Abstract
CBFβ is the non-DNA binding subunit of the core binding factors (CBFs). Mice with reduced CBFβ levels display profound, early defects in T-cell but not B-cell development. Here we show that CBFβ is also required at very early stages of natural killer (NK)–cell development. We also demonstrate that T-cell development aborts during specification, as the expression of Gata3 and Tcf7, which encode key regulators of T lineage specification, is substantially reduced, as are functional thymic progenitors. Constitutively active Notch or IL-7 signaling cannot restore T-cell expansion or differentiation of CBFβ insufficient cells, nor can overexpression of Runx1 or CBFβ overcome a lack of Notch signaling. Therefore, the ability of the prethymic cell to respond appropriately to Notch is dependent on CBFβ, and both signals converge to activate the T-cell developmental program.
Introduction
T-cell development begins with colonization of the thymus by rare circulating bone marrow (BM)–derived progenitors,1 which expand to generate a population of early T lineage progenitors (ETPs). ETPs then give rise in an orderly fashion to cells expressing both CD4 and CD8 (double positive (DP)) via several intermediate CD4/CD8 double negative (DN) stages (DN2, DN3, and DN4).2 The earliest intrathymic stages of T lineage development, proliferation, and survival require signaling through Notch, c-kit, and the IL-7 receptor, plus the activity of several transcription factors, including core binding factors (CBFs), Gata3, E2A, c-myb, Ikaros, TCF/LEF, and Ets family members.2 Notch and the CBFs were shown to interact genetically in other contexts, although their genetic hierarchy during T-cell development is unknown.
Natural killer (NK) cells develop in multiple sites, including liver (fetal and adult), BM, spleen, and thymus.3,4 The first NK lineage committed progenitors can be identified through their expression of the interleukin-2 (IL-2)/IL15Rβ chain (CD122) and the absence of lineage-specific and mature NK cell markers.3 The differentiation of NK progenitors (NKPs) into immature and mature NK cells is exquisitely dependent on IL-15 signaling.3 NK-cell development does not require signaling through the c-kit receptor,3 and sustained Notch signaling inhibits NK-cell differentiation.5-7 Transcription factors required for NK-cell development include Ets-1, MEF, Id2, TCF/LEF, and members of the Ikaros family.3
CBFs are heterodimeric transcription factors consisting of a DNA binding subunit (Runx1, Runx2, or Runx3) and a non-DNA binding CBFβ subunit that increases the affinity of the Runx subunits for DNA. Homozygous disruption of Runx1 results in a failure of hematopoietic stem cell (HSC) emergence in the conceptus,8 and in the adult Runx1 is required for megakaryocyte, B-cell, and T-cell development.9,10 Conditional deletion of Runx1 in BM progenitors using Mx1-Cre blocked T-cell development at the DN2 to DN3 transition,9,10 whereas deletion in DN3 cells with Lck-Cre modestly impaired the formation of DN4 and intermediate single positive cells.11 Runx1 deletion in DP cells with Cd4-Cre reduced the number of mature CD4+ cells and eliminated a specialized subset of T cells with NK markers.11,12 However, an earlier, collective role for CBFs in T-cell development was revealed by a hypomorphic Cbfb allele (Cbfbrss) that, when carried over a nonfunctional Cbfb allele, caused an 85% reduction in CBFβ protein levels.13 Although HSCs emerged in Cbfbrss/− fetuses and B cells were generated, there was a profound defect in T-cell development with what appeared to be consecutive partially penetrant blocks in the generation of ETPs, DN2, and DN3 cells, and an almost complete absence of DN4 and DP cells.13
Notch proteins (Notch1-4) are transmembrane receptors that, on binding the cell surface ligands Delta-like or Jagged, undergo 2 proteolytic cleavages to release the Notch intracellular domain (ICN).14 ICN translocates into the nucleus, where it binds to the CSL/RBP-J (CBF1/RBP-J, Suppressor of Hairless, Lag-1) transcription factor, displacing corepressors and recruiting coactivators of the Mastermind-like (MAML) family. Disruption of Notch signaling either by conditional deletion of Notch1, by conditional deletion of CSL, or through expression of a truncated, dominant negative form of MAML1 completely blocks T-cell development and results in the generation of intrathymic B cells.14 Conversely, Notch is sufficient to drive T-cell development because overexpression of a constitutively active form of Notch (ICN) leads to T-cell at the expense of B-cell development at extrathymic sites.14 Exposure of hematopoietic progenitors to plate- or cell-bound Notch ligands of the Delta-like family can drive T lineage development in culture.14-16
In 2 well-characterized examples in hematopoiesis, Notch signaling was shown to function genetically upstream of the CBFs. Inactivation of Notch or its ligand Serrate in Drosophila caused the loss of Lozenge (a Runx homolog) expression in hemocyte progenitors,17 and mutations in Notch, Serrate, or Lozenge itself resulted in a failure to generate the subset of hemocytes called crystal cells.17,18 In mice, both Notch1 signaling and Runx1 are required for hematopoietic cell emergence from the aorta/gonad/mesonephros (AGM) region.8,19-22 Notch1 signaling defects in mice and zebrafish impair Runx1 expression in the AGM region,19,20,23 and overexpression of Runx1 in Notch signaling mutants can rescue the emergence of hematopoietic cells from the AGM region, demonstrating that Runx1 is, at least in part, genetically downstream of Notch signaling.23,24
Here we characterized the molecular mechanism underlying the T-cell defect caused by insufficient CBFβ levels. We show that T-cell specification does not occur, as its multiple early markers (Gata3, Tcf7, Cd3e) fail to be expressed. Notch signaling is not impaired; and although IL-7 signaling is decreased, it is not solely responsible for the T-cell defect. Finally, we show that reduced CBF levels cause an early and profound block in NK-cell development, which is the first demonstration that the CBFs play an essential role in the NK-cell lineage.
Methods
Mice
Transplant analyses
C57BL/6 (B6.SJL-Ptprc < a > Pep3 < b > /BoyJ) x 129S1/SVImJ F1 mice (Ly5.1+/Ly5.2+) were subjected to 2 split doses of 550 cGy 3 to 4 hours apart. Each recipient received donor fetal liver (FL) and competitor BM cells (2 × 105 cells of each) via tail vein injection. All donor fetuses were of a mixed C57BL/6J and 129S1/SVImJ background and expressed the Ly5.2 (CD45.2) haplotype. Whole BM competitor cells were prepared from C57BL/6 (B6.SJL-Ptprc < a > Pep3 < b > /BoyJ) (Ly5.1+) mice.
Flow cytometry and cell sorting
Flow cytometric analyses were performed on a dual-laser FACSCalibur, FACSCanto, or on a 4-laser LSRII (BD Biosciences, San Jose, CA). The following antibodies were purchased from BD PharMingen (San Diego, CA), eBiosciences (San Diego, CA), or BioLegend (San Diego, CA): CD3ϵ (145-2C11), CD4 (RM4-5), CD8α (53-6.7), CD11b (M1/70), CD11c (HL3), CD19 (1D3), CD25 (7D4), CD27 (LG.7F9), CD44 (IM7), CD45 (30-F11), B220 (RA3-6B2), Gr-1 (RB6-8C5), CD127/IL-7Rα (A7R34), T-cell receptorβ (TCRβ; H57-597), TCRγ (GL3), NK1.1 (PK136), CD49b (DX5), CD122 (TM-b1), CD132 (4G3), NKG2D (CX5), CD45.1 (A20), CD45.2 (104), Thy1.2 (53.2.1), c-kit (2B8), and pStat5 (47). On the LSRII platform, doublets were excluded through their FSC-W and SSC-W characteristics, and 4,6 diamidino-2-phenylindole was used for dead cell exclusion. The data were analyzed using FlowJo (version 6.1.1, TreeStar, San Carlos, CA). Cells were sorted on a FACSAria (BD Biosciences).
Cell-cycle analysis
Fetal thymocytes (17.5 days postcoitus (dpc)) were stained with fluorescein isothiocyanate-conjugated antibodies (CD8, TCRβ, TCRγ, CD11b, Ter119, B220) and subsequently cultured in Dulbecco modified Eagle medium with 10% fetal bovine serum (FBS) and pulsed with bromodeoxyuridine (BD Biosciences) for 1 hour. Cells were harvested and stained with anti-bromodeoxyuridine allophycocyanin, CD45 PerCP-Cy5.5, and 7-amino-actinomycin D.
In vitro IL-7 stimulation and intracellular staining for TCRβ, TCRγ, and pStat5
Fetal thymocytes (17.5 dpc) were stained with fluorescein isothiocyanate-conjugated lineage antibodies (CD8, CD3, TCRβ, TCRγ, CD11c, B220, NK1.1, Mac1, Ter119) as described elsewhere.26,27 Thymocytes were then incubated at 37°C for 20 minutes in Dulbecco modified Eagle medium with 10% FBS, then treated with 0, 1, and 5 ng/mL IL-7 (PeproTech, Rocky Hill, NJ) for an additional 20 minutes. At the end of the stimulation, cells were immediately fixed with 1.6% formaldehyde at room temperature for 10 minutes and permeabilized in ice-cold methanol for 20 minutes. Cells were stained with phycoerythrin-conjugated anti-phospho-STAT-5 (Tyr694) plus antibodies to CD44, CD25, and CD45. The analysis of intracellular TCR staining was performed using the same protocol minus the incubation and cytokine stimulation.
Enrichment of FL progenitors and OP9 cocultures
Lineage negative (Lin−) FL cells (E14.5-E17.5) were isolated by depletion of Lin+ (CD19, Gr-1, Ter119, F4/80) cells using MACS LS columns (Miltenyi Biotec, Auburn, CA). An anti–IL-7Rα biotin-labeled antibody was included in the lineage cocktail in experiments in which Notch signaling was inhibited by γ-secretase inhibitor (GSI).
OP9 and OP9-DL1 cells16 were cultured in Minimum Essential Medium Alpha supplemented with 20% FBS. Cocultures were performed in 24-well plates by adding 1 to 5 × 105 Lin− cells to confluent OP9-DL1 monolayers along with 5 ng/mL human Flt3L and 1 ng/mL murine IL-7 (PeproTech). IL-6 (1 ng/mL) and 25 ng/mL IL-15 were included in the cultures to enhance the generation of T, B, and NK cells. Various concentrations of GSI (InSolution, EMD Biosciences, San Diego, CA) in dimethyl sulfoxide were added to the cultured cells. Cocultured cells were harvested and analyzed weekly unless otherwise indicated.
Retroviral infection of hematopoietic progenitors
cDNAs encoding Runx1 (AML1b), the CBFβ heterodimerization domain (aa 1-141), full-length CBFβ, Stat5a, and Stat5aF were subcloned into the bicistronic MigR1 retrovirus.28 MigR1 expressing the Notch ICN was previously described.29 Retroviruses were produced in Phoenix cells. One milliliter of viral supernatant, polybrene (2 ng/mL), and cytokines (IL-7 and Flt3L) were added to overnight cocultures of Lin− FL cells or thymocytes on OP9-DL1 in 24-well plates and spinoculated at 1400g at room temperature for 2 hours. The media was changed 24 hours after spinoculation and the coculture continued.
Quantitative RT-PCR
Total RNA was extracted from sorted or unsorted cells using the RNeasy Mini Kit and DNase I treatment (Qiagen, Valencia, CA). RNA quality was assessed on agarose gels and quantified by Nano-Drop1000 (Nano-Drop, Wilmington, DE). First-strand cDNA was generated using reverse transcriptase SuperscriptIII (Invitrogen) and oligo (dT)20 primers. Real-time polymerase chain reaction (PCR) was performed in triplicate on Applied Biosystems' 7500 Real-Time PCR System (Foster City, CA). Either Taqman probes or SYBR-Green (Applied Biosystems) were used to detect gene expression. The following premade mixture of primers and Taqman probes were used: Runx1 (Mm00486762_m1); Runx3 (Mm00490666_m1); Cbfb (Mm00491551_m1); Jak1 (Mm00600614_m1); Jak3 (Mm00439962_m1); Stat3 (Mm00456961_m1); Stat5a (Mm00839861_m1); Stat5b (Mm00839861_m1); and Hprt1 (Mm00-446968_m1).
The following primers were used for SYBR Green detection: Dtx1 For TGAGGATGTGGTTCGGAGGT, Rev CCCTCATAGCCAGATGCTGTG; Hprt For CTCCTCAGACCGCTTTTTGC, Rev TAACCTGGTTCATCATCGCTAATC; Notch1 For CAGCTTGCACAACCAGACAGAC, Rev ACGGAGTACGGCCCATGTT. Primers for Pu.1, Cd3g, and Cd3e were described previously.30,31 Absolute quantification of each gene was calculated by the standard curve method using 10-fold dilutions of a positive control (spleen cell cDNA). Expression of individual genes was normalized to Hprt expression.
Western blot analysis
Green fluorescent protein (GFP)+ cells were sorted from OP9-DL1 cocultures (purity > 99.9%) and resuspended at 105 cells per milliliter in lysis buffer (150 mM of NaCl, 50 mM of Tris, pH 8.0, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate (SDS), 0.2 mM of ethylenediaminetetraacetic acid, 2.0 mM of ethyleneglycoltetraacetic acid plus 1 μg/mL of pepstatin A, 1 μM of Pefablock, 2 μg/mL of leupeptin, 2 μg/mL of aprotinin). Lysates were boiled in SDS loading buffer, resolved by SDS-PAGE through 4% to 12% Bis-Tris gels (Invitrogen), proteins transferred to nitrocellulose, and the blot probed with a mouse monoclonal antibody to CBFβ (β141.2).25 The blots were developed with enhanced chemiluminescence reagents (Pico Kit; Pierce Chemical, Rockford, IL).
Results
The T-cell defect exhibited by Cbfbrss/− cells is recapitulated ex vivo
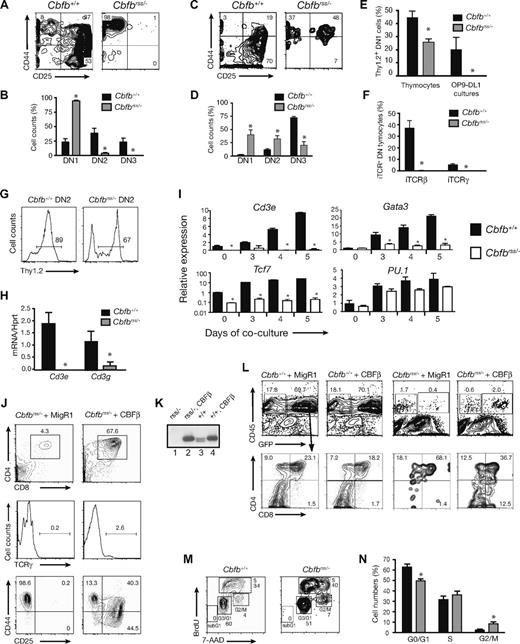
We previously assessed the collective role of core binding factors in hematopoiesis using a hypomorphic allele of the common non-DNA binding CBFβ subunit gene (Cbfbrss). Cbfbrss/− FL cells (expressing ∼15% of normal CBFβ levels) contained HSCs that could contribute to the formation of myelo-erythroid and B lineage cells, but not to DN4 or DP T cells.13 To further investigate the molecular basis of this defect, we cultured Lin− FL progenitors from 14.5 to 17.5 dpc mice on OP9 stromal cells expressing the Notch ligand Delta-like1 (OP9-DL1 cells) to induce T-cell differentiation ex vivo.16 Within 2 weeks after establishing the OP9-DL1 cocultures, Cbfb+/+ progenitors underwent extensive proliferation (Figure 1A,B), and approximately half became CD4/CD8 DP (Figure 1C,D) and gave rise to clear populations of TCRβ+ (not shown) and TCRγ+ cells (Figure 1E,F). In contrast, the ex vivo expansion of Cbfbrss/− cells was significantly depressed (Figure 1A,B). Cbfbrss/− progenitors generated very few DP cells (Figure 1C,D), TCRγ+ cells (Figure 1E,F), or TCRβ+ cells (not shown), and the DN cells contained no detectable intracellular TCRβ (Figure 1G). Cbfbrss/rss Lin− FL cells, which have higher CBFβ levels than Cbfbrss/− cells,13 also produced fewer cells and a significantly smaller percentage of TCRγ+ cells (Figure 1B,F); however, the defects were not as pronounced as those of Cbfbrss/− cells, indicating that T-cell development was affected in a dose-dependent manner ex vivo.
T-cell differentiation is blocked in a CBFβ dosage-dependent manner. (A) Proliferation of Lin− (Ter119, CD19, Gr-1, F4/80) 17.5 dpc FL cells cocultured on OP9-DL1 in the presence of Flt3L and IL-7, counted at weekly intervals. Each data point is averaged from at least 9 fetal livers. Error bars in all panels represent SEM. (B) Bar graph depicting the fold expansion of Lin− FL cells from an allelic Cbfb dosage reduction series, after 14 days of OP9-DL1 coculture. The percentage of CBFβ protein relative to wild-type levels is indicated below the bars.13 The number of independent animals analyzed was: Cbfb+/+ = 13; Cbfb+/− = 7; Cbfbrss/rss = 5; Cbfbrss/− = 10. *indicates significant (P < .05) difference relative to Cbfb+/+ animals (unpaired, 2-tailed Student t test). (C) CD4 and CD8 expression after 14 days of coculture on OP9-DL1. (D) Mean (± SEM) percentages of CD4+CD8+ (DP) cells. Cbfb+/+ = 17; Cbfb+/− = 7; Cbfbrss/rss = 5; Cbfbrss/− = 16. *Significant (P < .05) difference relative to Cbfb+/+ animals. **indicates significant (P < .05) difference relative to Cbfbrss/rss animals. (E) Cell surface TCRγ expression in the bulk cocultured cells. (F) Mean (± SEM) percentages of TCRγ+ cells after 14 days of culture. Cbfb+/+ = 17; Cbfb+/− = 7; Cbfbrss/rss = 5; Cbfbrss/− = 16. (G) Intracellular TCRβ staining in DN cells (DN = CD8−Gr-1− because CD4 expression is inappropriately unregulated in Cbfbrss/− DN cells13,72 ) after 6 and 9 days of OP9-DL1 culture. Cbfb+/+ = 4; Cbfbrss/− = 3.
T-cell differentiation is blocked in a CBFβ dosage-dependent manner. (A) Proliferation of Lin− (Ter119, CD19, Gr-1, F4/80) 17.5 dpc FL cells cocultured on OP9-DL1 in the presence of Flt3L and IL-7, counted at weekly intervals. Each data point is averaged from at least 9 fetal livers. Error bars in all panels represent SEM. (B) Bar graph depicting the fold expansion of Lin− FL cells from an allelic Cbfb dosage reduction series, after 14 days of OP9-DL1 coculture. The percentage of CBFβ protein relative to wild-type levels is indicated below the bars.13 The number of independent animals analyzed was: Cbfb+/+ = 13; Cbfb+/− = 7; Cbfbrss/rss = 5; Cbfbrss/− = 10. *indicates significant (P < .05) difference relative to Cbfb+/+ animals (unpaired, 2-tailed Student t test). (C) CD4 and CD8 expression after 14 days of coculture on OP9-DL1. (D) Mean (± SEM) percentages of CD4+CD8+ (DP) cells. Cbfb+/+ = 17; Cbfb+/− = 7; Cbfbrss/rss = 5; Cbfbrss/− = 16. *Significant (P < .05) difference relative to Cbfb+/+ animals. **indicates significant (P < .05) difference relative to Cbfbrss/rss animals. (E) Cell surface TCRγ expression in the bulk cocultured cells. (F) Mean (± SEM) percentages of TCRγ+ cells after 14 days of culture. Cbfb+/+ = 17; Cbfb+/− = 7; Cbfbrss/rss = 5; Cbfbrss/− = 16. (G) Intracellular TCRβ staining in DN cells (DN = CD8−Gr-1− because CD4 expression is inappropriately unregulated in Cbfbrss/− DN cells13,72 ) after 6 and 9 days of OP9-DL1 culture. Cbfb+/+ = 4; Cbfbrss/− = 3.
Cbfbrss/− cells fail to undergo T lineage specification
We examined the DN populations to more precisely define the T-cell developmental defect caused by reduced CBFβ dosage. After 7 days of culture on OP9-DL1, most Cbfb+/+Lin− FL cells had progressed to the DN2 and DN3 stages, whereas the majority of Cbfbrss/− cells were arrested before the DN2 stage (Figure 2A,B). Thymocytes from 16.5 to 17.5 dpc Cbfbrss/− fetuses also exhibited an early T-cell developmental arrest, but a higher percentage of thymocytes appeared to progress to the DN2 and DN3 stages (Figure 2C,D). We suspect that the lower numbers of phenotypic DN2 and DN3 cells generated ex vivo may be caused by differences in the quality or intensity of signaling provided by OP9-DL1 cells compared with thymic stromal cells.
T-cell specification fails to occur in Cbfbrss/− cells. (A) Lin− FL cells cocultured on OP9-DL1 in the presence of Flt3L and IL-7 for 7 days. CD8−Gr-1−GFP− cells (GFP gating was used to eliminate OP9-DL1) were analyzed for CD44 and CD25 expression. (B) Percentages of DN1, DN2, and DN3 cells (from panel A) averaged from 12 independent experiments. Error bars represent SEM. *indicates significant difference (P ≤ .01) between the percentages of Cbfb+/+ and Cbfbrss/− cells. (C) CD45+ Lin− (Lin = CD8, CD3, TCRβ, TCRγ, CD11c, B220, Mac1, NK1.1, Ter119) 17.5 dpc fetal thymocytes analyzed for CD44 and CD25 expression. (D) Data from 6 animals of each genotype (from panel C). *indicates significant difference (P ≤ .03) between the percentages of Cbfb+/+ and Cbfbrss/− cells. (E) Expression of cell surface Thy1.2 on 17.5 dpc DN1 (Lin−CD45+CD44+CD25−) thymocytes, and on DN1 cells (CD8−Gr-1−CD45+CD44+CD25−) from day 6 OP9-DL1 cultures. The thymocyte data were averaged from 4 Cbfb+/+ and 4 Cbfbrss/− fetuses, and the culture data were averaged from 4 Cbfb+/+ and 3 Cbfbrss/− fetuses. *indicates significant difference (P = .01) between Cbfb+/+ and Cbfbrss/− samples. (F) Percentage of 17.5 dpc DN thymocytes (CD45+ Lin−, Lin = CD8, CD3, CD11c, B220, Mac-1, Ter119) expressing intracellular TCRβ (iTCRβ) and iTCRγ. Data are averaged from 3 Cbfb+/+ and 3 Cbfbrss/− fetuses. (G) Percentage of Thy1.2+ DN2 cells after 7 days of OP9-DL1 culture. (H) Cd3e and Cd3g expression by qRT-PCR in DN2 cells sorted from day 7 OP9-DL1 cultures (culture conditions and staining as described in panel A). The purity of postsort populations was more than 95%. Expression of individual genes was normalized to Hprt. Data were derived from triplicate amplifications from 3 independent samples. *indicates significant difference (P≤ .01) between Cbfb+/+ versus Cbfbrss/− values. (I) Lin− FL cells cultured on OP9-DL1 cells, harvested at indicated time points. DN1 cells (Lin−CD45+CD44+CD25−) were isolated by cell sorting (purity > 95%). The expression of individual genes was normalized to Hprt (note log scale for Tcf7) and displayed as relative to day 0 values of Cbfb+/+ DN1 cells. Values are averaged from triplicate samples isolated from 3 independent experiments. *indicates significant difference (P ≤ .05) between Cbfb+/+ versus Cbfbrss/− values. (J) Rescue of T-cell development with CBFβ in 17.5 dpc Cbfbrss/− FL cells. Lin− FL cells (E17.5) were transduced with the indicated retroviruses and cocultured on OP9-DL1. Cells were harvested after 2 weeks and GFP+CD45+ cells analyzed for CD4 and CD8 expression (top panels) and TCRγ expression (middle panels). The bottom 2 plots are DN cells analyzed after 1 week of coculture. Gated GFP+Lin−CD45+ cells were analyzed for expression of CD44 and CD25 (nexperiments = 11). (K) Western blot showing CBFβ (p22) protein levels resulting from retroviral expression relative to endogenous protein levels, in whole cell extracts prepared from GFP+CD45+ cells purified from the OP9-DL1 cultures of Lin− FL cells (CD45+ cell purity ≥ 99.9%). The blot was probed with a monoclonal antibody to CBFβ. The 2 endogenous CBFβ isoforms generated as a result of alternative splicing (p21.5 and p22) are both visible on this gel. Samples were normalized for actin expression, and the relative amounts of CBFβ determined from a dilution series (not shown). CBFβ levels in retrovirally transduced cells were 5-fold higher than endogenous levels. Lane 1, Cbfbrss/− + MigR1; lane 2, Cbfbrss/− + CBFβ; lane 3, Cbfb+/+ + MigR1; lane 4, Cbfb+/+ + CBFβ. (L) Inefficient rescue of T-cell development on restoring CBFβ expression in 17.5 dpc Cbfbrss/− thymocytes. Thymocytes were transduced with the indicated retroviruses and cocultured for 7 days on OP9-DL1. Gated GFP+CD45+ cells were analyzed for CD4 and CD8 expression in the plots below. GFP+CD45− cells are predominantly OP9-DL1 cells.16 (M) Cell-cycle status of CD45+ Lin− (Lin = CD8, TCRβ, TCRγ, CD11b, Ter119, B220) 17.5 dpc fetal thymocytes. (N) Summary of cell-cycle data from 5 individual samples per genotype. The differences between Cbfb+/+ and Cbfbrss/− cells in G0/G1 and G2/M were significant at P≤ .01.
T-cell specification fails to occur in Cbfbrss/− cells. (A) Lin− FL cells cocultured on OP9-DL1 in the presence of Flt3L and IL-7 for 7 days. CD8−Gr-1−GFP− cells (GFP gating was used to eliminate OP9-DL1) were analyzed for CD44 and CD25 expression. (B) Percentages of DN1, DN2, and DN3 cells (from panel A) averaged from 12 independent experiments. Error bars represent SEM. *indicates significant difference (P ≤ .01) between the percentages of Cbfb+/+ and Cbfbrss/− cells. (C) CD45+ Lin− (Lin = CD8, CD3, TCRβ, TCRγ, CD11c, B220, Mac1, NK1.1, Ter119) 17.5 dpc fetal thymocytes analyzed for CD44 and CD25 expression. (D) Data from 6 animals of each genotype (from panel C). *indicates significant difference (P ≤ .03) between the percentages of Cbfb+/+ and Cbfbrss/− cells. (E) Expression of cell surface Thy1.2 on 17.5 dpc DN1 (Lin−CD45+CD44+CD25−) thymocytes, and on DN1 cells (CD8−Gr-1−CD45+CD44+CD25−) from day 6 OP9-DL1 cultures. The thymocyte data were averaged from 4 Cbfb+/+ and 4 Cbfbrss/− fetuses, and the culture data were averaged from 4 Cbfb+/+ and 3 Cbfbrss/− fetuses. *indicates significant difference (P = .01) between Cbfb+/+ and Cbfbrss/− samples. (F) Percentage of 17.5 dpc DN thymocytes (CD45+ Lin−, Lin = CD8, CD3, CD11c, B220, Mac-1, Ter119) expressing intracellular TCRβ (iTCRβ) and iTCRγ. Data are averaged from 3 Cbfb+/+ and 3 Cbfbrss/− fetuses. (G) Percentage of Thy1.2+ DN2 cells after 7 days of OP9-DL1 culture. (H) Cd3e and Cd3g expression by qRT-PCR in DN2 cells sorted from day 7 OP9-DL1 cultures (culture conditions and staining as described in panel A). The purity of postsort populations was more than 95%. Expression of individual genes was normalized to Hprt. Data were derived from triplicate amplifications from 3 independent samples. *indicates significant difference (P≤ .01) between Cbfb+/+ versus Cbfbrss/− values. (I) Lin− FL cells cultured on OP9-DL1 cells, harvested at indicated time points. DN1 cells (Lin−CD45+CD44+CD25−) were isolated by cell sorting (purity > 95%). The expression of individual genes was normalized to Hprt (note log scale for Tcf7) and displayed as relative to day 0 values of Cbfb+/+ DN1 cells. Values are averaged from triplicate samples isolated from 3 independent experiments. *indicates significant difference (P ≤ .05) between Cbfb+/+ versus Cbfbrss/− values. (J) Rescue of T-cell development with CBFβ in 17.5 dpc Cbfbrss/− FL cells. Lin− FL cells (E17.5) were transduced with the indicated retroviruses and cocultured on OP9-DL1. Cells were harvested after 2 weeks and GFP+CD45+ cells analyzed for CD4 and CD8 expression (top panels) and TCRγ expression (middle panels). The bottom 2 plots are DN cells analyzed after 1 week of coculture. Gated GFP+Lin−CD45+ cells were analyzed for expression of CD44 and CD25 (nexperiments = 11). (K) Western blot showing CBFβ (p22) protein levels resulting from retroviral expression relative to endogenous protein levels, in whole cell extracts prepared from GFP+CD45+ cells purified from the OP9-DL1 cultures of Lin− FL cells (CD45+ cell purity ≥ 99.9%). The blot was probed with a monoclonal antibody to CBFβ. The 2 endogenous CBFβ isoforms generated as a result of alternative splicing (p21.5 and p22) are both visible on this gel. Samples were normalized for actin expression, and the relative amounts of CBFβ determined from a dilution series (not shown). CBFβ levels in retrovirally transduced cells were 5-fold higher than endogenous levels. Lane 1, Cbfbrss/− + MigR1; lane 2, Cbfbrss/− + CBFβ; lane 3, Cbfb+/+ + MigR1; lane 4, Cbfb+/+ + CBFβ. (L) Inefficient rescue of T-cell development on restoring CBFβ expression in 17.5 dpc Cbfbrss/− thymocytes. Thymocytes were transduced with the indicated retroviruses and cocultured for 7 days on OP9-DL1. Gated GFP+CD45+ cells were analyzed for CD4 and CD8 expression in the plots below. GFP+CD45− cells are predominantly OP9-DL1 cells.16 (M) Cell-cycle status of CD45+ Lin− (Lin = CD8, TCRβ, TCRγ, CD11b, Ter119, B220) 17.5 dpc fetal thymocytes. (N) Summary of cell-cycle data from 5 individual samples per genotype. The differences between Cbfb+/+ and Cbfbrss/− cells in G0/G1 and G2/M were significant at P≤ .01.
We examined Cbfbrss/− DN cells for molecular markers of T-cell differentiation. Thy1.2+ DN1 thymocytes were present in Cbfbrss/− 17.5 dpc fetuses, but in significantly reduced numbers (Figure 2E). Cbfbrss/− DN thymocytes contained no intracellular TCRβ or TCRγ chains, which are normally found in DN3 and DN4 cells, respectively,32 indicating that development arrested before the commitment stage (Figure 2F). The ex vivo defect was more pronounced, as Thy1.2+ cells were absent in the DN1 population (Figure 2E), although most thymocytes that progressed to the DN2 stage did express Thy1.2 (Figure 2G). Cd3e expression, which can normally be found in both DN1 and DN2 cells,33 was essentially undetectable in Cbfbrss/− DN2 cells purified from the OP9-DL1 cultures, and Cd3g mRNA levels were significantly reduced (Figure 2H). Gata3 and Tcf7 expression progressively increased in wild-type DN1 (Figure 2I) and DN2 (not shown) cells over a 5-day culture period30 but remained low and unchanged in Cbfbrss/− DN1 and DN2 cells. PU.1 expression was not elevated in the Cbfbrss/− DN1 population, suggesting that respecification into myeloid lineage cells had not occurred.30
To determine whether Cbfbrss/− fetuses contained functional thymic progenitors, we attempted to rescue T-cell development by reintroducing CBFβ into Cbfbrss/− thymocytes. Although we could successfully rescue the formation of DP, TCRγ+, and DN2-DN4 cells after retroviral transduction of CBFβ into Lin−Cbfbrss/− FL progenitors (Figure 2J,K), we could not efficiently rescue T-cell development from Cbfbrss/− fetal thymocytes (Figure 2L). We could transduce wild-type thymocytes with GFP-expressing retroviruses and recover GFP+CD45+ and GFP+DP cells; thus, our retroviral transductions were effective. However, we could recover only very few GFP+CD45+ cells from Cbfbrss/− thymocytes transduced with a bicistronic virus expressing both CBFβ and GFP, although these included DP cells (Figure 2L). Cbfbrss/− DN thymocytes were actively cycling (Figure 2M,N); thus, it is unlikely that our failure to rescue T-cell development is the result of a lack of cells permissive for retroviral transduction. We conclude that Cbfbrss/− thymi contain very few functional T-cell progenitors. Altogether, these data indicate that Cbfbrss/− cells have a global, early defect in T-cell specification.
IL-7 signaling and c-kit expression in Cbfbrss/− cells
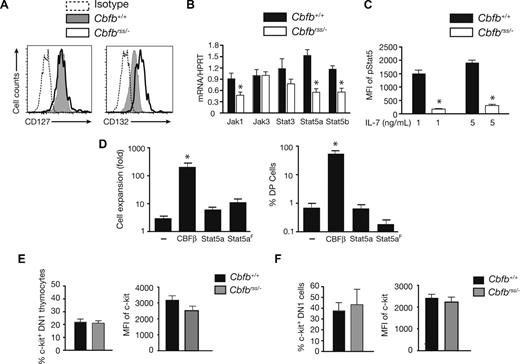
We determined whether CBFβ insufficiency affected the expression or activity of one or more signaling pathways required at early stages of T-cell development. IL-7 signaling is essential at the DN1 and DN2 stages, and cell surface IL-7Rα levels become elevated at the DN2 stage.34-38 CBFs directly activate the Il7ra (IL-7Rα) gene at later stages of T-cell differentiation, in DP and CD4+ cells.11,39 We found that the mean fluorescence intensity (MFI) of IL-7Rα (CD127) staining was comparable in Cbfb+/+ and Cbfbrss/− DN2 thymocytes, and that of the common gamma chain (γc/IL2RG/CD132) was approximately 2.5-fold higher (Figure 3A). However, the levels of mRNAs encoding several IL-7 signaling molecules, including Jak1, Stat5a, and Stat5b, were significantly (∼2 fold) lower in Cbfbrss/− DN2 cells (Figure 3B) and intracellular pStat5 levels were decreased by approximately 7-fold (Figure 3C). Decreased JAK/STAT signaling, however, cannot be solely responsible for the T-cell expansion or differentiation defects, as neither could be reversed on expressing a wild-type or constitutively active form of Stat5a40 (Figure 3D).
JAK/STAT and c-kit signaling in Cbfbrss/− T cells. (A) Expression of IL-7Rα (CD127) and γc (IL2RG/CD132) on the DN2 thymocytes from Figure 2C (17.5 dpc). Data are representative of 4 experiments. (B) DN2 cells were sorted from OP9-DL1 cultures (culture conditions and staining as described in Figure 2A). The purity of postsort populations was more than 95%. Expression of individual genes was normalized to Hprt. Values are averaged from triplicate samples from 3 independent experiments. *indicates significant difference (P≤ .05). (C) pStat5 levels in 17.5 dpc DN2 thymocytes on ex vivo stimulation with 0, 1, and 5 ng/mL IL-7 for 20 minutes. Shown is the MFI of the difference between pStat5 under IL-7–stimulated and nonstimulated conditions. The data are averaged from 5 Cbfb+/+ and 5 Cbfbrss/− fetuses. Differences are significant at P < .001. (D) Rescue of T-cell development from Cbfbrss/−Lin− FL cells with CBFβ, but not wild-type or constitutively active Stat5a (S711F; Stat5aF), or MigR1 alone (−) (n = 5). Cells were harvested after 2 weeks. The expansion of Cbfbrss/− cells (left graph) is calculated by dividing the number of GFP+ cells on day 14 by that on day 7 of culture. Only CBFβ expression significantly (*P≤ .001) increased cell numbers compared with MigR1-transduced Cbfbrss/− cells. The right-hand graph shows the percentage of GFP+ cells expressing CD4 and CD8. (E) Expression of cell surface c-kit on 17.5 dpc DN1 cells. Bar graph on left is the percentage of c-kit+ DN1 (Lin− as in Figure 2B; CD45+CD44+CD25−) thymocytes, and on right is the MFI of c-kit staining on c-kit+ DN1 thymocytes. The data are averaged from 4 Cbfb+/+ and 4 Cbfbrss/− fetuses. (F) Percentage of DN1 (CD44+CD25−CD45+ Lin−) cells expressing surface c-kit and MFI of c-kit staining on c-kit+ DN1 cells after 6 days of OP9-DL1 culture. The data are averaged from 4 Cbfb+/+ and 3 Cbfbrss/− fetuses.
JAK/STAT and c-kit signaling in Cbfbrss/− T cells. (A) Expression of IL-7Rα (CD127) and γc (IL2RG/CD132) on the DN2 thymocytes from Figure 2C (17.5 dpc). Data are representative of 4 experiments. (B) DN2 cells were sorted from OP9-DL1 cultures (culture conditions and staining as described in Figure 2A). The purity of postsort populations was more than 95%. Expression of individual genes was normalized to Hprt. Values are averaged from triplicate samples from 3 independent experiments. *indicates significant difference (P≤ .05). (C) pStat5 levels in 17.5 dpc DN2 thymocytes on ex vivo stimulation with 0, 1, and 5 ng/mL IL-7 for 20 minutes. Shown is the MFI of the difference between pStat5 under IL-7–stimulated and nonstimulated conditions. The data are averaged from 5 Cbfb+/+ and 5 Cbfbrss/− fetuses. Differences are significant at P < .001. (D) Rescue of T-cell development from Cbfbrss/−Lin− FL cells with CBFβ, but not wild-type or constitutively active Stat5a (S711F; Stat5aF), or MigR1 alone (−) (n = 5). Cells were harvested after 2 weeks. The expansion of Cbfbrss/− cells (left graph) is calculated by dividing the number of GFP+ cells on day 14 by that on day 7 of culture. Only CBFβ expression significantly (*P≤ .001) increased cell numbers compared with MigR1-transduced Cbfbrss/− cells. The right-hand graph shows the percentage of GFP+ cells expressing CD4 and CD8. (E) Expression of cell surface c-kit on 17.5 dpc DN1 cells. Bar graph on left is the percentage of c-kit+ DN1 (Lin− as in Figure 2B; CD45+CD44+CD25−) thymocytes, and on right is the MFI of c-kit staining on c-kit+ DN1 thymocytes. The data are averaged from 4 Cbfb+/+ and 4 Cbfbrss/− fetuses. (F) Percentage of DN1 (CD44+CD25−CD45+ Lin−) cells expressing surface c-kit and MFI of c-kit staining on c-kit+ DN1 cells after 6 days of OP9-DL1 culture. The data are averaged from 4 Cbfb+/+ and 3 Cbfbrss/− fetuses.
c-kit signaling is also critical for the proliferation and differentiation of DN1 and DN2 cells.38,41 We found that neither the percentage of c-kit+ DN1 thymocytes nor the MFI of c-kit staining was altered in Cbfbrss/− fetuses (Figure 3E). The percentage of c-kit+ cells and staining intensity was also similar in Cbfb+/+ and Cbfbrss/− DN1 cells derived from OP9-DL1 cultures (Figure 3F). Therefore, the T-cell defect does not appear to be caused by reduced cell surface c-kit expression, although deficiencies in c-kit signaling were not examined and cannot be ruled out.
Notch signaling is active in Cbfbrss/− cells
To examine the ability of Cbfbrss/− progenitors to receive Notch signals, we purified the DN1 population from OP9-DL1 cocultures at days 3, 4, and 5 and quantified the expression of Notch1 and several of its target genes by quantitative RT-PCR (qRT-PCR; Figure 4A). Notch1 was expressed in Cbfbrss/− cells at all time points examined, although its levels were reduced approximately 2-fold in freshly isolated “DN1” (ie, Lin−CD45+ CD44+CD25−) FL cells (day 0) and in DN1 cells isolated after 3 days of culture on OP9-DL1 cells, and slightly more than 2-fold at days 4 and 5 of culture. Expression of the Notch1 target genes Hes1 and Hes5 was reduced less than or equal to 2-fold in Cbfbrss/− DN1 cells at all time points examined (Figure 4A). Deltex1 (Dtx1) expression is known to be a very sensitive indicator of Notch signaling42-44 and was reduced almost 10-fold in Cbfbrss/− DN1 cells by days 4 and 5 of culture. Although this is a substantial decrease, Dtx1 levels did indeed increase in Cbfbrss/− DN1 cells approximately 100-fold during the culture period. Thus, although there were significant reductions in the expression of Notch1 and several of its target genes, the fact that their expression was not completely absent and was significantly induced after exposure to Notch ligands suggested that Notch signaling was active in Cbfbrss/− cells. Differences in the initial composition of the DN1 population that could change further over the culture period could account for the relative decreases in the levels of Notch1 and its targets because of enrichment of the Cbfb+/+ (but not the Cbfbrss/−) DN1 population for T lineage–specified cells.
Notch signaling is active in Cbfbrss/− cells and NK-cell development is defective. (A) Lin− FL cells cultured on OP9-DL1 cells, harvested at indicated time points, and stained with antibodies as described in Figure 2A. DN1 cells (Lin−CD45+CD44+CD25−) were isolated by cell sorting (purity > 95%). The expression of individual genes was normalized to Hprt (note log scale for Dxt1) and displayed as relative to day 0 Cbfb+/+ DN1 cells. Values are averaged from 9 samples (triplicate samples from 3 independent experiments). *indicates significant difference (P≤ .05) between Cbfb+/+ versus Cbfbrss/− values. (B) Flow cytometric analysis of Lin− FL cells cultured on OP9-DL1 (+ Flt3L, IL-7, IL-6, IL-15) in the absence and presence of the gamma secretase inhibitor (GSI) inhibitor X for 7 days. GFP−CD45+ cells were analyzed for expression of NK1.1 (NK cells) and CD19 (B cells). nexperiments = 7. (C) Lin−Cbfb+/+ 14.5 dpc FL cells transduced with either MigR1 or a retrovirus expressing the Notch1 intracellular domain (ICN) and cultured for 7 days on OP9 stromal cells in the presence of Flt3L and IL-7. CD8−CD19−Gr-1− cells were gated for GFP expression in the left-hand panels, and GFP+ cells analyzed for CD44 and CD25 expression in the right-hand panels. (D) Lin−Cbfbrss/− FL cells transduced with MigR1 or ICN, and analyzed as in panel C.
Notch signaling is active in Cbfbrss/− cells and NK-cell development is defective. (A) Lin− FL cells cultured on OP9-DL1 cells, harvested at indicated time points, and stained with antibodies as described in Figure 2A. DN1 cells (Lin−CD45+CD44+CD25−) were isolated by cell sorting (purity > 95%). The expression of individual genes was normalized to Hprt (note log scale for Dxt1) and displayed as relative to day 0 Cbfb+/+ DN1 cells. Values are averaged from 9 samples (triplicate samples from 3 independent experiments). *indicates significant difference (P≤ .05) between Cbfb+/+ versus Cbfbrss/− values. (B) Flow cytometric analysis of Lin− FL cells cultured on OP9-DL1 (+ Flt3L, IL-7, IL-6, IL-15) in the absence and presence of the gamma secretase inhibitor (GSI) inhibitor X for 7 days. GFP−CD45+ cells were analyzed for expression of NK1.1 (NK cells) and CD19 (B cells). nexperiments = 7. (C) Lin−Cbfb+/+ 14.5 dpc FL cells transduced with either MigR1 or a retrovirus expressing the Notch1 intracellular domain (ICN) and cultured for 7 days on OP9 stromal cells in the presence of Flt3L and IL-7. CD8−CD19−Gr-1− cells were gated for GFP expression in the left-hand panels, and GFP+ cells analyzed for CD44 and CD25 expression in the right-hand panels. (D) Lin−Cbfbrss/− FL cells transduced with MigR1 or ICN, and analyzed as in panel C.
To determine whether Notch signaling was functionally intact, we treated OP9-DL1 cocultures with a GSI to block Notch signaling. When Notch signaling is blocked in wild-type progenitors, B cells will develop at the expense of T cells.45,46 Because OP9-DL1 cocultures of Cbfbrss/−Lin− cells did not contain CD19+ B cells (Figure 4B), nor were B cells detected in thymi of mice transplanted with Cbfbrss/− FL cells,13 this suggested that Notch signaling was actively repressing B-cell development. As expected, inhibition of Notch signaling with 1 μM and 3 μM GSI resulted in the differentiation of CD19+ B cells from Cbfb+/+Lin− cells (Figure 4B). GSI even more efficiently induced the formation of B cells from Cbfbrss/−Lin− cells, confirming that Notch1 signaling was sufficiently active in Cbfbrss/− cells to repress B-cell fate (Figure 4B). An increase in NK1.1+ NK cells was also observed in Cbfb+/+ cocultures treated with GSI, but not in Cbfbrss/− cocultures. Thus, Cbfbrss/− cells had a defect in NK-cell development, which will be discussed in more detail later.
Although Notch signaling in Cbfbrss/− cells was adequate to suppress B-cell development, higher levels of Notch signaling are required to drive T-cell development.7 Therefore, to confirm that the lower levels of Notch1 or other defects in Notch signaling were not the underlying cause of the T-cell developmental defect, we transduced Cbfbrss/−Lin− FL cells with a retrovirus expressing the constitutively active intracellular form of Notch1 (ICN). Cbfb+/+ cells expressing high levels of ICN (based on the surrogate GFP marker) efficiently differentiated into DN2 and DN3 cells after 7 days of culture on OP9 cells (Figure 4C). On the other hand, Cbfbrss/− progenitors expressing ICN grew poorly, giving rise to a low percentage of GFP+ cells (1.7% ± 0.5% vs 66.6% ± 15.0% for Cbfb+/+ cells), and only a few of those cells differentiated to and past the DN2 stage (Figure 4D). Thus, constitutively high levels of Notch signaling did not rescue T-cell development but instead selected against the growth or survival of Cbfbrss/− cells. Because high levels of Notch signaling efficiently block B- and NK-cell development,47 and T-cell development from Cbfbrss/− progenitors could not be rescued by ICN, the ICN-expressing Cbfbrss/− cells could not differentiate into the T-, NK-, or B-cell lineages and appeared to be selected against, thus yielding relatively few GFP+ cells in the cultures. Therefore, the moderate decrease in Notch1 expression, or potential defects in ligand binding or cleavage of the Notch receptor are unlikely to be responsible for the Cbfbrss/− early T-cell defect but indeed may be selectively enriched to permit survival of the Cbfbrss/− cells.
Runx1, Runx3, and CBFβ expression are not dependent on Notch signaling
Runx1 expression is activated by Notch signaling during hematopoietic stem cell development19,20,23,24 ; thus, we determined whether Runx1, Runx3, or Cbfb expression in T cells also required Notch signaling. Addition of 1 μM and 3 μM GSI to Cbfb+/+ OP9-DL1 cocultures impaired the differentiation of DN2 and DN3 cells in a dose-dependent manner (Figure 5A). Expression of the Notch1 targets Hes1 and Dtx1 was significantly decreased in DN1 cells, as was Gata3 and Tcf7 (Figure 5B), demonstrating that Notch signaling and T-cell development were indeed inhibited at the 3 μM GSI concentration. However, Runx1, Runx3, and Cbfb expression did not significantly change in DN1 cells in the presence of GSI, consistent with the conclusion that none of these genes is a downstream activated target of Notch1 in early T lineage progenitors (Figure 5B). Because heterogeneity of the DN1 population could obscure modest changes in gene expression, we purified c-kit+CD27+CD25− lymphoid progenitors from the DN1 population30 and assessed Runx1 expression (Figure 5C). Inhibition of Notch signaling with 3 μM GSI resulted in a small but significant increase in Runx1 mRNA levels in c-kit+CD27+CD25− cells, confirming that Runx1 expression in lymphoid progenitors does not require Notch signaling. Moreover, retroviral overexpression of neither Runx1 (Figure 5D) nor CBFβ (not shown) in Cbfb+/+ progenitors treated with 3 μM GSI could rescue the perturbation in T-cell development caused by inhibition of Notch signaling, supporting the hypothesis that blocking Notch signaling does not impair T-cell development by affecting Runx1, Runx3, or Cbfb expression. In summary, the activity of the Notch pathway does not depend on the CBFs, nor does CBF expression require Notch activity. Rather, Notch and CBF expression and activity are independently regulated in uncommitted thymic progenitors and converge to specify the T-cell lineage.
Inhibition of Notch signaling does not affect Runx1, Runx3, or Cbfb expression. (A) Flow cytometric analysis of Lin− FL cells cultured on OP9-DL1 in the absence and presence of the indicated concentrations of GSI for 7 days. CD8−CD19−Gr-1−GFP− cells were analyzed for CD44 and CD25 expression (nexperiments = 8). (B) Gene expression profile of DN1 cells sorted from day 7 OP9-DL1 cocultures treated with dimethyl sulfoxide (□), 1.0 μM GSI ( ), or 3.0 μM GSI (■). Cell sorting, RNA preparation, and real-time PCR were performed as described for Figure 4A. Taqman probes were used for the quantification of Runx1, Runx3, Cbfb, and Hprt, and SYBR green was used for the remainder of the genes. Expression of each gene was quantified in comparison to a standard curve prepared with dilutions of spleen cDNA. The expression of individual genes is displayed relative to Hprt. Data are averaged from 4 independent experiments. Error bars represent SEM. *indicates significant difference (P≤ .05) between GSI-treated and untreated cells. (C) Runx1 expression in lymphoid progenitors (c-kit+CD27+CD25−) isolated from day 3 OP9-DL1 cultures in the absence and presence of 3 μM GSI (averaged from triplicate samples from 3 independent experiments). The increase in Runx1 expression in GSI-treated cultures was significant at P < .01. (D) Ectopic expression of Runx1 in Cbfb+/+ FL cells cultured on OP9-DL1 in the absence and presence of GSI at 3 μM. Analysis was performed as in panel A.
), or 3.0 μM GSI (■). Cell sorting, RNA preparation, and real-time PCR were performed as described for Figure 4A. Taqman probes were used for the quantification of Runx1, Runx3, Cbfb, and Hprt, and SYBR green was used for the remainder of the genes. Expression of each gene was quantified in comparison to a standard curve prepared with dilutions of spleen cDNA. The expression of individual genes is displayed relative to Hprt. Data are averaged from 4 independent experiments. Error bars represent SEM. *indicates significant difference (P≤ .05) between GSI-treated and untreated cells. (C) Runx1 expression in lymphoid progenitors (c-kit+CD27+CD25−) isolated from day 3 OP9-DL1 cultures in the absence and presence of 3 μM GSI (averaged from triplicate samples from 3 independent experiments). The increase in Runx1 expression in GSI-treated cultures was significant at P < .01. (D) Ectopic expression of Runx1 in Cbfb+/+ FL cells cultured on OP9-DL1 in the absence and presence of GSI at 3 μM. Analysis was performed as in panel A.
Inhibition of Notch signaling does not affect Runx1, Runx3, or Cbfb expression. (A) Flow cytometric analysis of Lin− FL cells cultured on OP9-DL1 in the absence and presence of the indicated concentrations of GSI for 7 days. CD8−CD19−Gr-1−GFP− cells were analyzed for CD44 and CD25 expression (nexperiments = 8). (B) Gene expression profile of DN1 cells sorted from day 7 OP9-DL1 cocultures treated with dimethyl sulfoxide (□), 1.0 μM GSI ( ), or 3.0 μM GSI (■). Cell sorting, RNA preparation, and real-time PCR were performed as described for Figure 4A. Taqman probes were used for the quantification of Runx1, Runx3, Cbfb, and Hprt, and SYBR green was used for the remainder of the genes. Expression of each gene was quantified in comparison to a standard curve prepared with dilutions of spleen cDNA. The expression of individual genes is displayed relative to Hprt. Data are averaged from 4 independent experiments. Error bars represent SEM. *indicates significant difference (P≤ .05) between GSI-treated and untreated cells. (C) Runx1 expression in lymphoid progenitors (c-kit+CD27+CD25−) isolated from day 3 OP9-DL1 cultures in the absence and presence of 3 μM GSI (averaged from triplicate samples from 3 independent experiments). The increase in Runx1 expression in GSI-treated cultures was significant at P < .01. (D) Ectopic expression of Runx1 in Cbfb+/+ FL cells cultured on OP9-DL1 in the absence and presence of GSI at 3 μM. Analysis was performed as in panel A.
), or 3.0 μM GSI (■). Cell sorting, RNA preparation, and real-time PCR were performed as described for Figure 4A. Taqman probes were used for the quantification of Runx1, Runx3, Cbfb, and Hprt, and SYBR green was used for the remainder of the genes. Expression of each gene was quantified in comparison to a standard curve prepared with dilutions of spleen cDNA. The expression of individual genes is displayed relative to Hprt. Data are averaged from 4 independent experiments. Error bars represent SEM. *indicates significant difference (P≤ .05) between GSI-treated and untreated cells. (C) Runx1 expression in lymphoid progenitors (c-kit+CD27+CD25−) isolated from day 3 OP9-DL1 cultures in the absence and presence of 3 μM GSI (averaged from triplicate samples from 3 independent experiments). The increase in Runx1 expression in GSI-treated cultures was significant at P < .01. (D) Ectopic expression of Runx1 in Cbfb+/+ FL cells cultured on OP9-DL1 in the absence and presence of GSI at 3 μM. Analysis was performed as in panel A.
CBFβ is required for NK-cell development
Although Cbfbrss/− Lin− cells were capable of producing B cells, we observed a defect in NK-cell development when Notch signaling was inhibited (Figure 4B). We examined this more directly by culturing Lin− FL cells on OP9 stromal cells in the presence of Flt3L, IL-7, and IL-15. CD122 (IL2Rβ) is a subunit of the IL-15 receptor and is regulated by the CBFs.48 Cbfb+/+Lin− FL cells generated a large number of CD122+ cells, including NK1.1+CD122+ NK cells and NK1.1−CD122+ NKPs. Cbfbrss/− Lin− FL cells, on the other hand, produced very few CD122+ cells (Figure 6A,B) that would be capable of responding to IL-15 signaling. These data indicate that ex vivo NK-cell development from fetal liver cells was blocked very early, before the NKP stage.
CBFβ is required at an early stage of NK-cell development in vivo. (A) Representative scatter plots of Lin− 15.5 dpc FL cells grown on OP9 cells in the presence of Flt3L, IL-7, and IL-15. Gr-1−CD19− cells were analyzed for CD122 and NK1.1 expression. (B) Percentage and absolute number of CD122+ cells (± SD) harvested from OP9 cultures established from 3 Cbfb+/+ and 4 Cbfbrss/− fetuses (nexperiments = 3). (C) NK cells in 15.5 dpc fetal thymi. (D) Total number of thymic NK1.1+ and NK1.1+CD122+ cells averaged from 3 Cbfb+/+ and 3 Cbfbrss/− fetuses (nexperiments = 3). (E-G) Lethally irradiated CD45.1 × CD45.2 F1 recipients were reconstituted with wild-type CD45.1+ BM and CD45.2+ FL cells from Cbfb+/+, Cbfbrss/rss, or Cbfbrss/− fetuses (expressing 100%, 30%, and 15% of normal CBFβ levels, respectively). (E) Bone marrow of recipient mice analyzed 10 months after reconstitution. The data are from gated FL-derived (CD45.2+) progenitors. A representative example is shown in each group (Cbfb+/+, n = 4; Cbfbrss/rss, n = 4; Cbfbrss/−, n = 5). Bone marrow from a 10-week-old C57BL/6 (B6) mouse is shown as control. CD19−CD3−TCRβ−NK1.1+DX5+ cells are mature NK cells, whereas immature NK lineage cells (iNK) have a NK1.1+Dx5− phenotype. (F) The spleen of recipient mice analyzed in a similar way, illustrating the absence of immature and mature NK cells among cells derived from Cbfbrss/− progenitors. (G) Primitive NK lineage committed progenitors (NKP) (CD122/IL2Rβ+NKG2D+NK1.1−Dx5−LinT−) (LinT = CD3CD4CD8αTCRβTCRγ) are preserved in the spleen of recipient mice. NKP express the IL-2/IL-15Rβ chain and have an NK1.1−Dx5−CD19−CD3−CD4−CD8a−TCRβ−TCRγ− phenotype. A fraction of these cells expresses NKG2D.3 (H) Expression Runx1, Runx3, and Cbfb by qRT-PCR in LSK (Lin = CD4, CD8, TCRβ, TCRγ, DX5, CD19, Mac-1, Ter119), iNK (CD4−CD8−TCRβ−TCRγ−CD19−CD49b−NK1.1+CD122+) and mature NK (CD4−CD8−TCRβ−TCRγ−CD19−CD49b+NK1.1+CD122+) cells sorted from BM of 8- to 12-week-old wild-type mice. The purity of post-sort populations was more than 98%. Expression of individual genes was normalized to Hprt. * Significant difference (P≤ .01) from LSK cell values.
CBFβ is required at an early stage of NK-cell development in vivo. (A) Representative scatter plots of Lin− 15.5 dpc FL cells grown on OP9 cells in the presence of Flt3L, IL-7, and IL-15. Gr-1−CD19− cells were analyzed for CD122 and NK1.1 expression. (B) Percentage and absolute number of CD122+ cells (± SD) harvested from OP9 cultures established from 3 Cbfb+/+ and 4 Cbfbrss/− fetuses (nexperiments = 3). (C) NK cells in 15.5 dpc fetal thymi. (D) Total number of thymic NK1.1+ and NK1.1+CD122+ cells averaged from 3 Cbfb+/+ and 3 Cbfbrss/− fetuses (nexperiments = 3). (E-G) Lethally irradiated CD45.1 × CD45.2 F1 recipients were reconstituted with wild-type CD45.1+ BM and CD45.2+ FL cells from Cbfb+/+, Cbfbrss/rss, or Cbfbrss/− fetuses (expressing 100%, 30%, and 15% of normal CBFβ levels, respectively). (E) Bone marrow of recipient mice analyzed 10 months after reconstitution. The data are from gated FL-derived (CD45.2+) progenitors. A representative example is shown in each group (Cbfb+/+, n = 4; Cbfbrss/rss, n = 4; Cbfbrss/−, n = 5). Bone marrow from a 10-week-old C57BL/6 (B6) mouse is shown as control. CD19−CD3−TCRβ−NK1.1+DX5+ cells are mature NK cells, whereas immature NK lineage cells (iNK) have a NK1.1+Dx5− phenotype. (F) The spleen of recipient mice analyzed in a similar way, illustrating the absence of immature and mature NK cells among cells derived from Cbfbrss/− progenitors. (G) Primitive NK lineage committed progenitors (NKP) (CD122/IL2Rβ+NKG2D+NK1.1−Dx5−LinT−) (LinT = CD3CD4CD8αTCRβTCRγ) are preserved in the spleen of recipient mice. NKP express the IL-2/IL-15Rβ chain and have an NK1.1−Dx5−CD19−CD3−CD4−CD8a−TCRβ−TCRγ− phenotype. A fraction of these cells expresses NKG2D.3 (H) Expression Runx1, Runx3, and Cbfb by qRT-PCR in LSK (Lin = CD4, CD8, TCRβ, TCRγ, DX5, CD19, Mac-1, Ter119), iNK (CD4−CD8−TCRβ−TCRγ−CD19−CD49b−NK1.1+CD122+) and mature NK (CD4−CD8−TCRβ−TCRγ−CD19−CD49b+NK1.1+CD122+) cells sorted from BM of 8- to 12-week-old wild-type mice. The purity of post-sort populations was more than 98%. Expression of individual genes was normalized to Hprt. * Significant difference (P≤ .01) from LSK cell values.
To assess whether there was also an in vivo NK-cell defect, we examined thymi from 15.5 dpc Cbfbrss/− fetuses for NK cells. Bipotent T/NK progenitors and mature NK cells in the fetal thymus both express NK1.1.49 Cbfbrss/− fetal thymi contained very few NK1.1+ (or NK1.1+CD122+) cells (Figure 6C,D), indicating that almost no mature NK cells or bipotent T/NK progenitors were present. We also examined mice transplanted with Cbfb+/+, Cbfbrss/rss, and Cbfbrss/− 17.5 dpc FL cells13 for the presence of donor-derived NK cells. As expected, there were no or very few donor-derived T cells in the BM or spleen of mice transplanted with Cbfbrss/− FL cells (Figure 6E,F). Donor-derived mature NK cells were present in the peripheral blood (not shown), BM (Figure 6E), spleen (Figure 6F), and thymus (not shown) of transplant recipients of Cbfb+/+ and Cbfbrss/rss FL cells. However, Cbfbrss/− donor-derived mature and immature NK cells (NK and iNK) were completely absent in recipients reconstituted with Cbfbrss/− FL cells (Figure 6E,F). We also observed a decreased contribution of Cbfbrss/− FL cells to CD19+ B cells in the BM, but this decrease was not observed in the spleen, suggesting peripheral compensation for a partial defect in BM B-cell development (Figure 6E,F). The precipitous decline in NK-cell development that occurred when the CBFβ concentration dropped from 30% (Cbfbrss/rss) to 15% (Cbfbrss/−) of normal levels mirrored the abrupt and profound T-cell defects we observed at this same transition,13 indicating that both early T- and NK-cell development requires a similar threshold level of CBFβ. We also examined the transplant recipients for committed NKP (Figure 6G). The percentage of donor-derived NK cells and their precursors in both BM and spleen of the transplant recipients was lower than in the C57BL/6 (B6) control, perhaps because of the more advanced age of the transplant recipients. This precluded evaluation of donor-derived NKP in the BM; however, we were able to detect NKP in the spleen. Mice transplanted with Cbfb+/+, Cbfbrss/rss, and Cbfbrss/− FL cells all contained a detectable population of NKP in their spleen (Figure 6G). Therefore, the NK- cell deficiency in transplanted adult mice occurred at the NKP to iNK transition, whereas in the fetal liver and thymus it preceded the emergence of NKP.
We (and others) examined the expression of core binding factor genes in iNK and NK cells purified from wild-type mice to ascertain which members of the Runx family might be required in NK cells.48 Runx1, Runx3, and Cbfb mRNA were all found in Lin−Sca-1+c-kit+ BM progenitors, iNK, and NK cells, and both Runx3 and Cbfb mRNA levels increased during NK-cell differentiation (Figure 6H). Runx2 mRNA levels were very low in Lin−Sca-1+c-kit+, iNK, and NK cells (not shown). Based on expression levels, we predict that Runx1 and/or Runx3 are the Runx subunits most likely to be involved in NK-cell development.
Discussion
The CBFs and Notch cooperate in early T-cell development
We endeavored to define the molecular mechanisms underlying the requirement for CBFs in early T-cell development by examining the expression of T-cell specific genes, and signaling pathways important for T-cell development in mice with reduced CBFβ levels. A small percentage of Cbfbrss/− cells reached the DP stage of T-cell development when cultured on OP9-DL1 stromal cells, although the absolute number of cells that progressed past the DN1 stage was extremely low. The majority of Cbfbrss/− cells expressed very little, if any, Cd3e, Gata3, or Tcf7 mRNA, and no intracellular TCRγ or TCRβ chains were present. Thus, Cbfbrss/− cells lacked some of the earliest and most critical mediators of T-lineage development, indicating that T-lineage specification had essentially failed to occur. Although Cbfbrss/− cells were competent to receive Notch signals and by virtue of that were restrained in their ability to undergo B-cell development, they could not integrate the Notch signal to fully activate the T-lineage program. Thus, CBFs are needed to establish a T-lineage inducible state that is competent to respond to Notch signaling.
The mechanisms by which the CBFs and Notch signaling converge to specify T cells are unknown. One possibility is that they integrate at the level of common target genes in ETPs and at subsequent stages of DN thymocyte development. A few bona fide direct Notch targets are known in mammals, the best examples of which are the Hairy/Enhancer of SPLIT (HES) genes.50 Although no HES genes have been implicated as direct CBF targets, the mammalian HES-1 protein has been shown to physically interact with both Runx1 and Runx2, and HES-1 can potentiate Runx2 transactivation in reporter assays.51 Thus, one mechanism by which Notch and CBFs may integrate their function is through the direct interaction of Runx and HES proteins. Genes, such as Il2ra, which encode the CD25 molecule, could be directly and coordinately regulated by Notch and the CBFs.39,43,52
Our observations during T-cell development contrast with the initial stages of hematopoiesis in vertebrates, where Runx1 expression is dependent on Notch1 signaling (Figure 7), and overexpression of the mammalian or zebrafish Runx1 protein could rescue hematopoiesis from the AGM region in Notch mutant animals.19,20,23,24,53 In contrast, neither Runx1 nor CBFβ expression was decreased, nor could overexpression of Runx1 or CBFβ restore T-cell development when Notch signaling was inhibited.
Genetic interactions between the CBFs and Notch signaling in the specification of HSCs, T cells, and NK cells in the fetus. Notch receptors and ligands are expressed on the aortic endothelial cells in the AGM region that give rise to HSCs.20 Notch signaling is required for Runx1 expression in endothelial cells and for the formation of HSCs19,20,23,24 ; thus, Notch is genetically upstream of Runx1 in HSC formation. — represent molecular interactions; ---- represent cell migrations. PB indicates peripheral blood. HSCs and progenitors are released into the circulation from their sites of formation and colonize the fetal liver. We speculate that Runx1 and/or Runx3 plus CBFβ are required to generate NKP from either a bipotent T/NK or another progenitor in the fetus, and perhaps in T/NK progenitors themselves. CBFs are also required for the NKP to iNK transition, based on data from the adult. Circulating progenitors expressing all 3 CBF complexes colonize the thymus where they encounter high levels of Notch ligands. CBFs confer on these progenitors the ability to respond to Notch signaling, which results in T-cell specification and progression to the ETP/DN2 stage accompanied by the expression of a suite of T cell–specific genes. Not shown are NKT cells, which differentiate from DP T cells in a Runx1-dependent manner.
Genetic interactions between the CBFs and Notch signaling in the specification of HSCs, T cells, and NK cells in the fetus. Notch receptors and ligands are expressed on the aortic endothelial cells in the AGM region that give rise to HSCs.20 Notch signaling is required for Runx1 expression in endothelial cells and for the formation of HSCs19,20,23,24 ; thus, Notch is genetically upstream of Runx1 in HSC formation. — represent molecular interactions; ---- represent cell migrations. PB indicates peripheral blood. HSCs and progenitors are released into the circulation from their sites of formation and colonize the fetal liver. We speculate that Runx1 and/or Runx3 plus CBFβ are required to generate NKP from either a bipotent T/NK or another progenitor in the fetus, and perhaps in T/NK progenitors themselves. CBFs are also required for the NKP to iNK transition, based on data from the adult. Circulating progenitors expressing all 3 CBF complexes colonize the thymus where they encounter high levels of Notch ligands. CBFs confer on these progenitors the ability to respond to Notch signaling, which results in T-cell specification and progression to the ETP/DN2 stage accompanied by the expression of a suite of T cell–specific genes. Not shown are NKT cells, which differentiate from DP T cells in a Runx1-dependent manner.
Defects in both early T-cell and NK-cell development implicate pathways essential for both lineages
Although we could restore T-cell development when we overexpressed CBFβ in Cbfbrss/− Lin− FL cells, we were unable to efficiently restore proliferation or T-cell differentiation from Cbfbrss/− thymocytes. One possible explanation for this failure is that the defect may originate in a prethymic T-cell progenitor, and as a result very few of them emigrate to the thymus or they fail to proliferate or survive once they arrive. The majority of T-cell progenitors in the fetal liver and thymus have both T- and NK-cell potential,54-58 and a defect within this population could also account for the lack of NK cells we observed in OP9 cultures of fetal liver cells and in the fetal thymus. Both the T- and NK-cell defects became pronounced when CBFβ levels were reduced from 30% to 15% of normal levels, consistent with the hypothesis that they be ultimately traced, in part, to perturbations in the expression of genes required in both lineages, or to a common progenitor. Preliminary data indeed suggest that 12.5 dpc Cbfbrss/− fetal livers contained 6- to 7-fold fewer phenotypic (c-kit+CD127+PIR-A+) trilineage T-, NK-, and dendritic cell (T/NK/DC) progenitors55 (not shown). Furthermore, fetal thymi contained only very low numbers of NK1.1+ cells, which include all the bipotent T/NK progenitors and mature NK cells.49 The small number of Cbfbrss/− T/NK progenitors that do colonize the thymus may, in addition, be functionally defective, perhaps because they have undergone irreversible epigenetic changes or were unresponsive to the cytokines (Flt3L and IL-7) that we used in the transduction cocktail.
The NK-cell defect in the transplanted adult mouse appeared to occur somewhat later, at the transition between NKP and iNK cells. NKP differentiate from the HSC through intermediate precursors that have not been unambiguously identified, but may include the early and common lymphoid progenitors (ELP and CLP), both of which are distinct from the fetal liver T/NK progenitor.3 Because B-cell development in Cbfbrss/− mice is only mildly affected, the ELP and CLP are presumably present, and are apparently able to give rise to NKP.
Other transcription factors are necessary for both T- and NK-cell development, including members of the Ikaros, ETS, Id, and TCF7/LEF-1 families.59-68 However, mutations in none of these genes selectively and profoundly affected both T- and NK- but not B-cell development in mice. The profound T- and NK-cell defects we observed in Cbfbrss/− mice most closely resemble those associated with the T−B+NK− forms of severe combined immune deficiency syndrome in humans, which are caused by mutations that affect both the IL-7 and IL-15 signaling pathways.69 Molecules in both pathways are regulated by the CBFs. The CBFs regulate CD122 (Il2rb) expression in NK1.1+ cells, and Runx1 is required for full activation of the Il7ra gene in DN2, DN3, and DP (TCRβhi CD69+) thymocytes and in Foxp3−CD4+ cells.11,48 We found essentially no CD122+ cells in ex vivo cultures of Cbfbrss/− Lin− cells or in the thymus. IL-7Rα levels were unchanged in Cbfbrss/− DN2 cells, but IL-7 signaling was nonetheless compromised, as levels of phosphorylated Stat5 were lower. However, defective JAK/STAT signaling is unlikely to be the only defect in Cbfbrss/− T and NK cells. Neither wild-type nor constitutively active Stat5a could rescue the T-cell abnormalities, indicating that defective JAK/STAT signaling alone cannot explain the T-cell defect. Egawa et al11 similarly concluded that defective IL-7 signaling was not the only factor limiting the number of Runx1-deficient CD4+ T cells. Furthermore, the defects in Cbfbrss/− cells are manifested before the IL-7–dependent stage of T-cell growth, as their inability to express Cd3e, Gata3, or Tcf7 is consistent with a profound regulatory defect in T-cell specification. In NK cells CBFs appear to regulate multiple genes, including the Ly49 family in mouse and the killer cell Ig-like receptors in human.70,71 In the future, it will be interesting to explore whether abnormalities in Runx/CBFβ function might account for a subset of SCID patients with as of yet no identified mutations.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Juan Carlos Zún̋iga-Pflücker for providing the OP9-DL1 cells; Michael Chen for the Runx1 virus; Richard Moriggl for the Stat5a cDNA; and Caroline Speck, Michael Chen, and Brandon Zeigler for technical assistance.
This work was supported by National Institutes of Health (NIH) grant RO1CA075611 (N.A.S.) and Damon Runyon Cancer Research Foundation grant DRG-102-05 (I.M.). Core services were supported in part by the Norris Cotton Cancer Center (NIH grant CA23108).
National Institutes of Health
Authorship
Contribution: Y.G., S.C., and I.M. performed experiments; N.A.S., I.M, and Y.G. created the figures; all authors analyzed the results, designed the research, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nancy A. Speck, Department of Biochemistry, HB7200, Dartmouth Medical School, Hanover, NH 03755; e-mail: nancy.speck@dartmouth.edu.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal