Abstract
To characterize genetic contributions toward aberrant splicing of the hyaluronan synthase 1 (HAS1) gene in multiple myeloma (MM) and Waldenstrom macroglobulinemia (WM), we sequenced 3616 bp in HAS1 exons and introns involved in aberrant splicing, from 17 patients. We identified a total of 197 HAS1 genetic variations (GVs), a range of 3 to 24 GVs/patient, including 87 somatic GVs acquired in splicing regions of HAS1. Nearly all newly identified inherited and somatic GVs in MM and/or WM were absent from B chronic lymphocytic leukemia, nonmalignant disease, and healthy donors. Somatic HAS1 GVs recurred in all hematopoietic cells tested, including normal CD34+ hematopoietic progenitor cells and T cells, or as tumor-specific GVs restricted to malignant B and plasma cells. An in vitro splicing assay confirmed that HAS1 GVs direct aberrant HAS1 intronic splicing. Recurrent somatic GVs may be enriched by strong mutational selection leading to MM and/or WM.
Introduction
Splicing of pre-mRNA contributes to protein diversity in humans. Pre-mRNA splicing is regulated by cis- and trans-splicing elements, involving a complex repertoire of splicing factors, with spliceosome assembly directed by splicing motifs in the DNA template.1 Evidence is accumulating for an association of defective gene splicing with susceptibility to and progression of cancer.2,3 Genetic variations underlying cancer are linked to altered splicing.4-6 Genetic changes in malignant cells can alter the genomic context of splice sites by activating otherwise weak or cryptic splice sites, leading to aberrant exon skipping, intron retention, or both.1 Inherited polymorphisms and somatic (acquired) mutations underlie aberrant splicing in cancer.4-8 Most aberrantly spliced products involve loss of tumor suppressor activity.3,6,7 However, 2 genes, cyclin D14,9 and hyaluronan synthase 1 (HAS1),10-12 undergo aberrant splicing to generate proteins with new functions or localizations that may directly promote cancer.
Hyaluronan synthases have been implicated in malignant transformation.13-16 HAS1 overexpression was described in patients with multiple myeloma (MM),10 Waldenstrom macroglobulinemia (WM),11 and colon,17 ovarian,18,19 bladder,20,21 and endometrial carcinomas.22 We identified a family of aberrant HAS1 splice variants, termed HAS1Va, HAS1Vb, and HAS1Vc10 in MM and WM that are undetectable in B-chronic lymphocytic leukemia (B-CLL) and healthy donors (HDs).10,11 Aberrant intronic splicing of HAS1 pre-mRNA, spanning exons 3 to 5, correlates with significantly reduced survival in MM.10 In WM, up-regulation of HAS1 intronic splice variants occurs in a majority of CD20+ B-lineage cells, detected by single-cell reverse transcriptase–polymerase chain reaction (RT-PCR).11 HAS1Va has been detected in bladder cancer.20
Aberrant splicing results from genetic variations (GVs), including substitutions, deletions, and insertions, detected in the sequence of classical splicing elements and within exons and introns.23-30 These mutations include missense mutations, which change amino acids and consequently protein function, and nonsense or silent mutations leading to frame-shifting and novel protein production.27,31-33 “Deep” intronic mutations may cause aberrant splicing of disease-related genes by creating or strengthening cryptic splicing elements.34,35 GVs promote aberrant splicing in genes encoding MLH1, MLH2,8 CHEK2,6 RB1,36,37 p53,5 NF1,28 BRCA1,38,39 PTEN,40 and the cystic fibrosis transmembrane conductance regulator.41
Here, the contributions of germ line origin or malignant-cell specific GVs to aberrant HAS1 splicing were determined by extensive sequencing of HAS1 gene segments from buccal epithelial cells (BECs), hematopoietic progenitor cells (HPCs), T cells, B cells, and plasma cells (PCs) obtained from patients with MM and WM. BECs represent the germ line “host” genotype, whereas HPCs, T cells, B cells, and PCs represent normal and malignant components of the hematopoietic lineage in patients with MM and WM. We found a total of 197 GVs in 17 patients with MM and WM. Nearly all of these GVs were absent from 23 control subjects, including 4 with B-CLL, 11 with monoclonal gammopathy of undetermined significance (MGUS), and 8 HDs. In addition to 12 known HAS1 polymorphisms, in 17 patients we found 46 novel germ line GVs, 87 somatic GVs, and 52 unclassified GVs among all tested cell types. Among somatic GVs, defined as those that were absent from autologous BECs, 26 were found in presumptively normal HPCs and T cells from patients with MM and WM, and 61 tumor-specific GVs were found only in B cells and PCs of patients with MM or WM or both. An in vitro splicing assay confirmed that a combination of germ line and somatic GVs leads to aberrant HAS1 splicing.
Methods
Patients and controls
The present study includes samples from 10 patients with MM (age range, 41-85 years; mean, 64 years), 7 patients with WM (age range, 44-76 years; mean, 63 years), and, as control subjects, 4 with B-CLL, 11 with MGUS (age range, 61-84 years), and 8 HDs (age range, 50-69 years, with the exception of 1 HD who was 30 years of age). Patients with MM included 5 with IgG myeloma and 5 with IgA myeloma; 8 had detectable bone lesions. There are no known familial forms of disease, and no familial relationships among the patients analyzed with MM and WM. The patients with MM and WM expressed at least one HAS1 splice variant. Approximately 60% of patients have such variants.10,11 Patients were recruited independently at the Cross Cancer Institute and the University of Alberta Hospital (Edmonton, AB), and the Dana-Farber Cancer Institute (Boston, MA). All patients were diagnosed according to the recommendations of the International Myeloma Working Group,42 and the consensus guidelines from the 2nd International Workshop on Waldenstrom Macroglobulinemia.43 Peripheral blood (PB), bone marrow (BM), and BEC samples were taken at the time of diagnosis or at follow-up, after approval from the University of Alberta and the Alberta Cancer Board Institutional Review Boards or the Dana-Farber Institutional Review Board; informed consent in accordance with the Declaration of Helsinki was obtained from all subjects. All patients and HDs included in this study were of white origin.
Tissue and sample preparation
Tissue and sample preparation and cell sorting were conducted as previously described.44 gDNA samples from sorted cells and unfractionated peripheral blood mononuclear cells (PBMCs) or BM cells (BMCs) were isolated using QIAamp DNA Blood mini kit (QIAGEN, Mississigua, ON) according to the manufacturer's instructions. For 17 patients with MM and WM sequencing of all indicated HAS1 gene segments and HAS1 minigenes was performed on populations of 6 sorted PB B cells, 3 sorted BM B cells, 2 sorted PCs, 6 sorted T cells, 8 sorted CD34+ HPCs, 2 unfractionated PBMCs, 3 unfractionated BMCs, and 8 BECs. Among the 23 control subjects, HAS1 gene segments were analyzed from purified B cells of 4 B-CLL (intron 4), unfractionated PBMCs from 11 MGUS, sorted B and T cells from 2 healthy donors and unfractionated PBMCs from 6 HDs.
Cloning and sequencing
Cloning and sequencing of HAS1 gene segments used gDNA isolated from BECs, purified cell subsets, BMCs, or PBMCs obtained from patients with MM, patients with WM, or HDs and PBMCs from MGUS and purified B cells from B-CLL. Patients with MM and WM expressed HAS1 and its splice variants. The HAS1 genomic segments from exon 3 to exon 5 were amplified, cloned, and sequenced using a series of primer sets (Figure 1; Table 1) spanning the region involved in the observed HAS1 aberrant splicing (exons 3-4 and introns 3-4). The reverse and forward primers used in these PCR reactions were significantly overlapped to evaluate the accuracy of PCR and sequencing in multiple sequencing reactions (Figure 1). PCR products were cloned, and 3 to 19 subclones were sequenced in both directions for each HAS1 gene segment from each subset of cells, using the ABI3130xl DNA capillary analysis system (Applied Biosystems, Foster City CA). For one small segment of intron 4 from one patient, only 2 subclones were sequenced. Because of primer overlap for HAS1 gene segments, we obtained up to 60 sequencing reactions for each sample, which included the approximately 40 to 100 bp range of exon-intron boundaries of the HAS1 gene.
Strategies for sequencing the HAS1 gene segments. Described are 2 strategies used to amplify the HAS1 gene segments. Overlapping reverse and forward primers were designed to anneal with exons and introns of the HAS1 gene to identify genetic variations that may contribute to aberrant HAS1 splicing in patients with WM. Minigene sequencing in strategy 2 was used to determine whether the recurrent mutations were clustered. gDNA PCR and cloning were carried out as described in “Methods.” We picked 24 to 48 subclones to screen and sequence inserts (gDNA PCR product of the HAS1 gene segment) in the TOPO TA plasmid using the appropriate primer sets for each segment. While using the first strategy of amplifying HAS1 gene segments from the patients or HDs, we cloned 7 or 3 segments from the exon 3 to exon 5 region of the HAS1 gene using A or B primer sets, respectively. For each segment, 3 to 10 positive subclones were selected, and, for each cell subset, more than 50 plasmids were isolated and sequenced both directions using M13 and T7 sequencing primers. Using strategy 2, we cloned 30 HAS1 minigene plasmids from MM and WM, and 33 minigenes from B-CLL and MGUS. B-CLL minigenes encompassed only intron 4. MGUS minigenes encompassed exon 3 to exon 5. Each plasmid was sequenced using overlapping HAS1 gene-specific A and B primer sets. Because we used overlapping primers either in gDNA PCR (strategy 1) or for sequencing (strategy 2), we analyzed 50 to 60 sequencing reactions for exon-intron spanning segments of the HAS1 gene. The HAS1 gene segments of 2 HDs (B and T cells) were sequenced using strategy 1. The genetic variations identified in patients with MM and WM were assessed, based on a total of 4119 sequencing reactions.
Strategies for sequencing the HAS1 gene segments. Described are 2 strategies used to amplify the HAS1 gene segments. Overlapping reverse and forward primers were designed to anneal with exons and introns of the HAS1 gene to identify genetic variations that may contribute to aberrant HAS1 splicing in patients with WM. Minigene sequencing in strategy 2 was used to determine whether the recurrent mutations were clustered. gDNA PCR and cloning were carried out as described in “Methods.” We picked 24 to 48 subclones to screen and sequence inserts (gDNA PCR product of the HAS1 gene segment) in the TOPO TA plasmid using the appropriate primer sets for each segment. While using the first strategy of amplifying HAS1 gene segments from the patients or HDs, we cloned 7 or 3 segments from the exon 3 to exon 5 region of the HAS1 gene using A or B primer sets, respectively. For each segment, 3 to 10 positive subclones were selected, and, for each cell subset, more than 50 plasmids were isolated and sequenced both directions using M13 and T7 sequencing primers. Using strategy 2, we cloned 30 HAS1 minigene plasmids from MM and WM, and 33 minigenes from B-CLL and MGUS. B-CLL minigenes encompassed only intron 4. MGUS minigenes encompassed exon 3 to exon 5. Each plasmid was sequenced using overlapping HAS1 gene-specific A and B primer sets. Because we used overlapping primers either in gDNA PCR (strategy 1) or for sequencing (strategy 2), we analyzed 50 to 60 sequencing reactions for exon-intron spanning segments of the HAS1 gene. The HAS1 gene segments of 2 HDs (B and T cells) were sequenced using strategy 1. The genetic variations identified in patients with MM and WM were assessed, based on a total of 4119 sequencing reactions.
Primer sets used for PCR and/or sequencing reactions
| HAS1 primers . | Sequence 5′ to 3′ . |
|---|---|
| 5′ exon 3 | GGGGTCTGTGCTGATCCTGG |
| 3′ exon 3 | GCTTCCAGTTTTATCCCATC |
| 5′ intron 3 | CTTCCACTGTGTATCCTGCATC |
| 3′ intron 3 | AACTGCTGCAAGAGGTTATTCC |
| 5′ exon 4 | TGGGGTTGGAACTGGAGATG |
| 3′ exon 4 | CATGCACACACGCTAGGATA |
| 5′ intron 4a | GCTCAGCATGGGTTATGCTA |
| 3′ intron 4a | GTATCCCCGCAGCTTAAACA |
| 5′ intron 4b | TTGGGATAATCCAGGGGAAT |
| 3′ intron 4b | CAAGATGGGTGTGGTTGCTA |
| 5′ intron 4c | GGTAGCAACCACACCCATCT |
| 3′ intron 4c | AGGAATGAGGGCATCATCG |
| 5′ exon 5a | CTCGCCCCCGTGCAGGTACA |
| 3′ exon 5a | AGGCCCCCAAGCAGCAGCAGCGC |
| 5′ exon 3-exon 4* | ATGGGATAGGCTTGGAGTCA* |
| 3′ exon 3-exon 4* | CCCATCCAAAACCCACTGCA* |
| 5′ exon 4-intron 4 (a)* | GGAGACCAAGGTAGCACAGT* |
| 3′ exon 4-intron 4 (a)* | GTCTCTTGCCCTTCCTACTT* |
| 5′ exon 4-intron 4 (b)* | CAAGGGTGGTGGATAGGAAGTT* |
| 3′ exon 4-intron 4 (b)* | CCTCAGGCACTCCACTTAACAC* |
| 5′ intron 4-exon 5 (a)* | GGGCACGATCATGGCTCACT* |
| 3′ exon 4-intron 4 (a)* | GAGGTAGGGGGATCACTTGA* |
| 5′ intron 4-exon 5 (b)* | CCCCAGGGAGCACGCGATGA* |
| 3′ exon 4-intron 4 (b)* | GCACGGGGGCGAGGAATGAG* |
| 5′ minigene | CTTCCACCTTACAGGTCTGTGACT |
| 3′ minigene | CCACTCTGGTTCATGGTGACTA |
| 5′intron 4c (T) | GCAAATACTATCTACTGGTCCTAAGC |
| 3′intron 4c (T) | TGTGTAGAATAGGGTGGATAATGGT |
| M13R | CAGGAAACAGCTATGAC |
| T7 | TAATACGACTCACTATAGGG |
| HAS1 primers . | Sequence 5′ to 3′ . |
|---|---|
| 5′ exon 3 | GGGGTCTGTGCTGATCCTGG |
| 3′ exon 3 | GCTTCCAGTTTTATCCCATC |
| 5′ intron 3 | CTTCCACTGTGTATCCTGCATC |
| 3′ intron 3 | AACTGCTGCAAGAGGTTATTCC |
| 5′ exon 4 | TGGGGTTGGAACTGGAGATG |
| 3′ exon 4 | CATGCACACACGCTAGGATA |
| 5′ intron 4a | GCTCAGCATGGGTTATGCTA |
| 3′ intron 4a | GTATCCCCGCAGCTTAAACA |
| 5′ intron 4b | TTGGGATAATCCAGGGGAAT |
| 3′ intron 4b | CAAGATGGGTGTGGTTGCTA |
| 5′ intron 4c | GGTAGCAACCACACCCATCT |
| 3′ intron 4c | AGGAATGAGGGCATCATCG |
| 5′ exon 5a | CTCGCCCCCGTGCAGGTACA |
| 3′ exon 5a | AGGCCCCCAAGCAGCAGCAGCGC |
| 5′ exon 3-exon 4* | ATGGGATAGGCTTGGAGTCA* |
| 3′ exon 3-exon 4* | CCCATCCAAAACCCACTGCA* |
| 5′ exon 4-intron 4 (a)* | GGAGACCAAGGTAGCACAGT* |
| 3′ exon 4-intron 4 (a)* | GTCTCTTGCCCTTCCTACTT* |
| 5′ exon 4-intron 4 (b)* | CAAGGGTGGTGGATAGGAAGTT* |
| 3′ exon 4-intron 4 (b)* | CCTCAGGCACTCCACTTAACAC* |
| 5′ intron 4-exon 5 (a)* | GGGCACGATCATGGCTCACT* |
| 3′ exon 4-intron 4 (a)* | GAGGTAGGGGGATCACTTGA* |
| 5′ intron 4-exon 5 (b)* | CCCCAGGGAGCACGCGATGA* |
| 3′ exon 4-intron 4 (b)* | GCACGGGGGCGAGGAATGAG* |
| 5′ minigene | CTTCCACCTTACAGGTCTGTGACT |
| 3′ minigene | CCACTCTGGTTCATGGTGACTA |
| 5′intron 4c (T) | GCAAATACTATCTACTGGTCCTAAGC |
| 3′intron 4c (T) | TGTGTAGAATAGGGTGGATAATGGT |
| M13R | CAGGAAACAGCTATGAC |
| T7 | TAATACGACTCACTATAGGG |
Primer set intron 4c (T) was used to verify common motif sequences 1st T, 2nd T, and TTTA stretches. Primer sequences were used in sequencing reactions only.
For each subject, we also generated up to 15 minigenes of 3503 bp (chromosomal location 56912272-56908769) in length, which spanned gDNA of HAS1 exon 3 to exon 5. A minigene is the product of a gDNA PCR reaction. Usually, this segment includes exons and introns of any given gene, preserving the linkages among genetic variations on the DNA strand that was copied. In our case, HAS1 minigenes include the genomic sequence of HAS1 exons 3, 4, and 5 and introns 3 and 4. The main purpose for using minigenes in this study was to determine whether recurrent GVs are expressed as clusters and if they belong to the same allele. Each minigene was sequenced in both directions using HAS1 gene primer sets shown in Figure 1. Sequencing of HAS1 minigenes allowed us to determine whether the recurrent GVs detected in patients occurred as a cluster of mutations. The regions of HAS1 that were sequenced included exons 3 and 4, introns 3 and 4, and part of exon 5.
Overall, more than 4000 sequencing reactions were performed and analyzed. Sequencing runs included a defined patient sample to validate our sequencing analysis. In addition, cloning and sequencing were conducted by 7 different persons over the course of this study, and sequence analysis was performed by 3 different persons, all with consistent results. As a control, some samples were blinded and sequenced more than once, with consistent results. All PCR reactions were performed using High Fidelity Taq polymerase that has proofreading capability. For randomly chosen reactions, direct sequencing was done to verify the subcloning analysis.
Conditions for gDNA PCR were as follows: 50 μL PCR reaction mix contained 50 ng gDNA, 5 μL of 1XPCR buffer, 2 mM MgSO4, 0.2 mM dNTPs, 0.4 mM HAS1 primer set (Figure 1), and 0.5 U High Fidelity Platinum Taq (Invitrogen, Carlsbad, CA). The PCR cycling parameters were as follows: denaturation for 5 minutes at 94°C, followed by denaturation for 30 seconds at 94°C, annealing for 40 seconds at 60°C, and extension at 68°C for 5 minutes for 35 cycles, with a final extension period of 10 minutes at 72°C. The HAS1 PCR products were cloned into the pCR2.1 TOPO TA cloning system (Invitrogen) and sequenced using BigDye V1.1 and V1.1 chemistry (Applied Biosystems) according the manufacturer's instructions.
Sequencing analysis
Obtained sequences were analyzed with the use of sequencing analysis software, whereas alignment was done with the use of SeqScape software; both software packages were provided by Applied Biosystems. Obtained sequences were compared with the HAS1 reference sequence reported in the National Center for Biotechnology Information (NCBI) database.45 For the analysis reported here, GVs from patients with MM and WM were included if they were present on at least 20% of the subclones sequenced for that sample or were identified in more than one patient. HAS1 GVs were identified as recurrent if they were detected in more than one patient. GVs were categorized as hematopoietic/germ line origin if no BECs were available for sequencing. GVs were categorized as confirmed germ line only if BEC sequences were available for comparison with hematopoietic cells. Other categories were as reported in “Results.” HAS1 sequences from 23 control subjects were screened for the presence, or not, of GVs previously identified in patients with MM or WM or both.
In vitro splicing assay
A HAS1 minigene, a segment of HAS1 gDNA, extending from exon 3 to exon 5 (NC_000049) was amplified from gDNA of a patient with WM who expressed HAS1Vb transcripts. HAS1 minigene includes sequence of the HAS1 exons 3, 4, and 5 and introns 3 and 4. The amplified HAS1 minigene was joined to the upstream sequence of HAS1 cDNA, including exon 1 and 2, at SmaI site located within exon 3 to generate a hybrid HAS1 cassette composed of cDNA linked to a gDNA fragment (Figure 3F). Four subclones of the hybrid fragments were cloned into pcDNA3 (Invitrogen), yielding pcDNA3HAS1-g3-4-5 constructs. RNA splicing was analyzed by transfecting the construct into HeLa cells using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. Cells were harvested 24 hours after transfection for RT-PCR. We made a plasmid construct containing full-length HAS1 cDNA, pcDNA3-HAS1-FL (A.G., H.K., and L.M.P., manuscript submitted), for use as a positive control in the transfection experiments.
Total RNA was isolated from transfected HeLa cells using a standard Trizol isolation method (Invitrogen). RT-PCR reactions were conducted following a standard protocol (Invitrogen). PCR primer sequences used in the RT-PCR reactions are as follows: HAS1FL sense primer, 5′-GCGGTCCTCTAGGCCTATATAGGA-3′; HAS1FL antisense primer, 5′-CTGGAGGTGTACTTGGTAGCATAA-3′; HAS1Vb sense primer, 5′-GCGGTCCTCTAGAATCCTGCCCAG-3′; and HAS1Vb antisense primer, 5′-CTGGAGGTGTACCTGCACGGGGGC-3′.
Bioinformatic analysis
To evaluate recurrent GVs leading to aberrant HAS1 splicing in patients, we first identified classical splicing sites (5′ and 3′ SS, BP, PPT) and splicing elements (putative exonic and intronic enhancers and suppressors: ESE, ISE, ESS, and ISS) located within the alternatively spliced exons and introns of this gene using “ESE finder” web interface (release 2.0). In addition, cis-splicing elements (exonic and intronic enhancers [ESE, ISE] and suppressors [ISE, ISS], splicing branch point [BP] and polypyrimidine tract [PPT]) of wild-type and mutated HAS1 gene segments were mapped and evaluated using the publicly available bioinformatic software, Splicing Signal Analysis tools (http://www.ebi.ac.uk/asd-srv/wb.cgi). Next, GVs detected through cloning and sequencing were mapped with the splicing elements identified by the above-mentioned methods. Each GV was evaluated alone and in combination to determine whether any of these variations or clusters of GVs had a predicted effect on the activation of cryptic splice sites in the HAS1 gene. Activation of cryptic splice sites was determined using the Splice Site Prediction tool (http://www.cbs.dtu.dk/biolinks/pserve2.php).
Results
Sequencing analysis of HAS1 gene segments in MM and WM
Because of their involvement in the aberrant splicing of HAS1 pre-mRNA, we sequenced genomic segments (exons and introns) or minigenes or both of HAS1 from exon 3 to exon 5 (3616 bp or 3503 bp, chromosomal location at 56912295-56908679 bp or 56912272-56908769 bp). We obtained samples from 10 patients with MM and 7 patients with WM expressing HAS1 splice variant transcripts and sequenced genomic HAS1 from defined cell subsets. To avoid false detection of GVs, 3 to 19 subclones or minigenes were sequenced in both directions for each cell subset. Only those GVs present in 20% or more of the subclones for a given sample are reported here.
GVs were defined as recurrent if they were detected in at least 2 patients. Sequencing analysis identified 50 recurrent and 147 unique HAS1 GVs that include substitutions, insertions, and deletions (Tables 2,Table 3–4; Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). We identified 46 novel germline and 87 somatic HAS1 GVs (acquired in hematopoietic or tumor cells but absent from BECs), as well as 52 unclassified novel GVs and 12 frequent NCBI single nucleotide polymorphisms (SNPs). A median of 23 GVs/patient were identified (range, 3-24 GVs). The 50 recurrent HAS1 GVs included 28 inherited (16 germ line origin GVs and 12 NCBI-SNPs), 10 acquired hematopoietic origin, and 7 acquired tumor-specific GVs (Tables 2,3; classification of GVs is described in “Classification of GVs detected in patients with MM and WM”). We also detected 5 GVs that were classified as hematopoietic/germ line origin, because no BECs were available to confirm germ line or hematopoietic origin for the patients in whom these GVs were detected. Each category of recurrent HAS1 GVs was detected in 2 to 15 of the 17 patients with MM and WM analyzed (Table 4). Both inherited and acquired sets included GVs recurrent only in MM (7 of 50; ∼14%), only in WM (10 of 50; ∼20%), or shared by MM and WM (34 of 50; ∼68%) (Table 3).
Distribution of HAS1 GVs on HAS1 exons and introns
| Classification of GVs . | HAS1 gene . | |||||||
|---|---|---|---|---|---|---|---|---|
| Exon 3 . | Intron 3 . | Exon 4 . | Intron 4 . | |||||
| U . | R . | U . | R . | U . | R . | U . | R . | |
| Patients with MM and WM (n = 17) | ||||||||
| Tumor specific | 8 | 2 | 6 | 1 | 0 | 0 | 40 | 4 |
| Hematopoietic origin | 1 | 2 | 0 | 1 | 0 | 2 | 15 | 5 |
| Hematopoietic/germline origin | 9 | 1 | 11 | 2 | 1 | 0 | 26 | 2 |
| Germline origin | 4 | 5 | 9 | 1 | 0 | 0 | 17 | 10 |
| NCBI-SNP | 0 | 1 | 0 | 3 | 0 | 0 | 0 | 8 |
| Total | 22 | 11 | 26 | 8 | 1 | 2 | 98 | 29 |
| Controls (n = 23) | ||||||||
| Tumor specific | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Hematopoietic origin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Hematopoietic/germline origin | 2 | 0 | 0 | 0 | 0 | 0 | 5 | 0 |
| Germline origin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 |
| NCBI-SNP | 0 | 1 | 0 | 3 | 0 | 0 | 0 | 8 |
| Total | 2 | 1 | 0 | 3 | 0 | 0 | 6 | 12 |
| Classification of GVs . | HAS1 gene . | |||||||
|---|---|---|---|---|---|---|---|---|
| Exon 3 . | Intron 3 . | Exon 4 . | Intron 4 . | |||||
| U . | R . | U . | R . | U . | R . | U . | R . | |
| Patients with MM and WM (n = 17) | ||||||||
| Tumor specific | 8 | 2 | 6 | 1 | 0 | 0 | 40 | 4 |
| Hematopoietic origin | 1 | 2 | 0 | 1 | 0 | 2 | 15 | 5 |
| Hematopoietic/germline origin | 9 | 1 | 11 | 2 | 1 | 0 | 26 | 2 |
| Germline origin | 4 | 5 | 9 | 1 | 0 | 0 | 17 | 10 |
| NCBI-SNP | 0 | 1 | 0 | 3 | 0 | 0 | 0 | 8 |
| Total | 22 | 11 | 26 | 8 | 1 | 2 | 98 | 29 |
| Controls (n = 23) | ||||||||
| Tumor specific | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Hematopoietic origin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Hematopoietic/germline origin | 2 | 0 | 0 | 0 | 0 | 0 | 5 | 0 |
| Germline origin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 |
| NCBI-SNP | 0 | 1 | 0 | 3 | 0 | 0 | 0 | 8 |
| Total | 2 | 1 | 0 | 3 | 0 | 0 | 6 | 12 |
Distribution of unique (U) and recurrent (R) of GVs on HAS1 gene exon 3 to 4 and introns 3 to 4 from patients with MM and WM. We have detected a total of 196 unique and recurrent GVs in 17 patients with MM and WM. Among these GVs, 61 are tumor specific, 26 are hematopoietic origin, 52 are hematopoietic/germline origin, 46 are germline origin, and 11 are NCBI SNPs. Some of the GVs detected in hematopoietic cells were classified as hematopoietic/germline origin because no BECs were available for these patients to confirm germ line or hematopoietic origin. We also detected sporadic substitutions, each in only one subclone from one individual HD, that were not detected in patients with MM or WM and are not reported in the NCBI database.
Recurrent GVs and their distribution in patients with MM and WM
| . | Chromosomal location . | Nucleotide change . | AA changes . | Type of GVs . | Recurrence of GVs in MM and WM* . |
|---|---|---|---|---|---|
| Tumor specific GVs | |||||
| Exon 3 | 56912068 | a>T | Tyr>Phe | 1TS | 2 WM |
| Exon 3 | 56912051 | t>A | Cys>Ser | 2TS | 1 MM and 1 WM |
| Intron 3 | 56911759† | a>T | 3TS | 1 WM | |
| Intron 3 | 56911759† | a>G | 3TS | 1 WM | |
| Intron 4 | 56909899 | a>G | 4TS | 2 WM | |
| Intron 4 | 56909573 | del.-c | 5TS | 1 MM and 1 WM | |
| Intron 4 | 56909521 | t>C | 6TS | 1 MM and 1 WM | |
| Intron 4 | 56909482 | t>C | 7TS | 2 WM | |
| Hematopoietic origin GVs | |||||
| Exon 3 | 56912079 | t>C | Ala>Ala | 1HO | 2 MM and 1 WM |
| Exon 3 | 56912077 | g>A | Cys>Try | 2HO | 1 MM and 1 WM |
| Intron 3 | 56911668 | a>G | 3HO | 2 WM | |
| Exon 4 | 56911348 | g>C | Arg>Pro | 4HO | 1 MM and 3 WM |
| Exon 4 | 56911346 | a>T | Met>Leu | 5HO | 1 MM and 3 WM |
| Intron 4 | 56910198 | a>C | 6HO | 2 MM | |
| Intron 4 | 56910041 | g>T | 7HO | 1 MM and 4 WM | |
| Intron 4 | 56909423 | t>C | 8HO | 2 MM | |
| Intron 4 | 56909270 | a>G | 9HO | 1 MM and 1 WM | |
| Intron 4 | 56909186 | a>G | 10HO | 2 MM | |
| Germline origin | |||||
| Exon 3 | 56912080 | c>T | Ala>Val | 1GO | 1 MM and 1 WM |
| Exon 3 | 56912058 | t>C | Cys>Cys | 2GO | 1 MM and 1 WM |
| Exon 3 | 56912056 | t>C | Val>Ala | 3GO | 3 MM and 1 WM |
| Exon 3 | 56912051 | t>C | Cys>Arg | 4GO | 3 MM and 1 WM |
| Exon 3 | 56912041 | g>A | Gly>Asp | 5GO | 1 MM and 1 WM |
| Intron 3 | 56911526 | g>A | 6GO | 2 WM | |
| Intron 4 | 56910856 | t>C | 7GO | 1 MM and 1 WM | |
| Intron 4 | 56910811 | a>G | 8GO | 2 WM | |
| Intron 4 | 56910410 | a>G | 9GO | 2 MM | |
| Intron 4 | 56910219 | a>G | 10GO | 2 MM | |
| Intron 4 | 56909762 | ins(Ts) | 11GO | 6 MM and 7 WM | |
| Intron 4 | 56909764 | ins (Ts) | 12GO | 3 MM and 2 WM | |
| Intron 4 | 56909589 | del or ins (T)s | 13GO | 7 MM and 7 WM | |
| Intron 4 | 56909447 | del or ins (TTTA)s | 14GO | 6 MM and 5 WM | |
| Intron 4 | 56909315 | c>T | 15GO | 4 MM | |
| Intron 4 | 56909217 | c>T | 16GO | 4 MM and 1 WM | |
| Hematopoietic/germline origin | |||||
| Exon 3 | 56912197 | a>G | Asp>Gly | 1H/G | 1 MM and 1 WM |
| Intron 3 | 56911983 | a>G | 2H/G | 2 MM | |
| Intron 3 | 56911977 | a>G | 3H/G | 1 MM and 1 WM | |
| Intron 4 | 56910790 | a>C | 4H/G | 2 WM | |
| Intron 4 | 56909253 | g>A | 5H/G | 2 WM | |
| NCBI-SNPs | |||||
| Exon 3 | 56912163 | c>T | Asp>Asp | rs 11084111 | 7 MM and 2 WM |
| Intron 3 | 56911889 | g>A | rs 11084110 | 8 MM and 2 WM | |
| Intron 3 | 56911831 | g>A | rs 11084109 | 8 MM and 7 WM | |
| Intron 3 | 56911750 | t>A | rs 11669079 | 8 MM and 7 WM | |
| Intron 4 | 56910770 | g>C | rs 11667974 | 3 MM and 1 WM | |
| Intron 4 | 56910738 | t>G | rs 11667949 | 8 MM and 7 WM | |
| Intron 4 | 56910711 | g>A | rs 7254072 | 8 MM and 7 WM | |
| Intron 4 | 56910493 | g>C | rs 4802850 | 4 MM and 3 WM | |
| Intron 4 | 56910155 | c>A | rs 4802849 | 5 MM and 4 WM | |
| Intron 4 | 56910154 | c>G | rs 4802848 | 5 MM and 4 WM | |
| Intron 4 | 56909763 | c>T | rs 8104157 | 8 MM and 7 WM | |
| Intron 4 | 56909604 | ins T | rs 11438660 | 8 MM and 7 WM | |
| . | Chromosomal location . | Nucleotide change . | AA changes . | Type of GVs . | Recurrence of GVs in MM and WM* . |
|---|---|---|---|---|---|
| Tumor specific GVs | |||||
| Exon 3 | 56912068 | a>T | Tyr>Phe | 1TS | 2 WM |
| Exon 3 | 56912051 | t>A | Cys>Ser | 2TS | 1 MM and 1 WM |
| Intron 3 | 56911759† | a>T | 3TS | 1 WM | |
| Intron 3 | 56911759† | a>G | 3TS | 1 WM | |
| Intron 4 | 56909899 | a>G | 4TS | 2 WM | |
| Intron 4 | 56909573 | del.-c | 5TS | 1 MM and 1 WM | |
| Intron 4 | 56909521 | t>C | 6TS | 1 MM and 1 WM | |
| Intron 4 | 56909482 | t>C | 7TS | 2 WM | |
| Hematopoietic origin GVs | |||||
| Exon 3 | 56912079 | t>C | Ala>Ala | 1HO | 2 MM and 1 WM |
| Exon 3 | 56912077 | g>A | Cys>Try | 2HO | 1 MM and 1 WM |
| Intron 3 | 56911668 | a>G | 3HO | 2 WM | |
| Exon 4 | 56911348 | g>C | Arg>Pro | 4HO | 1 MM and 3 WM |
| Exon 4 | 56911346 | a>T | Met>Leu | 5HO | 1 MM and 3 WM |
| Intron 4 | 56910198 | a>C | 6HO | 2 MM | |
| Intron 4 | 56910041 | g>T | 7HO | 1 MM and 4 WM | |
| Intron 4 | 56909423 | t>C | 8HO | 2 MM | |
| Intron 4 | 56909270 | a>G | 9HO | 1 MM and 1 WM | |
| Intron 4 | 56909186 | a>G | 10HO | 2 MM | |
| Germline origin | |||||
| Exon 3 | 56912080 | c>T | Ala>Val | 1GO | 1 MM and 1 WM |
| Exon 3 | 56912058 | t>C | Cys>Cys | 2GO | 1 MM and 1 WM |
| Exon 3 | 56912056 | t>C | Val>Ala | 3GO | 3 MM and 1 WM |
| Exon 3 | 56912051 | t>C | Cys>Arg | 4GO | 3 MM and 1 WM |
| Exon 3 | 56912041 | g>A | Gly>Asp | 5GO | 1 MM and 1 WM |
| Intron 3 | 56911526 | g>A | 6GO | 2 WM | |
| Intron 4 | 56910856 | t>C | 7GO | 1 MM and 1 WM | |
| Intron 4 | 56910811 | a>G | 8GO | 2 WM | |
| Intron 4 | 56910410 | a>G | 9GO | 2 MM | |
| Intron 4 | 56910219 | a>G | 10GO | 2 MM | |
| Intron 4 | 56909762 | ins(Ts) | 11GO | 6 MM and 7 WM | |
| Intron 4 | 56909764 | ins (Ts) | 12GO | 3 MM and 2 WM | |
| Intron 4 | 56909589 | del or ins (T)s | 13GO | 7 MM and 7 WM | |
| Intron 4 | 56909447 | del or ins (TTTA)s | 14GO | 6 MM and 5 WM | |
| Intron 4 | 56909315 | c>T | 15GO | 4 MM | |
| Intron 4 | 56909217 | c>T | 16GO | 4 MM and 1 WM | |
| Hematopoietic/germline origin | |||||
| Exon 3 | 56912197 | a>G | Asp>Gly | 1H/G | 1 MM and 1 WM |
| Intron 3 | 56911983 | a>G | 2H/G | 2 MM | |
| Intron 3 | 56911977 | a>G | 3H/G | 1 MM and 1 WM | |
| Intron 4 | 56910790 | a>C | 4H/G | 2 WM | |
| Intron 4 | 56909253 | g>A | 5H/G | 2 WM | |
| NCBI-SNPs | |||||
| Exon 3 | 56912163 | c>T | Asp>Asp | rs 11084111 | 7 MM and 2 WM |
| Intron 3 | 56911889 | g>A | rs 11084110 | 8 MM and 2 WM | |
| Intron 3 | 56911831 | g>A | rs 11084109 | 8 MM and 7 WM | |
| Intron 3 | 56911750 | t>A | rs 11669079 | 8 MM and 7 WM | |
| Intron 4 | 56910770 | g>C | rs 11667974 | 3 MM and 1 WM | |
| Intron 4 | 56910738 | t>G | rs 11667949 | 8 MM and 7 WM | |
| Intron 4 | 56910711 | g>A | rs 7254072 | 8 MM and 7 WM | |
| Intron 4 | 56910493 | g>C | rs 4802850 | 4 MM and 3 WM | |
| Intron 4 | 56910155 | c>A | rs 4802849 | 5 MM and 4 WM | |
| Intron 4 | 56910154 | c>G | rs 4802848 | 5 MM and 4 WM | |
| Intron 4 | 56909763 | c>T | rs 8104157 | 8 MM and 7 WM | |
| Intron 4 | 56909604 | ins T | rs 11438660 | 8 MM and 7 WM | |
Details are included of recurrent GVs that were detected in MM and WM, in exons and introns. TS indicates tumor specific GVs; HO, hematopoietic origin GVs; GO, germline origin GVs; H/G, hematopoietic/germline GVs; AA, amino acid; ins, insertion; del, deletion. GVs are numbered according to their position on exons and introns.
The numbers represent the number of patients with MM or WM or both in whom the GV was recurrent. A total of 49 chromosomal positions in HAS1 harbored GVs.
Two different mutated alleles were detected, yielding an aggregate of 50 recurrent GVs.
Number of persons in whom recurrent HAS1 GVs were detected
| Type . | Tumor . | Hematopoietic . | Germline . | Germline/hematopoietic . | NCBI-SNP . |
|---|---|---|---|---|---|
| MM and WM Patients (n = 17) | |||||
| Percentage of patients with the indicated type of HAS1 GVs (n/n) | 59 (10/17) | 65 (11/17) | 94 (16/17) | 41 (7/17) | 88 (15/17) |
| No. of patients in whom HAS1 GVs were recurrent | 2-3 | 2-5 | 2-14 | 2-4 | 4-15 |
| Controls (n = 23) | |||||
| Percentage of controls with recurrent HAS1 GVs | 0 | 0 | 4-39 | 0 | 39-83 |
| No. of controls in whom HAS1 GVs were recurrent | 0 | 0 | 1-6 | 0 | 2-14 |
| Type . | Tumor . | Hematopoietic . | Germline . | Germline/hematopoietic . | NCBI-SNP . |
|---|---|---|---|---|---|
| MM and WM Patients (n = 17) | |||||
| Percentage of patients with the indicated type of HAS1 GVs (n/n) | 59 (10/17) | 65 (11/17) | 94 (16/17) | 41 (7/17) | 88 (15/17) |
| No. of patients in whom HAS1 GVs were recurrent | 2-3 | 2-5 | 2-14 | 2-4 | 4-15 |
| Controls (n = 23) | |||||
| Percentage of controls with recurrent HAS1 GVs | 0 | 0 | 4-39 | 0 | 39-83 |
| No. of controls in whom HAS1 GVs were recurrent | 0 | 0 | 1-6 | 0 | 2-14 |
A patient or control was counted as having the indicated type of GV if at least one such GV was detected in the samples that were sequenced.
The presence or not of GVs from the 17 patients with MM and WM was determined for the NCBI HAS1 gene sequence and the HAS1 exons and introns from 23 control subjects (Table 2). HAS1 segments were sequenced from 4 B-CLL, 11 MGUS, and 8 HDs. Most of the control subjects expressed the NCBI SNPs found in MM and WM, although 4 of the HDs expressed only major alleles. Four germ line GVs were detected in B-CLL and a subset of MGUS, which were found in only a minority of subclones or were absent from the HDs; these may be as yet unreported polymorphisms. Of 147 unique GVs, 8 were detected sporadically in HDs or MGUS, in only a minority of subclones per sample, and, with one exception, each GV was found in only 1 of the 23 control subjects. Seven of the 8 unique GVs found in control subjects were classified as germline/hematopoietic origin (Table 3). This indicates that the majority of the HAS1 GVs reported here for patients with MM and WM, including all of the recurrent acquired GVs, appear restricted to MM and WM.
Classification of GVs detected in patients with MM and WM
GVs were classified as follows: (1) GVs detected only in B cells and PCs from patients were classified as “tumor-specific.” These GVs were absent from T cells, HPCs, and BECs, as well as from control subjects; the stage of disease during which these were acquired is unknown. (2) GVs identified in hematopoietic cell populations but absent from BECs were defined as being of “hematopoietic-origin.” They were found in all hematopoietic populations tested from MM and WM, including HPCs and T cells (nonmalignant) and B cells and PCs (malignant). Their absence from BECs indicates that these are somatic GVs acquired by presumptively normal HPCs and transmitted to their T- and B-lineage progeny. Hematopoietic-origin GVs were absent from cell subsets of control subjects. (3) Newly identified HAS1 GVs identified in all cell populations, including BECs, were classified as “germline-origin,” 4 of which were detected in control subjects and may be unreported SNPs. In MM and WM, these substitutions were frequently homozygous, defined by their presence in every subclone sequenced for a given patient. We also identified a high frequency of the mutated alleles for 12 NCBI HAS1 SNPs. The mutated alleles of these SNPs were present in most patients with MM and WM and were also detectable in control subjects (Table 2).
GVs detected in MM and WM
Tumor cells from MM and WM had 4 or more tumor-specific GVs in their B cells, PCs or both, but the majority of these tumor-specific GVs were unique (Figure 2B). In addition, 10 (59%) of 17 patients with MM and WM carried recurrent tumor-specific HAS1 GVs in B cells and /PCs, whereas 11 (65%) had recurrent hematopoietic-origin GVs and 16 (94%) had recurrent germline-origin GVs (Table 4). The type, genomic location, and the distribution of recurrent GVs in MM and WM are reported in Table 3. None of the recurrent somatic GVs were found in control subjects.
Distribution of HAS1 GVs. (A) Cell type distribution of HAS1 GVs detected in patients with MM and WM. Mutations identified in various types of cells from patients with MM were classified as tumor specific, hematopoietic and germline–origin based on their occurrence in these cells. HPCs indicates CD34+45low HPCs from mobilized blood of patients with MM or bone marrow aspirates from patients with WM; BECs indicate buccal epithelial cells. (B) Distribution of GVs in genomic HAS1. This figure shows relative distribution of GVs detected in patients with MM and WM. Recurrent NCBI-SNPs are absent from this figure. The inserts detail sets of GVs located at the boundary of exon 3 and exon 4, respectively. GVs are represented by yellow rings. On the figure, the first break on intron 4 represents 180 nucleotides, whereas the second break represents 100 nucleotides. 1st “T,” 2nd “T,” and “TTTA” are the common motif detected in patients with MM and WM. The spaces between mutations are arranged according to a scale of 50 bp = 4 mm.
Distribution of HAS1 GVs. (A) Cell type distribution of HAS1 GVs detected in patients with MM and WM. Mutations identified in various types of cells from patients with MM were classified as tumor specific, hematopoietic and germline–origin based on their occurrence in these cells. HPCs indicates CD34+45low HPCs from mobilized blood of patients with MM or bone marrow aspirates from patients with WM; BECs indicate buccal epithelial cells. (B) Distribution of GVs in genomic HAS1. This figure shows relative distribution of GVs detected in patients with MM and WM. Recurrent NCBI-SNPs are absent from this figure. The inserts detail sets of GVs located at the boundary of exon 3 and exon 4, respectively. GVs are represented by yellow rings. On the figure, the first break on intron 4 represents 180 nucleotides, whereas the second break represents 100 nucleotides. 1st “T,” 2nd “T,” and “TTTA” are the common motif detected in patients with MM and WM. The spaces between mutations are arranged according to a scale of 50 bp = 4 mm.
Among the 7 recurrent tumor-specific (TS) HAS1 GVs, 4 were detected only in WM and 3 were shared by WM and MM. Among these GVs 2 (no. 1TS and no. 2TS) are missense transversions and one, no. 5TS, is a deletion. Other recurrent GVs on intron 3 and intron 4 are transitions (Table 3). Furthermore, groups of tumor-specific GVs are coexpressed in the same patients. For example, exon 3 no. 1TS, intron 3 no. 3TS, and intron 4 no. 6TS are coexpressed together; a second coexpressed group included no. 2TS on exon 3 and no. 7TS on intron 4.
For 10 recurrent GVs of hematopoietic origin (HO), distributed across 65% of patients, 6 (50%) recur in both MM and WM, whereas 3 HO GVs (40%) are specific to MM and 1, no. 3HO (10%), is specific to WM (Table 3). Acquired hematopoietic–origin GVs were detected in as many as 5 different patients.
Of 16 recurrent germline origin (GO) HAS1 GVs (defined by their presence in BECs), the majority are on exon 3 or intron 4. Most novel germline GVs (11 of 16) are shared by patients with MM and WM, with 3 (no. 9GO, no. 10GO, no. 15GO) recurring only in MM and 2 (no. 6GO, no. 8GO) recurring only in WM (Table 3). These may be candidate SNPs not yet reported. The sequencing analysis also detected mutated alleles of 12 NCBI-SNPs. These HAS1 SNPs are found in 88% of the patients with MM and WM analyzed. Coding GVs detected on exon 3 and 4 (inserts in Figure 2B) were all shared by patients with MM and patients with WM.
Five recurrent GVs were provisionally classified as “hematopoietic/germline-origin” because no BECs were available for the patients analyzed. Of these 5 GVs, 2 were shared by MM and WM, one was specific to MM, and 2 were specific to WM (Table 3).
As indicated above, 3 tumor-specific and 6 hematopoietic-origin HAS1 GVs were shared by MM and WM. The existence of these shared, somatically acquired GVs suggests that there may exist fundamental similarities in the events that underlie development and progression of MM and WM
Linked clusters of GVs are detected in HAS1 minigenes
Our sequencing analysis showed that recurrent GVs are distributed as linked clusters, as distinct from random locations (Table 5). The linked clusters of GVs described in Table 5 are distributed in the vicinity of splicing elements. HAS1 minigene sequencing identified 3 distinct clusters of GVs that include 3 NCBI-SNPs and germline/hematopoietic origin GVs that are detected within a specific sequence stretch of intron 4 (first “T” stretch, second “T” stretch, and TTTA repeats; shown in Figure 2B), near splicing elements at the 3′ end of intron 4, where partial intron retention leads to HAS1Vb transcripts. These GVs and NCBI-SNPs were present in every cluster detected in patients with MM and WM, and will be referred to as a “common motif.” The common motif appears to cluster with other recurrent HAS1 GVs (Figure 2B).
Clusters of GVs detected in HAS1 gene exons and introns from patients with MM and WM
| . | Nucleotide changes . | Chromosomal location . | Type . | Effects on the protein . | Cluster* . |
|---|---|---|---|---|---|
| GVs clusters detected in MM patients | |||||
| Exon 3 | c>T | 56912163 | NCBI rs 1108411 | 1, 2, 3 | |
| Exon 3 | t>C | 56912056 | Germline origin | Val>Ala | 1 |
| Intron 4 | g>C | 56910770 | NCBI rs 11667974 | 1 | |
| Intron 4 | c>T | 56109315 | Germline origin | 2 | |
| Intron 4 | c>T | 56109217 | Germline origin | 2 | |
| Intron 4 | inst T | 56909764 | Germline origin | 2 | |
| Exon 3 | t>C | 56912051 | Germline origin | Cys>Arg | 3 |
| Intron 4 | g>C | 56910493 | NCBI rs 4802850 | 3 | |
| Intron 4 | c>A | 56910155 | NCBI rs 4802849 | 3 | |
| Intron 4 | c>G | 56910154 | NCBI rs 4802848 | 3 | |
| GVs clusters detected in WM patients | |||||
| Exon 3 | a>T | 56912068 | Tumor specific | Tyr>Phe | 1 |
| Intron 3 | a>G | 56911668 | Tumor specific | 2 | |
| Exon 4 | a>T | 56911346 | Hematopoietic origin | Met>Leu | 2 |
| Exon 4 | g>C | 56911348 | Hematopoietic origin | Arg>Pro | 2 |
| Intron 4 | g>t | 56910041 | Hematopoietic origin | 2 | |
| Intron 4 | t>C | 56909252 | Hematopoietic/germline origin | 3 | |
| Intron 4 | c>G | 56910154 | SNP-NCBI rs 4802848 | 3 | |
| Intron 4 | c>A | 56910155 | SNP-NCBI rs 4802849 | 3 | |
| Intron 4 | g>C | 56910493 | SNP-NCBI rs 4802850 | 3 | |
| Common motifs | |||||
| Intron 3 | t>A | 56911750 | SNP-NCBI rs 11669079 | ||
| Intron 3 | g>A | 56911831 | SNP-NCBI rs 11084109 | ||
| Intron 3 | g>A | 56911889 | SNP-NCBI rs 11084110 | ||
| Intron 4 | inst T | 56909604 | SNP-NCBI rs 11438660 | ||
| Intron 4 | inst (TTTA)s | 56909447 | Germline origin | ||
| Intron 4 | del (TTTA)s | 56909447 | Germline origin | ||
| Intron 4 | inst/del (T)s | 56909589 | Germline origin | ||
| Intron 4 | inst (Ts) | 56909762 | Germline origin | ||
| Intron 4 | c>T | 56909763 | SNP-NCBI rs 8104157 | ||
| Intron 4 | g>A | 56910711 | SNP-NCBI rs 7254072 | ||
| Intron 4 | t>G | 56910738 | SNP-NCBI rs 11667949 | ||
| . | Nucleotide changes . | Chromosomal location . | Type . | Effects on the protein . | Cluster* . |
|---|---|---|---|---|---|
| GVs clusters detected in MM patients | |||||
| Exon 3 | c>T | 56912163 | NCBI rs 1108411 | 1, 2, 3 | |
| Exon 3 | t>C | 56912056 | Germline origin | Val>Ala | 1 |
| Intron 4 | g>C | 56910770 | NCBI rs 11667974 | 1 | |
| Intron 4 | c>T | 56109315 | Germline origin | 2 | |
| Intron 4 | c>T | 56109217 | Germline origin | 2 | |
| Intron 4 | inst T | 56909764 | Germline origin | 2 | |
| Exon 3 | t>C | 56912051 | Germline origin | Cys>Arg | 3 |
| Intron 4 | g>C | 56910493 | NCBI rs 4802850 | 3 | |
| Intron 4 | c>A | 56910155 | NCBI rs 4802849 | 3 | |
| Intron 4 | c>G | 56910154 | NCBI rs 4802848 | 3 | |
| GVs clusters detected in WM patients | |||||
| Exon 3 | a>T | 56912068 | Tumor specific | Tyr>Phe | 1 |
| Intron 3 | a>G | 56911668 | Tumor specific | 2 | |
| Exon 4 | a>T | 56911346 | Hematopoietic origin | Met>Leu | 2 |
| Exon 4 | g>C | 56911348 | Hematopoietic origin | Arg>Pro | 2 |
| Intron 4 | g>t | 56910041 | Hematopoietic origin | 2 | |
| Intron 4 | t>C | 56909252 | Hematopoietic/germline origin | 3 | |
| Intron 4 | c>G | 56910154 | SNP-NCBI rs 4802848 | 3 | |
| Intron 4 | c>A | 56910155 | SNP-NCBI rs 4802849 | 3 | |
| Intron 4 | g>C | 56910493 | SNP-NCBI rs 4802850 | 3 | |
| Common motifs | |||||
| Intron 3 | t>A | 56911750 | SNP-NCBI rs 11669079 | ||
| Intron 3 | g>A | 56911831 | SNP-NCBI rs 11084109 | ||
| Intron 3 | g>A | 56911889 | SNP-NCBI rs 11084110 | ||
| Intron 4 | inst T | 56909604 | SNP-NCBI rs 11438660 | ||
| Intron 4 | inst (TTTA)s | 56909447 | Germline origin | ||
| Intron 4 | del (TTTA)s | 56909447 | Germline origin | ||
| Intron 4 | inst/del (T)s | 56909589 | Germline origin | ||
| Intron 4 | inst (Ts) | 56909762 | Germline origin | ||
| Intron 4 | c>T | 56909763 | SNP-NCBI rs 8104157 | ||
| Intron 4 | g>A | 56910711 | SNP-NCBI rs 7254072 | ||
| Intron 4 | t>G | 56910738 | SNP-NCBI rs 11667949 | ||
The first GV cluster of MM includes the common motif plus a recurrent hematopoietic origin missense mutation t>C (CH56912056) detected on exon 3 and NCBI-SNP rs 1667974. The second MM GV cluster comprises the common motif and 2 additional germline GVs, both on intron 4. The third MM GV cluster includes the common motif with an additional one germline origin missense t>C substitution (Cys>Arg) on exon 3 and 3 NCBI-SNPs on intron 4. All patients with MM were homozygous for mutated alleles of NCBI-SNPs included in the third GV cluster. WM GV cluster 1 includes the common motif plus a recurrent tumor-specific missense mutation a>T (Tyr>Phe, CH6912068) detected on exon 3. The second WM GV cluster includes the common motif and 4 additional hematopoietic origin GVs, missense mutations, a>T (CH56911346) and g>C (CH56911348) in exon 4, that lead to amino acid changes Met>Leu and Arg >Pro, respectively, one tumor specific in intron 4 (CH56910041), and one recurrent tumor-specific transition (CH5611668) in intron 3. The third WM GV cluster includes the common motif with an additional 4 GVs all detected in intron 4, 1 hematopoietic origin, and 3 NCBI-SNPs. Similar to patients with MM, all patients with WM were homozygous for mutated alleles of NCBI-SNPs included in any clusters.
The GV clusters in WM were compared with those in MM, referred to as “WM clusters” or “MM clusters,” respectively, both including the common motif. Interestingly, GVs comprising MM GV cluster no. 3 and WM GV cluster no. 3 are identical with the exception of a germline origin mutation in MM exon 3 (CH56912051) instead of the hematopoietic/germline GV in intron 4 (CH56909252) for WM cluster no. 3. Two other HAS1 GV clusters from patients with MM or WM include the common motif plus other recurrent GVs (Table 5). MM and WM clusters appear to harbor abnormalities that may accompany early stages of malignancy or characterize progression events or both.
Recurrent mutations detected in intron 4 promote HAS1 gene aberrant splicing
Bioinformatic analysis predicted that HAS1 GVs lead to splicing events that generate HAS1Vb, the variant most significantly correlated with poor outcome (Figure 3A-E; S.A., manuscript in preparation.).10 We constructed a HAS1 minigene splicing cassette derived from gDNA of a patient with WM whose cells expressed HAS1Vb transcripts. This HAS1 cassette was used in an in vitro splicing assay to verify that HAS1 GVs from WM could direct the splicing of HAS1Vb. The splicing construct incorporating the HAS1 gDNA segment from exon 3 to exon 5 was transfected into HeLa cells, which do not otherwise express full-length HAS1 or the aberrant HAS1 splice variants (Figure 3F). Sequencing analysis identified 2 unique substitutions in exon 3 and 2 in intron 4, as well as the common motif on HAS1 minigene cassettes. After transfection of HeLa cells with the HAS1 minigene cassettes, we detected aberrantly spliced HAS1Vb transcripts and normally spliced full-length HAS1 (Figure 3F). Sequencing of the HAS1Vb PCR product confirmed its identity as HAS1Vb. This indicates that HeLa cells conserve the trans-splicing elements (small ribonuclear proteins) required for aberrant splicing of HAS1 pre-mRNA, but the HAS1Vb transcript is spliced only when cis-elements of the WM HAS1 template are introduced by the splicing construct.
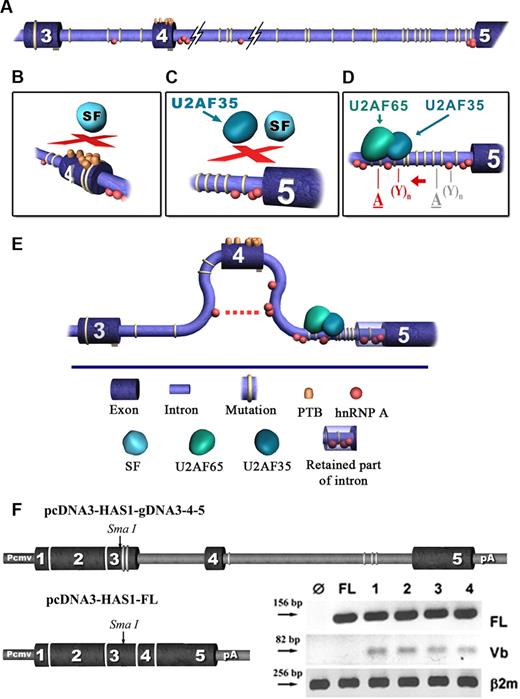
Bioinformatic model. Clusters of recurrent GVs facilitate aberrant splicing of HAS1 gene in patients with MM to create the intronic HAS1Vb splice variants. In this analysis we used the web-based bioinformatic tool ESE finder V2. Results were evaluated using ESE V3 (http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi). For more detailed analysis we used ASD (The Alternative Splicing Database; workbench bioinformatics tools). Using these tools, we evaluated the distribution of splicing elements in HAS1 exons 3 and 4 and introns 3 and 4 of wild-type and mutated sequences. (A) Relative distribution of recurrent mutations detected in patients with MM and WM is shown, and the accumulation of 2 important splicing cofactors, hnRNP I (PTB) and hnTNP A, is shown in exon 4 and introns 3 and 4. (B-D) The location on the HAS1 gene where the aberrations occur. (E) Predicts the effect of recurrent GVs on HAS1 splicing. (D), The red letters “A” and “Y” represent activated splicing branch point (BP) and polypyrimidine tract (PPT) of splicing, respectively, and gray letters “A” and “Y” represent native BP and PPT. Description of the model. No differences were found between wild-type and mutated exon 3 with respect to the accumulation of hnRNPs which bind manly splicing suppressors and promote exon exclusion. However, in mutated exon 4, compared with wild-type exon 4 and in mutated exon 3, bioinformatic analysis predicted a massive accumulation of hnRNPs, including hnRNP I (PTB, polypyrimidine tract binding protein), which is distributed across the entire mutated exon 4 (A,B). As suggested in the diagram, the binding of PTBs at several sites of an exon could cause a loopout of this exon, and subsequently these types of exons become inaccessible for the assembly of the spliceosome (B,C,E). The analysis did not predict any significant differences between wild-type and mutated intron 3 with respect to Serin/Arginine-rich proteins (SRs) or the distribution of hnRNP binding motifs. In addition, no significant difference was found when BP and PPT were mapped on wild-type and mutated intron 3. However, for mutated intron 4, the existence of alternative splicing branch points were predicted. These alternative BPs are located upstream of the alternative PPT (D). In addition, splicing element analysis of wild-type and mutated intron 4 showed an accumulation of a significant number of SR and hnRNP binding motifs in mutated intron 4. Among them, the most significant predicted difference that contributes to intronic splicing of HAS1 is recruitment of U2AF65 protein by the alternative PPTs (D). These predicted PPT sequences overlap with the 1st and 2nd “T” stretches and TTTA repeats of mutated intron 4 (the common motif) where the MM clusters of GVs are located. The protein U2SF65 is known to be responsible for the recruitment of SFs to splicing BP. Subsequently, this protein acts as a “bridge” between BP and PPT and stabilizes the spliceosomal complex necessary for the first stage of the splicing reaction. In addition, our analysis of wild-type and mutated intron 4 predicted the loss of a significant number of binding motifs for hnRNP proteins from mutated intron 4 compared with wild type. However, mutated intron 4 maintained ability to recruit hnRNP, a protein which most likely contributes to the exclusion of exon 4 through its ability to dimerise with other molecules of hnRNP A located within and on adjacent introns (E). (F) Splicing of aberrant HAS1 Vb transcripts in transfected HeLa cells. The diagram shows the expression cassettes for HAS1 minigene constructs. HAS1 sequences were flanked by a mammalian CMV promoter at the 5′ end of HAS1 gene and the bovine growth hormone polyadenylation signal, poly A, at the 3′ end. mRNA splicing was analyzed by transfecting HeLa cells with HAS1 minigene cassettes. RT-PCR was performed 24 hours after transfection, using specific primers for HAS1 full-length (FL), HAS1Vb or B2m (β-2 microglobulin). On the gel, Ø indicates the result obtained from the cells transfected with cassette without HAS1 gene; FL, the result obtained from the cells transfected with pcDNA3-HAS1-FL cDNA construct, which is already spliced; Lanes 1 to 4 represent Hela cells transfected with pcDNA3-HAS1-g3–4-5 construct. We tested 4 subclones of the HAS1 minigene cassette; transfection of all 4 subclones gave identical results. Product identity was confirmed by sequencing. For this experiment, Ø and FL were used as controls to verify specificity of the in vitro splicing assay.
Bioinformatic model. Clusters of recurrent GVs facilitate aberrant splicing of HAS1 gene in patients with MM to create the intronic HAS1Vb splice variants. In this analysis we used the web-based bioinformatic tool ESE finder V2. Results were evaluated using ESE V3 (http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi). For more detailed analysis we used ASD (The Alternative Splicing Database; workbench bioinformatics tools). Using these tools, we evaluated the distribution of splicing elements in HAS1 exons 3 and 4 and introns 3 and 4 of wild-type and mutated sequences. (A) Relative distribution of recurrent mutations detected in patients with MM and WM is shown, and the accumulation of 2 important splicing cofactors, hnRNP I (PTB) and hnTNP A, is shown in exon 4 and introns 3 and 4. (B-D) The location on the HAS1 gene where the aberrations occur. (E) Predicts the effect of recurrent GVs on HAS1 splicing. (D), The red letters “A” and “Y” represent activated splicing branch point (BP) and polypyrimidine tract (PPT) of splicing, respectively, and gray letters “A” and “Y” represent native BP and PPT. Description of the model. No differences were found between wild-type and mutated exon 3 with respect to the accumulation of hnRNPs which bind manly splicing suppressors and promote exon exclusion. However, in mutated exon 4, compared with wild-type exon 4 and in mutated exon 3, bioinformatic analysis predicted a massive accumulation of hnRNPs, including hnRNP I (PTB, polypyrimidine tract binding protein), which is distributed across the entire mutated exon 4 (A,B). As suggested in the diagram, the binding of PTBs at several sites of an exon could cause a loopout of this exon, and subsequently these types of exons become inaccessible for the assembly of the spliceosome (B,C,E). The analysis did not predict any significant differences between wild-type and mutated intron 3 with respect to Serin/Arginine-rich proteins (SRs) or the distribution of hnRNP binding motifs. In addition, no significant difference was found when BP and PPT were mapped on wild-type and mutated intron 3. However, for mutated intron 4, the existence of alternative splicing branch points were predicted. These alternative BPs are located upstream of the alternative PPT (D). In addition, splicing element analysis of wild-type and mutated intron 4 showed an accumulation of a significant number of SR and hnRNP binding motifs in mutated intron 4. Among them, the most significant predicted difference that contributes to intronic splicing of HAS1 is recruitment of U2AF65 protein by the alternative PPTs (D). These predicted PPT sequences overlap with the 1st and 2nd “T” stretches and TTTA repeats of mutated intron 4 (the common motif) where the MM clusters of GVs are located. The protein U2SF65 is known to be responsible for the recruitment of SFs to splicing BP. Subsequently, this protein acts as a “bridge” between BP and PPT and stabilizes the spliceosomal complex necessary for the first stage of the splicing reaction. In addition, our analysis of wild-type and mutated intron 4 predicted the loss of a significant number of binding motifs for hnRNP proteins from mutated intron 4 compared with wild type. However, mutated intron 4 maintained ability to recruit hnRNP, a protein which most likely contributes to the exclusion of exon 4 through its ability to dimerise with other molecules of hnRNP A located within and on adjacent introns (E). (F) Splicing of aberrant HAS1 Vb transcripts in transfected HeLa cells. The diagram shows the expression cassettes for HAS1 minigene constructs. HAS1 sequences were flanked by a mammalian CMV promoter at the 5′ end of HAS1 gene and the bovine growth hormone polyadenylation signal, poly A, at the 3′ end. mRNA splicing was analyzed by transfecting HeLa cells with HAS1 minigene cassettes. RT-PCR was performed 24 hours after transfection, using specific primers for HAS1 full-length (FL), HAS1Vb or B2m (β-2 microglobulin). On the gel, Ø indicates the result obtained from the cells transfected with cassette without HAS1 gene; FL, the result obtained from the cells transfected with pcDNA3-HAS1-FL cDNA construct, which is already spliced; Lanes 1 to 4 represent Hela cells transfected with pcDNA3-HAS1-g3–4-5 construct. We tested 4 subclones of the HAS1 minigene cassette; transfection of all 4 subclones gave identical results. Product identity was confirmed by sequencing. For this experiment, Ø and FL were used as controls to verify specificity of the in vitro splicing assay.
Discussion
Sequencing of the HAS1 gene in patients with MM and WM evaluated genetic contributions to the aberrant intronic splicing of HAS1 pre-mRNA that correlates with significantly reduced overall survival.10 We report the existence of novel inherited (germline) and acquired (somatic) GVs in regions of the HAS1 gene involved in aberrant splicing events. Somatic HAS1 GVs appear to accumulate throughout hematopoietic development, thereby leaving a mutational “trace” in nonmalignant HPCs and T cells as well as in malignant B cells and PCs, with the largest number of GVs occurring in malignant MM and WM cells. In patients with MM and WM, we identified 3 categories of genetic change: inherited germline origin, acquired hematopoietic origin, and acquired tumor-specific GVs. All types included recurrent HAS1 GVs, defined by their detection in 2 or more patients with MM or patients with WM. The majority of germline GVs and all recurrent acquired HAS1 GVs were absent from the 23 control subjects.
Of the 197 novel GVs reported here, 50 were recurrent and 147 were unique. We anticipate that when larger cohorts are analyzed, some of the GVs currently classified as unique will prove to be recurrent. Recurrent somatic GVs are restricted to MM, restricted to WM, or shared by both MM and WM; none are detected in the 23 control subjects. The majority of recurrent GVs (both inherited and acquired), including 3 recurrent tumor-specific GVs, are shared between MM and WM. Although global gene expression profiling suggests that WM has more in common with B-CLL than with MM,46 genomic analysis of HAS1 indicates that MM and WM, but not B-CLL, share a very close genetic relationship. The acquisition of recurrent somatic changes in the HAS1 gene, particularly in the noncoding intron 4, suggests that the HAS1 gene undergoes hypermutation and that strong selective pressures enrich these GVs in MM and WM.
Somatic mutations with the potential to alter splicing are frequent in some cancers.28,36,44,47,48 In patients with MM and WM we detected 87 somatic GVs, including 61 tumor-specific and 26 of hematopoietic-origin on HAS1 exons and introns (Table 3). MM and WM are B-lineage cancers. In malignant B cells, aberrant somatic hypermutation affects genes outside of the immunoglobulin variable region,49-54 which may occur before neoplastic transformation.51 The frequency of hypermutated genes such as BCL-6 and PAX5 ranges from approximately 0.2 to 0.6/100 bp,49-54 consistent with the degree of hypermutation detected within the HAS1 gene in MM and WM (∼0.1-0.5/100 bp).
Somatic GVs were found in all subsets of differentiated hematopoietic cells (PCs, PB B and T cells) and in purified CD34+ HPCs. Somatically mutated HPCs were found in G-CSF–mobilized blood autografts or bone marrow from all 8 patients from whom HPCs were available. This implies that somatic HAS1 GVs may be acquired at the earliest stages of hematopoietic development. HPCs harboring somatic HAS1 GVs have normal generative capabilities and are clearly nonmalignant, because their nonmalignant T-cell progeny carry the same somatic GVs. Our previous work shows that HAS1Vb transcripts are found in MM and WM cells but are undetectable in nonmalignant MM HPCs or MM T cells and normal B cells.10,12 Because at birth HPCs must, by definition, have the same genotype as BECs, our work suggests that in persons who are destined for MM and WM, an undefined mechanism may enrich those HPCs that have acquired somatic HAS1 GVs.
The pattern of germline GVs suggests that MM and WM, but not B-CLL, inherit recurrent germline GVs that are necessary but not sufficient for progression to malignancy. Acquisition of recurrent, somatic HAS1 GVs in otherwise healthy HPCs from patients with MM and WM appears to further increase the risk of MM or WM. This idea is supported by our demonstration that transfection of a HAS1-splicing construct from a patient with WM directs the aberrant splicing of HAS1Vb (in vitro splicing assay). The B-cell stage, at which acquisition of critical tumor-specific GVs occurs, may determine whether a person develops MM or WM. Our work suggests that, similar to leukemias, genetic changes leading to myelomagenesis may first accumulate in HPCs during the nonmalignant or premalignant stages of hematopoietic differentiation.55-57 The presence of tumor-specific recurrent GVs restricted to MM B cells or PCs, coupled with expression of clinically predictive aberrant HAS1Vb transcripts by B cells,10 supports the concept that transforming events may occur at the B-cell stage,44,58 and that malignant B cells transfer mutated HAS1 alleles to their MM PC progeny.
Provocatively, clusters of recurrent GVs are localized at sites of the HAS1 gene that control pre-mRNA splicing, particularly in intron 4, as shown in Figures 2B and 3. NCBI HAS1 SNPs appear to be significant predisposing elements in oncogenesis, as evidenced by their presence in combination with the common motif detected on the HAS1 gene. In silico analysis predicted that this common motif contributes to aberrant HAS1 splicing. To support the idea that HAS1 GVs play a role in HAS1 aberrant splicing, we conducted an in vitro splicing assay using a HAS1 splicing cassette genetically engineered from gDNA of a patient with WM. The successful in vitro splicing of HAS1Vb verified bioinformatic predictions that the GVs (particularly aberrations in the common motif region of first and second T and TTTA stretches of the HAS1 cassette) would direct splicing of HAS1Vb after transfection with the WM HAS1 cassette of host cells that do not otherwise express HAS1 or HAS1Vb transcripts.
The HAS1 GVs described here and the aberrant splicing reported previously10 are probably important in malignancy. The aberrant HAS1 splice variants synthesize HA in ex vivo MM cells10 and in transfectants (A.G., H.K., and L.M.P., manuscript submitted). In ex vivo MM cells from patients, HAS1Va correlates with synthesis of extracellular HA and HAS1Vb is associated with synthesis of intracellular HA,10 suggesting a contribution to, respectively, malignant spread59 and altered mitosis.60 In HAS1 transfectants, normally spliced HAS1 (HAS1-FL) has a short half-life and localizes to the plasma membrane, whereas the HAS1 splice variants have a prolonged half-life and localize to the cytoplasm (A.G., H.K., and L.M.P., manuscript submitted). The aberrant splice variants form hetero-multimers with HAS1-FL and each other, thereby prolonging the half-life of HAS1-FL and providing a potential mechanism for promoting cancer (A.G., H.K., and L.M.P., manuscript submitted). Finally, as a single agent of an intronic splice variant, HAS1Vc is aggressively transforming in vitro and forms tumors in vivo (A.G., H.K., and L.M.P., manuscript submitted). It seems likely that HAS1 family members will act synergistically among themselves and with other molecules, for example RHAMM,60-63 to confer malignant characteristics. Work is in progress to evaluate this.
If verified in large-scale studies, the recurrent HAS1 GVs identified here may have the potential to identify persons at risk of MM or WM. Persons with germline HAS1 GVs are predicted to have some degree of risk of developing MM or WM. Persons who, in addition to inheriting HAS1 germ line GVs have acquired HAS1 GVs, may have a greatly increased risk of developing MM (or WM), perhaps requiring closer monitoring. The possibility exists that early detection of acquired HAS1 mutations may identify cryptic early stages of MM or WM. Overall, this work shows that inherited and acquired HAS1 GVs may contribute to the development of overt disease or disease progression in MM and WM by directing the aberrant intronic splicing of HAS1.
The online version of this article contains a data supplement.
Presented in abstract form at the 49th annual meeting of the American Society of Hematology, Atlanta, GA, December 2007.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Carolyn McQuarrie, Erin Strachan, Juanita Wizniak, and Sarah Motz for skilled technical assistance, and Daniel Santos for his help in obtaining WM samples.
This work was supported by grants from the Canadian Institutes of Health Research, the Alberta Cancer Research Institute, the International Waldenstrom Macroglobuli-nemia Foundation, and the Alberta Cancer Board Research Initiatives Program. S.A. was supported by an AHFMR Studentship. T.R. is an AHFMR Clinical Investigator. L.M.P. is the Canada Research Chair in Biomedical Nano-technology and this work was supported in part by the Chairs Program.
Authorship
Contribution: S.A. conceived the study, designed and executed experiments, performed data analysis and interpretation, and wrote the manuscript; A.A.R., H.K., A.G., and J.J.H. acquired and analyzed data; P.M.P. assisted in data analysis; J.K. designed and executed in vitro splicing experiments; T.R., M.J.M., S.P.T. and A.R.B. assisted in experimental design and writing the manuscript; and L.M.P. directed the research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Linda M. Pilarski, 11560 University Ave, Cross Cancer Institute, Edmonton, AB, Canada T6G 1Z2; e-mail: lpilarsk@ualberta.ca.