | 14/21 (31) | .006 |
| GA | 19/64 (30) | 16/64 (22) | 23/64 (62) | |||
| AA | 11/41 (18) | 8/41 (20) | 13/41 (67) | |||
| Donor | ||||||
| GG | 5/23 (19) | NS | 13/23 (53) | .001 | 14/23 (38) | .003 |
| GA | 15/55 (23) | 10/55 (18) | 19/55 (64) | |||
| AA | 13/38 (33) | 8/38 (19) | 13/38 (63) | |||
| NLRP3 | ||||||
| 10925027 | ||||||
| Patient | ||||||
| TT | 9/18 (44) | .12 | 4/18 (23) | NS | 8/18 (52) | NS |
| TC | 17/61 (25) | 13/61 (20) | 20/61 (66) | |||
| CC | 1/49 (23) | 16/49 (31) | 24/49 (50) | |||
| Donor | ||||||
| TT | 16/26 (53) | .001 | 4/26 (12) | NS | 12/26 (52) | NS |
| TC | 10/48 (18) | 15/48 (30) | 17/48 (63) | |||
| CC | 8/43 (17) | 12/43 (28) | 17/43 (59) | |||
| 10754558 | ||||||
| Patient | ||||||
| GG | 6/24 (27) | NS | 7/24 (30) | NS | 12/24 (47) | NS |
| GC | 17/68 (22) | 17/68 (23) | 25/68 (62) | |||
| CC | 14/34 (35) | 8/34 (24) | 13/34 (59) | |||
| Donor | ||||||
| GG | 3/27 (12) | .002 | 10/27 (37) | NS | 13/27 (52) | NS |
| GC | 14/56 (22) | 17/56 (29) | 21/56 (61) | |||
| CC | 20/40 (42) | 7/40 (15) | 18/40 (53) | |||
| CARD8 | ||||||
| 11665831 | ||||||
| Patient | ||||||
| TT | 14/64 (19) | .05 | 16/64 (23) | NS | 23/64 (63) | NS |
| TC | 13/42 (25) | 13/42 (30) | 18/42 (55) | |||
| CC | 10/19 (53) | 4/19 (21) | 10/19 (43) | |||
| Donor | ||||||
| TT | 16/53 (25) | NS | 14/53 (26) | NS | 24/53 (54) | NS |
| TC | 12/47 (26) | 15/47 (31) | 19/47 (56) | |||
| CC | 6/17 (30) | 3/17 (12) | 4/17 (74) |
NS indicates not significant.
Cumulative Incidence at 1 year.
Cumulative incidence of relapse according to donor NLRP3 rs10925027 genotype with death as competing event. N = 113, TT 54% versus TC/CC 18% at 1 year (P = .001).
Cumulative incidence of relapse according to donor NLRP3 rs10925027 genotype with death as competing event. N = 113, TT 54% versus TC/CC 18% at 1 year (P = .001).
Multivariate analysis
| End point . | N . | Factor . | OR . | 95% CI . | P . |
|---|---|---|---|---|---|
| Relapse | 110 | Donor rs10925027 TT in NLRP3 | 6.3 | 3.1-12.5 | 1 × 10−7 |
| Patient rs11665831 CC in CARD8 | 2.6 | 1.3-5.3 | .007 | ||
| NRM | 114 | Donor rs1043684 GG in NLRP2 | 4.4 | 1.8-10.3 | 6 × 10−4 |
| Patient rs5862 AA in NLRP1 | 2.8 | 1.3-5.7 | .005 | ||
| OS | 114 | Donor rs1043684 GG in NLRP2 | 3.1 | 1.7-5.6 | .0001 |
| Advanced phase | 2.8 | 1.4-5.4 | .002 | ||
| Older age | 2.4 | 1.3-4.5 | .006 | ||
| Patient rs5862 AA in NLRP1 | 2.0 | 1.2-3.4 | .009 |
| End point . | N . | Factor . | OR . | 95% CI . | P . |
|---|---|---|---|---|---|
| Relapse | 110 | Donor rs10925027 TT in NLRP3 | 6.3 | 3.1-12.5 | 1 × 10−7 |
| Patient rs11665831 CC in CARD8 | 2.6 | 1.3-5.3 | .007 | ||
| NRM | 114 | Donor rs1043684 GG in NLRP2 | 4.4 | 1.8-10.3 | 6 × 10−4 |
| Patient rs5862 AA in NLRP1 | 2.8 | 1.3-5.7 | .005 | ||
| OS | 114 | Donor rs1043684 GG in NLRP2 | 3.1 | 1.7-5.6 | .0001 |
| Advanced phase | 2.8 | 1.4-5.4 | .002 | ||
| Older age | 2.4 | 1.3-4.5 | .006 | ||
| Patient rs5862 AA in NLRP1 | 2.0 | 1.2-3.4 | .009 |
NRM
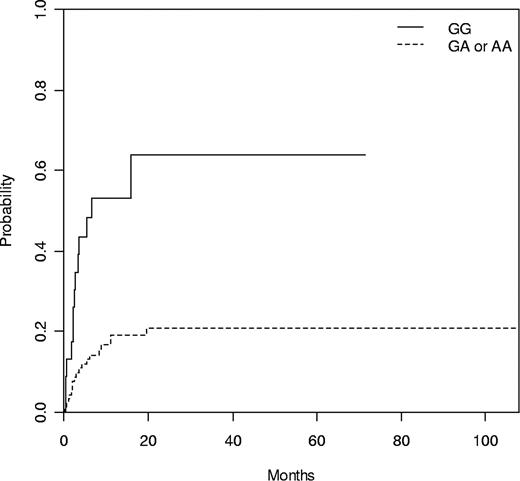
Thirty-six (27%) patients died while in remission. The only clinical risk factor with a trend to NRM was to receive a transplant in advanced phase (30% vs 16% at 1 year, P = .11). Three variants, 2 within the NLRP2 gene (rs1043673 and rs1043684) in both donor and patient and 1 within the NLRP1 gene (rs5862) in the patient, were associated with NRM (Table 3). Genotype GG at rs1043684 in NLRP2 was compared with the combination of GA and AA, both in donors (GG 53% vs GA/AA 19% at 1 year, P = .001; Figure 2) and patients (GG 44% vs GA/AA 21% at 1 year, P = .016). The following variables were included in the multivariate analysis: patient and donor rs1043684 genotype, patient rs5862 genotype, age at transplant, conditioning, and phase of the disease. Of them, only donor rs1043684 GG genotype in NLRP2 and patient rs5862 AA genotype in NLRP1 remained significant (Table 4). Interestingly, the most important prognostic factor for NRM was donor GG genotype at rs1043684 in NLRP2 (odds ratio = 4.4, P = .001).
Cumulative incidence of NRM according to donor NLRP2 rs1043684 genotype with relapse as competing event. N = 116, GG 53% versus GA/AA 19% at 1 year (P = .001).
Cumulative incidence of NRM according to donor NLRP2 rs1043684 genotype with relapse as competing event. N = 116, GG 53% versus GA/AA 19% at 1 year (P = .001).
Overall survival
Sixty-two (47%) patients died after the transplant. Median follow-up was 39 months (range, 1-118 months) for the surviving patients and 3 months (range, 0.2-31 months) for the patients who died. Clinical risk factors associated or with a trend for a worse OS were to receive a transplant in advanced phase (65% vs 42%, P = .003) and patients older than the median (56% vs 41%, P = .087). Those 3 variants associated with NRM (rs1043673 and rs1043684 in NLRP2 and rs5862 in NLRP1) were also associated with OS (Table 3). GG genotype at rs1043684 in NLRP2 was compared with the combination of GA and AA, both in donors (GG 15% vs GA/AA 57%, P = .001; Figure 3) and in patients (GG 18% vs GA/AA 56%, P = .002). The following variables were included in the multivariate analysis: donor and patient rs1043684 genotype, phase of the disease, age at transplant, and patient rs5862 genotype. Of them, all except patient rs1043684 genotype remained significant, being donor rs1043684 genotype the most important risk factor (Table 4).
Kaplan-Meier estimate, OS according to donor NLRP2 rs1043684 genotype. N = 116, GG 15% versus GA/AA 57% (P = .001).
Kaplan-Meier estimate, OS according to donor NLRP2 rs1043684 genotype. N = 116, GG 15% versus GA/AA 57% (P = .001).
Discussion
This study shows, for the first time, a strong association between common genetic variants in NALP genes and the outcome of HLA-identical allo-SCT. Interestingly, the variants studied affected the outcome of transplantation in different ways. Donor variants rs1043673 and rs1043684 in the NLRP2 gene were associated with NRM and OS, whereas donor rs10925027 and rs10754558 variants in the NLRP3 gene were associated with disease relapse. Variants rs1043673 and rs1043684 in the NLRP2 gene as well as variants rs10925027 and rs10754558 in the NLRP3 gene were in linkage disequilibrium, suggesting that their relationship with the clinical outcome found in this study may be part of a high-risk haplotype rather than the direct causal variants.
The NLRP1 gene is highly expressed in T cells and Langerhans cells. It encodes Nalp1 protein that has been involved in the assembly of Nalp1-inflammasome and the secretion of several caspase-1–dependent cytokines (IL-1β, IL-18, IL-33) in response to several endogenous or exogenous signals. Common variants in NLRP1 have been recently associated with vitiligo-associated autoimmune diseases,11 suggesting a role of such variants in the pro-IL1β processing activity. In this study, we have observed an association of patient rs5862 variant, located in the untranslated region 3′ of NLRP1, with NRM and OS.
The NLRP2 gene is expressed in several tissues, including monocyte/macrophage cells. It has been reported that lipopolysaccharide and several pro-inflammatory cytokines could increase NLRP2 expression, leading to caspase-1 activation and subsequent IL-1β release.5 Variants in NLRP2 might influence NRM and OS after HLA-identical allo-SCT by either modifying the incidence of severe aGVHD and/or by increasing the susceptibility to infections. Thus, in addition to its role in IL-1β secretion, Nalp2 also inhibits NF-κB signaling pathway,23 which is involved in the coordinated expression of a wide variety of genes involved in the control of the innate and adaptive immune responses.24 Similar to variants in NOD2 gene, which down-regulate several cytokines via the NF-κB pathway,25 variants in NLRP2 might promote severe aGVHD, as we have found in the univariate analysis. Nevertheless, the little number of patients dying of severe aGVHD and the strong impact of NLRP2 variants in survival suggest that the effect of such variants may be, at least in part, related to other transplant complications.
NLRP3 is highly expressed in polymorphonuclear cells.8 It encodes for Nalp3/Criopyrin protein, which constitute the Nalp3 inflammasome after its assembly with the adaptor molecules ASC and CARD8.26 Several pathogen-derived compounds (muramyl dipeptide, bacterial and viral RNA, microbial toxins) as well as internal nonmicrobial danger signals (adenosine triphosphate, monosodium urate, and calcium pyrophosphate crystals, and low intracellular potassium concentration) have been recognized as Nalp3 inflammasome activators, subsequently increasing its pro-IL1β processing activity.27 In the past, the NLRP3 gene and the caspase-1–dependent cytokines have been involved in the innate and adaptive immune response. With regard to the innate immune system, several autoinflammatory diseases, known as cryopyrinopathies, are a consequence of a deregulated inflammatory response. These diseases have been associated with gain-of-function mutations in NLRP3 gene resulting in an increased production of IL-1β and IL-18. Cryopyrinopathies have shown an excellent clinical and biochemical response to IL-1β blockade with anakinra.7-10 With regard to adaptive immune system, NLRP3-deficient mice showed an impaired contact hypersensitivity to the hapten trinitrophenylchloride, being the contact hypersensitivity a T cell–mediated immune response to repeated exposure to contact allergens.28 In the present study, a strong association between 2 variants in donor NLRP3 gene and relapse has been observed. Of note, both variants are located at both extremes of the gene (5′ and 3′ untranslated regions) and are in linkage disequilibrium, suggesting that other variants within the gene or around these variants may also be in linkage disequilibrium. We did not detect an underlying diagnosis most frequent among patients with the deleterious NLRP3 variants who relapsed than among those who did not relapse (data not shown), suggesting that this association may affect different hematologic malignancies. The mechanism of this association is at present unknown. A possible explanation would be that donor immune cells with the deleterious NLRP3 variants might have a defect in their ability to mount a correct adaptive immune response, which would impair a graft-versus-leukemia effect. However, further studies are needed to delineate the precise effects of these common variants in the NLRP3 gene on the production of caspase-1–dependent cytokines as well as in the development of an adequate adaptive immune response.
The association herein presented stresses the importance of common variants in NALP genes on the outcome of HLA-identical sibling donor allo-SCT. These results should be validated in larger series, which would also allow performing subgroup analysis to investigate in what kind of transplants these variants have a deeper impact. In addition, the mechanisms underlying these associations should be investigated, including the differential effects of NALP variants on transcription and translation. An increasing number of articles are reporting the association of polymorphisms of non-HLA genes with clinical outcome after allo-SCT. All these reported gene polymorphisms should be analyzed together in one large series, including thousands of patients, to identify among them which actually have a clinical impact in the outcome after allo-SCT. A multicenter and multinational study is warranted.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Silvia Pairet for her excellent technical assistance.
This work was supported by Instituto de Salud Carlos III (RTICC03/10, FISS 050209) and Fundació La Caixa (BM05-219-0). M.G. is supported by a grant of the Spanish Society of Hematology (AEHH).
Authorship
Contribution: M.G., A.U.-I., J.I.A., and J.Y. designed the study; A.U.-I., E.M., and M.M. obtained funding; M.G., A.G., C.T., F.F.-A., C.M., M.R., and E.C. obtained clinical data; A.P., A.N., S.J., M.J., and R.A. obtained genetic data; B.G. and C.R. performed the statistical analysis; M.G., A.U.-I., and J.I.A. drafted the manuscript; and all the authors reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Álvaro Urbano-Ispizua, Hospital Universitario Virgen del Rocío, Servicio de Hematología y Hemoterapia, Avda Manuel Siurot sn, 41013 Sevilla, Spain; e-mail: alvaro.urbano.sspa@juntadeandalucia.es.