Abstract
Inhibitors of DNA binding (Id) family members are key regulators of cellular differentiation and proliferation. These activities are related to the ability of Id proteins to antagonize E proteins and other transcription factors. As negative regulators of E proteins, Id proteins have been implicated in lymphocyte development. Overexpression of Id1, Id2, or Id3 has similar effects on lymphocyte development. However, which Id protein plays a physiologic role during lymphocyte development is not clear. By analyzing Id2 knock-out mice and retroviral transduced hematopoietic progenitors, we demonstrated that Id2 is an intrinsic negative regulator of B-cell development. Hematopoietic progenitor cells overexpressing Id2 did not reconstitute B-cell development in vivo, which resembled the phenotype of E2A null mice. The B-cell population in bone marrow was significantly expanded in Id2 knock-out mice compared with their wild-type littermates. Knock-down of Id2 by shRNA in hematopoietic progenitor cells promoted B-cell differentiation and induced the expression of B-cell lineage–specific genes. These data identified Id2 as a physiologically relevant regulator of E2A during B lymphopoiesis. Furthermore, we identified a novel Id2 function in erythroid development. Overexpression of Id2 enhanced erythroid development, and decreased level of Id2 impaired normal erythroid development. Id2 regulation of erythroid development is mediated via interacting with transcription factor PU.1 and modulating PU.1 and GATA-1 activities. We conclude that Id2 regulates lymphoid and erythroid development via interaction with different target proteins.
Introduction
Hematopoiesis, the process by which mature blood cells of distinct lineages are produced from multipotent hematopoietic stem cells (HSCs), is a highly orchestrated process, involving a hierarchy of progenitors with progressively restricted developmental potential.1,2 Transcription factors play a key role in hematopoietic lineage commitment, depending on their expression levels as well as their interactions.3-5 For example, the importance of E proteins in the regulation of lymphocyte development was firmly established in the studies of 2 independently produced E2A knock-out mouse strains, which display a block in B-cell development and perturbed T-cell development.6-8 During myeloid development, PU.1 and C/EBPα are up-regulated in the granulocyte/macrophage progenitors (GMPs) during granulocyte and macrophage development, and down-regulated in the megakaryocyte/erythrocyte progenitors (MEPs).4,9 They cooperate in the regulation of a number of myeloid-specific genes, such as the granulocyte/macrophage colony-stimulating factor receptor α (GM-CSFRα), macrophage CSF receptor (M-CSFR), and granulocyte CSF receptor (G-CSFR).10-13 In comparison, GATA-1 and its cofactor FOG-1, which are required for erythroid differentiation, are up-regulated in MEPs and down-regulated in GMPs.4,14,15 In addition, the antagonistic interaction between PU.1 and GATA-1 is critical in initiating the myeloid versus the erythroid program.16,17
Id2 is a member of the inhibitor of DNA binding protein (Id) family. Id proteins play important roles in regulating cell proliferation, differentiation, and apoptosis.18-20 Mechanistically, Id proteins act as dominant-negative regulators of other transcription factors and render them unable to bind DNA and regulate transcription. Id proteins bind to ubiquitously expressed bHLH transcription factors E proteins, and prevent E protein homodimerization or heterodimerization with tissue-restricted basic HLH (bHLH) proteins.21,22 In addition to their interaction with E proteins, Id proteins have also been shown to interact with other transcription factors, including transcription factors from the ETS family, Pax family, and retinoblastoma protein RB.23-25
As negative regulators of E proteins, Id proteins have been implicated in the lymphocyte proliferation and developmental progression.26,27 Overexpression of Id1, Id2, or Id3 has similar effects on lymphocyte development.28-31 However, which Id protein plays a physiologic role during lymphocyte development is not clear. In this study, by analyzing Id2 knock-out mice and retroviral transduced hematopoietic progenitors, we demonstrated that Id2 is an intrinsic negative regulator of B-cell development. Furthermore, we identified novel Id2 function in erythroid development. Id2 regulation of erythroid development is mediated by its interaction with PU.1. We conclude that Id2 regulates lymphoid and erythroid development via interaction with different target proteins.
Methods
Mice
Id2−/− mice of mixed genetic background (129/Sv × NMRI) were used in most of the experiments. C57BL/6-Ly5.2 (CD45.1) mice were obtained for experiments from the animal production area at NCI-Frederick (Frederick, MD). Animal care was provided in accordance with the National Institutes of Health's procedures for the care and use of laboratory animals.
Plasmids
For the MSCV-Id2-IRES-GFP plasmid, Id2 cDNA was amplified by reverse transcriptase–polymerase chain reaction (RT-PCR) using C57BL/6 mouse BM mRNA as template, and cloned between BglII and XhoI sites of murine stem cell virus (MSCV)–based retroviral vector MSCV-IRES-GFP. For the pTRE-Id2 plasmid, Id2 cDNA was amplified by RT-PCR using C57BL/6 mouse BM mRNA as template, and cloned into XbaI site of pTRE vector (BD Biosciences, San Jose, CA). For the pRetro-Id2-shRNA and pRetro-NS-shRNA plasmids, annealed DNA oligoes for expression of Id2-specific small hairpin RNA (target sequence: CATGAACGACTGCTACTCC) or nonspecific small hairpin RNA (target sequence: GTTCTCCGAACGTGTCACG) were cloned behind the human H1 RNA promoter between BamHI and XhoI sites of pRetro-H1G vector (Cellogenetics, Ijamsville, MD). All constructs were verified by DNA sequencing. pcDNA-GATA-1 and p4.2-Luc plasmids are gifts from Dr Stephen Brandt, Vanderbilt University (Nashville, TN). PU.1-pECE and M-CSFR-Luc plasmids are gifts from Dr Dong-Er Zhang, Scripps Research Institute (La Jolla, CA).
Cell lines
The stem cell factor (SCF)–dependent EML cell line was maintained in Iscove-modified Dulbecco medium (IMDM) supplemented with 20% horse serum, 8% conditioned medium from BHK/MKL cells. The U2OS cell line was maintained in complete Dulbecco-modified Eagle medium (cDMEM) with 10% fetal bovine serum. The U2OS-Id2 cell line that conditionally expresses Id2 was established by transfecting U2OS-Tet-On cell line (a gift from Dr Karen Vousden, Beatson Institute for Cancer Research, Glasgow, United Kingdom) with pTRE-Id2 and pTK-Hyg (BD Biosciences) plasmids. Stable U2OS-Id2 cell lines were cloned by limiting dilution in the presence of 400 μg/mL hygromycin (Calbiochem, San Diego, CA). The expression of Id2 was monitored 24 hours after doxycycline (Invitrogen, Carlsbad, CA) induction by Western blot using anti-Id2 antibody (Santa Cruz Biotechnology, Santa Cruz, CA).
Purification of hematopoietic progenitors and erythroblasts
Purification of hematopoietic progenitors was accomplished by staining bone marrow cells (BMCs) with purified rat antibodies specific for the following lineage markers: CD4, CD8, B220, TER119, Gr-1, and Mac-1 (BD PharMingen, San Diego, CA). Lin+ cells were removed with sheep anti–rat IgG–conjugated magnetic beads (Invitrogen), and the remaining cells were stained with PE-Cy5–conjugated anti–IL-7Rα, FITC-conjugated anti-CD34, PE-conjugated anti–c-Kit, APC-conjugated anti–Sca-1, and PE-Cy7–conjugated anti-FcγRII/III. LSK cells were sorted as Lin− IL-7Rα− Sca-1+ c-Kit+; common myeloid progenitors (CMPs) were sorted as Lin− IL-7Rα− Sca-1− c-Kit+ CD34+ FcγRII/IIIlo; GMPs were sorted as Lin− IL-7Rα− Sca-1− c-Kit+ CD34+ FcγRII/IIIhi; and MEPs were sorted as Lin− IL-7Rα− Sca-1− c-Kit+ CD34− FcγRII/IIIlo.
Purification of erythroblasts was accomplished by staining BMC from Id2+/+ or Id2−/− mice with PE-conjugated TER119 and FITC-conjugated CD71 antibodies. Ter119+CD71+ erythroblasts were purified by multicolor-based sorting.
Retroviral transduction of EMLs, CMPs, MEPs, and BMCs
MSCV-IRES-GFP and MSCV-Id2-IRES-GFP were transfected into Phoenix packaging cells (a gift from Dr Gary Nolan, Stanford University, Stanford, CA) with pCL-eco plasmid by FuGENE 6 (Roche Applied Science, Indianapolis, IN) to produce infectious ecotropic retrovirus. The viral supernatants were collected 48 hours after transfection and used to infect cells in the presence of 4 μg/mL Polybrene (Sigma-Aldrich, St Louis, MO). Briefly, BMCs were harvested from C57BL/6 mice 3 days after treatment with 150 mg/kg 5-fluorouracil (5FU), and cultured in IMDM supplemented with 10% fetal bovine serum, 100 ng/mL each of murine stem cell factor (mSCF), human Flt-3L (hFlt-3L), human thrombopoietin (hTPO), and 50 ng/mL murine IL-6 (mIL-6) for 24 hours. 5FU BMCs were infected with MSCV-Id2-IRES-GFP or MSCV-IRES-GFP control retroviral vectors 3 times over a 36-hour period, and then GFP+ cells were isolated by flow cytometry. CMPs and MEPs were purified from C57BL/6 mice, and cultured in IMDM supplemented with 10% fetal bovine serum, 100 ng/mL each of mSCF, hFlt-3L, hTPO, 50 ng/mL mIL-6, and 10 U/mL human erythropoietin (hEPO) for 24 hours. CMPs and MEPs were infected with MSCV-Id2-IRES-GFP or MSCV-IRES-GFP control retroviral vectors 3 times over a 36-hour period, and then GFP+ cells were isolated by flow cytometry. To induce erythroid development, CMPs and MEPs were cultured with mSCF (100 ng/mL), hTPO (100 ng/mL), and hEPO (40 U/mL) for 4 days. To induce myeloid development, CMPs and GMPs were cultured with mSCF (100 ng/mL), murine IL-3 (mIL-3, 30 ng/mL), and murine granulocyte monocyte–colony-stimulating factor (mGM-CSF 20 ng/mL) for 4 days. EML cells were infected with MSCV-Id2-IRES-GFP or MSCV-IRES-GFP, and then GFP+ cells were isolated by flow cytometry. To induce erythroid development, EML cells were cultured with mSCF (100 ng/mL) and hEPO (40 U/mL) for 5 days. To induce myeloid development, EML cells were cultured with mSCF (100 ng/mL), atRA (10 μM), and mIL-3 (30 ng/mL) for 5 days, followed by atRA (10 μM) and mGM-CSF (20 ng/mL) for 5 days.
BM transplantation and FACS analysis
BMCs (5 × 105) from Id2+/+ or Id2−/− mice (CD45.2) together with BMCs (5 × 105) from C57BL/6 (CD45.1) mice were transplanted into C57BL/6 (CD45.1) recipient mice exposed to 9.5 Gy (950 rad) from 137Cs source. Hematopoietic reconstitution was determined 4 months after transplantation by analyzing CD45.2-positive BMCs. Equal numbers of BMC-GFP and BMC-Id2-GFP, or BMC-NS-shRNA and BMC-Id2-shRNA, together with supporting BMCs from C57BL/6 mice, were transplanted into C57BL/6 recipient mice exposed to 9.5 Gy (950 rad) from 137Cs source. Hematopoietic reconstitution was determined 4 weeks or 16 weeks after transplantation by FACS analysis of GFP-positive BMCs. All lineage-specific antibodies and PerCP-Cy5.5–conjugated streptavidin were purchased from BD PharMingen. CD45.2 was FITC-conjugated antibody. B220, IgM, Gr-1, and TER119 were PE-conjugated antibodies. CD43, IgD, Mac-1, and CD71 were biotin-conjugated antibodies.
Real-time PCR
Total RNA was purified using the RNeasy kit (Qiagen, Valencia, CA) and converted to cDNA with iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). Real-time quantitative PCR was performed in triplicate on iCycler Thermal Cycler (Bio-Rad). All samples were normalized to GAPDH.
Luciferase reporter assays
For M-CSFR luciferase assay, 300 ng PU.1-pECE, 100 ng M-CSFR-Luc, and 25 ng phRL4.7 were cotransfected into inducible U2OS-Id2 cells. For p4.2 luciferase assay, 300 ng pcDNA-GATA-1, 500 ng PU.1-pECE, 100 ng p4.2-Luc, and 25 ng phRL4.7 were cotransfected into inducible U2OS-Id2 cells. Id2 expression was induced 24 hours after transfection. Luciferase activity was examined 24 hours after Id2 induction using the Dual-Glo Luciferase Assay Kit (Promega, Madison, WI) and normalized to renilla activity.
Immunoprecipitation
Whole-cell lysates from EML, EML-GFP, EML-Id2-GFP, EML-NS-shRNA, or EML-Id2-shRNA were prepared with the RIPA Lysis Buffer (Santa Cruz Biotechnology). Immunoprecipitation with anti-PU.1 antibody or rabbit IgG (Santa Cruz Biotechnology) was performed in RIPA buffer. Immuno-complexes were resolved on NuPAGE Novex 4-12% Bis-Tris Gel (Invitrogen), transferred to Immobilon-P membrane (Millipore, Billerica, MA), and probed with anti-Id2 antibody or anti–GATA-1 antibody (Santa Cruz Biotechnology).
Results
Id2 knock-out mice show enhanced B-cell development and impaired erythroid development
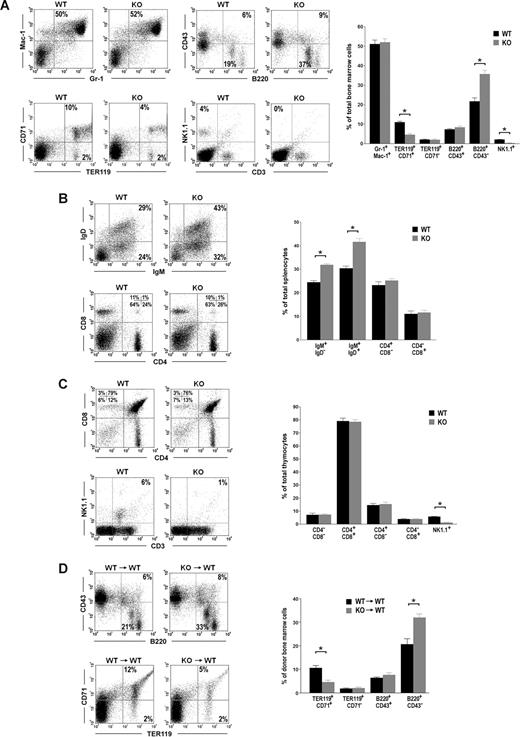
To understand Id2 function in hematopoiesis, we analyzed hematopoietic cells in the bone marrow (BM), spleens, and thymuses of Id2 knock-out mice. Consistent with previous reports, we observed natural killer cell defects in Id2−/− mice BM and thymuses. The percentages of natural killer cells (NK1.1+) in Id2+/+ mice BM and thymuses are 4% and 6%, respectively, whereas natural killer cells are absent in Id2−/− mice BM and thymuses (Figure 1A,C). In addition, we observed differences in B-cell population and erythrocyte population between Id2−/− mice and Id2+/+ mice. Compared with their Id2+/+ littermate controls, Id2−/− mice show a significantly increased percentage of B220+CD43− B cells in the BM, which include pre-B, immature, and mature B cells (Figure 1A). The percentages of IgM+IgD− immature B cells and IgM+IgD+ mature B cells in the spleen were also significantly increased in Id2−/− mice compared with their Id2+/+ littermate controls (Figure 1B). Id2−/− mice also show a significantly decreased percentage of TER119+CD71+ erythroid cells in the BM compared with their Id2+/+ littermate controls (Figure 1A). No difference in myeloid cells (Gr-1+Mac-1+ and Gr-1−Mac-1+) in the BM or T cells (CD4−CD8−, CD4+CD8−, CD4−CD8+, and CD4+CD8+) in the thymus was observed in Id2−/− and Id2+/+ mice (Figure 1A,C). These data suggest that Id2 functions in early B-cell development and early erythroid development.
Id2 knock-out mice show enhanced B-cell development and impaired erythroid development. (A-C) FACS analysis of hematopoietic lineages in Id2+/+ and Id2−/− BM, spleens, and thymuses. (D) BMCs (5 × 105) from Id2+/+ or Id2−/− mice (CD45.2) together with 5 × 105 BMCs from C57BL/6 (CD45.1) mice were transplanted into irradiated C57BL/6 (CD45.1) mice. B-cell and erythroid reconstitution were examined 4 months after transplantation by analyzing CD45.2-positive BMCs using B220, CD43, TER119, and CD71 antibodies. *P < .01.
Id2 knock-out mice show enhanced B-cell development and impaired erythroid development. (A-C) FACS analysis of hematopoietic lineages in Id2+/+ and Id2−/− BM, spleens, and thymuses. (D) BMCs (5 × 105) from Id2+/+ or Id2−/− mice (CD45.2) together with 5 × 105 BMCs from C57BL/6 (CD45.1) mice were transplanted into irradiated C57BL/6 (CD45.1) mice. B-cell and erythroid reconstitution were examined 4 months after transplantation by analyzing CD45.2-positive BMCs using B220, CD43, TER119, and CD71 antibodies. *P < .01.
To determine if the phenotypes of B cells and erythrocytes in Id2−/− are intrinsic to the hematopoietic stem cells, or due to the defect of the bone marrow microenvironment, we transplanted the Id2−/− or Id2+/+ bone marrow cells (BMCs) into lethally irradiated CD45.1 mice. After 4 months, hematopoietic reconstitution from CD45.2 donor was examined. We found that the percentage of B220+CD43− B cells in the BM was significantly increased in mice that underwent transplantation with Id2−/− BMCs compared with that in mice that underwent transplantation with Id2+/+ BMCs (Figure 1D). The percentage of erythroid cells in mice that underwent transplantation with Id2−/− BMCs was significantly lower than that in mice that underwent transplantation with Id2+/+ BMCs (Figure 1D). These data suggest that Id2 intrinsically regulates B-cell development and erythroid development.
Id2 is a negative regulator of early B-cell development
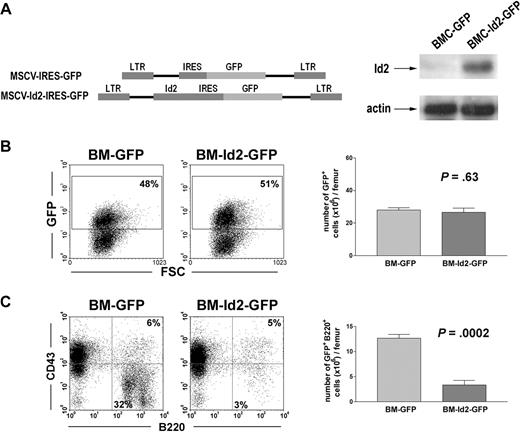
To further evaluate Id2 function in B-cell development, we infected BMCs from 5FU-treated mice with MSCV-Id2-IRES-GFP retrovirus or MSCV-IRES-GFP control retrovirus. Overexpression of Id2 in infected 5FU BMCs was verified by Western blot analysis (Figure 2A). GFP-positive cells, BMC-Id2-GFP or BMC-GFP, were purified by cell sorting and equal numbers of GFP-positive cells were transplanted into lethally irradiated mice with supporting BMCs. B-cell reconstitution was evaluated after 4 weeks and 16 weeks by analyzing GFP-positive cells in the BM of recipient mice. At 4 weeks after transplantation, in recipient BM, total GFP-positive cells were roughly 50% in mice that received transplants of BMC-GFP or BMC-Id2-GFP (Fig 2B). Mice that received transplants of BMC-Id2-GFP showed a significant decrease of the percentage and the total number of B220+CD43− B-cell population in BM compared with mice that received transplants of BMC-GFP (Figure 2C), suggesting that a high level of Id2 blocks B-cell development at the pro-B stage. This phenotype recapitulates the phenotype of the E2A−/− mouse.7 B-cell reconstitution was also examined 16 weeks after transplantation. Similar results were obtained as those of 4 weeks (data not shown).
Overexpression of Id2 blocks B-cell development. (A) Illustration of MSCV-IRES-GFP and MSCV-Id2-IRES-GFP retroviral vectors (left panel). Id2 expression levels in BMC-GFP and BMC-Id2-GFP were determined by Western blot analysis (right panel). (B) BMC-GFP and BMC-Id2-GFP were transplanted into lethally irradiated mice with supporting BMCs. A representative from 4 independent experiments is shown here. Eight mice were analyzed in each transplantation. Hematopoietic reconstitution in recipient BM was examined 4 weeks after transplantation by analyzing the percentage (left panel) and the total cellularity (right panel) of GFP-positive cells. (C) B-cell reconstitution in recipient BM was examined 4 weeks after transplantation by FACS analysis of GFP-positive BMCs using B220 and CD43 antibodies.
Overexpression of Id2 blocks B-cell development. (A) Illustration of MSCV-IRES-GFP and MSCV-Id2-IRES-GFP retroviral vectors (left panel). Id2 expression levels in BMC-GFP and BMC-Id2-GFP were determined by Western blot analysis (right panel). (B) BMC-GFP and BMC-Id2-GFP were transplanted into lethally irradiated mice with supporting BMCs. A representative from 4 independent experiments is shown here. Eight mice were analyzed in each transplantation. Hematopoietic reconstitution in recipient BM was examined 4 weeks after transplantation by analyzing the percentage (left panel) and the total cellularity (right panel) of GFP-positive cells. (C) B-cell reconstitution in recipient BM was examined 4 weeks after transplantation by FACS analysis of GFP-positive BMCs using B220 and CD43 antibodies.
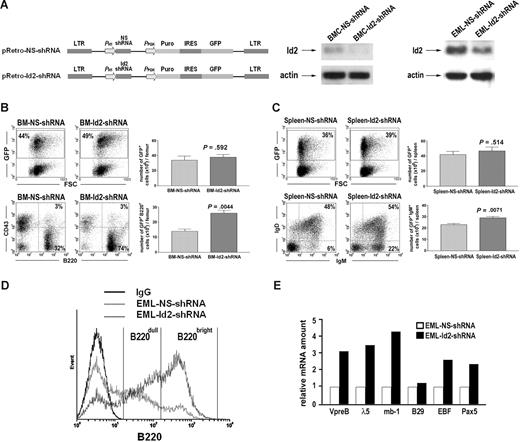
We further asked if knocking down Id2 can induce B-cell development. We infected BMCs from 5FU-treated mice with pRetro-Id2-shRNA retrovirus that expresses Id2-specific shRNA and GFP, or pRetro-NS-shRNA retrovirus that expresses nonspecific shRNA and GFP (Figure 3A). GFP-positive cells, BMC-Id2-shRNA or BMC-NS-shRNA, were purified by cell sorting and equal numbers of GFP-positive cells were transplanted into lethally irradiated mice with supporting BMCs. At 4 weeks after transplantation, we observed that mice that underwent transplantation with BMC-Id2-shRNA showed a significant increase of B220+CD43− B cells in BM, and a significant increase of IgM+IgD− immature B cells and IgM+IgD+ mature B cells in spleen compared with mice that underwent transplantation with BMC-NS-shRNA (Figure 3B,C), demonstrating that knocking down Id2 can induce B-cell development.
Knock-down of Id2 by shRNA induces B-cell development. (A) Illustration of pRetro-NS-shRNA and pRetro-Id2-shRNA retroviral vector (left panel). Id2 expression levels in BMC-NS-shRNA and BMC-Id2-shRNA were determined by Western blot analysis (middle panel). Id2 expression levels in EML-NS-shRNA and EML-Id2-shRNA were determined by Western blot analysis (right panel). (B) BMC-NS-shRNA and BMC-Id2-shRNA were transplanted into lethally irradiated mice with supporting BMCs. A representative from 3 independent experiments is shown here. Eight mice were analyzed in each transplantation. Hematopoietic reconstitution in recipient BM was examined 4 weeks after transplantation by analyzing the percentage and the total cellularity of GFP-positive cells (top panels). B-cell reconstitution was examined 4 weeks after transplantation in BM by FACS analysis using B220 and CD43 antibodies (bottom panels). (C) Hematopoietic reconstitution in recipient spleen was examined 4 weeks after transplantation by analyzing the percentage and the total cellularity of GFP-positive cells (top panels). B-cell reconstitution was examined 4 weeks after transplantation in spleen by FACS analysis using IgM and IgD antibodies (bottom panels). (D) FACS analysis of B cell–specific cell surface marker B220 in EML-NS-shRNA and EML-Id2-shRNA. A representative from 3 independent experiments is shown here. (E) Real-time PCR analysis of B cell–specific gene expression in EML-NS-shRNA and EML-Id2-shRNA. A representative from 3 independent experiments is shown here.
Knock-down of Id2 by shRNA induces B-cell development. (A) Illustration of pRetro-NS-shRNA and pRetro-Id2-shRNA retroviral vector (left panel). Id2 expression levels in BMC-NS-shRNA and BMC-Id2-shRNA were determined by Western blot analysis (middle panel). Id2 expression levels in EML-NS-shRNA and EML-Id2-shRNA were determined by Western blot analysis (right panel). (B) BMC-NS-shRNA and BMC-Id2-shRNA were transplanted into lethally irradiated mice with supporting BMCs. A representative from 3 independent experiments is shown here. Eight mice were analyzed in each transplantation. Hematopoietic reconstitution in recipient BM was examined 4 weeks after transplantation by analyzing the percentage and the total cellularity of GFP-positive cells (top panels). B-cell reconstitution was examined 4 weeks after transplantation in BM by FACS analysis using B220 and CD43 antibodies (bottom panels). (C) Hematopoietic reconstitution in recipient spleen was examined 4 weeks after transplantation by analyzing the percentage and the total cellularity of GFP-positive cells (top panels). B-cell reconstitution was examined 4 weeks after transplantation in spleen by FACS analysis using IgM and IgD antibodies (bottom panels). (D) FACS analysis of B cell–specific cell surface marker B220 in EML-NS-shRNA and EML-Id2-shRNA. A representative from 3 independent experiments is shown here. (E) Real-time PCR analysis of B cell–specific gene expression in EML-NS-shRNA and EML-Id2-shRNA. A representative from 3 independent experiments is shown here.
EML, an immortalized hematopoietic progenitor cell line, has the developmental potential for B cells, T cells, myeloid cells, and erythrocytes.32 We knocked down Id2 expression in EML cells by infecting cells with pRetro-Id2-shRNA retrovirus or pRetro-NS-shRNA retrovirus (Figure 3A). When maintained in growth medium containing mSCF, only 5% of the EML-NS-shRNA cells express a high level of B cell–specific surface marker B220 (B220bright), and 30% of the EML-NS-shRNA cells express a low level of B220 (B220dull). Knocking down Id2 in multipotent EML cells induces B220 expression, with over 45% of the EML-Id2-shRNA cells being B220bright and 35% of the EML-Id2-shRNA cells being B220dull (Figure 3D). Furthermore, real-time PCR analysis showed that knocking down Id2 in EML cells induced the expression of B cell–specific genes, including VpreB, λ5, mb-1, EBF, and Pax5 (Figure 3E), indicating that knocking down Id2 induces B-cell differentiation in multipotent EML cells.
Id2 promotes erythropoiesis of erythromyeloid progenitors
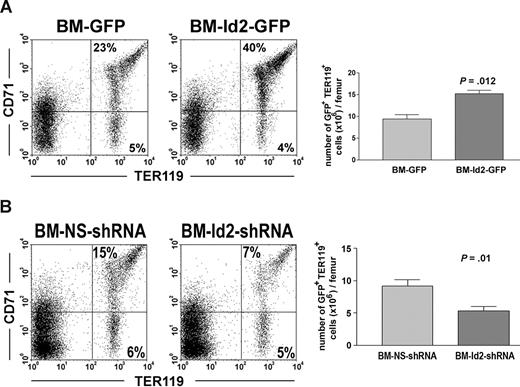
To further evaluate Id2 function in erythroid development, we examined the effects of Id2 overexpression and Id2 knock-down on erythroid reconstitution in mice that underwent transplantation with BMC-Id2-GFP, BMC-GFP, BMC-Id2-shRNA, or BMC-NS-shRNA. Mice that received transplants of BMC-Id2-GFP showed a significant increase of the percentage of TER119+CD71+ erythrocytes in BM compared with mice that received transplants of BMC-GFP (Figure 4A). To determine if Id2 directly affects erythroid development, we compared the total cellularity of erythrocytes in the BM. We found that the total cellularity of erythrocytes are higher in BMC-Id2-GFP recipients (Figure 4A), indicating that Id2 promotes erythroid development in addition to inhibiting B-cell development. Consistent with the phenotype of Id2 knock-out mice, we observed a significant decrease of TER119+CD71+ erythrocytes in mice that received transplants of BMC-Id2-shRNA compared with mice that received transplants of BMC-NS-shRNA (Figure 4B). This indicates that a decreased level of Id2 impairs normal erythroid development.
Id2 regulates erythroid development in vivo. (A) BMC-GFP and BMC-Id2-GFP were transplanted into lethally irradiated mice with supporting BMCs. A representative from 4 independent experiments was shown here. Eight mice were analyzed in each transplantation. Erythroid reconstitution in recipient BM was examined 4 weeks after transplantation by FACS analysis of GFP-positive cells using TER119 and CD71 antibodies. (B) BMC-NS-shRNA and BMC-Id2-shRNA were transplanted into lethally irradiated mice with supporting BMCs. A representative from 3 independent experiments is shown here. Eight mice were analyzed in each transplantation. Erythroid reconstitution was examined 4 weeks after transplantation in recipient BM by FACS analysis using TER119 and CD71 antibodies.
Id2 regulates erythroid development in vivo. (A) BMC-GFP and BMC-Id2-GFP were transplanted into lethally irradiated mice with supporting BMCs. A representative from 4 independent experiments was shown here. Eight mice were analyzed in each transplantation. Erythroid reconstitution in recipient BM was examined 4 weeks after transplantation by FACS analysis of GFP-positive cells using TER119 and CD71 antibodies. (B) BMC-NS-shRNA and BMC-Id2-shRNA were transplanted into lethally irradiated mice with supporting BMCs. A representative from 3 independent experiments is shown here. Eight mice were analyzed in each transplantation. Erythroid reconstitution was examined 4 weeks after transplantation in recipient BM by FACS analysis using TER119 and CD71 antibodies.
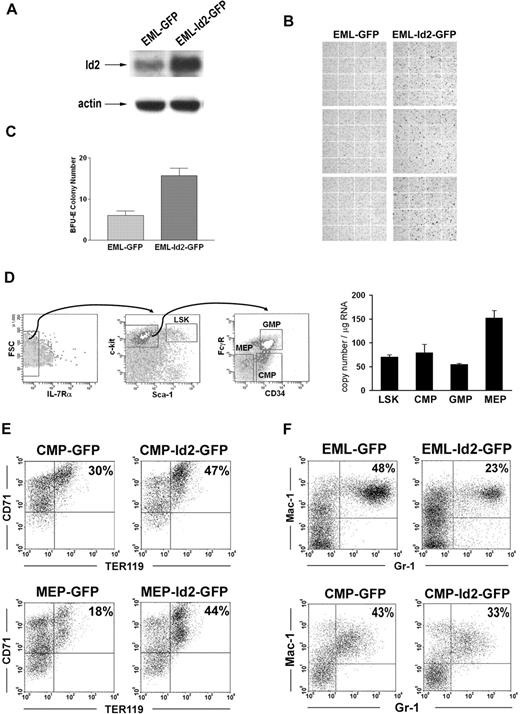
To further understand Id2 function in regulating the commitment of erythroid progenitors, we evaluated Id2 function in multipotent EML cells and purified normal hematopoietic progenitors. EML cells were transduced with MSCV-Id2-IRES-GFP retrovirus or MSCV-IRES-GFP control retrovirus. GFP-positive cells, EML-Id2-GFP or EML-GFP, were purified by cell sorting and then cultured in conditions that induce erythroid development. Overexpression of Id2 in EML cells was verified by Western blot analysis (Figure 5A). Erythroid differentiation was evaluated by Benzidine staining. After 5 days of culture, EML-GFP cells gave rise to 4% (± 1%) Benzidine-positive cells, whereas EML-Id2-GFP cells gave rise to 25% (± 5%) Benzidine-positive cells (Figure 5B). In addition, EML-Id2-GFP cells gave rise to more BFU-E colonies than EML-GFP cells in colony-forming assays (Figure 5C). These data suggest that Id2 promotes erythropoiesis in multipotent EML cells.
Id2 promotes erythropoiesis of erythromyeloid progenitors. (A) Id2 expression levels in EML-GFP and EML-Id2-GFP were determined by Western blot analysis. (B) EML-GFP or EML-Id2-GFP were induced for erythroid development in IMDM containing 10% FBS, 1 × 10−5 M 2-ME, 100 ng/mL mSCF, and 5 U/mL hEPO for 5 days. Erythrocytes were identified by Benzidine staining. (C) EML-GFP or EML-Id2-GFP were seeded at a density of 1 × 104 cells/mL in IMDM with 2.75% methylcellulose, 1 × 10−5 M 2-ME, 30% FBS, 100 ng/mL mSCF, 30 ng/mL mIL-3, and 5 U/mL hEPO. BFU-Es were determined after 10 days. (D) LSKs, CMPs, GMPs, and MEPs were purified from mouse BM according to their surface phenotypes, by multicolor-based sorting techniques. Total RNA was purified from LSKs, CMPs, GMPs, and MEPs, and converted to cDNA. Id2 expression levels in these purified progenitors were determined by real-time PCR. (E) MEP-GFP, MEP-Id2-GFP, CMP-GFP, or CMP-Id2-GFP were induced for erythroid development in IMDM containing 10% FBS, mSCF (100 ng/mL), hTPO (100 ng/mL), and hEPO (40 U/mL) for 4 days, and analyzed for TER119 expression by FACS analysis. (F) EML-GFP and EML-Id2-GFP were induced for myeloid development in IMDM containing 20% HS, mSCF (100 ng/mL), atRA (10 μM), and mIL-3 (30 ng/mL) for 5 days, followed by atRA (10 μM) and mGM-CSF (20 ng/mL) for 5 days, and analyzed for Gr-1 and Mac-1 expression by FACS analysis. CMP-GFP or CMP-Id2-GFP were induced for myeloid development in IMDM containing 10% FBS, mSCF (100 ng/mL), mIL-3 (30 ng/mL), and mGM-CSF (20 ng/mL) for 4 days, and analyzed for Gr-1 and Mac-1 expression by FACS analysis.
Id2 promotes erythropoiesis of erythromyeloid progenitors. (A) Id2 expression levels in EML-GFP and EML-Id2-GFP were determined by Western blot analysis. (B) EML-GFP or EML-Id2-GFP were induced for erythroid development in IMDM containing 10% FBS, 1 × 10−5 M 2-ME, 100 ng/mL mSCF, and 5 U/mL hEPO for 5 days. Erythrocytes were identified by Benzidine staining. (C) EML-GFP or EML-Id2-GFP were seeded at a density of 1 × 104 cells/mL in IMDM with 2.75% methylcellulose, 1 × 10−5 M 2-ME, 30% FBS, 100 ng/mL mSCF, 30 ng/mL mIL-3, and 5 U/mL hEPO. BFU-Es were determined after 10 days. (D) LSKs, CMPs, GMPs, and MEPs were purified from mouse BM according to their surface phenotypes, by multicolor-based sorting techniques. Total RNA was purified from LSKs, CMPs, GMPs, and MEPs, and converted to cDNA. Id2 expression levels in these purified progenitors were determined by real-time PCR. (E) MEP-GFP, MEP-Id2-GFP, CMP-GFP, or CMP-Id2-GFP were induced for erythroid development in IMDM containing 10% FBS, mSCF (100 ng/mL), hTPO (100 ng/mL), and hEPO (40 U/mL) for 4 days, and analyzed for TER119 expression by FACS analysis. (F) EML-GFP and EML-Id2-GFP were induced for myeloid development in IMDM containing 20% HS, mSCF (100 ng/mL), atRA (10 μM), and mIL-3 (30 ng/mL) for 5 days, followed by atRA (10 μM) and mGM-CSF (20 ng/mL) for 5 days, and analyzed for Gr-1 and Mac-1 expression by FACS analysis. CMP-GFP or CMP-Id2-GFP were induced for myeloid development in IMDM containing 10% FBS, mSCF (100 ng/mL), mIL-3 (30 ng/mL), and mGM-CSF (20 ng/mL) for 4 days, and analyzed for Gr-1 and Mac-1 expression by FACS analysis.
To determine if Id2 promotes erythropoiesis in normal hematopoietic progenitors, we purified Lin−Sca-1+c-Kit+ cells (LSKs), common myeloid progenitors (CMPs), GMPs, and MEPs from mouse BMCs by multicolor sorting. First, Id2 expression levels were examined in these hematopoietic progenitors by quantitive real-time RT-PCR. Id2 is expressed in LSKs and CMPs at similar levels. GMPs express lower levels of Id2, and MEPs express higher levels of Id2 (Figure 5D). This dynamic expression of Id2 in hematopoietic progenitors suggests that Id2 may play a role in the commitment of early erythromyeloid progenitors.
To evaluate Id2 effects on erythroid commitment in early erythromyeloid progenitors, we transduced purified CMPs and MEPs with MSCV-Id2-IRES-GFP retrovirus or MSCV-IRES-GFP control retrovirus, and cultured these cells in medium containing mSCF, hTPO, and hEPO to induce erythroid development. After 4 days, cells were analyzed for erythroid development by immunostaining for erythroid marker TER119, which is expressed in all erythroid precursors subsequent to the proerythroblast stage.33 CMP-Id2-GFP gave rise to 47% TER119-positive cells, while CMP-GFP gave rise to 30% TER119-positive cells (Figure 5E). The Id2 effect was even more significant in MEPs. MEP-Id2-GFP gave rise to 44% TER119-positive cells, while MEP-GFP gave rise to 18% TER119-positive cells (Figure 5E). This suggests that Id2 overexpression significantly enhanced erythropoiesis in purified CMPs and MEPs. Taken together, these data suggest that Id2 promotes erythroid commitment in hematopoietic progenitors.
Id2 promotes erythroid development in progenitors that also have myeloid potential, suggesting that Id2 may lead to bias between myeloid and erythroid development. Therefore, we evaluated the differentiation of EML-GFP, EML-Id2-GFP, CMP-GFP, or CMP-Id2-GFP cells in conditions that promote myeloid development. EML-GFP or EML-Id2-GFP cells were treated with atRA and IL-3 for 5 days, followed by atRA and GM-CSF treatment for 5 days to induce granulocyte development. CMP-GFP or CMP-Id2-GFP cells were cultured in medium containing mSCF, mIL-3, and mGM-CSF to induce myeloid development. Granulocyte differentiation was determined based on Gr-1 and Mac-1 expression by FACS. EML-Id2-GFP cells gave rise to fewer neutrophils than EML-GFP cells. CMP-Id2-GFP gave rise to fewer Gr-1+Mac-1+ neutrophils compared with CMP-GFP (Figure 5F).
Id2 regulates erythroid development by modulating PU.1 and GATA-1 activities
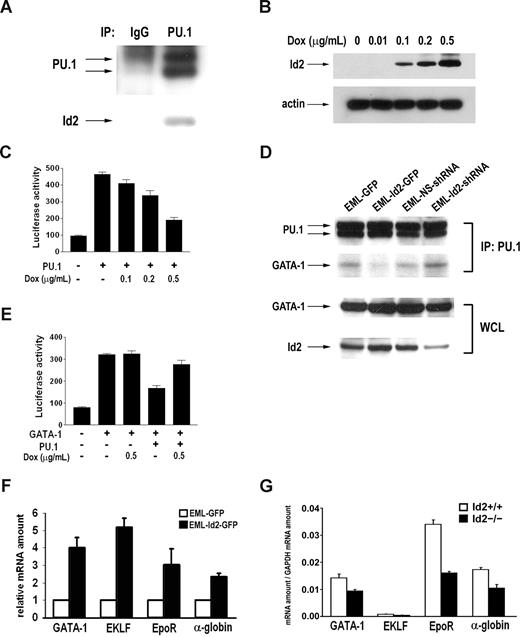
Previous studies indicate that Id proteins regulate lymphoid development through E proteins.21,34 This mechanism is further supported by the fact that Id2 overexpression resembles the phenotype of E2A knock-out mice (Figure 2C), and that knock-down of Id2 induces the E2A target genes (Figure 3E). However, the mechanism by which Id2 regulates erythroid development is completely unknown. Id proteins have been shown to bind and affect the transcriptional activity of Ets family transcription factors.24,35 PU.1, a member of the Ets family, plays an essential regulatory role in erythromyeloid development. Therefore, Id2 may affect erythroid development by interacting with PU.1. To determine if Id2 interacts with PU.1 in the hematopoietic cells, we performed coimmunoprecipitation of PU.1 and Id2 in EML cells, which express both proteins. Id2 protein was readily detected in the PU.1 immunocomplex, but not in the IgG control (Figure 6A). Id2-PU.1 interaction suggests that Id2 could affect PU.1 transcriptional activity. To test this, we examined Id2 effect on PU.1-mediated M-CSFR promoter activation in luciferase reporter assays. We cotransfected PU.1-pECE and M-CSFR-Luc into an inducible U2OS-Id2 cell line, in which Id2 protein expression can be induced by doxycycline in a dose-dependent manner (Figure 6B). Id2 expression was induced 24 hours after transfection of PU.1-pECE and M-CSFR-Luc, and luciferase activity was examined 24 hours after Id2 induction. PU.1 expression in U2OS-Id2 cells led to a 4- to 5-fold induction of M-CSFR promoter activity. PU.1-mediated M-CSFR promoter activity was inhibited by the expression of Id2 in a dose-dependent manner (Figure 6C), suggesting that Id2 can inhibit PU.1 transcriptional activity.
Id2 interacts with PU.1 and modulates PU.1 and GATA-1 activity. (A) Immunoprecipitation was performed using EML cell lysate with anti-PU.1 antibody and IgG. Id2 presence in the immunocomplex was examined by Western blot analysis. (B) Id2 expression was induced in dose-dependent manner by doxycycline in inducible U2OS-Id2 cells. (C) PU.1-pECE, M-CSFR-Luc, and phRL4.7 were cotransfected into U2OS-Id2 cells. At 24 hours after transfection, Id2 expression was induced by addition of doxycycline. Luciferase activity was examined and normalized to renilla activity. (D) Immunoprecipitation was performed in EML-GFP, EML-Id2-GFP, EML-NS-shRNA, and EML-Id2-shRNA with anti-PU.1 antibody. GATA-1 presence in the immunocomplex was examined by Western blot analysis (top panel). GATA-1 and Id2 levels in EML-GFP, EML-Id2-GFP, EML-NS-shRNA, and EML-Id2-shRNA whole-cell lysates were determined by Western blot analysis (bottom panel). (E) pcDNA-GATA-1, PU.1-pECE, p4.2-Luc, and phRL4.7 were cotransfected into U2OS-Id2 cells. At 24 hours after transfection, Id2 expression was induced by addition of doxycycline. Luciferase activity was examined and normalized to renilla activity. (F) Total RNA was purified from EML-GFP or EML-Id2-GFP, and converted to cDNA. GATA-1, ELKF, EpoR, and alpha-globin expression levels were determined by real-time PCR. (G) Ter119+CD71+ primary erythroblasts were purified from Id2+/+ and Id2−/− mice by multicolor-based sorting techniques. Total RNA was purified from sorted cells and converted to cDNA. GATA-1, ELKF, EpoR, and alpha-globin expression levels were determined by real-time PCR.
Id2 interacts with PU.1 and modulates PU.1 and GATA-1 activity. (A) Immunoprecipitation was performed using EML cell lysate with anti-PU.1 antibody and IgG. Id2 presence in the immunocomplex was examined by Western blot analysis. (B) Id2 expression was induced in dose-dependent manner by doxycycline in inducible U2OS-Id2 cells. (C) PU.1-pECE, M-CSFR-Luc, and phRL4.7 were cotransfected into U2OS-Id2 cells. At 24 hours after transfection, Id2 expression was induced by addition of doxycycline. Luciferase activity was examined and normalized to renilla activity. (D) Immunoprecipitation was performed in EML-GFP, EML-Id2-GFP, EML-NS-shRNA, and EML-Id2-shRNA with anti-PU.1 antibody. GATA-1 presence in the immunocomplex was examined by Western blot analysis (top panel). GATA-1 and Id2 levels in EML-GFP, EML-Id2-GFP, EML-NS-shRNA, and EML-Id2-shRNA whole-cell lysates were determined by Western blot analysis (bottom panel). (E) pcDNA-GATA-1, PU.1-pECE, p4.2-Luc, and phRL4.7 were cotransfected into U2OS-Id2 cells. At 24 hours after transfection, Id2 expression was induced by addition of doxycycline. Luciferase activity was examined and normalized to renilla activity. (F) Total RNA was purified from EML-GFP or EML-Id2-GFP, and converted to cDNA. GATA-1, ELKF, EpoR, and alpha-globin expression levels were determined by real-time PCR. (G) Ter119+CD71+ primary erythroblasts were purified from Id2+/+ and Id2−/− mice by multicolor-based sorting techniques. Total RNA was purified from sorted cells and converted to cDNA. GATA-1, ELKF, EpoR, and alpha-globin expression levels were determined by real-time PCR.
Since PU.1 and GATA-1 interact directly, and the antagonistic interaction between PU.1 and GATA-1 is critical in initiating the myeloid versus the erythroid program, we asked how Id2-PU.1 interaction affects PU.1-GATA-1 association and erythroid gene expression. We performed coimmunoprecipitation of PU.1 and GATA-1 in EML-GFP, EML-Id2-GFP, EML-NS-shRNA, and EML-Id2-shRNA cell lines. We found PU.1-GATA-1 association decreased in EML-Id2-GFP cells compared with EML-GFP cells, and increased in EML-Id2-shRNA cells compared with EML-NS-shRNA cells (Figure 6D), suggesting that Id2-PU.1 interaction interferes with PU.1-GATA-1 interaction. To determine if Id2 affects GATA-1 transcriptional activity, we cotransfected pcDNA-GATA-1, PU.1-pECE, and p4.2-Luc into U2OS-Id2 cells. Luciferase assays showed that GATA-1 protein induces the activation of the p4.2 promoter, and PU.1 protein antagonizes the induction. Id2 protein expression induced by doxycycline resulted in the release of PU.1-mediated GATA-1 inhibition (Figure 6E). To confirm the effect of Id2 on GATA-1 activity, we examined Id2 effect on the expression of GATA-1 target genes in EML cells and primary erythroblasts by real-time PCR. We found that overexpression of Id2 in EML cells induced the expression of GATA-1 target genes including GATA-1, ELKF, EpoR, and alpha-globin (Figure 6F). Conversely, the expression of GATA-1 target genes was lower in primary erythroblasts purified from Id2−/− mice compared with Id2+/+ mice (Figure 6G). These data indicate that Id2 can enhance the erythroid transcriptional activity of GATA-1.
Discussion
Our findings allow us to reach 2 conclusions. First, we demonstrated that Id2 is an intrinsic negative regulator of B-cell development. Id2 is the physiologically relevant regulator of E2A during B lymphopoiesis. Second, we demonstrated that Id2 plays a role in erythroid development via its interaction with PU.1 protein. Thus, Id2 regulates lymphoid and erythroid development via different targets.
It is well established that E proteins are pivotal in the regulation of lymphocyte development. E2A knock-out mice display a block in B-cell development and perturbed T-cell development.6-8 Id proteins act as dominant-negative regulators of E proteins. B-cell development is impaired at an early stage in transgenic mice constitutively expressing the Id1 gene. These Id1 transgenic mice have few B220+ IgM+ mature B cells or B220+ CD43− pre-B cells in the BM.28 Impaired B-cell development was also seen in transplantation of HSCs that overexpress Id1.36 In addition, we provide evidence that overexpression of Id2 also blocks early B-cell development at the pro-B stage. This raises the question of which Id protein plays a physiologic role during lymphocyte development. We found that B-cell population in BM was significantly expanded in Id2 knock-out mice compared with their wild-type littermates. Transplantation of BMCs from Id2 knock-out mice confirmed these results and showed that Id2 is an intrinsic negative regulator of B-cell development. In addition, knock-down of Id2 by shRNA in hematopoietic progenitor cells promoted B-cell differentiation and induced the expression of B-cell lineage–specific genes. These genes are regulated by E2A protein, suggesting that Id2 regulates B-cell development by regulating E2A function. In contrast, Id1 does not have an intrinsic effect on B-cell development based on the analysis of Id1 knock-out mice (H.C.S., M.J., J. Gooya, M. Lee, K.D.K., J.R.K., manuscript submitted May 2008). Our data suggest that Id2 is the physiologically relevant regulator of E2A during B lymphopoiesis. Id2 has also been implicated in the later stages of B-cell development. For example, Id2 negatively controls differentiation into mature B2 cells while allowing the commitment to MZ B cells. In the absence of Id2 control, the unregulated differentiation is directed toward the mature B2 population.37 Mature B cells lacking Id2 have increased E2A activity, which leads to specific enhancement of germ line transcription of the immunoglobulin epsilon locus. Thus, Id2-deficient B cells undergo class switch recombination to IgE at a much higher frequency than wild-type B cells.38 Therefore, Id2 regulates B-cell development at multiple stages. Consistent with the regulatory role of Id2 in normal B-cell development, aberrant expression of Id2 was observed in Hodgkin lymphoma, which may contribute to the loss of the B cell–specific gene expression in Hodgkin-Reed/Sternberg (HRS) cells.39,40 Our data show that knock-down of Id2 by shRNA in hematopoietic progenitor cells promoted B-cell differentiation and induced the expression of B-cell lineage–specific genes, suggesting the potential of Id2 as a therapeutic target of Hodgkin lymphoma.
Id1 was shown to inhibit erythroid differentiation in the erythroleukemia cell line MEL.41 However, little is known about the function of Id2 in erythroid differentiation. Here we evaluated Id2 effect in erythroid differentiation in vitro and in vivo. Our in vitro study in multipotent EML cells and purified normal hematopoietic progenitors indicates that Id2 enhances erythroid commitment. Consistent with what we observed in vitro, overexpression of Id2 led to increased erythropoiesis, and loss of Id2 function resulted in impaired erythropoiesis in vivo. Apparently, the function of Id2 in erythroid development is different from Id1, which inhibits erythroid differentiation in the erythroleukemia cell line.41 However, it is worth noting that we evaluated the function of Id2 during early erythropoiesis in normal hematopoietic progenitors, a system quite different from the erythroleukemia cell line.
Id proteins were initially identified as negative regulators of bHLH transcription factors. Later studies indicated that Id proteins interact with transcription factors from many other protein families, such as Ets, Pax, and Rb.23-25 Here, we showed that Id2 interacts with PU.1 and represses PU.1 transcription activity. Furthermore, we found that Id2 and PU.1 interaction interferes with GATA-1 and PU.1 interaction. It has been demonstrated that GATA-1 and PU.1 interaction is mediated via the PU.1 ETS domain and the C-terminal finger region of GATA-1.42 Although Id2 and PU.1 interaction was not characterized in detail, previous studies showed that ETS domain binds to Id proteins. In fact, among Id family members, Id2 binds ETS domain with the highest efficiency.24 Thus, Id2 competes with GATA-1 for PU.1 binding, which leads to release of PU.1-mediated repression of GATA-1 activity. The antagonistic interaction between PU.1 and GATA-1 is critical in initiating the myeloid versus the erythroid program.16,17 Based on our data, we suggest the following model for Id2 function: high levels of Id2 decrease GATA-1 and PU.1 interaction, release PU.1-mediated repression of GATA-1 activity, and promote the erythoid program; low levels of Id2 increase GATA-1 and PU.1 interaction, allow stronger GATA-1 repression, and favor the myeloid program. Based on our model, correlations between Id2, PU.1, and GATA-1 expression would be expected. Indeed, quantitive real-time PCR showed that Id2 expression levels are up-regulated in MEPs and down-regulated in GMPs, which is positively correlated with GATA-1 level and negatively correlated with PU.1 level.4
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Steve Stull for technical assistance in BM transplantation, and Kathleen Noer, Roberta Matthai, and Samatha Bauchiero for technical assistance in FACS. We also thank Drs Francis W. Ruscetti and Kristbjorn Orri Gudmundsson for critical review of the manuscript.
This project was funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health (contract number NO1-CO-12 400). This research was supported in part by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
National Institutes of Health
Authorship
M.J. designed and performed research, analyzed data, and wrote the paper; H.L., H.-C.S., and K.K. performed research and analyzed data; Y.Y. contributed vital new reagents or analytical tools; J.R.K. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jonathan R. Keller, Basic Research Program, SAIC-Frederick, National Cancer Institute at Frederick, Bldg 560, Rm 12–03, Frederick, MD 21702; e-mail: kellerj@ncifcrf.gov.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal