Abstract
Commitment of hematopoietic progenitor cells to B-lymphoid cell fate has been suggested to coincide with the development of PAX5-expressing B220+CD19+ pro–B cells. We have used a transgenic reporter mouse, expressing human CD25 under the control of the B-lineage–restricted Igll1 (λ5) promoter to investigate the lineage potential of early progenitor cells in the bone marrow. This strategy allowed us to identify a reporter expressing LIN−B220−CD19−CD127+FLT3+SCA1lowKITlow population that displays a lack of myeloid and a 90% reduction in in vitro T-cell potential compared with its reporter-negative counterpart. Gene expression analysis demonstrated that these lineage-restricted cells express B-lineage–associated genes to levels comparable with that observed in pro–B cells. These data suggest that B-lineage commitment can occur before the expression of B220 and CD19.
Introduction
The development of mature blood cells can be viewed as a process where multipotent progenitors gradually lose lineage potential as they progress through maturation to become functional hematopoietic cells. One of the first steps in the progression of lymphoid differentiation involves the development of lineage marker (LIN)–negative SCA1+KIT+CD34+FLT3+ lymphoid primed multipotent progenitor (LMPP) cells that display a reduced megakaryocyte and erythroid lineage potential but a preserved ability to differentiate into myeloid and lymphoid cells.1,2 A subfraction of the LMPPs express a number of lymphoid-associated genes, including Rag1 and Rag2 as well as Dntt (TdT) and CD127 (Il7rα),2 and although the expression of a Rag1 or Rag2 reporter gene in early progenitors does not mark complete loss of myeloid lineage potential,3,4 such cells are primed to lymphoid development and further differentiation into common lymphoid progenitors (CLPs).4 The surface phenotype of the CLP largely resembles that of the LMPP,1 however, with lower expression of SCA1 and KIT and robust expression of CD127.5 CLPs have been shown to support B-, T-, and NK-cell development, although their myeloid potential is largely reduced compared with more immature cells.5-7 The regulation of B-lymphoid fate has been suggested to depend on an interplay between transcription factors including PU.1 (Sfpi1), early B-cell factor 1 (EBF1), and E2A,6,8,9 with extracellular signals such as those mediated by FLT3-ligand (FL), interleukin 7 (IL7),10 and the ligands of the NOTCH signaling pathway.11,12 However, even if these factors are able to prime cells for or against development along the B-lymphoid pathway, the definitive lock of lineage potential has been suggested to depend on the expression of the transcription factor PAX5. Loss-of-function models have shown that in the absence of PAX5, development along the B-lymphoid pathway is initiated, but although the cells express a large number of B-lineage genes, such cells retain the ability to develop into alternative lymphoid or myeloid pathways.13-15 Hence B-cell commitment may occur at the B220+CD19+ stage, where robust expression of Pax5 can be detected.3,16 However, the finding that Pax5 is a direct target for EBF1,17 and vice versa,18 complicates the understanding of commitment in normal B-cell development. In addition, the fact that Ebf1 is expressed already at the CLP stage19,20 suggests that the issue of B-lineage commitment needs further investigations.
To investigate the temporal regulation of B-cell development and the link between functional activation of early B-lineage genes and lineage commitment, we have used a transgenic mouse where the expression of a human CD25 (hCD25) reporter gene is controlled by the EBF1-regulated Igll1 (λ5) promoter.21,22 Expression of this reporter could be detected on a fraction of the LIN−B220−CD19−CD127+FLT3+SCA1lowKITlow cells, commonly referred to as CLPs, with a highly reduced ability to develop into T cells and an inability to develop into myeloid cells. This was accompanied by increased expression of B-lymphoid–associated genes including Pax5, leading us to suggest that B-lineage commitment can occur before surface expression of B220 and CD19.
Methods
Quantitative RT-PCR
Quantitative reverse transcriptase–polymerase chain reaction (Q-RT-PCR) analysis of sorted cells was performed as previously described.1,2 Assays-on-Demand probes (Applied Biosystems, Foster City, CA) used were as follows: Cd19; Mm00515420_m1, CD79a (Mb1); Mm00432423_m1, Pax5; Mm00435501_m1, Hprt; Mm00446968_m1, Ebf1; and Mm00432948_m1. Sequences for the Assay-by-Design probe used for λ5 (Igll1) detection were as follows: fw, 5′-GGAACAACAGGCCTAGCTATGG; rev, 5′-CTCCCCGTGGGATGATCTG; and probe, 5′-CCGGCAGCTCCTGTTC. All experiments were performed in triplicates and differences in cDNA input were compensated by normalizing against Hprt expression levels.
Fluorescence-activated cell sorting of CLPs and B-lineage cells
Bone marrow (BM) and spleen cells were harvested from 10- to 15-week-old C57Bl/6 wild-type or heterozygous hCD25 (λ5) transgenic mice21 (on C57Bl/6 background) and single-cell suspensions were prepared.
For CLP isolation, BM cells were subjected to magnetic-activated cell sorting (MACS) column (Miltenyi Biotec, Bergisch Gladbach, Germany) enrichment of KIT+ cells using anti-CD117 immunomagnetic beads (Miltenyi Biotec). KIT+ cells were subsequently stained with Fc-block (CD16/CD32, 2.4G2) followed by CD19 (6D5) fluorescein isothiocyanate (FITC), FLT3 (A2F10) phycoerythrin (PE), CD11b (M1/70) PE-cyanin-5 (PE-Cy5), GR1 (RB6-8C5) PE-Cy5, TER119 (Ter119) PE-Cy5, CD3 (145-2c11) PE-Cy5, B220 (RA3-6B2) PE-Cy7, hCD25 (BC96) allophycocyanin (APC), KIT (2B8) APC-Alexa750, SCA1 (D7) PacificBlue, IL7r (A7R34) biotin (visualized with streptavidin QDot605 or streptavidin QDot655), and propidium iodide (PI).
For isolation of B-lineage cells, BM/spleen cells were stained with purified TER119 (Ter119), GR1 (RB6-8C5), CD11c (M1/70) CD4 (GK1.5), and CD8a (53-6.7) (visualized with goat antirat QDot605). Cells were subsequently stained with Fc-block (CD16/CD32, 2.4G2) followed by IgM (R6-60.2) FITC, CD43 (S7) PE, B220 (RA3-6B2) APC, CD19 (1D3) PE-Cy7, AA4.1 (AA4.1) biotin (visualized with streptavidin QDot655), and PI.
Analysis and cell sorting was done on a BD FACSAria (BD Biosciences, San Jose, CA).
Affymetrix gene expression and data analysis
RNA was extracted from purified adult BM subsets as described for Q-RT-PCR. RNA was labeled and amplified according to the Affymetrix GeneChip Expression Analysis Technical Manual.23 Chips were scanned using a GeneChip Scanner 3000 (Affymetrix, Santa Clara, CA) and scaled to a median intensity of 100. RNA from cells sorted on separate occasions was separately hybridized for each investigated population. Probe level expression values were calculated using robust multichip average (RMA) and further analysis was done using dChip (Dana-Farber Cancer Institute and Harvard School of Pubic Health, http://www.dchip.org). Array data used are accessible through the Gene Expression Omnibus (GEO; GSE7302 and GSE11110).24
In vitro evaluation of B- and T-lymphoid potential by OP9/OP9DL1 coculture
For evaluating B-cell and T-cell potential, cells were clone sorted (using a FACSAria) directly into 96-well plates containing preplated (2000 cells/well) OP9 (supplemented with 10 ng/mL FLT3L and IL7) and OP9DL1 (supplemented with 10 ng/mL FLT3L and IL7 or only 10 ng/mL FLT3L) stroma layers, respectively. Cultures were substituted with new cytokines every 7 days. OptiMEM supplemented with 10% fetal calf serum, 50 μg/mL gentamicin, and 50 μM β-mercaptoethanol was used for maintaining the OP9/OP9DL1 stroma cell lines as well as for cocultures.
Cocultures were evaluated by flow cytometry staining with CD19 (1D3) PE, B220 (RA3-6B2) APC, and PI for OP9 cocultures and with CD25 (7D4) FITC, CD19 (1D3) PE, CD90.2/Thy1.2 (53-2.1) APC, and PI for OP9DL1 cocultures.
OP9 and OP9DL1 cocultures of CLPs were in initial experiments evaluated at day 14 and day 21, respectively, and in later experiments at days 7 to 8 and days 14 to 15, respectively, with indistinguishable results. Samples were analyzed on a BD FACSCalibur (BD Biosciences).
In vitro evaluation of myeloid potential
Myeloid potential was evaluated as previously described1,2 but with a modified cytokine combination. In brief, 150 cells were sorted in 3 mL medium (OptiMEM supplemented with 10% fetal calf serum, 50 μg/mL gentamicin, 50 μM b-mercaptoethanol, 25 ng/mL KIT ligand [KL], 25 ng/mL fms-like tyrosine kinase 3 ligand [FLT3L], 5 ng/mL interleukin 3 [IL-3], 5 ng/mL colony-stimulating factor 1 [CSF1], 5 ng/mL colony stimulating factor 2 [CSF2], and 10 ng/mL colony-stimulating factor 3 [CSF3]) and 20 μL was plated into each well of 2 60-well plates (Nunc Minitrays; Rochester, NY). Wells were scored after 6 days, with an inverted light microscope, for clonal growth and size of the clones. At least 600 cells per population were plated in 2 independent experiments and at least 16 clones picked randomly from each population for evaluation by RT-PCR. Nested RT-PCRs were essentially done as described for single-cell RT-PCRs (for primers used, see “Gene expression analysis of single cells by multiplex RT-PCR” and Mansson et al2 ). For morphologic evaluation, May-Grünwald Giemsa–stained cytospin preparations were made from 3 to 4 pooled clones to make 16 slides per population from 2 independent experiments.
Gene expression analysis of single cells by multiplex RT-PCR
Multiplex single-cell RT-PCR analysis was performed as previously described.1,2,25 Primers used were as follows: Hprt: (1) 5′-GGGGGCTATAAGTTCTTTGC; (2) 5′-GTTCTTTGCTGACCTGCTGG; (3) 5′-TGGGGCTGTACTGCTTAACC; (4) 5′-TCCAACACTTCGAGAGGTCC. Rag1: (1) 5′-CCAAGCTGCAGACATTCTAGCACTC; (2) 5′-CAGACATTCTAGCACTCTGG; (3) 5′-GCTTGACTTCCCATCAGCATGGA; (4) 5′-CAACATCTGCCTTCACGTCGATCC. Il7r: (1) 5′-CTCTCTCAGAATGATGGC; (2) 5′-TGACTCAGACATCAACACAGC; (3) 5′-AAGATACATGCGTCCAGTTGC; (4) 5′-GTCGTAGTTTTCTCTGTGG. Pax5: (1) 5′-CTACAGGCTCCGTGACGCAG; (2) 5′-ATGGCCACTCACTTCCGGGC; (3) 5′-GTCATCCAGGCCTCCAGCCA; (4) 5′-TCTCGGCCTGTGACAATAGG. IgII1 (λ5): (1) 5′-AGTTCTCCTCCTGCTGCTGC; (2) 5′-GGGTCTAGTGGATGGTGTCC; (3) 5′-CAAAACTGGGGCTTAGATGG; (4) 5′-CCCACCACCAAAGACATACC. Ebf1: (1) 5′-CCCTCTTATCTGGAACATGC; (2) 5′-CTACTCCCTGTATCAAAGCC; (3) 5′-TGTACGACAGTGTGACTTCC; (4) 5′-TAAGGATCACTTCCTTTGGC. Mb-1: (1) 5′-CCTCCTCTTCTTGTCATACG; (2) 5′-AAACAATGGCAGGAACCC; (3) 5′-TGATGATGCGGTTCTTGG; (4) 5′-GAACAGTCATCAAGGTTCAGG. Primers 1 plus 4 and 2 plus 3 are outer and inner primer pairs, respectively.
Institutional review board approval
Animal procedures were performed with consent from the local ethics committee at Lund Univeristy (Lund, Sweden) and Linköping University (Linköping, Sweden).
Results
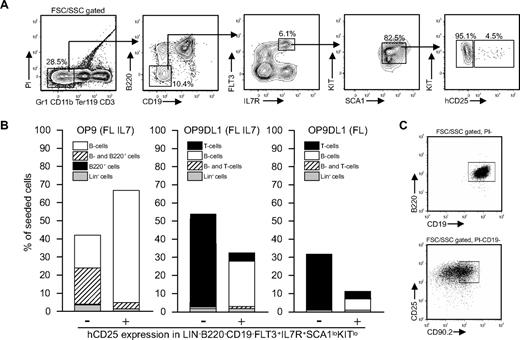
Although CLPs have been reported to express significant amounts of B-lineage–associated transcripts,3,26 it has not been reported that this is associated with B-cell commitment. Rather, such transcripts have been assigned to lineage priming, associated with the activation of chromatin, making progenitor cells permissive for development along the B-lymphoid pathway. However, single-cell PCR has suggested that approximately one-third of CLPs coexpress the B-lineage genes Igll1 and Pax5 without the expression of the T-lineage genes Pta or CD3e,26 a finding that might suggest that the CD19−B220− CLP compartment contains a B cell–committed subpopulation. To investigate if the early lineage marker–negative compartment contains committed progenitors, we adopted an approach using an Igll1 (λ5) promoter–controlled human CD25 reporter gene (hCD25)21 to trace cells with an active B-lineage–associated transcription program. We speculated that the use of this reporter mouse should allow us to identify cells that have initiated the expression of B-lineage–restricted genes, possibly reflecting lineage commitment. The fact that the Igll1 promoter is a direct target for EBF122 and that functional expression of this transcription factor is crucial for the expression of the Igll127 gene make it useful as a reporter for functional EBF1 activity. As expected, the reporter gene was active in a majority of the B220+CD19+ bone marrow cells (data not shown and Hu et al25 ), but we could also detect a minor fraction (∼ 5%) of the LIN−B220−CD19−CD127+FLT3+SCA1lowKITlow cells (often referred to as CLPs) that expressed hCD25 on the surface (Figure 1A). This would correspond to a population size comparable with that of the CD34−FLT3− LSK compartment1 (in the range of 1/20 000 bone marrow cells). To verify that the reporter gene expression overlapped with that of the endogenous Igll1 gene, we performed Q-PCR analysis of sorted CD25+ and CD25− cells, revealing that all the detectable Igll1 expression was found in the CD25+ progenitor fraction, supporting the notion that the reporter gene provides correct information about expression of the endogenous gene (Figure 3C).
The B220−CD19− compartment contains lineage-restricted B-cell progenitors marked by transgenic expression of hCD25. (A) Representative FACS plots and purification scheme used to identify hCD25+ and hCD25− LIN−B220−CD19−CD127+FLT3+SCA1lowKITlow fractions. Numbers indicate percentage of gated cells. (B) The frequency (%) of cells composing colonies generated by coculture of single cells on OP9/OP9DL1 cells in the presence of indicated cytokines. The data are collected from 2 or more independent experiments. (C) FACS plots of the output from representative clones generated on OP9 (top) and OP9DL1 (bottom).
The B220−CD19− compartment contains lineage-restricted B-cell progenitors marked by transgenic expression of hCD25. (A) Representative FACS plots and purification scheme used to identify hCD25+ and hCD25− LIN−B220−CD19−CD127+FLT3+SCA1lowKITlow fractions. Numbers indicate percentage of gated cells. (B) The frequency (%) of cells composing colonies generated by coculture of single cells on OP9/OP9DL1 cells in the presence of indicated cytokines. The data are collected from 2 or more independent experiments. (C) FACS plots of the output from representative clones generated on OP9 (top) and OP9DL1 (bottom).
Knowing that the LIN−B220−CD19−CD127+FLT3+SCA1lowKITlow fraction contains a subpopulation of cells with a functional transcription factor network that allows for the activation of the Igll1 gene, we wanted to investigate whether the hCD25+ progenitors were functionally different from the hCD25− LIN−B220−CD19−CD127+FLT3+SCA1lowKITlow cells. To gain the highest possible resolution and study lineage potential at a single-cell level, we sorted hCD25+ or hCD25− LIN−B220−CD19−CD127+FLT3+SCA1lowKITlow single cells and cultured them on OP9 stroma cells (Figure 1B). Of 294 hCD25+ cells plated, 181 (62%) developed into CD19+B220+ B-lineage progenitors (B cells, Figure 1B), whereas 3% of the wells contained a portion of cells with only B220 and no CD19 expression, indicating an incomplete differentiation process. Plating of hCD25− cells revealed that 18% of the 180 analyzed wells contained only B220+CD19+ cells, whereas 20% contained a portion of cells lacking the expression of CD19, indicating that these cells develop with a slower kinetics than the hCD25+ cells. To investigate this further, we seeded hCD25− or hCD25+ LIN−B220−CD19−CD127+FLT3+SCA1lowKITlow cells on OP9 feeders and investigated the cellular output after 3, 4, 5, 6, and 10 days after initiation of the cultures (Figure S1). At day 3, the cultures initiated with hCD25+ cells contained more than 95% CD19+ cells, whereas the cultures from the reporter-negative cells contained mainly CD19− cells at this time after seeding. The overall cell numbers were modestly different, but because the fraction of CD19+ cells was approximately 5 times higher in cells from the hCD25+ cells, these data support the idea that hCD25+ cells are more potent as short term B-cell progenitors. This difference was somewhat reduced at later time points after seeding, and after 10 days of differentiation the contents of the cultures were comparable although the number of cells generated from hCD25− cells were higher (Figure S1). We also investigated the presence of hCD25+ cells in these cultures, revealing that a low number of B220− hCD25+ cells were generated from the hCD25− cells and the LMPPs (Figure S2). This rapid kinetics makes in vivo analysis complicated due to the short time given for expansion and the short time between radiation and analysis. However, to ensure that the hCD25+ progenitors were able to give rise to CD19+ cells in vivo, we transplanted 1000 hCD25+ or hCD25− progenitors into irradiated mice (800 rad). Analysis of spleen and bone marrow suggested that both populations gave rise to a comparable amount of CD19+ cells 7 days after transplantation (Figure S3). The apparent lack of functional differences in vivo could reflect the in vitro finding that the difference in B-cell output is less apparent already after 6 days. We were also able to detect a low amount of lineage-negative cells selectively in the spleens of mice that received a transplant of hCD25− CLPs, indicating that residual progenitors were present at the time of analysis. However, we have been unable to investigate in vivo reconstitution at earlier time points, and we are therefore limited to the conclusion that the reporter-positive cells have in vivo B-cell potential. This suggests that the LIN−B220−CD19−CD127+FLT3+SCA1lowKITlow (CLP) progenitor population contains a subpopulation of cells prone to development into B-lineage cells in vitro.
To investigate if this increased B-cell potential was associated with any changes in T-cell potentials, we sorted single cells on OP9DL1 cells known to be highly efficient to stimulate development of T-lineage cells (Figure 1B). Thirty-two percent of the hCD25+ cells generated clones under these conditions, but these were composed mainly of B220+CD19+ cells (26%), and only 4% of the 192 plated cells generated T-cell progenitors (CD25+CD90.2+; Figure 1B). In contrast 51% of the 192 seeded hCD25− cells gave rise to colonies composed of CD25+CD90.2+ cells, and only 1% of the cells gave rise to B220+CD19+ cells. The protocol used for OP9DL1 cocultures includes the addition of IL7, known to be a highly potent stimulator of B-lineage development. Thus, to investigate the role of IL7 in this apparent loss of T-lineage potential in hCD25+ LIN−B220−CD19−CD127+FLT3+SCA1lowKITlow cells, we incubated the OP9DL1 cocultures with FL alone (Figure 1B). Under these conditions, the cloning frequency dropped so that only 11% of the 213 plated hCD25+ cells developed into detectable clones. Among these, 7% were composed of B-lineage and 3.5% of T-lineage cells. Of the 288 plated hCD25− cells, 31% developed into T-lineage cells, whereas only one clone contained B-lineage cells. These data suggest that expression of the Igll1 promoter–controlled reporter is linked to a reduced ability of CD19−B220− cells to develop into T-lymphoid cells, supporting that B- versus T-lineage restriction may occur already at this lineage marker–negative stage.
Expression of hCD25 on LIN−B220−CD19−CD127+FLT3+SCA1lowKITlow cells marks a loss of residual myeloid potential in early progenitors
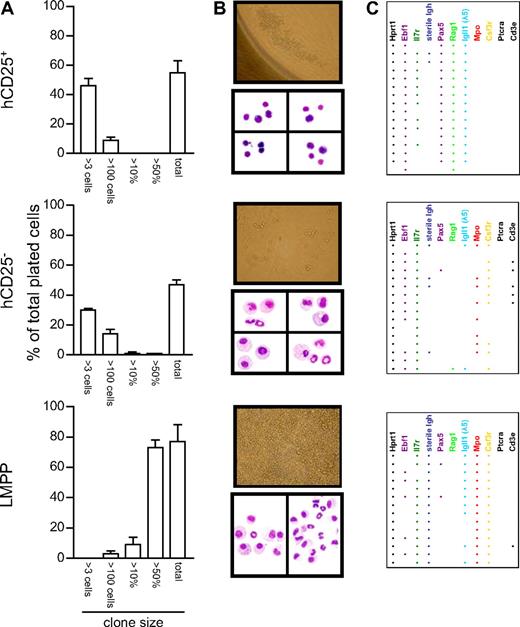
Although the previously defined CLPs, which functionally have a pronounced preference to develop into lymphoid cells,5,7 evidence suggesting that these cells maintain a myeloid potential3,28 (which may be lost as late as after the B- versus T-lineage choice2 ) has been presented. Therefore, to investigate whether we could reveal a myeloid potential in LIN−B220−CD19−CD127+FLT3+SCA1lowKITlow compartment of cells, we plated hCD25+ and hCD25− LIN−B220−CD19−CD127+FLT3+SCA1lowKITlow cells into 60-well plates and incubated them for 6 days in a cytokine cocktail (KL, FL, IL3, CSF1, CSF2, and CSF3) to promote the development of myeloid cells. As a control population, we sorted LMPPs and cultured them under the same conditions for 6 days. Analysis of the colonies generated suggested that although the general cloning frequency was comparable among the hCD25− and hCD25+ populations, the hCD25− cells generated a few large clones of cells resembling those generated by the LMPPs (Figure 2A). An even more striking difference was seen in the distribution of the cells in the wells and in the morphology of the cells. The clones generated from the hCD25+ cells were small, focused at a small area of the well, and the cells were rounded and nonadherent (Figure 2B). In contrast, a majority of the hCD25− cells gave rise to colonies composed of a mix of cells that were spread over the well, attached to the plastic, and also in some cases appeared to have pseudopodias (Figure 2B). To investigate this further, we randomly pooled the content of 3 to 4 wells to perform cytospins and May-Grünwald Giemsa stainings. Investigating the morphology of the cells revealed that all 16 slides obtained from 2 independent experiments using hCD25+ progenitors contained only rounded cells with a small cytoplasm, indicative that these cells are lymphoid related (Figure 2B). hCD25− cells did, however, generate a mix of cells on all of the 16 slides investigated. Among these were granular cells with large cytoplasm, a morphology clearly indicative of myeloid cells (Figure 2B). To obtain molecular support for the morphologic analysis, we picked cells from at least 16 randomly chosen clones and performed nested RT-PCR in search for myeloid and lymphoid transcripts in the generated cells (Figure 2C). Using cells generated from hCD25+ LIN−B220−CD19−CD127+FLT3+SCA1lowKITlow progenitors, 15 of 16 samples contained detectable amounts of both Pax5 and Igll1 transcripts, whereas none of the samples contained either Mpo or Csf3r message normally found in myeloid cells. This was in contrast to the cells obtained from the LIN−B220−CD19−CD127+FLT3+SCA1lowKITlow hCD25− progenitors, because 12 of the 16 samples analyzed contained transcripts encoding either Mpo, Csf3r, or both. Igll1 expression could be detected in 2 of the samples and Pax5 in 1 sample. We were also able to detect CD3ϵ expression, indicative of T-lineage cells, in 5 of these samples. As expected LMPPs gave rise mainly to myeloid cells and all 16 samples contained Mpo and Csf3r message, although lymphoid transcripts could be detected in some samples. These data indicate that although the hCD25− LIN−B220−CD19−CD127+FLT3+SCA1lowKITlow cells had lost most of their potential to generate large number of myeloid cells, the conditions we provided allowed them to reveal a residual myeloid potential that was lost in the hCD25+ cells. Thus, hCD25+ LIN−B220−CD19−CD127+FLT3+SCA1lowKITlow cells display a dramatically reduced ability to adopt other cell fates than B-lineage even under highly permissive conditions, supporting the idea that B-lineage commitment may occur before the expression of B220 and CD19.
Activation of the Igll1 promoter is associated with a reduced myeloid potential in progenitor cells. (A) The cloning frequency and the size of the colonies obtained after culture of single cells in a mix of myeloid cytokines. Error bars represent SEM. (B) Colonies detected (50× magnification) and the morphology of the cells generated during in vitro incubation with cytokines as visualized by May-Grünwald Giemsa staining (MGG; 500× magnification). Cells displayed as one picture are collected from the same slide, but the images of individual cells have been moved to fit the picture format. Images of tissue culture plate wells were taken with an Olympus IX70 microscope (with a LCPlanF1 20×/0.40 Ph1 objective; Olympus Optical, Tokyo, Japan) using a Nikon E4500 camera (Nikon Nordic AB, Solna, Sweden). Images of MGG-stained slides were acquired with an Olympus BX51 microscope (with a UPlanF1 100×/0.30 oil objective; Olympus Optical) using an Olympus DP70 camera and acquisition software (DP Controller 1.1.1.65; Olympus, Tokyo, Japan). (C) The number of PCR-positive samples generated after analysis of 16 randomly chosen clones by multiplex RT-PCR. Each horizontal line of dots indicates the gene expression pattern observed in a single investigated clone. Data presented are collected from 2 independent experiments.
Activation of the Igll1 promoter is associated with a reduced myeloid potential in progenitor cells. (A) The cloning frequency and the size of the colonies obtained after culture of single cells in a mix of myeloid cytokines. Error bars represent SEM. (B) Colonies detected (50× magnification) and the morphology of the cells generated during in vitro incubation with cytokines as visualized by May-Grünwald Giemsa staining (MGG; 500× magnification). Cells displayed as one picture are collected from the same slide, but the images of individual cells have been moved to fit the picture format. Images of tissue culture plate wells were taken with an Olympus IX70 microscope (with a LCPlanF1 20×/0.40 Ph1 objective; Olympus Optical, Tokyo, Japan) using a Nikon E4500 camera (Nikon Nordic AB, Solna, Sweden). Images of MGG-stained slides were acquired with an Olympus BX51 microscope (with a UPlanF1 100×/0.30 oil objective; Olympus Optical) using an Olympus DP70 camera and acquisition software (DP Controller 1.1.1.65; Olympus, Tokyo, Japan). (C) The number of PCR-positive samples generated after analysis of 16 randomly chosen clones by multiplex RT-PCR. Each horizontal line of dots indicates the gene expression pattern observed in a single investigated clone. Data presented are collected from 2 independent experiments.
Increased level of Ebf1 expression is associated with the activation of a B-lymphoid regulatory program in CD19−B220− progenitor cells
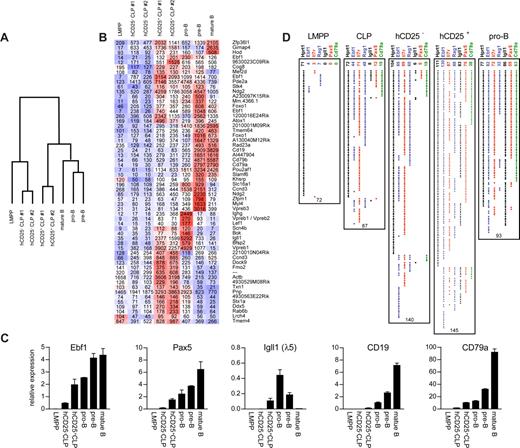
Although our data strongly support the idea that the expression of the Igll1-controlled hCD25 reporter in early progenitors marks B-lineage commitment, we wanted to investigate molecular changes associated with this event. To this end, we performed microarray experiments to analyze gene expression patterns in hCD25+ and hCD25− LIN−B220−CD19−CD127+FLT3+SCA1lowKITlow cells. To put the data into context, expression patterns were compared with those in LMPPs, CD19+AA4.1+CD43low pro-B cells,29 CD19+B220+CD43−IgM− pre-B cells, and IgM+CD19+ mature spleen B cells. This revealed a significant difference in overall expression patterns: the hCD25+ cells clustered with the committed B-lineage cells, whereas the hCD25− cells rather resembled the more immature LMPP cells (Figure 3A). This was a result mainly of a reduction in the expression of a large set of genes, including Notch1, comparably low in the hCD25+ cells (Figure S4). hCD25+ cells also expressed higher levels of Igll1, Vpreb1, Vpreb3, Cd19, Cd79a, Cd79b, and Ebf1 (Figure 3B), all indicative of B-lineage cells. We could also see an increased expression of several B-lineage–associated transcription factors such as Ocab, Lef1, and interestingly Foxo1, possibly linking Ebf1 directly into the network of Fox proteins that play an important role in early B-cell development.30 To verify some of these data, we analyzed the expression of a set of these genes by Q-RT-PCR (Figure 3C) and compared the relative expression levels with those observed in IgM−B220+CD43lowCD19+ pro-B cells,29 IgM−B220+CD43− CD19+ pre-B cells, and IgM+CD19+ splenic B cells. The hCD25+ cells expressed 4 times as much Ebf1 message as the hCD25− cells. This level of Ebf1 expression was comparable with that observed in the pro-B cells and only 2-fold lower than in the pre-B cells. Igll1 transcripts were not detectable in the hCD25− cells, whereas levels comparable with those in the pro-B cell could be found in the hCD25+ cells, providing evidence that the reporter gene expression is perfectly correlated to the expression of the endogenous Igll1 gene. In addition, both Pax5 and Mb1 message levels were increased to levels comparable with that in CD19+B220+ pro-B cells in the hCD25+ cells. These cells also expressed significant amounts of Cd19 mRNA, and although this level was low compared to what was observed in the pro-B cells, it was higher than in the hCD25− cells. LMPPs expressed low or undetectable levels of all the analyzed transcripts (Figure 3C). These data support the idea that the expression of the reporter gene allows us to identify two molecular and functionally distinct LIN−B220−CD19−CD127+FLT3+SCA1lowKITlow cell populations in the mouse bone marrow.
Expression of the Igll1 promoter–controlled hCD25-transgenic marker in LIN− progenitors is associated with induced expression of genes linked to B-cell development. (A) A diagram of the relative relationship between the hCD25+ and hCD25− LIN−B220−CD19−CD127+FLT3+SCA1lowKITlow progenitors to LMPPs and B-lineage cells. The diagram is based on genes differentially expressed between the hCD25+ and hCD25− cells. The complete analysis is presented in Figure S4 (available on the Blood website; see the Supplemental Materials link at the top of the online article). (B) A dChip analysis of hCD25+ and hCD25− LIN−B220−CD19−CD127+FLT3+SCA1lowKITlow cells in the context of LMPPs and other stages of B-cell development. Clustering shows genes being expressed in either pro-B cells or LMPPs (100 + expression units in either) and being up-regulated 1.5-fold from hCD25− to hCD25+. Red represents high; white, intermediate; and blue, low expression of the gene indicated to the far right. Superimposed values show RMA-modeled array expression values. (C) Q-RT-PCR data from CD25+ and CD25− cells as well as control cell populations as indicated (data from one representative experiment). The error bars indicate standard deviation. (D) The collected result of multiplex single-cell PCR on LMPPs, CLPs, pro-B cells, and hCD25+/− LIN−B220−CD19−CD127+FLT3+SCA1lowKITlow cells as indicated. Each horizontal line of dots indicates the gene expression pattern observed in a single investigated cell. The values on top of the data panel indicate the number of positive cells and the values below the panels indicate the total number of cells analyzed. Error bars indicate standard deviation.
Expression of the Igll1 promoter–controlled hCD25-transgenic marker in LIN− progenitors is associated with induced expression of genes linked to B-cell development. (A) A diagram of the relative relationship between the hCD25+ and hCD25− LIN−B220−CD19−CD127+FLT3+SCA1lowKITlow progenitors to LMPPs and B-lineage cells. The diagram is based on genes differentially expressed between the hCD25+ and hCD25− cells. The complete analysis is presented in Figure S4 (available on the Blood website; see the Supplemental Materials link at the top of the online article). (B) A dChip analysis of hCD25+ and hCD25− LIN−B220−CD19−CD127+FLT3+SCA1lowKITlow cells in the context of LMPPs and other stages of B-cell development. Clustering shows genes being expressed in either pro-B cells or LMPPs (100 + expression units in either) and being up-regulated 1.5-fold from hCD25− to hCD25+. Red represents high; white, intermediate; and blue, low expression of the gene indicated to the far right. Superimposed values show RMA-modeled array expression values. (C) Q-RT-PCR data from CD25+ and CD25− cells as well as control cell populations as indicated (data from one representative experiment). The error bars indicate standard deviation. (D) The collected result of multiplex single-cell PCR on LMPPs, CLPs, pro-B cells, and hCD25+/− LIN−B220−CD19−CD127+FLT3+SCA1lowKITlow cells as indicated. Each horizontal line of dots indicates the gene expression pattern observed in a single investigated cell. The values on top of the data panel indicate the number of positive cells and the values below the panels indicate the total number of cells analyzed. Error bars indicate standard deviation.
To investigate the modulation in gene expression patterns at the single-cell level, we sorted hCD25− and hCD25+ LIN−B220−CD19−CD127+FLT3+SCA1lowKITlow cells and analyzed their gene expression patterns by multiplex RT-PCR analysis (Figure 3D). To put the data into context, we also sorted LIN−KIT+SCA1+FLT3+ LMPPs, LIN−KITLowSCA1lowCD127+FLT3+ CLPs, as well as CD19+AA4.1+CD43low pro-B cells. Among the LMPP cells for which we obtained HPRT signal, 4 (6%) contained detectable levels of Ebf1 transcripts, whereas 3 (4%) expressed Il7rα (CD127) transcripts. Seven (10%) expressed Rag1 transcripts, whereas none of the cells contained detectable levels of Pax5, Igll1, or Mb1 (CD79α) mRNA. In the CLP compartment, we were able to detect Rag1 expression in 48 (55%) of the 87 analyzed cells and Ebf1 expression in more than 51% of the analyzed cells, whereas Pax5, Igll1, and Mb1 transcripts were found in 14%, 9%, and 17% of the cells, respectively. Notably, the majority of cells expressed Pax5, Igll1, and Mb1 in a coordinated manner, supporting the idea that the lineage program is initiated already at the level of the currently defined CLP. A similar gene expression pattern was seen upon analysis of hCD25− LIN−B220−CD19−CD127+FLT3+SCA1lowKITlow cells. Of 140 analyzed cells, 85 (61%) expressed Ebf1 message, whereas in 99 cells (71%) we were able to detect Il7rα, and in 60 (43%) we detected Rag1 transcripts. Six cells (4%) expressed Igll1; 5 (4%), Pax5; and 19 (14%), Mb1. In the hCD25+ LIN−B220−CD19−CD127+FLT3+SCA1lowKITlow cells, a similar pattern was seen when analyzing the expression of Il7rα (60%) and Rag1 (61%) in the 145 investigated cells. A minor difference could be observed in the percentage of Ebf1-positive (90%) and Mb1-positive (26%) cells, whereas the number of Igll1 and Pax5-positive cells increased to 57% and 52%, respectively. This was more comparable with the data obtained using the pro-B cells: 88% expressed detectable levels of Ebf1 message, whereas 55 (66%) of 93 contained Pax5 mRNA. Igll1 was detected in 92% of the cells; Mb-1, in 68%; and Rag-1, in 34% of the pro-B cells. These data support the idea that B-cell commitment occurs already in the phenotypically defined CLP compartment and indicate that the expression of Ebf1 is a precommitment event, not directly linked to the expression of target genes or lineage restriction. Furthermore, because the expression level is increased 4-fold in the hCD25+ progenitors compared with the hCD25− fraction, although the number of cells with detectable expression is increased only from 68% to 92%, this taken together suggests an increase of Ebf1 mRNA at the single-cell level. Thus, investigations of transcriptional programs at a single-cell level support the idea that B-lineage fate can be determined before the expression of B220 and CD19.
Discussion
We here report data suggesting that B-lineage commitment of hematopoietic progenitors can occur at an early progenitor stage before the expression of B220 and CD19 in LIN−CD127+FLT3+SCA1lowKITlowhCD25+ B-lymphoid committed progenitor (BLCP). These finding does not support the currently dominating view that commitment occurs at a later stage of development with surface expression of both CD19 and B220. The idea that CD19 expression would coincide with commitment is to a substantial degree based on data from Pax5-deficient mice or cell lines where robust expression of Pax5 has been shown to result in expression of Pax5 target genes such as CD19 and repression of alternative cell fates.13-16 This view is also supported by studies in wild-type mice where CD19−B220− CLPs have been reported to give rise to CD11+ myeloid cells as well as lymphoid cells, whereas B220+ cells maintain their T-cell potential and only B220+CD19+ cells appear to have undergone lineage commitment.3 However, our experiments using a transgenic reporter provide a possibility to investigate these phenomena with a higher resolution. Our data do not contradict the idea that PAX5 is involved in lineage commitment because Pax5 expression is increased in the BLCP. However, they add new information concerning the temporal regulation of events suggested from these previous observations. Several transgenic mice models have been used to identify lineage-committed or -restricted progenitors in the early compartments, and although the use of Rag1 reporter mice4 or staining for TDT31 has allowed for a dramatic enrichment of cells primed for lymphoid development, the cells committed to B-lymphoid fate have been identified mainly in the lineage marker–positive fractions in the bone marrow.3 Experiments using a hCD25 reporter under the regulation of the T-lineage–restricted pTα promoter have allowed for the identification of lineage-restricted T-cell progenitors in the blood,32 whereas reporter-positive cells in the bone marrow appeared to retain a substantial B-lineage potential.33 To our knowledge, there are no previous reports of a committed B220−CD19− cell, however, our data are supported by observations in other transgenic models. First, Rag expression and DJ recombination of the immunoglobulin heavy chain locus are dramatically impaired in CLPs from mice lacking the B-lineage–restricted E-Rag enhancer.34,35 This suggests that B-lineage–restricted control elements may be active already in the CLP. Second, mice carrying a GFP reporter gene under the control of the Pax5 locus present a population of GFP+ cells within the LIN−CD127+SCA1lowKITlow compartment.16 The lineage potential of these cells was not investigated in the published report, but the size of this population is comparable with that of the hCD25+ progenitor population and may well represent the same cells. Furthermore, multiplex single-cell PCR analysis of sorted CLPs suggested that approximately one third of the cells contained only B-lineage–associated transcripts.26 Thus, we believe that our data find support in the existing literature.
In addition to an increased insight into the temporal regulation of B-lineage commitment, our results allow for alternative models for the molecular regulation of lineage choices in the early progenitors. Single-cell PCR analysis suggests that, as predicted from transgenic models, the transcription of lymphoid-associated genes such as CD127 or Rag1 is not linked to B- versus T-lineage decision event.4 This also appears to be true for the expression of Ebf1 because the number of progenitors that express EBF1 in hCD25+ and hCD25− cells are comparable. However, in hCD25+ cells, the level of Ebf1 message is increased 4-fold and the expression of EBF1 target genes is induced in a coordinated manner. The extremely low transcript levels (Figure 3C) and the low number of cells with detectable levels of transcripts from lineage-restricted genes in the noncommitted progenitors (Figure 3D) argue against a lineage priming model. Rather, our data suggest that the transcripts found in the CLP compartment are generated from B-lineage–restricted cells. This is also supported by the finding that the levels of Ebf1, Pax5, and Mb1 mRNA found in the BLCPs are fully comparable with those found in IgM−B220+CD43lowCD19+ pro-B cells. The finding that a reporter gene controlled by the Igll1 promoter—a direct target gene for EBF1—allows for tracing of B-lineage commitment points to a direct role of functional EBF1 activity in lineage restriction events, as previously suggested from overexpression studies.36 The finding that Ebf1 message can be detected in the majority (61%) of the CD127+FLT3+SCA1lowKITlow hCD25− cells able to efficiently generate T-cell clones, and before the detectable expression of either Pax5 or Igll1, suggests that the activation of the Ebf1 gene is a precommitment event but that further increased levels of Ebf1 have a striking effect on target gene activation and lineage commitment. This is also in line with the finding that Pax5-deficient pro-B cells could be made B-lineage restricted by ectopic expression of EBF, suggesting that the key role of PAX5 in lineage commitment is to enhance EBF expression.37 Although EBF1 has been proposed to act in a dose-dependent manner17 and the effect observed may be a direct response to increased level of EBF1, other posttranscriptional regulatory mechanisms including reduction of EBF1 activity by active NOTCH signaling38 might be involved.
Although our data do not directly contradict the idea that PAX5 is the crucial factor in B-lineage commitment, the low level of CD19 transcripts in the BLCPs argues against the idea that these cells have established a fully developed PAX5 regulatory network. We rather believe that our findings fit a model where lineage restriction initially is mediated by EBF1, whereas the role of PAX5 is to ensure the establishment of a regulatory circuit involving the control of EBF1 expression levels in the committed B-lineage cells.37 Although we currently are unable to say how large a part of the B cells that develop through a BLCP pathway, as opposed to those that undergo commitment at a CD19+B220+ stage,3 our findings suggest that this process can occur in lineage-negative cells—presenting new possibilities to study early events in B-cell development.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank A. Cumano and J. C. Zuniga-Pflücker for providing the OP9 and OP9DL1 stromal cell lines; Liselotte Lenner and Gerd Sten for expert advice and technical assistance; and the SweGene Affymetrix facility for its assistance with the microarrays.
These studies were generously supported by grants from the Swedish Cancer Society, the Swedish Research Council, Tobias Stiftelsen, the Swedish Childhood Cancer Foundation, and Swedish Foundation for Strategic Research (all Stockholm, Sweden). The Lund Stem Cell Center is supported by a Center of Excellence grant from the Swedish Foundation for Strategic Research.
Authorship
Contribution: R.M. and S.Z. performed the majority of the in vitro differentiation experiments and single-cell PCR; D.B. performed cell sorting experiments; K.A. performed and analyzed Giemsa stainings; M.S. performed Q-PCR experiments; R.M., S.Z., D.B., S.E.W.J., and M.S. were involved in the design of the experiments; I.-L.M. provided crucial reagents; all authors have read and given comments to the paper written by R.M. and M.S.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mikael Sigvardsson, Faculty of Health Sciences, Linköping University Lab1, Level 13, Linköping, Sweden; e-mail: mikael.sigvardsson@ibk.liu.se.