Abstract
Despite advances in the curative treatment of acute myeloid leukemia (AML), recurrence will occur in the majority of cases. At diagnosis, acquisition of segmental uniparental disomy (UPD) by mitotic recombination has been reported in 15% to 20% of AML cases, associated with homozygous mutations in the region of loss of heterozygosity. This study aimed to discover if clonal evolution from heterozygous to homozygous mutations by mitotic recombination provides a mechanism for relapse. DNA from 27 paired diagnostic and relapsed AML samples were analyzed using genotyping arrays. Newly acquired segmental UPDs were observed at relapse in 11 AML samples (40%). Six were segmental UPDs of chromosome 13q, which were shown to lead to a change from heterozygosity to homozygosity for internal tandem duplication mutation of FLT3 (FLT3 ITD). Three further AML samples had evidence of acquired segmental UPD of 13q in a subclone of the relapsed leukemia. One patient acquired segmental UPD of 19q that led to homozygosity for a CEBPA mutation 207C>T. Finally, a single patient with AML acquired segmental UPD of chromosome 4q, for which the candidate gene is unknown. We conclude that acquisition of segmental UPD and the resulting homozygous mutation is a common event associated with relapse of AML.
Introduction
Dramatic advances have been made during the past 40 years in the treatment of acute myeloid leukemia (AML) such that 40% to 50% of patients presenting younger than 60 years can be cured with initial therapy.1-3 Complete remission, determined clinically, morphologically, immunologically, and sometimes at the molecular level, is reliably reported to be achieved in up to 80% of patients in this age group.3 Despite this success, reemergence of leukemia is the rule rather than the exception; at least 50% of younger patients and 80% of those older than 60 years will see their leukemia return within 5 years.4,5 Data from our institution similarly show that two-thirds who achieve first complete remission, subsequently relapse. Although a second remission may be achieved, it is rarely sustained, even when allogeneic stem cell transplantation is successful, such that only 20% of adult patients survive at 4 years.6 Hence, there is a compelling urgency to determine mechanisms of resistance to identify appropriate alternative treatment strategies.
Cytogenetic abnormalities are the most important biologic prognostic factors for risk of relapse, with the occurrence of rearrangements involving inv(16), t(8;21), and t(15;17) associated with a better prognosis, and patients with AML with deletions involving chromosomes 7q, 5q, and abnormalities of 3q are among those having the poorest prognosis.4 Three case series have reviewed cytogenetic changes at diagnosis and relapse.7-9 No consistent cytogenetic abnormalities were acquired at relapse; therefore, it is not clear what role acquired karyotypic abnormalities play in the pathogenesis of relapse.
Resistance to chemotherapy is intrinsic to relapsed leukemic cells. The presence of a chemoresistant population of leukemic cells is shown by the detection of minimal residual disease after chemotherapy, which increases the risk of relapse for some subtypes of AML.10-12 However, the underlying pathologic mechanisms leading to relapse are poorly understood.
Single gene mutations are an additional biologic risk factor for relapse, with FLT3 internal tandem duplication (ITD) being an important predictor of poor prognosis.13 When FLT3 ITD is detected at diagnosis, the same mutation is noted at relapse in 75% of cases.14 Some of these have been found to have an increased dosage of mutant FLT3 at disease recurrence, suggesting a clonal outgrowth of a cell with a homozygous or hemizygous mutation.14 Homozygous single gene mutations are known to occur in AML, and these are associated with mitotic recombination or possibly a nondisjunction event leading to acquired uniparental disomy (UPD), also known as acquired isodisomy.15 Studies of acquired UPD in diagnostic AML samples give a frequency of 15% to 20%,16,17 with the largest study to date having analyzed 454 samples.18 A possible mechanism for relapse in AML is one in which a heterozygous mutation is followed by a recombination event leading to homozygosity and clonal evolution. To evaluate this hypothesis, 27 paired diagnosis and relapse AML samples were studied with the use of array-based genome-wide mapping to identify regions of segmental UPD and gene-specific mutation analysis.
Methods
Sample selection
Sufficient paired diagnostic and relapse blood and bone marrow samples for analysis were available from 27 patients with AML. Ethical approval was obtained from the local research ethics committee, and written informed consent was obtained from all patients in accordance with the Declaration of Helsinki. Ethics approval was obtained from the East London and City Research Ethics Committee (ref P1/03/310). Samples were stored in the tissue bank of Barts and the London School of Medicine and Dentistry (Human Tissue Authority deemed license 12199). Because samples were chosen based on their availability, there was a bias toward samples with relatively high white blood cell counts. Patients presented between April 1983 and September 2004. Patient demographics are shown in Table 1. Median age was 51 years (range, 35-68 years); 18 were men and 9 were women. The median number of days to relapse was 179 days (range, 88-758 days). The median overall survival of this group of patients was 1.02 years. Standard cytogenetic analysis was performed with bone marrow or peripheral blood. Fluorescent in situ hybridization was performed according to the manufacturer's protocol (Abbott Molecular, Abbott Park, IL). Patient karyotypes were described according to the International System for Human Cytogenetic Nomenclature.19 By criteria from the Medical Research Council,4 at diagnosis 2 patients had favorable risk cyto-genetics, 20 had intermediate risk cytogenetics, 1 had poor risk cytogenetics, and 2 were unknown because there was no sample available for cytogenetic analysis.
Demographic data
| Patient . | Sex . | Age at diagnosis, y . | FAB type . | WBC count at presentation, ×109/L . |
|---|---|---|---|---|
| 1 | F | 25 | M5 | 89.0 |
| 2 | M | 47 | M1 | 182.0 |
| 3 | F | 60 | M2 | 10.4 |
| 4 | M | 27 | M1 | 65.9 |
| 5 | M | 35 | M4 | 79.7 |
| 6 | F | 67 | M5 | 127.2 |
| 7 | M | 61 | M4 | 11.3 |
| 8 | F | 47 | M2 | 53.1 |
| 9 | F | 47 | M4 | 88.1 |
| 10 | F | 41 | M1 | 9.0 |
| 11 | M | 47 | M1 | 90.0 |
| 12 | M | 68 | M2 | 19.8 |
| 13 | M | 59 | M1 | 88.4 |
| 14 | M | 64 | M4 | 31.2 |
| 15 | M | 48 | M5 | 33.2 |
| 16 | F | 36 | M4 | 30.7 |
| 17 | F | 65 | M1 | 4.4 |
| 18 | M | 52 | M4 | 5.5 |
| 19 | M | 52 | M2 | 11.3 |
| 20 | M | 46 | M4 | 127.2 |
| 21 | M | 43 | M4 | 11.4 |
| 22 | M | 61 | M2 | 29.1 |
| 23 | M | 50 | M4 | 5.7 |
| 24 | F | 65 | M1 | 6.2 |
| 25 | M | 25 | M5 | 96.8 |
| 26 | M | 21 | M5 | 51.8 |
| 27 | M | 43 | M5 | 43.6 |
| Patient . | Sex . | Age at diagnosis, y . | FAB type . | WBC count at presentation, ×109/L . |
|---|---|---|---|---|
| 1 | F | 25 | M5 | 89.0 |
| 2 | M | 47 | M1 | 182.0 |
| 3 | F | 60 | M2 | 10.4 |
| 4 | M | 27 | M1 | 65.9 |
| 5 | M | 35 | M4 | 79.7 |
| 6 | F | 67 | M5 | 127.2 |
| 7 | M | 61 | M4 | 11.3 |
| 8 | F | 47 | M2 | 53.1 |
| 9 | F | 47 | M4 | 88.1 |
| 10 | F | 41 | M1 | 9.0 |
| 11 | M | 47 | M1 | 90.0 |
| 12 | M | 68 | M2 | 19.8 |
| 13 | M | 59 | M1 | 88.4 |
| 14 | M | 64 | M4 | 31.2 |
| 15 | M | 48 | M5 | 33.2 |
| 16 | F | 36 | M4 | 30.7 |
| 17 | F | 65 | M1 | 4.4 |
| 18 | M | 52 | M4 | 5.5 |
| 19 | M | 52 | M2 | 11.3 |
| 20 | M | 46 | M4 | 127.2 |
| 21 | M | 43 | M4 | 11.4 |
| 22 | M | 61 | M2 | 29.1 |
| 23 | M | 50 | M4 | 5.7 |
| 24 | F | 65 | M1 | 6.2 |
| 25 | M | 25 | M5 | 96.8 |
| 26 | M | 21 | M5 | 51.8 |
| 27 | M | 43 | M5 | 43.6 |
FAB indicates French-American-British; and WBC, white blood cell.
10K GeneChip assay
DNA was extracted with standard phenol-chloroform techniques, or from the organic phase of TRIzol (Invitrogen, Carlsbad, CA). The GeneChip mapping assay protocol (Affymetrix, Santa Clara, CA) was used to produce the 10K single-nucleotide polymorphism (SNP) array results and is described elsewhere.20,21 The protocol was adapted such that the purification of polymerase chain reaction (PCR) product was performed with the use of the Ultrafree-MC 30 000 NMWL filtration column (Millipore, Billerica, MA). Signal intensity data were analyzed by the GeneChip DNA analysis software, which uses a model algorithm, modified partitioning around medoids, to generate SNP calls.22,23 In brief, the relative allele signal (RAS) from each allele on the forward and reverse strands (RAS1 and RAS2, respectively) is used to determine heterozygosity or homozygosity (the call) for individual SNPs. The call is made based on whether the plotted RAS values lie within the boundary of zones previously determined from a training set of human samples.22 RAS values close to (0,0) and (1,1) are homozygous. No-calls are made when the RAS values are outside the call zones.
The data discussed in this publication have been deposited in the National Cancer Biotechnology Information (NCBI) Gene Expression Omnibus (GEO)24 and are accessible through GEO Series accession number GSE7210. Analysis of the call and copy number data was performed as previously described.16 Regions of acquired LOH were determined by comparing diagnostic and relapse samples. SNP copy number analysis identified these regions as being deleted, gained, or as being disomic.25 SNP and gene annotations used NCBI genome build 35.
Mutation analysis
Mutation screening was performed for the entire coding region of CEBPA26,27 and specific exons of FLT3 (exons 14-15 and 20),28,29 using standard primer and amplification conditions. PCR products were sequenced directly by use of an ABI 377 DNA sequencer (PE Applied Biosystems, Foster City, CA). Sequencing data were analyzed using 4Peaks software (Mekentosj, http://mekentosj.com/4peaks). Fragment analysis of FLT3 exon 14 to 15 PCR products was performed using the QIAGEN Multiplex PCR kit (Valencia, CA), and measured by capillary electrophoresis using an ABI Prism 3700 Genetic Analyzer (Applied Biosystems, Foster City, CA). Fluorescence signals were analyzed with Genotyper 3.7 software (Applied Biosystems).
Results
Matched paired samples were analyzed by SNP array and mutation analysis at presentation and at relapse for evidence of clonal evolution. Eighteen patients acquired copy number changes at relapse, and these correlated with karyotype data where available (Table 2). There were no consistently acquired karyotypic or copy number abnormalities at relapse, and only one patient acquired karyotypic changes at relapse that are reported to confer a poor prognosis at diagnosis (patient 17, deletions of chromosome 7q and 20q). Three patients (patients 7, 8, and 12) had acquired UPD at diagnosis. Patient 8 had UPD 13q with a homozygous FLT3 ITD mutation at diagnosis, and this was retained at relapse.
Chromosomal, copy number, and loss of heterozygosity (including uniparental disomy) changes between diagnosis and relapse
| Patient . | Diagnosis . | Relapse . | ||
|---|---|---|---|---|
| Cytogenetics . | Mapping array . | Cytogenetics . | Mapping array . | |
| 1 | 46,XX | No abnormality | No sample | LOH whole 13 |
| 2 | No sample | Gained whole 6 | No sample | Gained whole 6 |
| 3 | 46,XX | No abnormality | 47,XX,+11/46,XX | Gained whole 11 |
| 4 | 46,XY,t(6;9)(p23;q34)[10] | No abnormality | 46,XY,t(6;9)(p23;q34)[4]/46,XY[6] | Segmental UPD13q (262590909-term) |
| 5 | 46,XY,t(2;14)(q21;q32)/46,XY | No abnormality | 46,XY,t(2;14)(q21;q32)[8]/46,XY[2] | No abnormality |
| 6 | 46,XX,inv(16)(p13.1q22)[10] | No abnormality | nuc ish(CBFBx2)(5′CBFB sep 3′CBFBx1)[82/100] | Gained whole 22 |
| 7 | 46,XY | Segmental UPD 2p (45892803-66988762) | 46,XY[47] | Segmental UPD 2p (45892803-66988762) |
| Segmental UPD 5q (109245567-132912666) | Segmental UPD 5q (109245567-132912666) | |||
| LOH whole 13 | ||||
| 8 | 46,XX,del(16)(q12.1)/46,XX | Segmental UPD 13q (19029353-term) Del 16q (29142199-85631136) | 46,XX,del(11)(q13q22),del(16)(q12.1) [2]/46,idem,t(4;6)(q35;q15)[8] | Del 4q (67619803-93329691) |
| Del 11q (85287757-115209411) | ||||
| Segmental UPD 13q (19029353-term) | ||||
| Del 16q (29142199-85631136) | ||||
| 9 | 46,XX[25] | No abnormality | 46,XX[25] | Segmental UPD whole 13 |
| 10 | 46,XX | No abnormality | 46,XX | Segmental UPD 13q (19029353-term) |
| 11 | 46,XY | No abnormality | 46,XY | No abnormality |
| 12 | 45,X,-Y,t(8;21)(q22;q22)[10] | Segmental UPD 11 (44160735-62518998) | 45,X,-Y,t(8;21)(q22;q22) | Del 2q (198963363-240642812) |
| Segmental UPD 11 (44160735-62518998) | ||||
| 13 | 46,XY,del(7)(q11q21)/46,XY | No abnormality | No sample | No abnormality |
| 14 | 46,XY | No abnormality | 46,XY | LOH whole 13 |
| 15 | 46,XY[20] | No abnormality | 47,XX,+8,add(9)(q34)[8]/46,XY[2] | No abnormality |
| 16 | 46,XX [25] | No abnormality | No sample | Segmental UPD 19q (20533429-term) |
| Gained whole 21 | ||||
| 17 | 47,XX,+8[10] | No abnormality | 47,XX,del(6)(q13q21),der(7)t(7;17)(q11.1;q11.2),+8,add(17)(q11.2), del(20)(q13.1q13.3 or q11.2q13.1)[5]/47,idem,del(2)(q13q2?)[5] | Del 6q (64252743-101001308) |
| Del 7q (69512516-term) | ||||
| Segmental UPD whole 13 | ||||
| Del 20q (40318090-110881790) | ||||
| 18 | 46,XY[20] | No abnormality | 46,XY[20] | No abnormality |
| 19 | 46,XY,t(6;9)(p23;q34)[6]/46,XY[55] | No abnormality | 46,XY,t(6;9)(p23;q34)[10] | Segmental UPD 13q (18482385-term) |
| 20 | 46,XY[20] | Gained 1p (term-59198211) | 46,XY[20] | No abnormality |
| 21 | 46,XY | No abnormality | 46,XY | Segmental UPD 4q (87522823-term) |
| 22 | 46,XY,t(8;9)(q24;q32)[4] 45,sl,-Y,der(9)add(9)(p13)t(8;9)[11]45,sdl1,add(14)(q32)[3] | Del 9p (19871864-25253918) | No sample | Del 9p (19871864-25253918) |
| 23 | No sample | No abnormality | No sample | No abnormality |
| 24 | 46,XX[20] | No abnormality | 46,XX,t(1;5)(q4;q11.2),?t(11;17) (q13;q11.2),del(12)(p11.2p13)[3]46,XX[15] | No abnormality |
| 25 | 46,XY | No abnormality | 46,XY | Segmental UPD 13q (20860773-term) |
| 26 | 49,XY,+5,+8,+19[9]46,XY[1] | Gained whole 5 | 49,XY,+5,+8,+19[1]50,idem+mar[2]46, | Del 1p (5708940-38792696) |
| Gained whole 8 | Y,der(X),t(X;13)(q26;q12), add (7)(p11.2),-13,+mar[4] | Gained 3p (125054444-term) | ||
| Del 7p (term-61521113) | ||||
| 27 | 47,XY,+8/46,XY | Gained whole 8 | No sample | Gained whole 8 |
| Patient . | Diagnosis . | Relapse . | ||
|---|---|---|---|---|
| Cytogenetics . | Mapping array . | Cytogenetics . | Mapping array . | |
| 1 | 46,XX | No abnormality | No sample | LOH whole 13 |
| 2 | No sample | Gained whole 6 | No sample | Gained whole 6 |
| 3 | 46,XX | No abnormality | 47,XX,+11/46,XX | Gained whole 11 |
| 4 | 46,XY,t(6;9)(p23;q34)[10] | No abnormality | 46,XY,t(6;9)(p23;q34)[4]/46,XY[6] | Segmental UPD13q (262590909-term) |
| 5 | 46,XY,t(2;14)(q21;q32)/46,XY | No abnormality | 46,XY,t(2;14)(q21;q32)[8]/46,XY[2] | No abnormality |
| 6 | 46,XX,inv(16)(p13.1q22)[10] | No abnormality | nuc ish(CBFBx2)(5′CBFB sep 3′CBFBx1)[82/100] | Gained whole 22 |
| 7 | 46,XY | Segmental UPD 2p (45892803-66988762) | 46,XY[47] | Segmental UPD 2p (45892803-66988762) |
| Segmental UPD 5q (109245567-132912666) | Segmental UPD 5q (109245567-132912666) | |||
| LOH whole 13 | ||||
| 8 | 46,XX,del(16)(q12.1)/46,XX | Segmental UPD 13q (19029353-term) Del 16q (29142199-85631136) | 46,XX,del(11)(q13q22),del(16)(q12.1) [2]/46,idem,t(4;6)(q35;q15)[8] | Del 4q (67619803-93329691) |
| Del 11q (85287757-115209411) | ||||
| Segmental UPD 13q (19029353-term) | ||||
| Del 16q (29142199-85631136) | ||||
| 9 | 46,XX[25] | No abnormality | 46,XX[25] | Segmental UPD whole 13 |
| 10 | 46,XX | No abnormality | 46,XX | Segmental UPD 13q (19029353-term) |
| 11 | 46,XY | No abnormality | 46,XY | No abnormality |
| 12 | 45,X,-Y,t(8;21)(q22;q22)[10] | Segmental UPD 11 (44160735-62518998) | 45,X,-Y,t(8;21)(q22;q22) | Del 2q (198963363-240642812) |
| Segmental UPD 11 (44160735-62518998) | ||||
| 13 | 46,XY,del(7)(q11q21)/46,XY | No abnormality | No sample | No abnormality |
| 14 | 46,XY | No abnormality | 46,XY | LOH whole 13 |
| 15 | 46,XY[20] | No abnormality | 47,XX,+8,add(9)(q34)[8]/46,XY[2] | No abnormality |
| 16 | 46,XX [25] | No abnormality | No sample | Segmental UPD 19q (20533429-term) |
| Gained whole 21 | ||||
| 17 | 47,XX,+8[10] | No abnormality | 47,XX,del(6)(q13q21),der(7)t(7;17)(q11.1;q11.2),+8,add(17)(q11.2), del(20)(q13.1q13.3 or q11.2q13.1)[5]/47,idem,del(2)(q13q2?)[5] | Del 6q (64252743-101001308) |
| Del 7q (69512516-term) | ||||
| Segmental UPD whole 13 | ||||
| Del 20q (40318090-110881790) | ||||
| 18 | 46,XY[20] | No abnormality | 46,XY[20] | No abnormality |
| 19 | 46,XY,t(6;9)(p23;q34)[6]/46,XY[55] | No abnormality | 46,XY,t(6;9)(p23;q34)[10] | Segmental UPD 13q (18482385-term) |
| 20 | 46,XY[20] | Gained 1p (term-59198211) | 46,XY[20] | No abnormality |
| 21 | 46,XY | No abnormality | 46,XY | Segmental UPD 4q (87522823-term) |
| 22 | 46,XY,t(8;9)(q24;q32)[4] 45,sl,-Y,der(9)add(9)(p13)t(8;9)[11]45,sdl1,add(14)(q32)[3] | Del 9p (19871864-25253918) | No sample | Del 9p (19871864-25253918) |
| 23 | No sample | No abnormality | No sample | No abnormality |
| 24 | 46,XX[20] | No abnormality | 46,XX,t(1;5)(q4;q11.2),?t(11;17) (q13;q11.2),del(12)(p11.2p13)[3]46,XX[15] | No abnormality |
| 25 | 46,XY | No abnormality | 46,XY | Segmental UPD 13q (20860773-term) |
| 26 | 49,XY,+5,+8,+19[9]46,XY[1] | Gained whole 5 | 49,XY,+5,+8,+19[1]50,idem+mar[2]46, | Del 1p (5708940-38792696) |
| Gained whole 8 | Y,der(X),t(X;13)(q26;q12), add (7)(p11.2),-13,+mar[4] | Gained 3p (125054444-term) | ||
| Del 7p (term-61521113) | ||||
| 27 | 47,XY,+8/46,XY | Gained whole 8 | No sample | Gained whole 8 |
Each change is marked in bold. Regions are delineated by chromosome band or nucleotide position.
LOH indicates loss of heterozygosity; and UPD, uniparental disomy.
Clonal evolution of acquired UPD at relapse
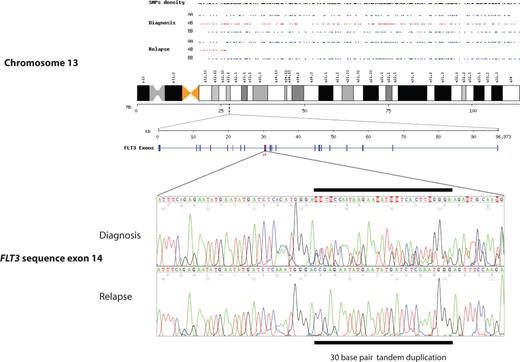
Newly acquired segmental UPD at disease recurrence was detected in 11 (41%) of 27 patients. The most common abnormality detected was segmental UPD of chromosome 13, which was observed in 6 (22%) of 27 patients (Table 2). The smallest size of UPD was approximately 88 Mb (megabase), including the telomere, and the largest involved the whole chromosome (114 Mb). As expected for UPD, copy number analysis and karyotype analysis showed 2 copies of chromosome 13 across the regions of LOH, excluding the possibility of a hemizygous deletion. All regions of UPD on chromosome 13 included the locus for FLT3. Sequencing of FLT3 showed an ITD mutation, which was heterozygous at diagnosis and homozygous at relapse in all 6 cases (Table 3; Figure 1). The mutations were identical at diagnosis and relapse in each case, showing that the second clone had evolved from the initial leukemia.
Associated mutation and acquired segmental uniparental disomy (UPD) or loss of call
| Patient . | Segmental UPD acquired at relapse . | Associated mutation status . | |
|---|---|---|---|
| Diagnosis . | Relapse . | ||
| 1 | Loss of call whole 13 | FLT3 ITD heterozygous | FLT3 ITD heterozygous |
| 4 | 13q | FLT3 ITD heterozygous | FLT3 ITD homozygous |
| 7 | Loss of call whole 13 | D835Y heterozygous | D835Y homozygous |
| 9 | 13q | FLT3 ITD heterozygous | FLT3 ITD homozygous |
| 10 | 13q | FLT3 ITD heterozygous | FLT3 ITD homozygous |
| 14 | Loss of call whole 13 | D835Y heterozygous | FLT3 wild type |
| 16 | 19q | CEBPA 207C>T heterozygous | CEBPA 207C>T homozygous |
| 17 | Whole 13 | FLT3 ITD heterozygous | FLT3 ITD homozygous |
| 19 | 13q | FLT3 ITD heterozygous | FLT3 ITD homozygous |
| 21 | 4q | Unknown | Unknown |
| 25 | 13q | FLT3 ITD heterozygous | FLT3 ITD homozygous |
| Patient . | Segmental UPD acquired at relapse . | Associated mutation status . | |
|---|---|---|---|
| Diagnosis . | Relapse . | ||
| 1 | Loss of call whole 13 | FLT3 ITD heterozygous | FLT3 ITD heterozygous |
| 4 | 13q | FLT3 ITD heterozygous | FLT3 ITD homozygous |
| 7 | Loss of call whole 13 | D835Y heterozygous | D835Y homozygous |
| 9 | 13q | FLT3 ITD heterozygous | FLT3 ITD homozygous |
| 10 | 13q | FLT3 ITD heterozygous | FLT3 ITD homozygous |
| 14 | Loss of call whole 13 | D835Y heterozygous | FLT3 wild type |
| 16 | 19q | CEBPA 207C>T heterozygous | CEBPA 207C>T homozygous |
| 17 | Whole 13 | FLT3 ITD heterozygous | FLT3 ITD homozygous |
| 19 | 13q | FLT3 ITD heterozygous | FLT3 ITD homozygous |
| 21 | 4q | Unknown | Unknown |
| 25 | 13q | FLT3 ITD heterozygous | FLT3 ITD homozygous |
ITD indicates internal tandem duplication.
Chromosome 13q segmental UPD acquired at relapse in patient 4, showing a change from heterozygosity (alleles AB in red) to homozygosity (alleles AA or BB in blue). An identical mutated sequence of FLT3 ITD is shown in the region of UPD at q12.2 at diagnosis and relapse, with loss of heterozygosity.
Chromosome 13q segmental UPD acquired at relapse in patient 4, showing a change from heterozygosity (alleles AB in red) to homozygosity (alleles AA or BB in blue). An identical mutated sequence of FLT3 ITD is shown in the region of UPD at q12.2 at diagnosis and relapse, with loss of heterozygosity.
Patient 16 acquired segmental UPD of 19q at relapse, and this was associated with a mutation of CEBPA located within the region of homozygosity. This mutation was also heterozygous at diagnosis and homozygous at relapse. The mutation, 207C>T (GenBank accession Y11525), created a stop codon that results in an N-terminal truncated protein.27 A further segmental UPD was acquired at 4q in patient 21. In the absence of an obvious candidate gene and in view of the large region involved (104 Mb), this was not investigated further.
Subclones with acquired UPD
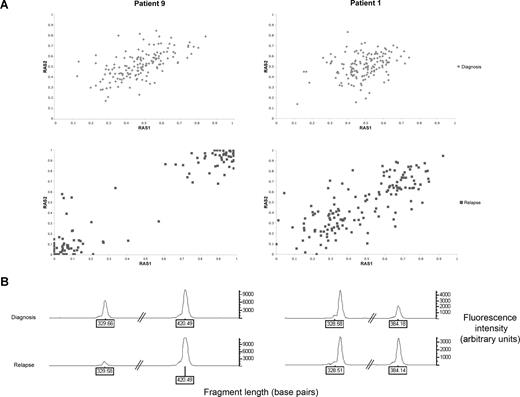
Three (patients 1, 7, and 14) of the 11 patients acquired UPD of chromosome 13 in subclones of the relapsed leukemic cell population. Figure 2A illustrates the shift in RAS values in patient 9, whereby the whole leukemic population acquires UPD of chromosome 13q, compared with patient 1, whereby only a subclone with UPD 13q is present. The allele call rates for all other chromosomes in each sample were greater than 90%, indicating that changes in RAS values were not due to a failure of hybridization and the changes were not due to normal germ line contamination because all samples contained greater than 70% blasts.30
Comparison between patients who have acquired UPD 13q at relapse in the whole leukemic clone (patient 9) or as a subclone (patient 1). (A) Change in relative allele signal (RAS) values between diagnosis and relapse, for SNPs heterozygous at diagnosis in the region of LOH on chromosome 13 for patients 1 and 9. The same SNPs that are heterozygous at diagnosis shift RAS values at relapse, consistent with LOH. (B) Electropherogram showing PCR fragment quantitation of FLT3 wild type, at 329 base pairs, and the larger ITD fragment in patients 1 and 9.
Comparison between patients who have acquired UPD 13q at relapse in the whole leukemic clone (patient 9) or as a subclone (patient 1). (A) Change in relative allele signal (RAS) values between diagnosis and relapse, for SNPs heterozygous at diagnosis in the region of LOH on chromosome 13 for patients 1 and 9. The same SNPs that are heterozygous at diagnosis shift RAS values at relapse, consistent with LOH. (B) Electropherogram showing PCR fragment quantitation of FLT3 wild type, at 329 base pairs, and the larger ITD fragment in patients 1 and 9.
PCR fragment analysis for FLT3 was performed on both patients 1 and 9, which was within the region of UPD (Figure 2B). Both patients had a reduction in the ratio of wild-type to mutant FLT3 at relapse, consistent with the presence of a homozygous FLT3 ITD mutation. Together with the array data, this is consistent with a leukemic cell subpopulation with UPD 13q and a homozygous FLT3 ITD mutation at disease recurrence in patient 1. Patient 7 had a FLT3 loop mutation (D835Y), which became homozygous at relapse. Patient 14 had a heterozygous D835Y mutation at diagnosis, but this was absent at relapse.
Discussion
This study shows that remarkably approximately 40% of patients with AML exhibit newly acquired UPD at relapse. Acquired segmental UPD frequently results in a homozygous mutation, most commonly involving chromosome 13 with FLT3 ITD. Interestingly, previous studies on diagnostic material have shown that loss of wild-type FLT3 in the presence of FLT3 ITD is a poor prognostic marker at presentation,31,32 and that a higher level of FLT3 ITD on PCR fragment analysis of genomic DNA increases the risk of relapse.33 Acquisition of UPD of chromosome 13 provides an explanation for the increase in gene dosage at diagnosis and at relapse.
Patient 7 had acquired homozygosity of the FLT3 D835Y loop mutation at relapse, and this finding lends further support to the hypothesis that increasing dosage of mutant leads to chemoresistance and relapse. The clinical relevance of FLT3 loop mutations is controversial; 2 studies suggest they confer a relatively good prognosis,34,35 although a smaller study showed an association with poor prognosis.36 Loss of a loop mutation without an ITD mutation was also associated with acquisition of UPD of 13q at relapse, suggesting there may be mutations of FLT3 outside of the juxtamembrane region or the activation loop as have been described,37 or, alternatively, another gene is mutated on 13q.
Tyrosine kinase inhibitors are currently undergoing trials in FLT3 ITD–positive AML.38,39 After acquisition of UPD of chromosome 13, these AML cells may be even more dependent on FLT3 signaling, which suggests inhibition of FLT3 may be an effective method of treating relapse in patients with AML with UPD 13q.
Biallelic mutations of CEBPA are well recognized in AML. They are occasionally homozygous,40 and these have been associated with acquired UPD of 19q.41 Although a biallelic mutation is not a prerequisite for AML, it is important in its development, as shown in familial cases of AML in which a germ line CEBPA mutation is followed after a long latency by a second CEBPA mutation and subsequently develops AML.42 Patients with AML with mutations of CEBPA have a relatively good prognosis.43 Although in this study, evolution of a homozygous CEBPA mutation was associated with relapsed AML, the numbers of patients with AML with homozygous CEBPA mutations are too few to ascertain whether they would have a poorer prognosis.
Mitotic recombination or nondisjunction leads to segmental or whole chromosomal UPD, respectively. It is unknown whether these events occur before, during, or after treatment. A rare chemoresistant subclone with UPD may be present in the initial population at diagnosis. In vitro studies on mouse fibroblasts and normal human lymphocytes have shown a physiologic background rate of mitotic recombination of approximately 1 in 100 000 cells,44 suggesting that rare subclones with acquired UPD could occur spontaneously in the developing leukemia. These rare subclones would not be detected, because a low proportion of homozygous cells cannot be observed with SNP array technology. In this study, 3 cases developed subclones with segmental UPD at detectable levels. The role of UPD may be underestimated, given our inability to detect low levels of homozygosity.
As expected, SNP arrays identified more abnormalities than did cytogenetic analysis alone. However, some cytogenetic abnormalities were not detected by SNP array; eg, patient 24 acquired a rare clone with a deletion of 12p at relapse, patient 15 acquired trisomy 8, and patient 13 had a deletion of 7q. We hypothesize that in these cases, the dominant leukemic clone had a normal karyotype, but it did not grow on short-term culture for karyotypic analysis.
Although the Affymetrix 10K arrays used in this study have been superseded by those at a much higher resolution, the study was designed to identify large regions of UPD as a result of mitotic recombination, for which the 10K arrays are quite adequate. However, higher-resolution arrays have the potential to identify much smaller recombinations of only a few thousand base pairs. In addition, with the small number of patients in this study, no significant correlations with clinical outcomes were possible. Recurrent AML defines itself as a poor prognostic group, and only 5 patients achieved a second remission, of which there is only 1 long-term survivor. The samples were selected retrospectively, based on the availability of both a diagnostic and a relapse leukemia sample. As a result, it was unavoidable that some subgroups were missed or underrepresented, eg, acute promyelocytic leukemias and complex karyotypes. Future studies should aim to use higher resolution arrays in a larger sample set to discover whether acquired UPD is of prognostic importance at relapse.
The frequency of acquired segmental UPD at diagnosis is 15% to 20%,16-18 and this study appears to show an increased frequency of UPD (41%) at relapse. In particular, the incidence of 13q UPD is only 6% at diagnosis.18 It is possible that mitotic recombination could occur more often after exposure to cytotoxic chemotherapy, perhaps because of an increased rate of DNA double-strand breaks.
This study provides the first data that mitotic recombination is an important mechanism responsible for recurrence of AML. Previously, 2 case reports of relapsed childhood AML have described a similar phenomenon, one involving FLT3 ITD,45 and another in which a homozygous WT1 mutation was acquired at relapse.46 In addition, several other cancers were shown to acquire UPD at diagnosis,47,48 suggesting mitotic recombination could be a general mechanism for cancer progression. Although UPD of 13q is the commonest abnormality described here, further studies into acquired segmental UPDs are likely to show novel targets in the treatment of AML and other cancers.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the Cancer Research UK and the Leukemia Research Fund for supporting this project. We also thank Sameena Iqbal and Finlay MacDougall for help in identifying samples and clinical data and John Amess and Michael Jenner for morphologic analysis.
Authorship
Contribution: M.R. designed and performed research, analyzed data and wrote the paper; L.-L.S., D.M.L., T.C., I.K., and G.M. performed research; C.C. and J.-B.C, analyzed data; J.D.C. designed research, and obtained samples; T.A.L. designed research, obtained samples, and wrote the paper; J.F. and B.D.Y. designed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Manoj Raghavan, Medical Oncology Unit, Centre for Medical Oncology, Barts and the London School of Medicine and Dentistry, Charterhouse Square, London EC1M 6BQ, United Kingdom; e-mail: manoj.raghavan@cancer.org.uk.