Abstract
Constitutive activation of the transcription factor STAT3 contributes to the pathogenesis of many cancers, including multiple myeloma (MM). Since STAT3 is dispensable in most normal tissue, targeted inhibition of STAT3 is an attractive therapy for patients with these cancers. To identify STAT3 inhibitors, we developed a transcriptionally based assay and screened a library of compounds known to be safe in humans. We found the drug nifuroxazide to be an effective inhibitor of STAT3 function. Nifuroxazide inhibits the constitutive phosphorylation of STAT3 in MM cells by reducing Jak kinase autophosphorylation, and leads to down-regulation of the STAT3 target gene Mcl-1. Nifuroxazide causes a decrease in viability of primary myeloma cells and myeloma cell lines containing STAT3 activation, but not normal peripheral blood mononuclear cells. Although bone marrow stromal cells provide survival signals to myeloma cells, nifuroxazide can overcome this survival advantage. Reflecting the interaction of STAT3 with other cellular pathways, nifuroxazide shows enhanced cytotoxicity when combined with either the histone deacetylase inhibitor depsipeptide or the MEK inhibitor UO126. Therefore, using a mechanistic-based screen, we identified the clinically relevant drug nifuroxazide as a potent inhibitor of STAT signaling that shows cytotoxicity against myeloma cells that depend on STAT3 for survival.
Introduction
Signal transducers and activators of transcription (STATs) are a family of transcription factors essential for the pathogenesis of many cancers.1 In normal cells, signaling initiated by cytokines and growth factors leads to the transient activation of STATs by tyrosine kinases, such as members of the Janus kinase (Jak) family. Activation involves phosphorylation of a tyrosine residue near the carboxyl terminus followed by dimerization of 2 STAT monomers. These activated STATs enter the nucleus, and regulate gene transcription by binding to specific DNA regulatory elements. STATs have long been known for their ability to up-regulate genes; however, emerging evidence demonstrates that STATs can also down-regulate key target genes.2,3 Soon after activation, STATs are typically dephosphorylated, ending the signaling process. In cancer, STAT activation is often constitutive.4 This can occur through a variety of mechanisms, such as positive signaling from mutated kinases, or by a lack of negative regulation by phosphatases or other feedback inhibitors. This continuously activated state leads to the inappropriate regulation of genes that are important for survival and proliferation, and that are known to contribute to the malignant phenotype.
STAT3 in particular has been found to be a critical mediator in the pathogenesis of many tumors. STAT3 is activated by several factors including IL-6, a cytokine essential for the survival of many cell lineages, including B lymphocytes and plasma cells. Multiple myeloma (MM) is a plasma cell malignancy often characterized by the constitutive activation of STAT3.5 In the bone marrow environment, cytokines such as IL-6 can be secreted by stromal cells or the myeloma cells themselves, and the presence of these cytokines can promote the continued survival and proliferation of MM cells.6 Thus, the pathogenesis of MM depends on STAT3 signaling, and interruption of this signaling pathway may lead to the death of these cancer cells.
To develop targeted inhibitors of STAT3 function for the treatment of MM and other forms of cancer, we developed a cell-based assay to measure STAT3-dependent transcriptional activity. To accelerate the identification of agents that could be used in clinical trials, we used this system to perform a high-throughput screen of compounds known to be bioactive, including many drugs presently in clinical use for a variety of diseases.
Methods
Cell culture
RPMI 8266, H929, and U266 myeloma cells were obtained from ATCC (Manassas, VA) and were grown in RPMI media containing 10% fetal calf serum. 293 human embryonic kidney cells (ATCC) were grown in DMEM containing 10% fetal calf serum. The IL-6–dependent INA6 myeloma cell line7 was obtained from Dr Renate Burger (University of Kiel, Kiel, Germany), and was grown in RPMI media containing 10% fetal calf serum supplemented with 1 ng/mL IL-6 (Peprotech, Rocky Hill, NJ). STAT1-null U3A cells and the parental 2FTGH cells were a gift from Dr George Stark (Cleveland Clinic, Cleveland, OH), and were grown in DMEM containing 10% fetal calf serum. T47D human mammary carcinoma cells (ATCC) were grown in DMEM containing 10% fetal calf serum. Bone marrow mononuclear cells from patients with untreated multiple myeloma were obtained through a Dana-Farber Cancer Institute IRB-approved protocol, and were cultured in RPMI media containing 10% fetal calf serum.
Antibodies
Antibodies recognizing STAT3 (sc-482), Jak2 (sc-278), Jak1 (sc-295), Tyk2 (sc-169), c-src (sc-18), and Mcl-1 (sc-819) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA); tyrosine-phosphorylated STAT3 (9131L), and pan-phospho-tyrosine (9411 and 9416) were from Cell Signaling Technology (Danvers, MA); epidermal growth factor receptor (EGFr) (2232), Parp (9542), p-STAT3 (9131L), and pan-phospho-tyrosine (9411 and 9416), p-src (2101), p-EGFr (2234), and p-Jak1 (3331) were from Cell Signaling Technology; anti–tyrosine 4G10 platinum (05-1050x) was from Millipore (Billerica, MA); tubulin was from Sigma (St Louis, MO); and serine-phosphorylated STAT3 was generated as described.8
Compounds
The Prestwick chemical library was obtained from Prestwick Chemical Company (Illkirch, France). Nifuroxazide (catalog no. 5227235) was obtained from Chembridge (San Diego, CA), and was dissolved in DMSO at a concentration of 150 mM. Jak inhibitor 1 (420099), WP1066 (573097), and AG490 (658401) were obtained from EMD (La Jolla, CA). The final concentration of DMSO in cellular experiments was 0.1%.
Generation of reporter cell lines
To measure STAT3-dependent luciferase activity, the STAT-luc/U3A cell line was used.9 A STAT1-responsive line (STAT-luc/2FTGH) was generated by cotransfecting pIRES-puro with a STAT-responsive luciferase plasmid10 into 2FTGH cells. A STAT5-responsive line (NCAM2-luc/T47D) was generated by cotransfecting pIRES-puro with a STAT5-responsive NCAM2-luciferase construct11 into T47D cells. An NF-κB–responsive cell line (NF-κB-luc/293) was generated by cotransfecting pIRES-puro with an NF-κB–responsive reporter plasmid (catalog no. 219078; Stratagene, La Jolla, CA) into 293 cells. After selection, individual clones were isolated. To assess reporter gene activity, STAT-luc/U3A cells were stimulated for 6 hours with 10 ng/mL IL-6; STAT-luc/2FTGH cells were stimulated with 500 units/mL IFNγ; NCAM2-luc/T47D cells were stimulated with 100 ng/mL prolactin; NF-κB-luc/293 cells were stimulated for 6 hours with 10 ng/mL TNFα. IL-6 and TNFα were obtained from Peprotech, whereas IFNγ and prolactin were obtained from R&D Systems (Minneapolis, MN).
Identification of STAT3 inhibitors
To identify STAT3 inhibitors, we screened the Prestwick Chemical Library, which contains 1120 bioactive compounds. Compounds were tested for their ability to modulate STAT3 activity using the STAT-luc/U3A cells. To eliminate compounds displaying nonspecific activity, a counter screen was performed using the NF-κB-luc/293 cells. Cells (2.5 × 103) were plated in triplicate in 30 μL media in an opaque 384-well dish. The following day, 100 nL (2 mg/mL) of each compound from the library was transferred to the cells. After one hour of incubation, cytokines were added at the indicated concentrations. For confirmation of inhibitory activity, and for testing compounds for STAT1 and STAT5 inhibitory activity, 104 cells were plated in opaque 96-well plates. The following day, drug was added for one hour, after which cytokines were added. Six hours later, luciferase activity was quantitated using the Bright-Glo Luciferase Assay System from Promega (Madison, WI) and a Luminoskan Ascent luminometer from Labsystems (Helsinki, Finland).
Western blotting and immunoprecipitation
Protein was isolated from myeloma cells by incubating cells on ice for 15 minutes in lysis buffer (50 mM Tris, pH 8.0, 250 mM NaCl, and 0.5% NP40) supplemented with sodium vanadate and Complete Protease Inhibitors (Roche, Indianapolis, IN). Lysates were cleared by centrifugation. Protein concentration was quantitated using the Bradford Reagent (Bio-Rad Laboratories, Hercules, CA). Lysates were mixed with 2× sample buffer (0.125 M Tris, pH 6.8, 4% SDS, 20% glycerol) containing 4% β-mercaptoethanol and boiled for 5 minutes. For STAT analysis, protein was resolved on an 8% acrylamide gel, and for Mcl-1 analysis, protein was resolved on a 12% gel, then transferred to nitrocellulose. Membranes were blocked in 5% milk in TBST (USB, Cleveland, OH) for 1 hour. Blots were washed in TBST and incubated with the appropriate horseradish peroxidase–labeled secondary antibody. Following washing in TBST, the blots were developed using Western Blot Chemiluminescence Reagent Plus (PerkinElmer, Boston, MA). For immunoprecipitations, cells were lysed on ice for 15 minutes in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 50 mM Tris, pH 7.4, 1% NP40, 0.5% sodium deoxycholate, and 0.1% SDS) supplemented with sodium vanadate and protease inhibitors. Lysates were cleared by centrifugation. The supernatant was incubated with the indicated antibody overnight. Protein A/G Plus beads (Santa Cruz Biotechnology) were added and incubated for 2 hours, after which the beads were washed 4 times in RIPA buffer. The beads were resuspended in 2× sample buffer containing 10% β-mercaptoethanol and boiled for 5 minutes. Quantitation of band intensity was performed using ImageJ software (National Institutes of Health, Bethesda, MD; http://rsb.info.nih.gov/ij/index.html).
Viability assays
Cells (104) were plated in opaque 96-well dishes. Compound or vehicle was added to yield a total volume of 100 μL. At the indicated times, cell viability was assessed using the CellTiter-Glo Luminescent Cell Viability Assay (Promega).
Bone marrow stromal cell culture
After IRB approval from the Dana-Farber Cancer Institute and informed consent in accordance with the Declaration of Helsinki, bone marrow specimens were obtained from patients with MM. Mononuclear cells (MNCs) separated by Ficoll-Hypaque density sedimentation were used to establish long-term bone marrow stromal cell (BMSC) cultures, as previously described,12 except no growth factors were added. When an adherent cell monolayer developed, cells were harvested in Hanks buffered saline solution containing 0.25% trypsin and 0.02% EDTA, washed, and collected by centrifugation. For coculture experiments, 5 × 105 BMSCs were seeded into a 25-cm2 flask and cultured overnight. The cells were washed 3 times with PBS, and U266 cells were added. The coculture was maintained for 48 hours after drug addition, after which, the U266 cells were collected for protein isolation.
Gene expression analysis
Cells were incubated for the indicated times with either compound or vehicle, and RNA was isolated using the RNeasy kit (QIAGEN, Valencia, CA). cDNA was generated using the Taqman reverse transcription kit (Applied Biosystems, Foster City, CA). Quantitative real-time polymerase chain reaction (PCR) was performed in triplicate using SYBR green master mix (Applied Biosystems) on a model 7500 real time PCR system (Applied Biosystems). Data are expressed as the mean fold change plus or minus SE of 3 replicates. Each assay was repeated at least 3 times. Primers used were Mcl-1, forward, GAGACCTTACGACGGGTT and reverse, TTTGATGTCCAGTTTCCG; HPRT, forward, GAACGTCTTGCTCGAGATGTG and reverse, CCAGCAGGTCAGCAAAGAATT.
Statistical analysis
Analysis of the effects of combinations of agents was performed using CalcuSyn software (Conservion, Ferguson, MO) using previously established parameters for determining synergy.13
Results
Identification of nifuroxazide as a STAT inhibitor
To assess the ability of compounds to inhibit STAT3 function, we used U3A cells stably expressing a luciferase reporter gene under the control of a STAT-dependent promoter.9 Basal luciferase expression is minimal in these cells. However, when they are treated with IL-6, robust and reproducible luciferase expression can be detected. Although STAT1 can also activate this promoter, U3A cells lack STAT1, and thus luciferase activity in these cells is completely dependent upon STAT3 function. We used this cell line to interrogate the Prestwick Library of 1120 bioactive compounds to identify clinically relevant drugs that specifically inhibit STAT3 function. To eliminate compounds that broadly inhibit gene expression, we performed a parallel counter screen using the NF-κB-luc/293 cell line in which luciferase expression is driven by the transcription factor NF-κB. Through this approach, we identified nifuroxazide as a potent inhibitor of STAT3 (Figure 1A). Nifuroxazide inhibited STAT3-dependent luciferase activity, with an EC50 of approximately 3 μM (Figure 1B). By contrast, at concentrations up to 10 μM, nifuroxazide had essentially no effect on NF-κB–dependent luciferase expression. Thus, nifuroxazide is a potent inhibitor of STAT3 function.
Nifuroxazide inhibits STAT3-dependent gene expression. (A) Chemical structure of nifuroxazide. (B) STAT3 and NF-κB reporter cells were pretreated with nifuroxazide at the indicated concentrations for 1 hour, after which cells were stimulated with IL-6 or TNF-α, respectively, for 6 hours. Luciferase activity was quantitated by luminometry.
Nifuroxazide inhibits STAT3-dependent gene expression. (A) Chemical structure of nifuroxazide. (B) STAT3 and NF-κB reporter cells were pretreated with nifuroxazide at the indicated concentrations for 1 hour, after which cells were stimulated with IL-6 or TNF-α, respectively, for 6 hours. Luciferase activity was quantitated by luminometry.
Nifuroxazide inhibits STAT3 tyrosine phosphorylation
Having shown that nifuroxazide inhibits STAT3 function, we next examined the mechanism for this effect. Since tyrosine phosphorylation is a critical event for STAT3 activation, we analyzed the effect of nifuroxazide on the constitutive STAT3 tyrosine phosphorylation found in U266 myeloma cells. When U266 cells were incubated with nifuroxazide, a significant dose-dependent decrease in STAT3 tyrosine phosphorylation was observed, reaching approximately 50% inhibition at a concentration of 10 μM (Figure 2A). This was comparable with the inhibition of STAT3-dependent reporter gene activity seen with this drug (Figure 1B). This inhibition of STAT3 tyrosine phosphorylation was rapid, occurring as early as one hour after treatment, and was sustained for at least 24 hours (Figure 2B). STAT3 can also be phosphorylated on a carboxy-terminal serine residue, serine 727, and this modification may modulate STAT3 transcriptional activity. U266 cells contain STAT3 that is constitutively phosphorylated on serine 727, but this modification is unaffected by nifuroxazide treatment (Figure 2B). Thus, nifuroxazide inhibits STAT3 activity through the inhibition of STAT3 tyrosine phosphorylation.
Nifuroxazide inhibits STAT3, Jak2, and Tyk2 tyrosine phosphorylation. (A) U266 cells were incubated with the indicated concentration of nifuroxazide for 24 hours, after which whole-cell lysates were prepared, and Western blot analysis was performed using antibodies specific for tyrosine-phosphorylated STAT3 (p-tyr STAT3) and total STAT3. (B) U266 cells were incubated for the indicated times with 10 μM nifuroxazide. Western blots were performed for tyrosine-phosphorylated STAT3, serine-phosphorylated STAT3 (p-ser STAT3), and total STAT3. (C) INA6 cells were incubated with 10 μM nifuroxazide or vehicle for 6 hours, after which immunoprecipitation was performed followed by Western blotting with the indicated antibodies. Quantitation of bands is shown for each panel.
Nifuroxazide inhibits STAT3, Jak2, and Tyk2 tyrosine phosphorylation. (A) U266 cells were incubated with the indicated concentration of nifuroxazide for 24 hours, after which whole-cell lysates were prepared, and Western blot analysis was performed using antibodies specific for tyrosine-phosphorylated STAT3 (p-tyr STAT3) and total STAT3. (B) U266 cells were incubated for the indicated times with 10 μM nifuroxazide. Western blots were performed for tyrosine-phosphorylated STAT3, serine-phosphorylated STAT3 (p-ser STAT3), and total STAT3. (C) INA6 cells were incubated with 10 μM nifuroxazide or vehicle for 6 hours, after which immunoprecipitation was performed followed by Western blotting with the indicated antibodies. Quantitation of bands is shown for each panel.
Nifuroxazide inhibits Jak kinase phosphorylation
Given these findings, we next determined whether an upstream tyrosine kinase is affected by nifuroxazide treatment. IL-6 autocrine and paracrine loops play a role in the pathogenesis of many cases of MM,14 and thus we focused on the Jak family kinases, Jak2, Tyk2, and Jak1, which are the central mediators of the effects of IL-6.15 To determine the effect of nifuroxazide on the activation of these kinases, we performed immunoprecipitations to determine their level of tyrosine phosphorylation, which reflects their kinase activity. IL-6–dependent INA6 MM cells were treated with 10 μM nifuroxazide for 6 hours, after which immunoprecipitations were performed to Jak2, Tyk2, Jak1, or STAT3. Western blotting was then performed using an antibody specific for phosphotyrosine to determine the activation state of these proteins. As expected, nifuroxazide led to a decrease in tyrosine phosphorylation of STAT3 by almost 70% (Figure 2C). In addition, the tyrosine phosphorylation of Jak2 and Tyk2 showed decreases of approximately 50%, whereas the low level of Jak1 phosphorylation was little changed. This suggested that nifuroxazide decreased STAT3 phosphorylation in these cells through an inhibition in upstream Jak kinases. To determine whether nifuroxazide inhibits tyrosine kinases broadly, we examined the effect of this drug on the EGF receptor tyrosine kinase (in HCC-827 cells) and Src (in A549 cells). Neither showed a significant change in activity (data not shown), indicating that nifuroxazide is relatively specific for Jak2 and Tyk2.
Having shown that nifuroxazide can inhibit Jak2 and Tyk2, we considered the possibility that nifuroxazide could inhibit the activation of other STAT family members that may be activated by these kinases. Using luciferase reporter systems dependent on the activation of STAT1 by interferon-γ or the activation of STAT5 in response to prolactin, we found that nifuroxazide could inhibit both of these reporter systems with a similar dose response as seen for STAT3 (data not shown). Thus, nifuroxazide can inhibit the activation of several STAT family members in Jak-dependent systems.
Nifuroxazide inhibits expression of the endogenous STAT3 target gene Mcl-1
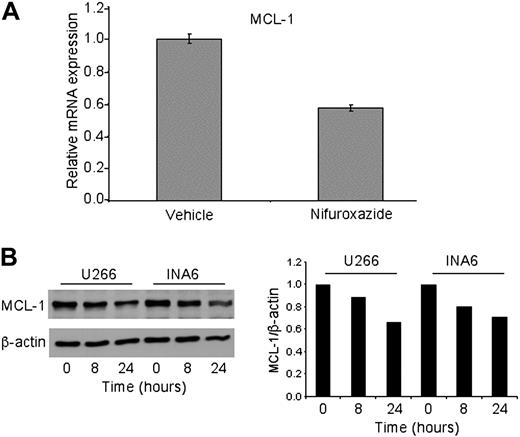
Having shown that nifuroxazide inhibits STAT3 phosphorylation and activation of a heterologous reporter gene, we next examined the effect of this drug on expression of an endogenous STAT3 target gene. We focused on Mcl-1, as this a key STAT3 target gene that is known to be critical for the aberrant survival and malignant phenotype of myeloma.16 Twenty-four–hour treatment of U266 cells with 10 μM nifuroxazide led to a 40% decrease in Mcl-1 mRNA expression when normalized to HPRT expression (Figure 3A). To determine whether this led to a loss of expression of Mcl-1 protein, Western analysis was performed on U266 and INA6 cells treated with nifuroxazide. The level of Mcl-1 protein was reduced at both 8 hours and 24 hours in both cell types (Figure 3B). Thus, nifuroxazide can decrease expression of a bona fide endogenous STAT3 target gene.
Nifuroxazide reduces the expression of Mcl-1. (A) U266 cells were treated with 10 μM nifuroxazide for 24 hours, and Mcl-1 mRNA expression was quantitated by quantitative reverse-transcription (RT)–PCR, using HPRT as an invariant control. (B) U266 and INA6 cells were treated with 10 μM nifuroxazide for the indicated times, and Western blot analysis was performed using antibodies to Mcl-1 or β-actin, which served as a loading control. Quantitation of the Western blot is shown at right.
Nifuroxazide reduces the expression of Mcl-1. (A) U266 cells were treated with 10 μM nifuroxazide for 24 hours, and Mcl-1 mRNA expression was quantitated by quantitative reverse-transcription (RT)–PCR, using HPRT as an invariant control. (B) U266 and INA6 cells were treated with 10 μM nifuroxazide for the indicated times, and Western blot analysis was performed using antibodies to Mcl-1 or β-actin, which served as a loading control. Quantitation of the Western blot is shown at right.
Nifuroxazide decreases viability of myeloma cells with activated STAT3
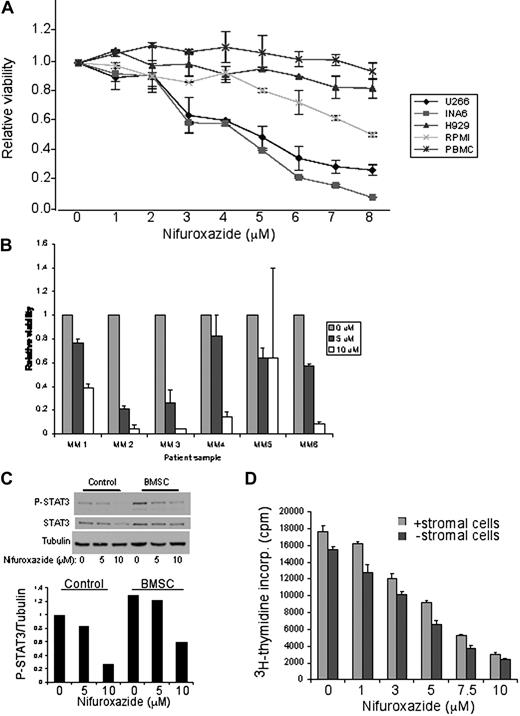
Given the evidence that STAT3 plays an essential role in the pathogenesis of myeloma, we next evaluated the effect of nifuroxazide on the viability of MM cell lines. We used U266 and INA6 myeloma cells that contain constitutive STAT3 phosphorylation, and RPMI 8226 and H929 cells that lack detectable STAT3 phosphorylation.17 Treatment of U266 or INA6 cells with nifuroxazide for 48 hours resulted in a dose-dependent loss of cell viability with an EC50 of approximately 4.5 μM in both cell types (Figure 4A). Notably, the MM cells lacking constitutive STAT3 activation showed little toxicity to nifuroxazide. Other known Jak inhibitors, including pyridone 6, AG490, and WP 1066, show similar effects in reducing INA6 viability (data not shown). Importantly, peripheral blood mononuclear cells from healthy donors showed essentially no toxicity to nifuroxazide treatment. These results suggest that nifuroxazide does not show nonspecific toxicity, and exerts a cytotoxic effect only on myeloma cells characterized by constitutive STAT3 activation.
Nifuroxazide inhibits the viability of MM cells containing constitutive STAT3 phosphorylation. (A) MM cells containing STAT3 phosphorylation (U266 and INA6), MM cells without STAT3 phosphorylation (RPMI 8226 and H929), and peripheral blood mononuclear cells (PBMCs) from healthy donors were treated with the indicated concentrations of nifuroxazide for 48 hours. Cell viability was measured using an ATP-dependent bioluminescence assay. (B) Bone marrow aspirates from MM patient samples were treated with the indicated concentrations of nifuroxazide for 72 hours. Cell viability was measured using an ATP-dependent bioluminescence assay. (C) U266 cells cultured in the presence or absence of BMSCs were treated with the indicated concentration of nifuroxazide for 6 hours, and then examined by Western blot analysis with the indicated antibodies. (D) U266 cells were cultured in the presence or absence of BMSCs for 48 hours with the indicated concentrations of nifuroxazide. 3H thymidine incorporation was measured during the final 8 hours of incubation to measure DNA synthesis.
Nifuroxazide inhibits the viability of MM cells containing constitutive STAT3 phosphorylation. (A) MM cells containing STAT3 phosphorylation (U266 and INA6), MM cells without STAT3 phosphorylation (RPMI 8226 and H929), and peripheral blood mononuclear cells (PBMCs) from healthy donors were treated with the indicated concentrations of nifuroxazide for 48 hours. Cell viability was measured using an ATP-dependent bioluminescence assay. (B) Bone marrow aspirates from MM patient samples were treated with the indicated concentrations of nifuroxazide for 72 hours. Cell viability was measured using an ATP-dependent bioluminescence assay. (C) U266 cells cultured in the presence or absence of BMSCs were treated with the indicated concentration of nifuroxazide for 6 hours, and then examined by Western blot analysis with the indicated antibodies. (D) U266 cells were cultured in the presence or absence of BMSCs for 48 hours with the indicated concentrations of nifuroxazide. 3H thymidine incorporation was measured during the final 8 hours of incubation to measure DNA synthesis.
It is notable that nifuroxazide causes a prominent decrease in viability of INA6 cells while causing only partial inhibition of STAT3 phosphorylation. To determine whether this was a characteristic of other Jak kinase inhibitors, we performed detailed quantitative dose-response analyses of both cellular viability and STAT3 phosphorylation in INA6 cells treated with nifuroxazide or 2 other Jak inhibitors, WP1066 and Jak inhibitor 1. At concentrations of these drugs that caused a 90% decrease in viability, nifuroxazide caused a 50% decrease in STAT3 phosphorylation (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Similarly, WP1066 induced a 60% decrease in STAT3 phosphorylation, whereas Jak inhibitor 1 led to a 90% decrease in STAT3 phosphorylation. Thus, even incomplete inhibition of STAT3 phosphorylation can be associated with inhibition of expression of key target genes (Figure 3) and substantial decreases in cellular viability.
Nifuroxazide leads to a loss of viability of primary myeloma cells
Although nifuroxazide shows considerable efficacy in decreasing the viability of multiple myeloma cell lines characterized by STAT3 activation, we wished to exclude the possibility that this was a feature unique to myeloma cells that had been selected for their ability to grow in vitro. We obtained bone marrow specimens from 6 untreated myeloma patients, and assessed the effect of nifuroxazide on the viability of these cells. When the cells were treated with nifuroxazide for 72 hours, the viability of all of the patient specimens decreased with a dose response comparable with that seen with U266 or INA6 cells (Figure 4B). Thus, nifuroxazide is active against primary myeloma samples.
Nifuroxazide overcomes the prosurvival effects of bone marrow stromal cells
Myeloma cells in vivo depend on survival and proliferation signals from bone marrow stromal cells, and thus it is essential for any effective treatment to overcome this effect.6 To assess the effects of nifuroxazide on MM cells in this environment, we first analyzed the effect of BMSC coculture on STAT3 activation of the myeloma cells. Consistent with the prosurvival effects of this environment, coculture of myeloma cells with BMSCs led to enhanced STAT3 tyrosine phosphorylation in the myeloma cells (Figure 4C). However, nifuroxazide led to a similar decrease in STAT3 phosphorylation in the presence or absence of BMSCs. Furthermore, as expected, the presence of bone marrow stromal cells also led to increased proliferation of U266 cells (Figure 4D). However, as with its effect on STAT3 phosphorylation, nifuroxazide reduced the proliferation of myeloma cells even in the presence of stroma, with a similar dose response as seen in monoculture. Thus, nifuroxazide is effective at decreasing the activation of STAT3 and reducing the viability of myeloma cells in an environment closely resembling that seen in vivo.
Consistent with the enhanced survival and proliferation seen when MM cells are cultured in the presence of BMSCs, phosphorylation of MAP kinase and Akt is also increased in U266 cells under coculture conditions (Figure S1). Interestingly, in contrast to the effect of nifuroxazide on STAT3 phosphorylation, nifuroxazide caused a further increase in both MAP kinase and Akt phosphorylation. Thus, the effects of nifuroxazide in decreasing survival and proliferation of myeloma cells are not mediated by inhibition of these pathways, and are more likely to be secondary to inhibition of STAT3.
The combination of nifuroxazide and the histone deacetylase inhibitor depsipeptide leads to enhanced toxicity of MM cells
There is evidence that the simultaneous inhibition of 2 signaling pathways is necessary to induce apoptosis of MM cells in the presence of bone marrow stromal cells.18 Thus we considered the possibility that a STAT3 inhibitor such as nifuroxazide might show enhanced cytotoxicity in combination with an agent that targeted a different mechanism. Neither doxorubicin nor bortezomib showed significant combinatorial effects with nifuroxazide (data not shown). However, increasing evidence has suggested that histone deacetylase (HDAC) inhibitors may show significant therapeutic effects in hematologic malignancies.19 We focused on depsipeptide, a natural product with extremely potent effects as an HDAC inhibitor.20 When INA6 cells were treated with the combination of nifuroxazide and picomolar concentrations of depsipeptide, nearly complete cytotoxicity could be induced (Figure 5A), with evidence of synergism with 3 μM nifuroxazide and 750 nM depsipeptide, and 5 μM nifuroxazide with either concentration of depsipeptide (combination index13 < 0.5). This combinatorial effect did not occur in H929 cells that lack activated STAT3 (data not shown). Nifuroxazide combined with depsipeptide in INA6 cells resulted in an enhanced reduction in STAT3 tyrosine phosphorylation, as well as an increase in Parp cleavage, indicating increased apoptosis (Figure 5B). Thus, the combination of STAT3 inhibition and HDAC inhibition may be a useful strategy for clinical trials for myeloma.
The combination of nifuroxazide and depsipeptide results in enhanced cytotoxicity of MM cells. (A) INA6 cells were incubated with the indicated doses of nifuroxazide and depsipeptide for 72 hours. Viability was measured using an ATP-dependent bioluminescence assay. (B) INA6 cells were incubated with the indicated doses of nifuroxazide and depsipeptide for 48 hours, after which Western analysis was performed with the indicated antibodies.
The combination of nifuroxazide and depsipeptide results in enhanced cytotoxicity of MM cells. (A) INA6 cells were incubated with the indicated doses of nifuroxazide and depsipeptide for 72 hours. Viability was measured using an ATP-dependent bioluminescence assay. (B) INA6 cells were incubated with the indicated doses of nifuroxazide and depsipeptide for 48 hours, after which Western analysis was performed with the indicated antibodies.
The combination of nifuroxazide and MEK inhibition leads to enhanced killing of MM cells
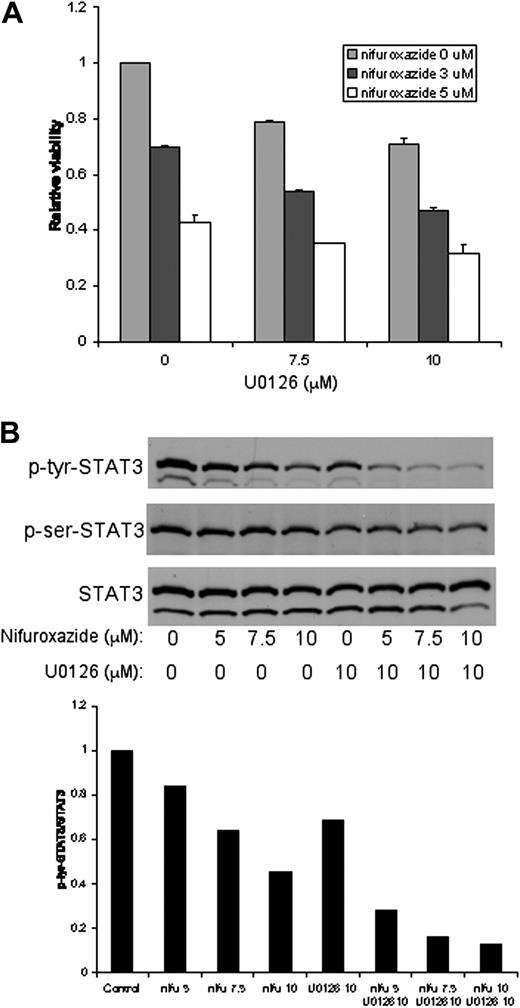
The MAP kinase pathway has been linked to survival signals for cancer cells, and inhibition of this pathway, together with the IL-6/STAT3 signaling pathway, leads to a loss of viability of MM cells.18 This suggested that combining a MAPK inhibitor with nifuroxazide might enhance the cytotoxicity of nifuroxazide. To determine whether inhibition of MAPK and STAT3 would result in enhanced cytotoxicity, we combined the MEK inhibitor UO126 with nifuroxazide and examined cellular viability. U266 cells treated with this combination showed enhanced cytotoxicity compared with either treatment alone (Figure 6A). Similar results were seen with INA6 cells (data not shown). H929 cells that have no activated STAT3 do not show this combinatorial effect (data not shown). These results suggest that clinically relevant MAPK inhibitors may be useful in combination with nifuroxazide for the treatment of patients with MM.
The combination of nifuroxazide and the MEK inhibitor UO126 results in enhanced cytotoxicity of MM cells and enhanced loss of STAT3 tyrosine phosphorylation. (A) U266 cells were incubated with the indicated concentrations of nifuroxazide and UO126 for 48 hours. Viability was measured using an ATP-dependent bioluminescence assay. (B) U266 cells were incubated with the indicated concentrations of nifuroxazide and UO126 for 24 hours. Western blots were performed for tyrosine-phosphorylated STAT3, serine-phosphorylated STAT3 (p-ser STAT3), and total STAT3.
The combination of nifuroxazide and the MEK inhibitor UO126 results in enhanced cytotoxicity of MM cells and enhanced loss of STAT3 tyrosine phosphorylation. (A) U266 cells were incubated with the indicated concentrations of nifuroxazide and UO126 for 48 hours. Viability was measured using an ATP-dependent bioluminescence assay. (B) U266 cells were incubated with the indicated concentrations of nifuroxazide and UO126 for 24 hours. Western blots were performed for tyrosine-phosphorylated STAT3, serine-phosphorylated STAT3 (p-ser STAT3), and total STAT3.
The MAPK pathway has been linked to the serine phosphorylation of STAT3, which can enhance its transcriptional activity.21 Since STAT3 shows constitutive phosphorylation of serine 727 in both U266 and INA6 cells (Figure 2 and data not shown), we considered the possibility that the combinatorial effect of nifuroxazide with U0126 may be due to the loss of both tyrosine and serine phosphorylation of STAT3. When U266 cells are treated with U0126, a slight reduction in serine phosphorylation of STAT3 is seen (Figure 6B). Nifuroxazide alone has no effect on serine phosphorylation of STAT3, and nifuroxazide does not enhance the inhibition of STAT3 phosphorylation induced by U0126. We next considered the possibility that these drugs might cooperate in inhibiting the tyrosine phosphorylation of STAT3. Whereas U0126 alone shows a modest reduction in STAT3 tyrosine phosphorylation, the combination of nifuroxazide and UO126 leads to nearly complete inhibition of this modification (Figure 6B). This suggests that the enhanced cytotoxicity of nifuroxazide combined with UO126 may be due to increased inhibition of tyrosine phosphorylation of STAT3.
Discussion
Although STAT3 is a critical mediator of the oncogenic phenotype of many cancers, it is not required for the function of most normal cells.1 Thus, finding effective inhibitors of STAT3 may provide useful targeted agents for cancer therapy. Although much effort has gone into the development of STAT3 inhibitors, it has been difficult to introduce these agents into clinical trials. To circumvent this problem, we sought to identify drugs known to be safe in humans that might act as effective STAT3 inhibitors. Starting with a library of known bioactive agents, we identified the antidiarrheal agent nifuroxazide as a potent inhibitor of STAT3. Nifuroxazide reduces STAT3 tyrosine phosphorylation, and inhibits the viability of both primary myeloma cells and myeloma cell lines containing constitutive STAT3 activation. Nifuroxazide also showed combinatorial effects on the viability of myeloma cells when used in conjunction with the histone deacetylase inhibitor depsipeptide, as well as with the MEK inhibitor UO126. Given that nifuroxazide showed no effect on normal hematopoietic cells, this drug may be useful in clinical trials for the treatment of patients with multiple myeloma.
Although not currently available for clinical use in the United States, nifuroxazide is available in many countries worldwide. In randomized double-blind control trials, nifuroxazide has shown benefit in adults with acute diarrhea, and is extremely well tolerated.22 Since it is generally given orally, its systemic bioavailability is unclear, and little information is available on the parenteral administration of nifuroxazide. However, it is chemically related to nitrofurantoin, which does show good oral absorption, and is used widely for the treatment of urinary tract infections. In addition, several analogs of nifuroxazide have been synthesized,23,24 and one or more of these may show STAT3 inhibitory activity as well.
Nifuroxazide appears to inhibit STAT3 phosphorylation through inhibition of the Jak family kinases Jak2 and Tyk2. Given that the constitutive activation of STATs in many tumors is due to aberrant activation of Jaks,25,26 nifuroxazide may be useful for the treatment of these other tumors as well. Nifuroxazide is not a nonspecific tyrosine kinase inhibitor, as it showed no effects against the EGF receptor kinase or Src. Although the full spectrum of kinases inhibited by nifuroxazide is not yet known, increasing evidence supports the utility of multikinase inhibitors in oncology.27 Thus, nifuroxazide may have broader efficacy as well.
At active concentrations, nifuroxazide inhibits STAT3 tyrosine phosphorylation by approximately 50%, and decreases STAT3-dependent gene expression by 50%. This is consistent with findings from other approaches to inhibit STAT3 that there is likely a threshold effect at which STAT3-dependent gene products promote malignant cellular behavior. For example, partial depletion of constitutively active STAT3 leads to prominent effects on viability in lung cancer cells.28 Similarly, in prostate cancer cells, inhibition of STAT3 phosphorylation with low concentrations of the Jak inhibitor AG490 (25-50 μM) leads to partial inhibition of STAT3 activation, but significant effects on cellular viability.29 Thus, even partial inhibition of STAT3 is likely to confer therapeutic benefit, although these findings can also reflect effects of these drugs on additional targets. In this context, it is also notable that nifuroxazide showed no inhibition of Akt and MAP kinase phosphorylation. This provides evidence that the effects of this drug on myeloma cell survival and proliferation are more likely mediated by its inhibition of STAT signaling than one of these other pathways.
Since many STAT3 target genes are prosurvival genes, it is not surprising that STAT3 inhibition may show combinatorial effects when paired with other modalities of therapy. We have shown an additive effect of nifuroxazide on the viability of myeloma cells when used in combination with the MEK inhibitor U0126. It has been suggested that MAP kinases can mediate the serine phosphorylation of STAT3,30 and thus the combination of a MEK inhibitor and an inhibitor of STAT3 tyrosine phosphorylation might show synergy based on this mechanism. Inhibition of MAPK showed only a slight reduction in the constitutive serine phosphorylation of STAT3 found in myeloma cells. However, the combination of U0126 with nifuroxazide caused a pronounced decrease in STAT3 tyrosine phosphorylation. Although the mechanism for this is unclear, this finding shows another level of cross talk between the STAT3 and MAPK pathways, and suggests that the mechanism of the combinatorial effect of nifuroxazide and U0126 is likely through their convergent effects on STAT3 tyrosine phosphorylation.
Nifuroxazide also shows enhanced cytotoxicity to multiple myeloma cells when combined with the HDAC inhibitor depsipeptide, a drug currently in clinical trials for the treatment of hematologic malignancies. The exact mechanism by which HDAC inhibitors exert their therapeutic effect is unclear, but may involve enhanced acetylation of lysine residues in histones, transcription factors, and other cellular proteins.31 This can lead to changes in gene expression, though depsipeptide may also cause cell death through caspase activation.32 Thus, it is possible that the decrease in prosurvival genes mediated by STAT3 inhibition may cooperate with caspase activation mediated by depsipeptide to enhance cell killing. We demonstrate that depsipeptide and nifuroxazide show a combinatorial effect on STAT3 activation, with a greater reduction in STAT3 phosphorylation compared with cells treated with either drug alone. In addition, this drug combination results in enhanced apoptosis as measured by Parp cleavage. Given the known safety characteristics of both nifuroxazide and depsipeptide, these drugs may be useful in clinical trials for the treatment of myeloma.
In conclusion, the development of a nonbiased functional screen to identify STAT3 inhibitors in conjunction with a chemical library biased to drugs known to be safe in humans has allowed the identification of nifuroxazide as an effective inhibitor of STAT3. Nifuroxazide decreases the viability of multiple myeloma cells with constitutive activation of STAT3, while showing minimal effects on nontransformed and normal cells. In addition, this drug shows enhanced effects with agents that target other cellular pathways. Given that STAT3 activation is a common event in a wide range of human cancers, and this drug has been given safely to humans for many years, it is possible that clinical trials exploiting this approach could be developed in a rapid time frame.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the National Cancer Institute and the Initiative for Chemical Genetics (Bethesda, MD), who provided support for this publication, and the Chemical Biology Platform of the Broad Institute of Harvard and MIT (Boston, MA) for their assistance in this work. The collection of tissue samples was supported by the Ted and Eileen Pasquarello Tissue Bank for Hematologic Malignancies (Dana-Farber Cancer Institute). We thank Roberto Bellucci for preparing and providing normal primary cells.
This work was supported by the Multiple Myeloma Research Foundation (Norwalk, CT) and the Brent Leahey Fund (Dana-Farber Cancer Institute).
National Institutes of Health
Authorship
Contribution: E.A.N. designed the research, performed experiments, and wrote the paper; S.R.W. designed the research and performed experiments; A.K., L.B.G., H.I., and T.H. performed experiments; D.C. and K.C.A. designed the research; and D.A.F. designed the research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: David A. Frank, Dana-Farber Cancer Institute, Department of Medical Oncology, Mayer 522B, 44 Binney Street, Boston, MA 02115; e-mail: david_frank@dfci.harvard.edu.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal