Abstract
Dissecting the molecular mechanisms used by developmental regulators is essential to understand tissue specification/differentiation. SCL/TAL-1 is a basic helix-loop-helix transcription factor absolutely critical for hematopoietic stem/progenitor cell specification and lineage maturation. Using in vitro and forced expression experimental systems, we previously suggested that SCL might have DNA-binding–independent functions. Here, to assess the requirements for SCL DNA-binding activity in vivo, we examined hematopoietic development in mice carrying a germline DNA-binding mutation. Remarkably, in contrast to complete absence of hematopoiesis and early lethality in scl-null embryos, specification of hematopoietic cells occurred in homozygous mutant embryos, indicating that direct DNA binding is dispensable for this process. Lethality was forestalled to later in development, although some mice survived to adulthood. Anemia was documented throughout development and in adulthood. Cellular and molecular studies showed requirements for SCL direct DNA binding in red cell maturation and indicated that scl expression is positively autoregulated in terminally differentiating erythroid cells. Thus, different mechanisms of SCL's action predominate depending on the developmental/cellular context: indirect DNA binding activities and/or sequestration of other nuclear regulators are sufficient in specification processes, whereas direct DNA binding functions with transcriptional autoregulation are critically required in terminal maturation processes.
Introduction
An important biologic process is the specification of tissue stem cells during development and their subsequent controlled differentiation to form the large number of specialized cell types. The molecular decisions that regulate these processes depend, in large part, on the establishment of specific transcriptional programs, mediated by the combined activities of ubiquitously expressed and tissue-specific transcription factors. Hematopoiesis provides a well-studied and accessible system to investigate transcriptional mechanisms involved in lineage specification and terminal differentiation. One of the critical hematopoietic-specific transcriptional regulators is the basic helix-loop-helix (bHLH) protein SCL.
SCL was discovered through its involvement in translocation events and is involved in up to 60% of cases in T-cell acute lymphoblastic leukemia.1 In normal hematopoiesis, it plays key roles in both specification of progenitors and terminal erythroid differentiation. Loss-of-function studies have shown that SCL is required for specification of the first primitive (embryonic) hematopoietic cells from a yolk sac mesodermal precursor at the origin of both blood and endothelium, the hemangioblast.2-4 As development proceeds, SCL is then required in the cellular processes leading to specification of definitive (adult) hematopoietic stem cells (HSCs) in the embryo proper.5-8 Functions for SCL in the maintenance and commitment of mouse short-term and human nonobese diabetic–severe combined immunodeficient mouse repopulating HSCs have been proposed,9-11 whereas SCL is not thought to be involved in mouse long-term HSC biology.12,13 Separately, SCL is necessary for proper development and maturation of erythroid and megakaryocytic progenitors, normal differentiation of mast cells, as well as in neural development.12,14-20
SCL belongs to the family of class II tissue-specific bHLH proteins. Similarly to the prototype of this class, MyoD, it binds specific DNA sequences, E-box (CANNTG) motifs, in cis-regulatory elements as a heterodimer with the E-proteins.21 Moreover, it interacts with multiple protein partners to regulate transcription. In erythroid cells, where its function has been most studied, it can positively as well as negatively modulate transcription of target genes by recruiting cofactors (activators or repressors) and chromatin remodelling proteins (Schuh et al22 and references therein).23
DNA binding has long been considered as a prerequisite for the transcriptional activity of bHLH proteins, as exemplified by muscle-specific proteins of this family. The DNA-binding domain of MyoD is crucial in conferring myogenic properties to a nonmyogenic cell line.24 Similarly, myogenin is unable to induce the myogenic program without DNA binding.25 We suggested that this might not be strictly true for SCL because some of its functions during hematopoietic differentiation can be mediated by DNA-binding–independent mechanisms.15 However, there are important caveats with these findings. The assays used were based on rescue as opposed to loss-of-function experiments. Importantly, SCL expression was forced at unregulated levels in in vitro embryonic stem (ES) cell differentiation assays and in the zebrafish mutant embryo cloche. Moreover, the consensus suggests that, although ES cell–derived hematopoiesis faithfully recapitulates primitive, yolk sac hematopoiesis, in vitro–differentiated ES cells do not give rise to definitive, adult hematopoietic stem cells and their progeny.26 Thus, to rigorously and systematically assess the DNA binding–dependent and -independent activities of SCL in developmental (primitive and definitive) hematopoiesis, an in vivo physiologic model was needed. We anticipated that this analysis would have broader implications for our understanding of how regulatory proteins achieve their functions.
To address this question, we have created a knockin mouse model expressing a mutant form of SCL that has no DNA-binding activity. In contrast to early lethality and complete absence of hematopoiesis in scl-null embryos, hematopoietic cells are specified in the DNA-binding homozygous mutant mice. Terminal erythroid cell maturation, however, is severely compromised and leads to lethality from embryonic day E14.5. This conclusively shows that SCL DNA-binding–independent activities are necessary and sufficient for hematopoietic stem/progenitor cell specification, whereas SCL direct DNA-binding activity is critically required in terminal erythroid maturation. We propose that the differential use of distinct functional domains is likely to be a general property of developmental regulators. In this context, dissection of the molecular mechanisms that regulatory proteins engage at different stages of development or in distinct cellular contexts should uncover novel transcriptional regulatory networks.
Methods
Generation of homologous recombinant ES clones and sclWT/RER mice
This study was conducted in compliance with the Home Office regulations at the University of Oxford. The targeting construct containing the mutations aimed at replacing the RER residues by alanines in SCL basic domain (details in Document S1, available on the Blood website; see the Supplemental Materials link at the top of the online article) was electroporated into mouse E14Tg2a ES cells maintained in BHK-21-Glasgow MEM (Invitrogen, Carlsbad, CA). On selection with G418 (0.3 mg/mL) and Gancyclovir (0.2 mM), resistant colonies were isolated and characterized by Southern blotting. To excise the floxed neomycin cassette, positive clones (sclWt/RERneo) were transiently transfected with a vector expressing a Cre recombinase/GFP fusion protein (gift from T. Enver, University of Oxford). Forty-eight hours after transfection, sorted GFP-positive cells were selected for G418 sensitivity. Excision of the neomycin cassette was further confirmed by Southern blotting. sclWt/RERΔneo ES cells with a normal karyotype were then injected into C57Bl/6 blastocysts and introduced into pseudo-pregnant C57Bl/6 females. Germ line transmission was obtained and heterozygotes were produced. The polymerase chain reaction (PCR)–based genotyping strategy relied on the presence of a PvuII restriction site created by the mutation introduced into the scl locus.
Selection of sclRER/RER ES cells and in vitro hemangioblast assay
Heterozygous sclWT/RERneo ES cells were grown in 1.8 mg/mL G418.27 Clones were picked and homozygosity was characterized by Southern blot analysis. The neomycin cassette was excised as described for sclWT/RERneo to generate sclRER/RER ES cells. Clones were analyzed by Southern blotting and allele-specific PCR as mentioned above. For in vitro hematopoietic differentiation, wild-type E14Tg2a, sclRER/RER, and scl−/−5 ES cells were differentiated into embryoid bodies (EBs) in IMDM as described.28 At day 3 of development, EBs were disaggregated by trypsin treatment, and cells were replated in 1% methylcellulose in VEGF-containing medium. Colonies (transitional, blast, and secondary EBs) were scored after 4 days of culture and individually expanded in liquid culture on Matrigel-coated wells in IMDM supplemented with cytokines that support the growth of both hematopoietic and endothelial lineages, as described.28 Hematopoietic colony-forming potential of the nonadherent cells was assayed on replating in 1% methylcellulose.28
In vitro progenitor assays
Cells from yolk sac (day E8.5), fetal liver (days E12.5 and 14.5), and adult bone marrow (total or common myeloid progenitor [CMP] and megakaryocyte-erythroid progenitor [MEP] populations) were subject to progenitor assays. At least 5 independent experiments were performed for each assay. Replating conditions are described in Document S1.
Isolation of CMP and MEP from adult bone marrow
Expansion and erythroid differentiation of day E12.5 fetal liver cells
Real-time PCR
RNA was extracted from day 0 and day 2 fetal liver populations using the RNAeasy Micro RNA isolation kit (Qiagen, Valencia, CA) and cDNA was synthesized using the Sensiscript kit (Qiagen). Ready-made primer and probe mixes from Applied Biosystems (ABI; Foster City, CA) were used for c-kit, gpa, eklf, pb4.2, and alpha globin. Primer sequences for beta globin and gata1 are in the Document S1. Samples were analyzed in duplicates using an ABI Prism 7000 sequence detection system.
For chromatin immunoprecipitation (ChIP) experiments, primers and 5′-6-carboxyfluorescein-3′-6-carboxy tetramethylrhodamine–labeled probes were selected from unique sequences in the pb4.2, eklf, and gpa loci and appropriate external controls using Primer Express (see Document S1). Input and immunoprecipitated material were analyzed in duplicates relative to a sequence in the gapdh locus as previously described.32
ChIP experiments
ChIP assays were performed as described.22
Image analysis
Hematopoietic colonies were imaged on an inverted IX51 Olympus microscope (Tokyo, Japan) with a Jenoptik C14 camera (Jena, Germany). The objectives used were 10×/0.3 NA (numeric aperture), 20×/0.4 NA, and 4×/0.13 NA; see figure legends for details). MGG/benzidine-stained cytospins were imaged on a BX60 Olympus microscope with a QImaging camera (Surrey, ON); all images were taken through a 40×/0.5 NA objective. Embryos were imaged on an SMZ1500 Nikon microscope (Tokyo, Japan) with a Nikon digital camera Dxm1200F. The Openlab (version 3; Improvision, Coventry, United Kingdom) software was used for image acquisition and images were exported into Adobe Photoshop (version CS2; Adobe Systems, San Jose, CA) for processing.
Results
Generation of SCL DNA-binding mutant mice
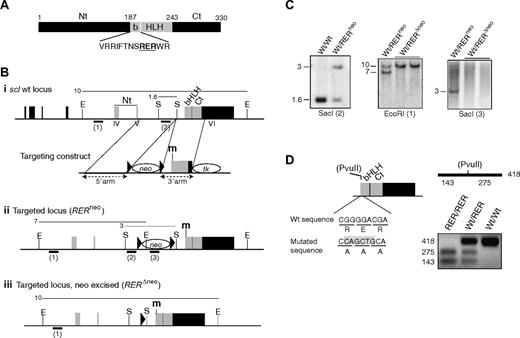
To abolish SCL DNA-binding activity, point mutations were introduced into exon VI of the scl genomic locus by homologous recombination in ES cells (targeting construct Figure 1Bi). These mutations substituted 3 highly conserved DNA-binding residues33 in SCL basic domain with alanine residues (amino acids RER at positions 195-197; Figure 1A). Southern blot analysis identified heterozygous sclWt/RERneo ES cell clones positive for homologous recombination (Figure 1Bii,C). On transient expression of a Cre recombinase, 2 ES subclones harboring the deletion of the floxed neomycin resistance gene (sclWt/RERΔneo; Figure 1Biii,C) were generated. Both clones had a normal karyotype (not shown), and the presence of the mutation in exon VI was confirmed by allele-specific digestion of PCR products (Figure 1D). One of the clones was then used for blastocyst injection, and mouse lines heterozygous for the SCL DNA-binding mutation (thereafter referred to as sclWt/RER) were generated.
Generation of sclRER/RER knockin mice. (A) Schematic representation of the SCL protein. In black are shown the N-terminal (Nt) and C-terminal (Ct) domains; in gray, the basic (b) helix-loop-helix (HLH) domain. Numbers correspond to amino acid positions. The amino acid sequence shows the 3 residues that are mutated in SCL basic domain (RER, bold and underlined). (B) Strategy for targeted insertion of the mutations into the scl locus. The scl wild-type genomic locus and targeting construct (i), the targeted locus resulting from homologous recombination before (ii, RERneo) and after (iii, RERΔneo) excision of the floxed neo cassette are shown. Solid boxes represent the exons (black indicates noncoding; gray, coding). The domains of SCL (Nt, bHLH, Ct) encoded by exons IV to VI are indicated. Neo, TK indicate neomycin and thymidine kinase selection cassettes, respectively; black triangles, LoxP sites; m: mutation introduced into the sequence coding for SCL basic domain. E, EcoRI; S, SacI. The thin lines above the loci represent the length (in kb) of the restriction fragments detected by Southern blotting. The bold bars under the loci show the position and name (in brackets) of the probes used in the Southern blot analyses. (C) Autoradiograms showing the genomic analysis of wild-type (Wt/Wt) and targeted heterozygous (Wt/RERneo and Wt/RERΔneo) ES clones. The restriction enzymes and probes used are indicated under each blot. (D) Allele-specific PCR analysis. (Left) Schematic representation of the exon coding for SCL bHLH and Ct domains. Homologous recombination introduced a PvuII site in the mutated scl allele. Nucleotide and amino acid sequences are indicated for wild-type and mutated alleles. The PvuII site created by the mutation in the targeted allele is highlighted. (Right) The 418-bp amplified fragment encompassing the mutation gives rise to 2 bands when digested by PvuII. An ethidium bromide–stained agarose gel shows a representative PCR analysis of DNA extracted from wild-type, heterozygous sclWt/RER, and homozygous sclRER/RER cells.
Generation of sclRER/RER knockin mice. (A) Schematic representation of the SCL protein. In black are shown the N-terminal (Nt) and C-terminal (Ct) domains; in gray, the basic (b) helix-loop-helix (HLH) domain. Numbers correspond to amino acid positions. The amino acid sequence shows the 3 residues that are mutated in SCL basic domain (RER, bold and underlined). (B) Strategy for targeted insertion of the mutations into the scl locus. The scl wild-type genomic locus and targeting construct (i), the targeted locus resulting from homologous recombination before (ii, RERneo) and after (iii, RERΔneo) excision of the floxed neo cassette are shown. Solid boxes represent the exons (black indicates noncoding; gray, coding). The domains of SCL (Nt, bHLH, Ct) encoded by exons IV to VI are indicated. Neo, TK indicate neomycin and thymidine kinase selection cassettes, respectively; black triangles, LoxP sites; m: mutation introduced into the sequence coding for SCL basic domain. E, EcoRI; S, SacI. The thin lines above the loci represent the length (in kb) of the restriction fragments detected by Southern blotting. The bold bars under the loci show the position and name (in brackets) of the probes used in the Southern blot analyses. (C) Autoradiograms showing the genomic analysis of wild-type (Wt/Wt) and targeted heterozygous (Wt/RERneo and Wt/RERΔneo) ES clones. The restriction enzymes and probes used are indicated under each blot. (D) Allele-specific PCR analysis. (Left) Schematic representation of the exon coding for SCL bHLH and Ct domains. Homologous recombination introduced a PvuII site in the mutated scl allele. Nucleotide and amino acid sequences are indicated for wild-type and mutated alleles. The PvuII site created by the mutation in the targeted allele is highlighted. (Right) The 418-bp amplified fragment encompassing the mutation gives rise to 2 bands when digested by PvuII. An ethidium bromide–stained agarose gel shows a representative PCR analysis of DNA extracted from wild-type, heterozygous sclWt/RER, and homozygous sclRER/RER cells.
Initial breeding produced sclRER/RER embryos at several developmental stages as well as sclRER/RER newborn mice (as shown in Tables 1 and 2, see next subsection). Before embarking on extensive breeding programs, we fully characterized the mutant SCLRER protein to ensure that substitution of the 3 residues (RER) in SCL basic domain had specifically inhibited DNA-binding activity. Inability to bind DNA, unperturbed levels of expression, and nuclear localization were confirmed from tissues isolated from homozygous embryos or adult mice (Figure S1A-C).
Survival at the yolk sac stage but fetal death
As shown in Table 1, mating of heterozygous sclWt/RER mice only produced 5% of homozygous sclRER/RER mice (as opposed to 25% for a mendelian ratio), suggesting embryonic lethality of approximately 80% of homozygotes. The surviving sclRER/RER mice developed to adulthood. To document time of death, timed matings were set up between homozygous males and heterozygous females. sclWt/RER and sclRER/RER embryos were observed at a mendelian frequency up to gestation day E12.5 (Table 2). Lethality was observed at day E14.5 and increased until day E17.5. Equivalent frequencies were observed in the progeny of heterozygous mice (data not shown).
Distribution of scl genotypes in newborn mice
| Genotypes . | sclWt/Wt . | sclWt/RER . | sclRER/RER . |
|---|---|---|---|
| Seen, % | 31 | 64 | 5 |
| Expected, % | 25 | 50 | 25 |
| Genotypes . | sclWt/Wt . | sclWt/RER . | sclRER/RER . |
|---|---|---|---|
| Seen, % | 31 | 64 | 5 |
| Expected, % | 25 | 50 | 25 |
The crosses were sclWt/RER × sclWt/RER. Newborn mice (n = 229) were analyzed. Genotyping was performed by PCR (see Figure 1D).
Expected indicates the expected mendelian frequency.
Distribution of scl genotypes in timed matings
| Genotypes . | sclWt/RER . | sclRER/RER . | |
|---|---|---|---|
| Alive . | Dead . | ||
| E8.5, % (n = 20) | 40 | 60 | |
| E9.5, % (n = 20) | 60 | 40 | |
| E12.5, % (n = 61) | 48 | 52 | |
| E14.5, % (n = 47) | 52 | 36 | 12 |
| E17.5, % (n = 10) | 50 | 20 | 30 |
| Expected, % | 50 | 50 | |
| Genotypes . | sclWt/RER . | sclRER/RER . | |
|---|---|---|---|
| Alive . | Dead . | ||
| E8.5, % (n = 20) | 40 | 60 | |
| E9.5, % (n = 20) | 60 | 40 | |
| E12.5, % (n = 61) | 48 | 52 | |
| E14.5, % (n = 47) | 52 | 36 | 12 |
| E17.5, % (n = 10) | 50 | 20 | 30 |
| Expected, % | 50 | 50 | |
Female sclWt/RER mice were crossed with male sclRER/RER mice. E8.5-E17.5 indicates embryonic days 8.5 to 17.5. The number of embryos analyzed are shown in the square brackets. Genotyping was performed by PCR (see Figure 1D). The first dead homozygous sclRER/RER embryos were observed at day E14.5.
Expected indicates the expected mendelian frequency.
In conclusion, we successfully generated a mouse model expressing a mutant form of SCL that was specifically impaired in its DNA-binding capacity. Contrary to the conventional SCL knockout embryos,2,4 the mutant homozygous sclRER/RER embryos survive at the yolk sac stage (day E9.5). The lethality observed at the fetal liver stage indicates a specific nonredundant requirement for SCL direct DNA-binding activity in definitive hematopoiesis.
Commitment of hematopoietic cells from the sclRER/RER hemangioblast
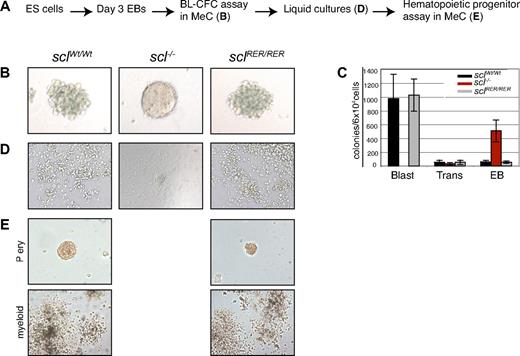
In vitro experiments have suggested that the scl−/− embryonic phenotype is due to failure of hematopoietic commitment from the hemangioblast.3 The fact that no lethality was observed at the yolk sac stage in the sclRER/RER embryos therefore suggested that the hemangioblast was specified and that its hematopoietic potential was expressed. To confirm this hypothesis, we used the ES cell in vitro differentiation system that gives easy access to this early embryonic hematopoietic compartment. The in vitro equivalent of the hemangioblast, called blast colony-forming cell (or BL-CFC), develops in ES-derived EBs from day 2.5 to day 4 of differentiation.28 We derived sclRER/RER ES cells from heterozygous clones (Figure S1D) and submitted them to in vitro differentiation, in parallel with wild-type and scl-null ES cells.5 At day 3 of EB development, the cells were replated in conditions favoring hemangioblast development (see Figure 2A for the culture steps involved in the blast colony assay). Remarkably, colonies that developed from sclRER/RER cultures were very similar in terms of number and morphology to those observed from wild-type cultures (Figure 2B right and left, C) and previously described as blast colonies (progeny of the BL-CFCs28 ). In contrast, scl-null EB-derived cells mainly gave rise to large numbers of secondary EBs (Figure 2B middle, C). To characterize their hematopoietic and vascular potential, blast colonies were transferred onto matrigel-coated plates. Both wild-type and sclRER/RER blast colonies developed into a typical layer of adherent (endothelial) cells surmounted by nonadherent (hematopoietic) cells (Figure 2D right and left). In contrast, the scl−/− EB structures generated an adherent layer and a compact inner core of cells (Figure 2D middle). The hematopoietic nature of the nonadherent wild-type and sclRER/RER cells was confirmed on replating in medium supplemented with hematopoietic growth factors (Figure 2E). In summary, the DNA-binding mutation did not affect SCL activity in blast colony-forming cells, and hematopoietic cells were specified from sclRER/RER hemangioblasts.
Hemangioblasts derived from sclRER/RER ES cells produce hematopoietic cells. (A) Steps involved in the BL-CFC colony assay. MeC indicates methylcellulose. (B) Representative blast colonies from wild-type (sclWt/Wt) and mutant (sclRER/RER) EB cells (left and right) and secondary EB from scl−/− cells (middle; original magnification, ×200). (C) Frequency of blast colonies (Blast), transitional colonies (Trans), and secondary EBs (EBs) in sclWt/Wt, scl−/−, and sclRER/RER cultures. Error bars indicate plus or minus 1 SD from 5 independent experiments. (D) When replated on Matrigel-coated wells, blast colonies gave rise to hematopoietic (cells in suspension) and endothelial (adherent cells) compartments (left and right). In contrast, the scl−/− EBs only gave rise to adherent cells and a core of cells (middle; original magnification, ×100). (E) The hematopoietic cells shown in panel D (left and right) were replated in methylcellulose. Primitive erythroid (Pery; original magnification, ×200) and myeloid (original magnification, ×40) colonies were observed.
Hemangioblasts derived from sclRER/RER ES cells produce hematopoietic cells. (A) Steps involved in the BL-CFC colony assay. MeC indicates methylcellulose. (B) Representative blast colonies from wild-type (sclWt/Wt) and mutant (sclRER/RER) EB cells (left and right) and secondary EB from scl−/− cells (middle; original magnification, ×200). (C) Frequency of blast colonies (Blast), transitional colonies (Trans), and secondary EBs (EBs) in sclWt/Wt, scl−/−, and sclRER/RER cultures. Error bars indicate plus or minus 1 SD from 5 independent experiments. (D) When replated on Matrigel-coated wells, blast colonies gave rise to hematopoietic (cells in suspension) and endothelial (adherent cells) compartments (left and right). In contrast, the scl−/− EBs only gave rise to adherent cells and a core of cells (middle; original magnification, ×100). (E) The hematopoietic cells shown in panel D (left and right) were replated in methylcellulose. Primitive erythroid (Pery; original magnification, ×200) and myeloid (original magnification, ×40) colonies were observed.
These data are therefore the first in vivo indication in a mammalian system that the direct DNA-binding activity of SCL is dispensable for its earliest role in hematopoietic development.
Anemia in sclRER/RER embryos at the yolk sac and fetal liver stages
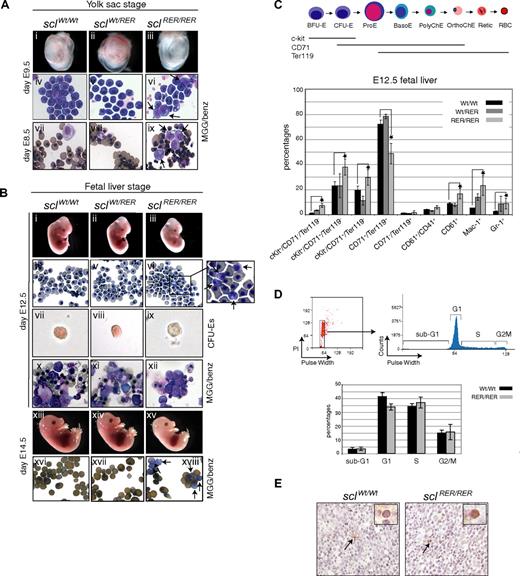
The first hematopoietic cells to emerge in the embryo are primitive erythroid progenitors in the yolk sac blood islands at day E7.5, that mature progressively after entering the circulation.34 To examine the effects of absence of SCL DNA-binding activity on primitive erythropoiesis, day E9.5 yolk sacs were dissected. Homozygous sclRER/RER yolk sacs were pale compared with wild-type controls and heterozygotes (Figure 3Ai-iii). Blood examination showed mature basophilic erythroblasts in control embryos (Figure 3Aiv,v), whereas the presence of immature proerythroblasts in the sclRER/RER blood suggested a block or a delay in maturation in mutant embryos (Figure 3Avi arrows). We then examined the differentiation potential of the erythroid progenitors in colony assays. Erythroid colonies derived from day E8.5 control yolk sacs showed the presence of mature, benzidine-positive, and enucleated erythrocytes (Figure 3Avii,viii). In contrast, mutant sclRER/RER colonies contained immature proerythroblasts and fewer fully differentiated hemoglobinized red cells (Figure 3Aix arrows). Together, these data indicate a requirement for SCL DNA-binding activity in the maturation of primitive erythroid cells.
Block in erythroid maturation at the yolk sac and fetal liver stages in sclRER/RER embryos. (A) Yolk sac erythropoiesis. Day E9.5 sclRER/RER yolk sacs (iii) are paler than wild-type and heterozygotes (i,ii). (iv-vi) MGG staining of yolk sac blood shows asynchronous maturation of primitive erythroid cells in mutant homozygous E9.5 embryos. Arrows show immature erythroid cells absent from wild-type and heterozygous control samples (original magnification, ×400). (vii-ix) MGG/benzidine staining of primitive erythroid colonies obtained on plating of day E8.5 sclWt/Wt, sclWt/RER, and sclRER/RER yolk sacs. Arrows point to immature erythroid cells in the mutant homozygous population that are absent from wild-type and heterozygous control samples (original magnification, ×400). (B) Fetal liver erythropoiesis. (i-iii) Day E12.5 embryos. Note the pale sclRER/RER embryo (iii). (iv-vi) MGG-stained E12.5 blood (original magnification, ×400). The inset shows the presence of immature cells in blood derived from sclRER/RER embryos (arrows); (vii-ix) representative CFU-E colonies from wild-type, sclWt/RER, and sclRER/RER E12.5 fetal liver replating (magnification, ×200). Note the poor hemoglobinization of the mutant CFU-E colony (ix); (x-xii) MGG/benzidine staining of CFU-E colonies. Note the lack of mature benzidine positive sclRER/RER erythrocytes (xii) (original magnification, ×400); (xii-xv) day E14.5 embryos. Note the pale sclRER/RER embryo in (xv). (xvi-xviii) Benzidine staining of E14.5 fetal blood (original magnification, × 400). The arrows show the presence of less mature, benzidine-negative erythroid cells in the mutant homozygous sample (xviii) that are absent from the wild-type and heterozygous samples (xvi,xvii). (C) Characterization of fetal liver hematopoietic cells. (Top) Schematic representation of the progressive maturation of erythroid cells and pattern of expression of the cell surface markers c-kit, CD71 and Ter119.37 ProE indicates proerythroblasts; basoE, polyChE, orthoChE, basophilic, polychromatic, and orthochromatic erythroid cells; retic, reticulocytes; RBC, red blood cells. (Bottom) Fetal liver cells derived from wild-type, heterozygous, and homozygous E12.5 embryos were analyzed by fluorescence-activated cell sorting (FACS). The percentages of fetal liver populations characterized by expression of various combinations of cell surface markers are indicated for each genotype. Error bars indicate plus or minus 1 SD from at least 3 independent experiments; *P < .01. (D) Cell-cycle analysis. (Top left) FACS analysis of PI-stained fetal liver cells representative of both wild-type and sclRER/RER E12.5 fetal liver samples. (Top right) The 3 cell cycle phases and the sub-G1 phase are visualized according to PI intensity and pulse width. (Bottom) The graph shows the proportion of fetal liver cells isolated from wild-type and sclRER/RER mice in sub-G1, G1, S, and G2/M phases. Error bars indicate plus or minus 1 SD from at least 3 independent experiments. (E) Apoptosis analysis. Cryosections of wild-type and sclRER/RER E12.5 fetal livers were analyzed by TUNEL assay. Arrowheads point to TUNEL-positive apoptotic cells shown at higher magnification in insets (upper right of both panels; original magnification, ×400).
Block in erythroid maturation at the yolk sac and fetal liver stages in sclRER/RER embryos. (A) Yolk sac erythropoiesis. Day E9.5 sclRER/RER yolk sacs (iii) are paler than wild-type and heterozygotes (i,ii). (iv-vi) MGG staining of yolk sac blood shows asynchronous maturation of primitive erythroid cells in mutant homozygous E9.5 embryos. Arrows show immature erythroid cells absent from wild-type and heterozygous control samples (original magnification, ×400). (vii-ix) MGG/benzidine staining of primitive erythroid colonies obtained on plating of day E8.5 sclWt/Wt, sclWt/RER, and sclRER/RER yolk sacs. Arrows point to immature erythroid cells in the mutant homozygous population that are absent from wild-type and heterozygous control samples (original magnification, ×400). (B) Fetal liver erythropoiesis. (i-iii) Day E12.5 embryos. Note the pale sclRER/RER embryo (iii). (iv-vi) MGG-stained E12.5 blood (original magnification, ×400). The inset shows the presence of immature cells in blood derived from sclRER/RER embryos (arrows); (vii-ix) representative CFU-E colonies from wild-type, sclWt/RER, and sclRER/RER E12.5 fetal liver replating (magnification, ×200). Note the poor hemoglobinization of the mutant CFU-E colony (ix); (x-xii) MGG/benzidine staining of CFU-E colonies. Note the lack of mature benzidine positive sclRER/RER erythrocytes (xii) (original magnification, ×400); (xii-xv) day E14.5 embryos. Note the pale sclRER/RER embryo in (xv). (xvi-xviii) Benzidine staining of E14.5 fetal blood (original magnification, × 400). The arrows show the presence of less mature, benzidine-negative erythroid cells in the mutant homozygous sample (xviii) that are absent from the wild-type and heterozygous samples (xvi,xvii). (C) Characterization of fetal liver hematopoietic cells. (Top) Schematic representation of the progressive maturation of erythroid cells and pattern of expression of the cell surface markers c-kit, CD71 and Ter119.37 ProE indicates proerythroblasts; basoE, polyChE, orthoChE, basophilic, polychromatic, and orthochromatic erythroid cells; retic, reticulocytes; RBC, red blood cells. (Bottom) Fetal liver cells derived from wild-type, heterozygous, and homozygous E12.5 embryos were analyzed by fluorescence-activated cell sorting (FACS). The percentages of fetal liver populations characterized by expression of various combinations of cell surface markers are indicated for each genotype. Error bars indicate plus or minus 1 SD from at least 3 independent experiments; *P < .01. (D) Cell-cycle analysis. (Top left) FACS analysis of PI-stained fetal liver cells representative of both wild-type and sclRER/RER E12.5 fetal liver samples. (Top right) The 3 cell cycle phases and the sub-G1 phase are visualized according to PI intensity and pulse width. (Bottom) The graph shows the proportion of fetal liver cells isolated from wild-type and sclRER/RER mice in sub-G1, G1, S, and G2/M phases. Error bars indicate plus or minus 1 SD from at least 3 independent experiments. (E) Apoptosis analysis. Cryosections of wild-type and sclRER/RER E12.5 fetal livers were analyzed by TUNEL assay. Arrowheads point to TUNEL-positive apoptotic cells shown at higher magnification in insets (upper right of both panels; original magnification, ×400).
Concomitant with the decline of this transient, first wave of erythropoiesis, definitive hematopoietic cells develop autonomously in the embryo proper and, by day E11.5, have colonized the fetal liver.35 At day E12.5, mutant sclRER/RER embryos were pale compared with controls (Figure 3Bi-iii). Circulating blood mainly consisted of primitive erythrocytes, as expected at that developmental stage34 (Figure 3Biv-vi). In agreement with yolk sac analyses, blood derived from homozygous sclRER/RER embryos contained cells of an immature phenotype (Figure 3Bvi arrows). At day E14.5, when lethality is first observed (Table 2), the surviving mutant sclRER/RER embryos were smaller and paler than their control littermates (Figure 3Bxiii-xv). At that stage, definitive erythrocytes have entered the bloodstream and outnumbered primitive red cells. MGG/benzidine staining of mutant blood films detected benzidine-negative cells and only few enucleated, benzidine-positive erythrocytes (compare Figure 3Bxviii with Figure 3Bxvi,xvii).
Block in erythroid maturation in E12.5 sclRER/RER embryos
We then performed further analyses on day E12.5 embryos, an age when lethality is not observed (Table 2) but when the phenotype is expressed and compensation mechanisms (as shown by the incomplete penetrance of the phenotype; Table 1) are less likely to mask the effects of loss of SCL DNA-binding activity.
Immunophenotypic characterization of the fetal liver hematopoietic compartments showed significant differences between mutant homozygote and wild-type samples (Figure 3C). Decrease in the homozygous mutant CD71+/Ter119+ compartment showed late defects in erythroid maturation. The increase in the mutant early erythroid compartments (corresponding to BFU-E, CFU-E, and proerythroblast stages) denotes either an accumulation of immature progenitors and a corresponding decrease in more mature CD71+/Ter119+ cells because of defective erythroid maturation or a response to the fast turnover of later precursors. Regarding the other cellular populations, an expansion of the sclRER/RER myeloid compartment was documented (see Document S1 for a quantitative analysis of the data).
The in vitro growth potential of E12.5 hematopoietic progenitors was then analyzed in colony assays. No difference in numbers of CFU-E, BFU-E, megakaryocyte, myeloid, and mixed colonies was observed between mutant and control samples (Figure S2A). On morphologic examination, mutant homozygous CFU-E colonies appeared poorly hemoglobinized (Figure 3Bvii-ix), contained an increased proportion of immature erythroblasts, and were devoid of terminally differentiated, benzidine-positive erythrocytes (Figure 3B compare ix and xii with vii,viii and x,xi).
Because SCL can have survival functions in red cells,36 and to further characterize the affected erythroid populations, E12.5 fetal liver cells were subject to cell-cycle and apoptosis analyses. No difference in cell-cycle progression (G1, S, G2/M phases) was observed between wild-type and mutant populations (Figure 3D). Presence of very few events in the sub-G1 population (Figure 3D) suggested low apoptotic rates in both mutant and wild-type samples. This was confirmed on analysis of fetal liver sections by transferase-mediated dUTP nick end labeling (TUNEL) assay. Very few apoptotic cells were detected in both sclRER/RER and wild-type control samples, that represented 0.13% of total fetal liver cells (Figure 3E; data not shown).
In summary, SCL DNA-binding mutant homozygous embryos are anemic and show late defects in erythroid maturation that do not seem to be due to impaired proliferative or survival capacities. These defects result in most homozygous fetuses dying from anemia and show a critical in vivo function for SCL DNA-binding activity in embryonic/fetal erythropoiesis. Analysis of heterozygous embryos showed phenotypes very similar to those described for wild-type embryos, indicating that haploinsufficiency does not obviously affect SCL activity.
sclRER/RER adult mice present with microcytic hypochromic anemia
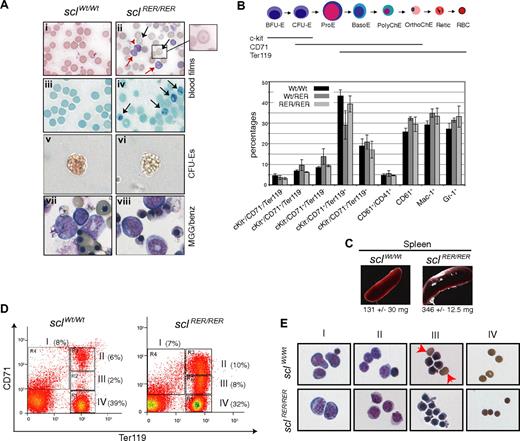
Surviving adult sclRER/RER mice did not present with obvious morphologic or behavioral abnormalities and showed no increase in death rate. Examination of blood variables, however, showed mild microcytic hypochromic anemia (Table 3). The reticulocyte count was increased. Red cell anisocytosis was shown by decreased mean corpuscular volume (MCV) and an increase in red cell distribution width (RDW). The homozygous sclRER/RER mice also showed decreased hemoglobin (HGB), hematocrit (HCT), and mean corpuscular hemoglobin (MCH). The rest of the peripheral blood count was normal (not shown). Examination of blood films confirmed the anemia in the sclRER/RER mice. May-Grünwald-Giemsa (MGG) staining showed the presence of reticulocytes and poorly hemoglobinized target cells compared with control wild-type cells (Figure 4Ai,ii) and Brilliant Cresyl Blue (BCB) staining confirmed a 4-fold increase in the number of reticulocytes in blood derived from the mutant mice (Figure 4Aiii,iv and data not shown). In summary, these blood analyses indicate defective erythropoiesis in the adult sclRER/RER mice.
Blood variables of sclWt/Wt, sclWt/RER, and sclRER/RER mice
| . | Reticulocytes, % . | RBC count, 106/mm3 . | HGB level, g/dL . | HCT, % . | MCV, μm3 . | MCH, pg . | RDW, % . |
|---|---|---|---|---|---|---|---|
| sclWt/Wt | 1.18 (0.35) | 10.07 (0.86) | 17.12 (1.47) | 51.26 (3.65) | 50.8 (2.59) | 17.0 (0.70) | 13.26 (0.72) |
| sclWt/RER | 0.9 (0.21) | 9.91 (0.60) | 15.76 (0.85) | 49.0 (2.08) | 49.33 (1.15) | 15.86 (0.42) | 15.1 (1.30) |
| sclRER/RER | 5.57 (1.47) | 10.26 (0.99) | 13.57 (1.16) | 42.5 (3.23) | 41.77 (2.11) | 13.27 (0.82) | 19.85 (1.82) |
| P | .001 | .71 | .002 | .002 | < .001 | < .001 | < .001 |
| . | Reticulocytes, % . | RBC count, 106/mm3 . | HGB level, g/dL . | HCT, % . | MCV, μm3 . | MCH, pg . | RDW, % . |
|---|---|---|---|---|---|---|---|
| sclWt/Wt | 1.18 (0.35) | 10.07 (0.86) | 17.12 (1.47) | 51.26 (3.65) | 50.8 (2.59) | 17.0 (0.70) | 13.26 (0.72) |
| sclWt/RER | 0.9 (0.21) | 9.91 (0.60) | 15.76 (0.85) | 49.0 (2.08) | 49.33 (1.15) | 15.86 (0.42) | 15.1 (1.30) |
| sclRER/RER | 5.57 (1.47) | 10.26 (0.99) | 13.57 (1.16) | 42.5 (3.23) | 41.77 (2.11) | 13.27 (0.82) | 19.85 (1.82) |
| P | .001 | .71 | .002 | .002 | < .001 | < .001 | < .001 |
Values in parentheses are the standard deviations from at least 5 independent experiments.
RBC indicates red blood cell; HGB, hemoglobin; HCT, hematocrit; MCV, mean corpuscular volume; MCH, mean corpuscular hemoglobin; and RDW, red cell distribution width.
The sclRER/RER adult surviving mice present with mild anemia. (A) (i,ii) Blood films obtained from 2-month-old wild-type and sclRER/RER mice were stained with MGG (original magnification, ×400). Black arrows indicate target cells; red arrows, reticulocytes. (iii,iv) BCB staining confirms the presence of reticulocytes in mutant sclRER/RER blood (arrows) (original magnification, ×400). (v,vi) Representative CFU-E colonies from wild-type and sclRER/RER total bone marrow replating (200× objective). Note the poor hemoglobinization of the mutant CFU-E colony (vi). (vii,viii) MGG/benzidine staining of CFU-E colonies (magnification, ×400). Note the lack of benzidine positive sclRER/RER erythrocytes (viii). (B) Characterization of adult bone marrow hematopoietic cells. (Top) See legend for Figure 3C. (Bottom) Bone marrow cells derived from wild-type, heterozygous, and homozygous 2-month-old mice were analyzed by FACS according to expression of various combinations of cell surface markers. The percentage of total bone marrow cells for each cellular population is indicated. Error bars indicate plus or minus 1 SD from at least 3 independent experiments. (C) Splenomegaly in sclRER/RER mice. (D) Splenocytes harvested from wild-type and sclRER/RER mice were analyzed for expression of the cell surface markers CD71 and Ter119 by FACS. Populations I to IV represent progressive maturation of erythroid cells. Representative FACS plots with the percentage of cells for each population are shown. (E) Cell populations as shown in panel D were sorted and stained with MGG/benzidine. Populations are mainly characterized by I, proerythroblasts; II, basophilic normoblasts; III, late normoblasts; and IV, enucleated red blood cells. Red arrowheads show enucleated cells frequently observed in wild-type population III but not in the corresponding mutant sample. Note the irregular shape of the mutant late normoblasts and the small size of the mutant enucleated cells (sclRER/RER, III and IV, respectively; original magnification, ×400).
The sclRER/RER adult surviving mice present with mild anemia. (A) (i,ii) Blood films obtained from 2-month-old wild-type and sclRER/RER mice were stained with MGG (original magnification, ×400). Black arrows indicate target cells; red arrows, reticulocytes. (iii,iv) BCB staining confirms the presence of reticulocytes in mutant sclRER/RER blood (arrows) (original magnification, ×400). (v,vi) Representative CFU-E colonies from wild-type and sclRER/RER total bone marrow replating (200× objective). Note the poor hemoglobinization of the mutant CFU-E colony (vi). (vii,viii) MGG/benzidine staining of CFU-E colonies (magnification, ×400). Note the lack of benzidine positive sclRER/RER erythrocytes (viii). (B) Characterization of adult bone marrow hematopoietic cells. (Top) See legend for Figure 3C. (Bottom) Bone marrow cells derived from wild-type, heterozygous, and homozygous 2-month-old mice were analyzed by FACS according to expression of various combinations of cell surface markers. The percentage of total bone marrow cells for each cellular population is indicated. Error bars indicate plus or minus 1 SD from at least 3 independent experiments. (C) Splenomegaly in sclRER/RER mice. (D) Splenocytes harvested from wild-type and sclRER/RER mice were analyzed for expression of the cell surface markers CD71 and Ter119 by FACS. Populations I to IV represent progressive maturation of erythroid cells. Representative FACS plots with the percentage of cells for each population are shown. (E) Cell populations as shown in panel D were sorted and stained with MGG/benzidine. Populations are mainly characterized by I, proerythroblasts; II, basophilic normoblasts; III, late normoblasts; and IV, enucleated red blood cells. Red arrowheads show enucleated cells frequently observed in wild-type population III but not in the corresponding mutant sample. Note the irregular shape of the mutant late normoblasts and the small size of the mutant enucleated cells (sclRER/RER, III and IV, respectively; original magnification, ×400).
Hematopoiesis in sclRER/RER adult mice
To survey the adult hematopoietic compartments, we conducted an immunophenotypic analysis of bone marrow committed progenitors and hematopoietic cells. The proportion of common myeloid CMP and megakaryocyte-erythroid MEP progenitors was not affected in mutant sclRER/RER mice (Figure S2B). In addition, and contrary to what we observed in fetal liver hematopoiesis, the proportions of sclRER/RER hematopoietic populations were not affected (Figure 4B).
We then studied the potential of bone marrow hematopoietic progenitors in colony assays. The homozygous mutant compartments (BFU-E, CFU-E, megakaryocytic, and myeloid) were not reduced compared with controls (Figure S2C). Qualitatively, however, as observed in fetal liver replating assays, mutant CFU-E colonies were poorly hemoglobinized and, on cytospins, deficient in mature benzidine-positive erythrocytes compared with wild-type controls (Figure 4Av-viii).
Abnormal splenic erythroid maturation
Compensatory mechanisms triggered by anemia often include an increase in erythropoiesis in spleen. Splenomegaly was observed in sclRER/RER mice (Figure 4C), indicative of an expanded extramedullary erythropoiesis. To confirm this, we analyzed the splenic erythroid compartment by flow cytometry based on expression of the cell surface markers CD71 and Ter119.37 Four populations (I to IV) corresponding to stages of progressive erythroid maturation were defined (Figure 4D). Analysis of cells isolated from sclRER/RER mice consistently showed an increased number of splenic red cells in regions II and III, reflecting perturbed maturation from early to late normoblasts. Morphologically, populations I (proerythroblasts) and II (early normoblasts) derived from sclRER/RER mice were similar to those derived from control wild-type mice (Figure 4E). The sclRER/RER mutant population III was composed of smaller, poorly hemoglobinized cells with abnormal membranes and devoid of enucleated erythrocytes (Figure 4E). Finally, the mutant population IV presented with microcytic red blood cells, in agreement with what has been observed in adult blood.
To further investigate the erythroid progenitor compartment in a stress situation, we induced anemia in the animals by phenylhydrazine injections or bleeding. Homozygote, heterozygote, and wild-type mice recovered similarly as assessed by the reestablishment of pretreatment hematocrit and reticulocyte count over a period of 17 days (Figure S2D; data not shown), thereby confirming the unaffected potentiality of the early splenic erythroid progenitor compartment in vivo.
We concluded that the increase in the early sclRER/RER splenic erythroid compartments was probably to compensate for impaired terminal differentiation.
SCL DNA-binding mutation differentially affects expression of erythroid genes
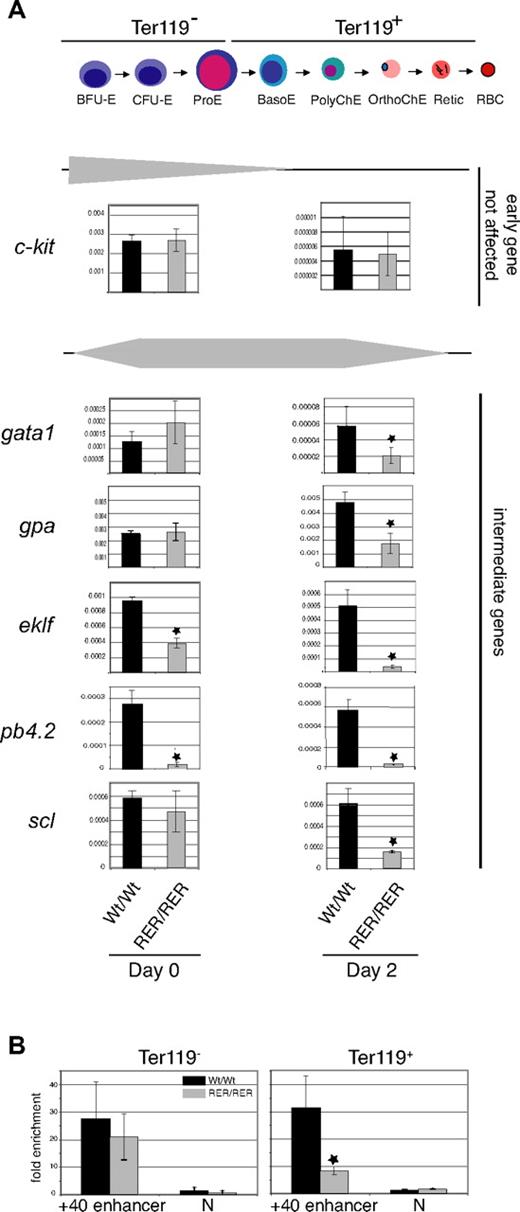
To characterize the molecular defects associated with SCL DNA-binding mutation in erythroid cells, we analyzed the expression levels of selected regulators of erythroid differentiation (known or potential SCL target genes) in material isolated from day E12.5 fetal livers. We used an in vitro differentiation system to expand c-kit+/CD71+/Ter119− erythroid precursors (day 0) that subsequently underwent differentiation to obtain CD71+Ter119+ erythrocytes after 48 hours (day 2; Figure S3).
Analysis in immature erythroid populations (day 0; Figure 5A; data not shown) showed genes whose expression was either not affected in mutant sclRER/RER cells (c-kit, gata1, gpa, alpha and beta globin, scl) or down-regulated (eklf and pb4.2). In more mature sclRER/RER erythroid cells (day 2; Figure 5A; data not shown), expression of the genes was either unchanged (c-kit, alpha and beta globin) or significantly decreased (gata1, gpa, eklf, pb4.2, and scl) compared with wild-type controls.
Levels of expression of erythroid-specific genes in SCL mutant erythroid cells. (A top) Schematic representation of the progressive maturation of erythroid cells. The BFU-E, CFU-E, and proE stages are Ter119−, whereas basoE, polyChE, orthoChE erythroid cells, retic, and RBC are Ter119+. (Bottom) Total RNA was prepared from Ter119− erythroid progenitors (day 0 population) or Ter119+ erythroid cells (day 2 population) derived from day E12.5 embryos. The genes chosen for analysis and indicated on the left side of the figure are grouped into 2 categories (early and intermediate) according to their normal pattern of expression during erythroid differentiation from the early BFU-E stage to mature erythrocytes22,32 (and this study). Levels of gene expression during normal erythroid maturation are schematized by gray triangles. The y-axis represents the enrichment in cDNA sequences as quantitated by real-time PCR normalized to 18S ribosomal gene control sequences for each gene. Error bars indicate plus or minus 1 SD from at least 3 independent experiments (*P < .01). (B) Chromatin derived from Ter119− and Ter119+ populations purified from sclWt/Wt and sclRER/RER fetal liver cultured cells was immunoprecipitated with α-SCL antibodies and the scl + 40 enhancer region analyzed by real-time PCR. The y-axis represents enrichment over input DNA, normalized to a control sequence in the gapdh gene. N indicates the transcriptionally inactive −16 region of the scl locus.
Levels of expression of erythroid-specific genes in SCL mutant erythroid cells. (A top) Schematic representation of the progressive maturation of erythroid cells. The BFU-E, CFU-E, and proE stages are Ter119−, whereas basoE, polyChE, orthoChE erythroid cells, retic, and RBC are Ter119+. (Bottom) Total RNA was prepared from Ter119− erythroid progenitors (day 0 population) or Ter119+ erythroid cells (day 2 population) derived from day E12.5 embryos. The genes chosen for analysis and indicated on the left side of the figure are grouped into 2 categories (early and intermediate) according to their normal pattern of expression during erythroid differentiation from the early BFU-E stage to mature erythrocytes22,32 (and this study). Levels of gene expression during normal erythroid maturation are schematized by gray triangles. The y-axis represents the enrichment in cDNA sequences as quantitated by real-time PCR normalized to 18S ribosomal gene control sequences for each gene. Error bars indicate plus or minus 1 SD from at least 3 independent experiments (*P < .01). (B) Chromatin derived from Ter119− and Ter119+ populations purified from sclWt/Wt and sclRER/RER fetal liver cultured cells was immunoprecipitated with α-SCL antibodies and the scl + 40 enhancer region analyzed by real-time PCR. The y-axis represents enrichment over input DNA, normalized to a control sequence in the gapdh gene. N indicates the transcriptionally inactive −16 region of the scl locus.
Autoregulation of scl expression
As shown above, SCL DNA-binding activity is necessary to achieve maximal expression of scl in late (day 2) erythroid cells, thereby showing either an autoregulatory mechanism or secondary effects of SCL DNA-binding mutation. To distinguish between these 2 possibilities, we performed ChIP from material isolated from both early (Ter119−) and late (Ter119+) erythroid populations and analyzed recruitment of SCL to its own + 40 enhancer. This enhancer is active in fetal liver red cells, contains E-box motifs, binds SCL in an erythroid cell line,38 and could therefore mediate transcriptional autoregulation. In sclRER/RER Ter119− cells, binding of SCL to + 40 enhancer was not affected, but it was impaired in more mature sclRER/RER Ter119+ cells (Figure 5B). This decrease in SCL binding correlated with the changes in scl expression levels. We concluded that maximal expression of scl in the late stages of erythroid maturation is likely to rely on an autoregulatory mechanism requiring direct DNA binding.
Loss of SCL DNA-binding and transcriptional activity marks on cis-acting elements of target genes in mutant erythroid cells
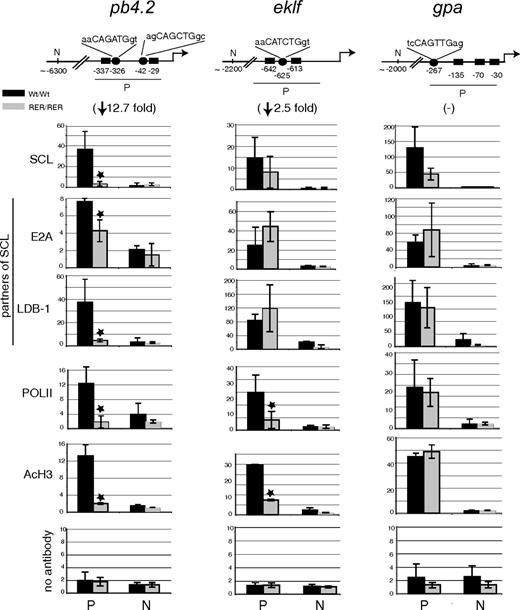
To directly link perturbed gene expression with impaired SCL DNA-binding activity, we analyzed the in vivo interaction of SCL with regulatory regions of genes whose expression was dysregulated in sclRER/RER cells. We performed ChIP from material isolated from Ter119− (day 0) and Ter119+ erythroid populations and focused our analyses on pb4.2, eklf, and gpa genes. The proximal promoter regions of these genes contain E-boxes and Gata motifs. These sequences are bound in hematopoietic cell lines by SCL and protein partners (E2A, Lim-only 2 [LMO2] and LIM domain binding protein 1 [LDB-1]), as well as GATA1 or GATA2, that drive gene expression in functional assays.39-41
When assayed for SCL recruitment in Ter119− cells, these promoters clearly showed different requirements for SCL DNA-binding activity in material isolated from wild-type versus mutant mice (Figure 6). In sclRER/RER cells, we observed a complete loss of SCL binding on the pb4.2 promoter. In contrast, there were only statistically insignificant or partial decreases in SCL recruitment on eklf and gpa promoters, respectively. These differences in SCL binding correlated with the extent in PolII recruitment, levels of acetylation of histone H3 (a mark of increased chromatin accessibility), the extent of reduction in gene expression (fold changes in brackets; Figure 6), and the binding of 2 members of the pentameric complex, E2A and LDB1. Of note, we observed unaffected weak binding of GATA1 on all 3 loci (data not shown).
Transcription factor binding and chromatin structure at the pb4.2, eklf, and gpa promoters in Ter119− cells. (Top) Schematic representation of the promoter (P) and distant upstream (N) regions in pb4.2, eklf, and gpa loci that were tested for transcription factor binding and chromatin modification. Numbers indicate the position of the cis-elements relative to the transcription start site (accession numbers are pb4.2, AF019074; eklf, AF033102; gpa, M26385). ■ indicates Gata sites; ●, E-box motifs and nucleotide sequence. In brackets under each locus is indicated the fold reduction in gene expression in the mutant cells compared with wild-type cells, as shown in Figure 5. (Below) Real-time PCR analysis of chromatin derived from Ter119− population purified from sclWt/Wt and sclRER/RER fetal liver cultured cells and immunoprecipitated using the antibodies indicated on the left. A no antibody control is also shown. The y-axis represents enrichment over input DNA, normalized to a control sequence in the gapdh gene. On the x-axis are shown the sequences that were analyzed; promoter areas (P) and negative points designed 5′ of the promoter regions (N). Error bars correspond to plus or minus 1 SD from at least 3 independent experiments; *P < .01.
Transcription factor binding and chromatin structure at the pb4.2, eklf, and gpa promoters in Ter119− cells. (Top) Schematic representation of the promoter (P) and distant upstream (N) regions in pb4.2, eklf, and gpa loci that were tested for transcription factor binding and chromatin modification. Numbers indicate the position of the cis-elements relative to the transcription start site (accession numbers are pb4.2, AF019074; eklf, AF033102; gpa, M26385). ■ indicates Gata sites; ●, E-box motifs and nucleotide sequence. In brackets under each locus is indicated the fold reduction in gene expression in the mutant cells compared with wild-type cells, as shown in Figure 5. (Below) Real-time PCR analysis of chromatin derived from Ter119− population purified from sclWt/Wt and sclRER/RER fetal liver cultured cells and immunoprecipitated using the antibodies indicated on the left. A no antibody control is also shown. The y-axis represents enrichment over input DNA, normalized to a control sequence in the gapdh gene. On the x-axis are shown the sequences that were analyzed; promoter areas (P) and negative points designed 5′ of the promoter regions (N). Error bars correspond to plus or minus 1 SD from at least 3 independent experiments; *P < .01.
We then repeated the ChIP experiments from material isolated from Ter119+ cells. In these cells however, a decrease in SCL recruitment in mutant sclRER/RER cells could not solely be attributed to the lack of DNA-binding activity but also to decreased SCL levels (Document S1; Figure S4A). In contrast with the observations in Ter119− cells, significant decrease in recruitment of SCL, E2A, LDB-1, and PolII, as well as in histone acetylation, was observed on the promoter region of each locus in sclRER/RER cells (Document S1; Figure S4B).
In summary, in early Ter119− erythroid progenitors, we report a strict dependence on SCL direct DNA-binding activity for the transcriptional regulation of pb4.2 promoter and a requirement for maximal expression of eklf. Importantly, in mature Ter119+ cells, scl expression is subject to an autoregulatory mechanism leading to decreased levels of the SCL protein in sclRER/RER cells.
Discussion
SCL direct DNA-binding activity is dispensable for specification of yolk sac and definitive hematopoiesis
In contrast to conventional scl knockout embryos, the striking observation that the DNA-binding mutant sclRER/RER embryos survive at days E9.5 and 10.5 but die at later stages is the first in vivo demonstration of the differential use of SCL's mechanisms of action during development.
Remarkably, direct DNA-binding activity of SCL is not required for development of the first yolk sac hematopoietic cells because primitive erythroid cells, absent in scl−/− embryos,2 were observed from sclRER/RER yolk sacs. Substantiating these findings, hematopoietic cells develop from sclRER/RER, but not scl-null, ES cell–derived hemangioblasts.
Previous loss-of-function studies have shown that SCL is required to establish definitive hematopoiesis (see “Introduction”). Moreover, analysis of conditional adult scl-null mice shows that SCL is absolutely necessary for the development of erythroid (BFU-E) and megakaryocyte progenitors (see “Introduction”). Our study shows that direct DNA binding is not required for the function of SCL in the establishment of definitive hematopoietic stem/progenitor cells in sclRER/RER mice, because similar numbers of wild-type and mutant hematopoietic colonies were observed in progenitor assays from day E12.5 fetal livers.
SCL DNA-binding mutation impairs erythroid maturation throughout development and at the adult stage
Anemia was documented in sclRER/RER embryos throughout development as well as in the surviving adult mutant mice. This was due to defective terminal erythroid maturation rather than to defective erythroid progenitor specification. These defects very probably resulted in the embryonic death observed from day E14.5.
As observed in days E8.5 and E9.5 embryos, SCL DNA-binding–dependent activity is required for terminal primitive erythroid cell maturation (this study and Juarez et al42 ).
At day E12.5, the maturation block in definitive erythropoiesis resulted in decreased numbers of mature CD71+/Ter119+ red cells and paucity of terminally differentiated erythrocytes. This therefore establishes that SCL's critical functions in fetal red cell maturation are mediated through direct DNA binding. No evidence of increased apoptosis or proliferation defects was observed that could have explained the decrease in erythroid precursors and the defect in maturation. Altogether, our data are in favor of a role for SCL in terminal maturation and hemoglobinization rather than control of survival or proliferation. Interestingly, expansion of the myeloid populations (CD61+, Mac-1+, Gr-1+; Figure 3C) suggests a possible role for SCL in repressing the macrophage/granulocyte program as myeloid precursors differentiate.
In our survey of gene expression, the gene most affected by the SCL mutation in fetal liver erythroid cells was that coding for protein band 4.2 (PB4.2). PB4.2 is a main component of the red blood cell membrane. The pb4.2 knockout mice have mild anemia.43 The decrease in pb4.2 mRNA levels observed in SCL mutant erythroid cells is therefore likely to contribute to the anemia. Decreased levels of the erythroid-specific transcription factor eklf and, possibly, of its target genes might have also contributed to the phenotype of the sclRER/RER mutant embryos.44 Importantly, the requirement of SCL direct DNA-binding for activation of its own expression in maturing erythroid cells confirms the autoregulatory mechanism previously hypothesized,38 and it suggests that one of the key functions of SCL DNA-binding activity might be to maintain its own expression in the terminal stages of red cell maturation. Finally, the erythroid phenotype may also result from the perturbation of expression of genes involved in other pathways important in erythropoiesis that remain to be characterized.
The long-term consequences of SCL deletion on adult erythropoiesis were recently analyzed in a conditional knockout model.16 Anemia, splenomegaly, and maturation defects in erythroid cell populations were reported. The phenotypic similarities between this model and ours suggest that most of SCL's functions in adult erythroid differentiation are mediated by its direct DNA-binding activity. Interestingly however, an expansion of the MEP compartment was documented in the scl-null model but was not observed in the sclRER/RER mice suggestive of an SCL DNA-binding independent function in this compartment.
Maturation versus specification
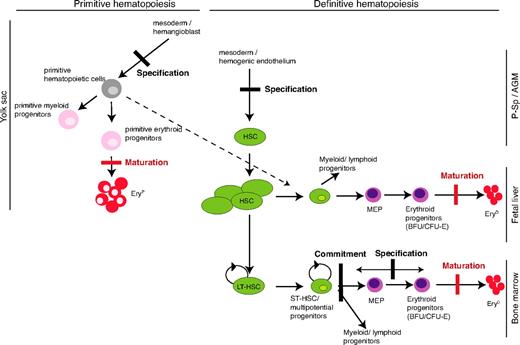
On the basis of the discussion above, we propose that SCL's critical functions in specification decisions are direct DNA binding–independent, whereas, for terminal maturation, its activity is critically mediated by direct DNA binding (Figure 7).
A model for SCL's mechanisms of action in developmental hematopoiesis. Comparative analyses of the hematopoietic defects observed in SCL−/− in vivo (conventional and conditional knockout, chimeras) and in vitro models (see “Introduction” for references) and in our DNA-binding mutant model allow to depict the steps in primitive and definitive hematopoiesis that do require SCL DNA-binding activity and, consequently, those that do not. The red thick bars represent the blocks in erythroid maturation that are documented in this report. The black thick bars represent blocks in specification or commitment that were reported in models in which SCL expression was totally abolished but not observed in this study. The dashed line shows the proposed contribution of primitive hematopoietic progenitors to definitive hematopoiesis.35
A model for SCL's mechanisms of action in developmental hematopoiesis. Comparative analyses of the hematopoietic defects observed in SCL−/− in vivo (conventional and conditional knockout, chimeras) and in vitro models (see “Introduction” for references) and in our DNA-binding mutant model allow to depict the steps in primitive and definitive hematopoiesis that do require SCL DNA-binding activity and, consequently, those that do not. The red thick bars represent the blocks in erythroid maturation that are documented in this report. The black thick bars represent blocks in specification or commitment that were reported in models in which SCL expression was totally abolished but not observed in this study. The dashed line shows the proposed contribution of primitive hematopoietic progenitors to definitive hematopoiesis.35
Why do different mechanisms of action of SCL predominate in different cellular contexts? The answer is likely to lie in the biologic function of the cells and the nature of the target genes/protein complexes.
We postulate that high DNA affinity mediated through direct binding ensures robust protein/DNA interactions in differentiating cells, probably to be required for tight regulation of expression of key target genes throughout maturation. This is exemplified by our data on the regulation of expression of pb4.2, important for terminally differentiating red cells. We show a strict dependence on SCL DNA-binding activity for its full expression, supported by the fact that 2 E-box motifs in pb4.2 regulatory sequences are functionally important.39 At eklf and gpa promoters, appreciable SCL binding is detected in sclRER/RER erythroid cells, suggesting that direct and indirect recruitment is also likely to occur in a dynamic process. Alternatively, it is possible that expression of only very few key genes requires SCL direct DNA-binding activity, such as pb4.2 in Ter119− cells and scl itself in Ter119+ cells. The positive autoregulation of scl expression, by engaging the cells in a forward momentum,45 could therefore be one of the key events driving terminal erythroid differentiation.
In cell fate decisions, at least 2 nonexclusive main mechanisms can account for SCL DNA-binding independent activities: (1) indirect recruitment to DNA through protein-protein interactions or (2) sequestration “off DNA” of other developmental regulators that mitigate against hematopoietic cell specification. In this context, note that functionally important residues are present at critical positions in loop domains of both SCL46 and the sequestering non–DNA-binding HLH-only protein, Id1.47 Regulation of expression of c-kit, a crucial hematopoietic cytokine, supports our first hypothesis. c-kit was previously shown to be regulated by SCL indirect DNA-binding activity,48 and, in agreement with this, its expression was not affected in sclRER/RER Ter119− erythroid progenitors. Of course, one cannot exclude the possibility that direct DNA binding might also occur in specification decisions but that this activity is not critical for hematopoietic development.
In early hematopoietic development, the recent demonstration that SCL-LMO2 interaction is required46,49 suggests that SCL is likely to nucleate the same multiprotein core complex in specification processes as it does in terminally differentiating erythroid cells.50 Whether this complex is recruited to DNA through other DNA-binding transcription factor(s) to regulate gene expression or fulfils its function off DNA by sequestering other nuclear regulators or both remains to be determined.
Concluding remarks
Other transcriptional regulators exert DNA binding–independent mechanisms of action. However, in contrast to what we describe here for SCL, these mechanisms are either associated to direct DNA-binding functions (as for GATA1 and PU.1 mutually antagonistic activities in lineage decisions51,52 ) or remain to be shown in a physiologic, loss-of-function model (as for the bHLH protein dHAND53 ). To our knowledge, the glucocorticoid receptor (GR) is the only other example in which specific abolishment of DNA-binding function in an in vivo setting has unveiled distinct mechanisms of action. However, the GR DNA binding–independent activities are not required for lineage specification, but participate in the control of physiologic pathways and in tissue organization.54
Thus, to date, SCL is the first example of a transcription factor exhibiting differential mechanisms of action in distinct developmental/cellular contexts, relying on DNA binding–independent functions in lineage specification. We propose that functional assessment of DNA binding, or other essential activities, of key proteins during development might show that differential use of functional domains is a property common to many transcriptional regulators.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Robert Sumner (Biomedical Services, University of Oxford) for mouse blastocyst injection and chimera production and Ann Atzberger (MRC Molecular Hematology Unit) for FACS sorting. We also thank Boris Guyot, Eduardo Anguita, and Douglas Vernimmen for advice on real-time PCR and ChIP assays; Jackie Sharpe and Jackie Sloane-Stanley for invaluable help; and Lauren Aronson for technical assistance. We thank Doug Higgs and Bill Wood for critical reading of the manuscript, and Roger Patient and Tariq Enver for helpful discussions.
This work was supported by the MRC and a grant from the Leukaemia Research Fund (M.T.K.). M.T.K. was the recipient of a Karim Rida Said foundation scholarship in the early stages of this work.
Authorship
Contribution: M.T.K. designed and performed research, analyzed the data, and wrote the paper; H.C. performed experiments; P.V. designed research and wrote the paper; and C.P. designed research, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Catherine Porcher, MRC Molecular Haematology Unit, Weatherall Institute of Molecular Medicine, John Radcliffe Hospital, Oxford OX3 9DS, United Kingdom; e-mail: catherine.porcher@imm.ox.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal