Abstract
CD9 has been reported to play a role in tumor metastasis suppression. However, it is not fully understood how CD9 affects the hematogenous spread of tumor cells. To clarify a new mechanism (or mechanisms), we generated HT1080 cells that had been transfected with a CD9-expressing plasmid. Ectopic expression of CD9 in HT1080 cells actually reduced their metastatic ability. CD9 expression reduced lung retention and platelet ag-gregation activity of the transfectants. Because HT1080 cells express the metastasis-promoting, platelet aggregation-inducing factor Aggrus/podoplanin on their surface, we examined the relationship between CD9 and Aggrus. We discovered that CD9 formed a complex with Aggrus via transmembrane domains 1 and 2 (TM1 and TM2) of CD9. Investigation of the interaction revealed that each CD9 and Aggrus interacted homophilically, and that they colocalized in low-density membrane fractions. Deleting TM1 and TM2 attenuated the ability of CD9 to interact homophilically or to localize in low-density membrane fractions. The expression of CD9–wild-type (WT), but not CD9 lacking TM1 and TM2, attenuated the platelet aggregation and metastasis induced by forced expression of Aggrus in CHO cells. Therefore, CD9 may act as a metastasis suppressor, at least in part, by neutralizing Aggrus-mediated platelet aggregation.
Introduction
Four-pass transmembrane proteins of the tetraspanin family regulate cell migration, fusion, and signaling events by functioning as organizers of a multimolecular membrane complex called the tetraspanin web or tetraspanin-enriched microdomains.1,2 The CD9 protein is a member of the tetraspanin family and has been identified as a suppressor of cancer spread.3 Numerous previous studies have shown reduced CD9 expression in various cancers, which correlated with the presence of distant metastasis and a poorer prognosis.4-11 The clinical importance of CD9 in the diagnosis, staging, and prognosis of tumors has been growing.12 Recently, it was reported that adenoviral transduction of CD9 inhibited lymph node metastasis in an orthotopic lung cancer model.13 The investigators evaluated metastatic growth in the mediastinal lymph nodes using gene delivery of CD9. These findings strongly suggest that CD9 plays indispensable roles in malignant tumor progression and shows the feasibility of gene therapy to prevent metastasis. However, we cannot say that the therapeutic application of CD9 is now available. Functions of tetraspanins, to a great extent, may depend on expression patterns of their counterparts or other molecules on the tetraspanin web. Actually, there are reports that overexpression of CD9 does not affect metastatic properties,14 or that CD9 expression is down-regulated at the primary site but reexpressed at the metastatic site.15 So, the precise mechanisms of the metastasis suppression should be elucidated more clearly.
Aggrus/podoplanin, a type-I transmembrane glycoprotein, has been shown to be up-regulated in a number of different cancers,16-23 suggesting that increased expression of Aggrus is associated with tumor malignancy and poor prognosis.24,25 We previously showed that Aggrus expression induced platelet aggregation with no requirement for plasma components,26 and we have discovered that Aggrus promoted pulmonary metastasis in mice.27 Platelet aggregation–inducing activity of Aggrus is directly associated with metastasis formation because introducing a point mutation that suppresses platelet aggregation or administering aspirin diminished the formation of pulmonary metastasis.27 It was recently reported that C-type lectin-like receptor 2 (CLEC-2) expressed on platelets was identified as one of the counterparts of Aggrus.28,29 When CLEC-2 binds to Aggrus expressed on tumor cells, it generates activation signals, intra-platelet phosphorylation, and eventually induces platelets to degranulate.30 Tumor cell–induced platelet aggregation is believed to facilitate large tumor-platelet aggregate formation, to embolize the microvasculature, and to promote tumor cell metastasis.31 Another survival advantage is tumor cell protection from immunologic assault and shear forces in the circulation.32,33 Indeed, mice without platelets, due to genetic elimination of Nf-E2, showed marked resistance to hematogenous metastasis formation.34 Therefore, Aggrus-mediated platelet aggregation may be a potential target for developing a hematogenous metastasis inhibitor.
In this study, we investigated how CD9 affected the hematogenous spread of tumor cells as a metastasis suppressor. Ectopic expression of CD9 in HT1080 cells reduced their metastatic ability without affecting their growth in vitro and in vivo. CD9 expression reduced platelet aggregation–inducing activity of HT1080 cells. Consistent with the results, ex vivo imaging of tumor cells revealed that CD9 expression reduced lung retention of HT1080 cells. Because HT1080 cells express the platelet aggregation–inducing factor Aggrus on their surfaces, we further examined the relationship between CD9 and Aggrus. We discovered that CD9 formed a complex with Aggrus, via transmembrane domains 1 and 2 (TM1 and TM2). The CD9-Δ6, lacking TM1 and TM2, could not form a complex with Aggrus. Moreover, CD9 and Aggrus interacted homophilically and localized in low-density membrane fractions (LMFs), while CD9-Δ6 could not interact homophilically and hardly existed in LMFs under Brij 97 treatment. Finally, we investigated the effects of CD9–wild-type (WT) or CD9-Δ6 on platelet aggregation and pulmonary metastasis in CHO cells overexpressing Aggrus. CD9-WT, but not CD9-Δ6, suppressed the abilities of Aggrus. Therefore, CD9 may act as a metastasis suppressor, at least in part, by neutralizing Aggrus-mediated platelet aggregation.
Methods
Cell culture conditions
HT1080 and CHO cells were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum (FBS; Sigma-Aldrich, St Louis, MO). LN319 cells were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% heat-inactivated FBS.
Animals
Female BALB/c mice, BALB/c-nu/nu mice, and C.B-17/Icr-scid mice were purchased from Charles River Japan (Kanagawa, Japan). All animal procedures were performed using protocols approved by the Japanese Foundation for Cancer Research Animal Care and Use Committee.
Plasmids and cell preparation
The human aggrus, CD9, and CD44 cDNAs were subcloned into a pcDNA3 vector. Substitution of the appropriate cysteine codons to serine codons in human CD9 cDNA was accomplished using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Deletion of the appropriate region in human CD9 cDNA was accomplished by polymerase chain reaction (PCR). HT1080 cells were transfected with vectors encoding nothing (mock), CD9-WT, or CD44 using the LipofectAMINE 2000 reagent (Invitrogen, Carlsbad, CA). Stable transfectants were selected by cultivating the cells in medium containing 0.4 mg/mL geneticin (Sigma-Aldrich). A pRetroQ-AcGFP-N1 retroviral vector was purchased from Clontech (Mountain View, CA). For ex vivo imaging, HT1080 cells were infected with the retroviral vector according to the manufacturer's instruction. Then, the established HT1080 (HT1080/AcGFP) was further transfected with plasmids using the LipofectAMINE 2000 reagent. After selection by cultivating the cells in the medium containing 0.5 mg/mL geneticin, stable transfectants were established. CHO cells were transfected with vectors encoding nothing (mock) or Aggrus using the LipofectAMINE 2000 reagent. Stable transfectants were selected by cultivating the cells in medium containing 1.0 mg/mL geneticin. Then the stable transfectants were transiently transfected with plasmids. To confirm the dependence on Aggrus expression in HT1080-induced platelet aggregation, cells were preincubated with rat anti–human Aggrus antibody (NZ-1) or control rat IgG for 1 hour at 4°C in Hanks balanced salt solution (HBSS). In some experiments, cells were transfected with aggrus-specific siRNA using the LipofectAMINE 2000 reagent. The coding strand of the siRNA-directed residues 183 through 201 of human aggrus mRNA was GUCUGGCUUGACAACUCUG. Nonsilencing control siRNA was purchased from Qiagen (Chatsworth, CA).
Flow cytometry
Cells were harvested by brief exposure to trypsin. After washing with phosphate-buffered saline (PBS), cells were treated with an anti–human CD9 monoclonal antibody (Sigma-Aldrich) or control mouse IgG, then were incubated with Oregon Green 488–conjugated antibody (Invitrogen). Flow cytometric analysis was performed using a Cytomics FC500 flow cytometry system (Beckman-Coulter, Miami, FL).
Experimental lung metastasis and ex vivo imaging of lung retention
Cells were harvested, washed, and resuspended in HBSS (HT1080, 106 cells/mL; CHO, 2.5 × 106 cells/mL). Then, 200 μL of the cell suspension were intravenously inoculated into lateral tail veins (HT1080, 8-week-old female C.B-17/Icr-scid mice; CHO, 8-week-old female BALB/c-nu/nu mice). After 28 days the mice were killed and surface lung metastatic foci were counted. For ex vivo imaging of lung retention, AcGFP-expressing HT1080 cells were resuspended in HBSS (106 cells/mL). Then 200 μL of the cell suspension were intravenously injected into lateral tail vein (8-week-old female BALB/c-nu/nu mice). After 5 minutes or 4 hours the mice were killed and the lungs were extirpated. The fluorescence was visualized with an IV100 imaging system (Olympus, Tokyo, Japan; 6×/0.14) and analyzed by FLUOVIEW software (Olympus); the fluorescence area was quantitated with NIH ImageJ software (National Institutes of Health, Bethesda, MD).
In vitro and in vivo proliferation assays
To assess in vitro cell proliferation, the MTT assay was used after 24, 48, or 72 hours of culture. For the in vivo proliferation assay, HT1080 cells were harvested, washed, and resuspended in HBSS. A total of 5 × 106 cells in 0.1 mL HBSS were subcutaneously injected into the backs of 8-week-old female C.B-17/Icr-scid mice. Tumor volume was calculated by the following formula: volume = W2 × L / 2, where W is the short diameter and L is the long diameter.
Platelet aggregation assay
Cells were harvested, washed, and resuspended in PBS (HT1080, 5 × 106 cells/mL; CHO, 107 cells/mL). Mouse platelet-rich plasma (PRP) was prepared from fresh, heparinized blood extracted from retired female BALB/c mice. A 200-μL aliquot of PRP was incubated at 37°C. A total of 10 μL cell suspension was added, and then light transmittance was monitored at 660 nm with an MCM HEMA TRACER 313M (SSR Engineering, Kanagawa, Japan). Platelet aggregation-inducing activity was calculated using the following formula: platelet aggregation–inducing activity = 1 / A, where A is the time from reaction-starting point to maximum value point. For the assay of forced platelet activation, 10 μL ADP (40 μM in PBS; WAKO, Osaka, Japan) and thrombin (50 units/mL in PBS; Invitrogen) were added to 200 μL PRP that had been preincubated with CHO/mock or CHO/CD9 cells for 10 minutes. For intraplatelet phosphorylation assay, mouse platelets were obtained by centrifugation of PRP with heparin and citrate-dextrose solution and resuspended in modified Tyrode buffer containing 1 mM GRGDS peptide. A total of 5 × 105 HT1080 cells were added to platelets, and reactions were terminated by the addition of 2× lysis buffer (2% Nonidet P-40, 20 mM Tris, 300 mM NaCl, 2 mM EDTA) after indicated times. The lysates were then electrophoresed and immunoblotted.
Immunoprecipitation and immunoblot analysis
Cells were harvested and solubilized in lysis buffer containing 1% detergent (Triton X, Brij 97, or CHAPS), 25 mM HEPES, 150 mM NaCl, 5 mM MgCl2, 1 mM phenylmethanesulfonyl fluoride, and 1 mM aprotinin. The FLAG-tagged proteins were immunoprecipitated with an anti-FLAG agarose (M2; Sigma-Aldrich). Endogenous CD9 and Aggrus were immunoprecipitated with protein G–Sepharose (Zymed Laboratories, South San Francisco, CA) that had been conjugated with antibodies to CD9 (M-L13; BD Biosciences, San Jose, CA) or Aggrus. Then, the immunoprecipitated proteins or the cell lysates were electrophoresed and blotted onto a nitrocellulose membrane. The membranes were incubated with antibodies to CD9, Aggrus, FLAG tag (Sigma-Aldrich), V5 tag (V5-10; Invitrogen), caveolin (BD Biosciences), Erk1/2 (Cell Signaling, Beverly, MA), phospho-Erk1/2 (Cell Signaling), or β-actin (Sigma-Aldrich). Subsequently, the membranes were incubated with a horseradish peroxidase–conjugated secondary antibody. After washing, the membranes were developed with an enhanced chemiluminescence (ECL) system.
Ultracentrifuge fractionation in sucrose gradients
CHO transfectants (107) were solubilized in 500 μL of lysis buffer containing 1% detergent (CHAPS or Brij 97), 25 mM MES, 150 mM NaCl, and 5 mM MgCl2. Lysates were then mixed with an equal volume of 90% sucrose. This 45% solution was overlaid onto 1.5 mL of 35% sucrose and 1 mL of 5% sucrose. Samples were centrifuged at 20 000g for 18 hours at 4°C, and 300 μL of each fraction were collected from the top of the gradient.35 The fractions were then electrophoresed and immunoblotted.
Immunostaining
LN319 cells were plated onto glass-bottomed dishes. After incubation for 20 hours, the cells were fixed and permeabilized with 4% paraformaldehyde and 0.2% Triton X in PBS for 10 minutes. Labeling was carried out by incubation for 1 hour with antibodies to Aggrus (NZ-1) or CD9 (DU-ALL-1), followed by incubation with Alexa Fluor 488–conjugated goat anti–rat IgG (for NZ-1) or Alexa Fluor 633–conjugated goat anti–mouse IgG (for DU-ALL-1) (Invitrogen). Subsequently, the cells were incubated with Hoechst 33342 (Invitrogen). After washing, the specimens were visualized using a fluorescence confocal microscope (FV1000; Olympus. 60×/1.42 oil objective) and analyzed by FLUOVIEW software (Olympus).
Invasion assay
Invasion of HT1080 cells was assessed by measuring their capacity to migrate across transwell inserts using BD Bio-Coat Matrigel invasion chamber in 6-well plates, 8-μm pore (BD Biosciences). A total of 2 × 105 HT1080 cells in serum-free RPMI 1640 medium were added to the upper compartment, and RPMI 1640 medium supplemented with 20% heat-inactivated FBS was added to the lower compartment. After incubation for 20 hours, invading HT1080 cells in the lower compartment were counted.
Gelatin zymography
Gelatin zymography was performed using a Gelatinzymo Electrophoresis Kit (Yagai, Yamagata, Japan). Supernatant was collected from HT1080 cells cultured in serum-free RPMI 1640 medium (with or without 50 ng/mL of phorbol 12-myristate 13-acetate; TPA) for 20 hours. The culture supernatants were concentrated with Centricon (Millipore, Bedford, MA) and underwent SDS-PAGE using a 7.5% gel containing 0.1% gelatin. After electrophoresis, the gel was soaked in 2.5% Triton X solution at 23°C with gentle shaking for 1 hour. After incubation for 40 hours in reaction buffer (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 10 mM CaCl2, and 0.01% Brij 35) at 37°C, the gels were stained with Coomassie Brilliant Blue (WAKO).
Statistical analysis
All data are shown as means plus or minus SD. The Student t test was performed. P values less than .05 were considered statistically significant. All statistical tests were 2-sided.
Results
CD9 suppressed pulmonary metastasis and lung retention of HT1080 cells
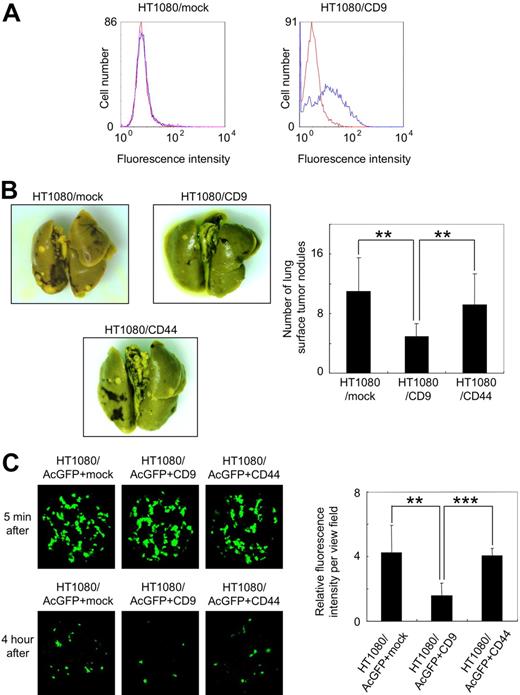
Forced expression of CD9 in tumor cells has been reported to reduce their metastatic ability.13,36-38 To confirm these reports, we prepared polyclonal HT1080 cells that had been stably transfected with vectors encoding nothing (HT1080/mock) or CD9 (HT1080/CD9). Although endogenous CD9 expression could not be detected in HT1080/mock cells, CD9 expression was clearly seen in HT1080/CD9 cells (Figure 1A). CD44 is a transmembrane glyco-protein involved in cell adhesion. We also established stably CD44-transfected polyclonal HT1080 cells (HT1080/CD44) as negative control cells. We then intravenously inoculated the HT1080/mock, HT1080/CD9, or HT1080/CD44 cells into C.B-17/Icr-scid mice. At 28 days after tumor inoculation, the number of lung surface nodules was counted, and the number in the HT1080/CD9-injected mice was significantly lower than in the HT1080/mock- or HT1080/CD44-injected mice (Figure 1B; **P < .005).
Ectopic expression of CD9 suppressed pulmonary metastasis and lung retention of HT1080 cells. (A) Stably transfected polyclonal HT1080/mock (left panel) and HT1080/CD9 (right panel) cells were treated with a control mouse IgG (red line) or an anti-CD9 antibody (blue line). After incubation with Oregon Green 488–conjugated second antibody, CD9 expression was estimated by flow cytometer. (B) HT1080/mock, HT1080/CD9, and HT1080/CD44 cells were intravenously injected into C.B-17/Icr-scid mice (n = 10). At 28 days after tumor inoculation, lung surface nodules were counted. Representative pictures of the lungs (left panels) and detailed numbers of the lung metastasis nodules in each mouse (right panel) are shown. Data are means plus or minus SD. **P < .005 by Student t test. (C) Stably transfected polyclonal HT1080/AcGFP+mock, HT1080/AcGFP+CD9, and HT1080/AcGFP+CD44 cells were intravenously injected into BALB/c-nu/nu mice (n = 6). After 5 minutes or 4 hours, the lungs were extirpated. The fluorescence was visualized using an IV100 imaging system, and the fluorescence area was quantitated. Representative pictures of the lungs (left panels) and detailed fluorescence area (right panels) are shown. Data are means plus or minus SD. **P < .005; ***P < .001 using the Student t test.
Ectopic expression of CD9 suppressed pulmonary metastasis and lung retention of HT1080 cells. (A) Stably transfected polyclonal HT1080/mock (left panel) and HT1080/CD9 (right panel) cells were treated with a control mouse IgG (red line) or an anti-CD9 antibody (blue line). After incubation with Oregon Green 488–conjugated second antibody, CD9 expression was estimated by flow cytometer. (B) HT1080/mock, HT1080/CD9, and HT1080/CD44 cells were intravenously injected into C.B-17/Icr-scid mice (n = 10). At 28 days after tumor inoculation, lung surface nodules were counted. Representative pictures of the lungs (left panels) and detailed numbers of the lung metastasis nodules in each mouse (right panel) are shown. Data are means plus or minus SD. **P < .005 by Student t test. (C) Stably transfected polyclonal HT1080/AcGFP+mock, HT1080/AcGFP+CD9, and HT1080/AcGFP+CD44 cells were intravenously injected into BALB/c-nu/nu mice (n = 6). After 5 minutes or 4 hours, the lungs were extirpated. The fluorescence was visualized using an IV100 imaging system, and the fluorescence area was quantitated. Representative pictures of the lungs (left panels) and detailed fluorescence area (right panels) are shown. Data are means plus or minus SD. **P < .005; ***P < .001 using the Student t test.
To assess the difference, we examined lung retention of tumor cells 5 minutes or 4 hours after inoculation. To visualize tumor cell retention, we first tried to generate AcGFP-expressing HT1080 cells. The established HT1080/AcGFP cells were further transfected with vectors encoding nothing (HT1080/AcGFP+mock), CD9 (HT1080/AcGFP+CD9), or CD44 (HT1080/AcGFP+CD44). Then, the established polyclonal HT1080/AcGFP+mock, HT1080/AcGFP+CD9, and HT1080/AcGFP+CD44 cells were intravenously injected into BALB/c-nu/nu mice. After 5 minutes or 4 hours the mice were killed and the lungs were extirpated. The fluorescence area was visualized and quantitated (Figure 1C). After 5 minutes, there was little difference among the 3 samples. After 4 hours, however, lung retention of HT1080/AcGFP+CD9 cells had drastically declined compared with others (Figure 1C; **P < .005; ***P < .001).
CD9 expression attenuated platelet aggregation induced by Aggrus without affecting cell growth
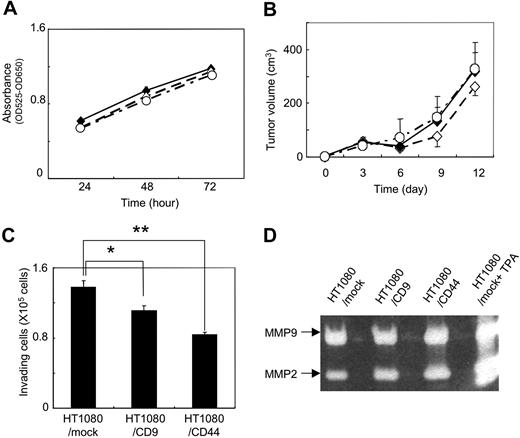
To identify the determinants associated with CD9-mediated metastasis suppression in vivo (Figure 1), we first estimated the growth of HT1080/mock, HT1080/CD9, and HT1080/CD44 cells by MTT assay. We also examined the in vivo growth of HT1080/mock, HT1080/CD9, and HT1080/CD44 cells that had been injected subcutaneously into the back of C.B-17/Icr-scid mice. We found almost no difference between in vitro and in vivo cell growth among HT1080/mock, HT1080/CD9, and HT1080/CD44 cells (Figure 2A,B). According to these results, CD9 expression might prevent ectopic dissemination without blocking tumor formation.
In vitro and in vivo growth and invasion of HT1080 transfectants. (A) Stably transfected polyclonal HT1080/mock (♦), HT1080/CD9 (◇), and HT1080/CD44 cells (○) were plated onto each well of 24-well plates. After incubation for 24, 48, or 72 hours, MTT assay was performed. Data are means plus or minus SD of triplicate determinations. (B) HT1080/mock (♦), HT1080/CD9 (◇), and HT1080/CD44 (○) cells were subcutaneously injected into the backs of C.B-17/Icr-scid mice (n = 5). Tumors were measured with calipers at 3, 6, 9, 12, and 15 days after injection. Tumor volume was calculated using the following formula: volume = W2 × L / 2, where W is the short diameter and L is the long diameter. Data are means plus or minus SD. (C) HT1080/mock, HT1080/CD9, and HT1080/CD44 cells in serum-free RPMI medium were added to the upper compartment of Matrigel-coated invasion chambers. Data are means plus SD of triplicate determinations. *P < .05; **P < .005 using the Student t test. (D) HT1080/mock, HT1080/CD9, and HT1080/CD44 cells were cultured in serum-free RPMI medium for 20 hours. In an experiment, HT1080/mock cells were stimulated with 50 ng/mL TPA (HT1080/mock+TPA). The culture supernatant was concentrated and electorophoresed. The gels were incubated for 40 hours in reaction buffer and stained with Coomassie Brilliant Blue.
In vitro and in vivo growth and invasion of HT1080 transfectants. (A) Stably transfected polyclonal HT1080/mock (♦), HT1080/CD9 (◇), and HT1080/CD44 cells (○) were plated onto each well of 24-well plates. After incubation for 24, 48, or 72 hours, MTT assay was performed. Data are means plus or minus SD of triplicate determinations. (B) HT1080/mock (♦), HT1080/CD9 (◇), and HT1080/CD44 (○) cells were subcutaneously injected into the backs of C.B-17/Icr-scid mice (n = 5). Tumors were measured with calipers at 3, 6, 9, 12, and 15 days after injection. Tumor volume was calculated using the following formula: volume = W2 × L / 2, where W is the short diameter and L is the long diameter. Data are means plus or minus SD. (C) HT1080/mock, HT1080/CD9, and HT1080/CD44 cells in serum-free RPMI medium were added to the upper compartment of Matrigel-coated invasion chambers. Data are means plus SD of triplicate determinations. *P < .05; **P < .005 using the Student t test. (D) HT1080/mock, HT1080/CD9, and HT1080/CD44 cells were cultured in serum-free RPMI medium for 20 hours. In an experiment, HT1080/mock cells were stimulated with 50 ng/mL TPA (HT1080/mock+TPA). The culture supernatant was concentrated and electorophoresed. The gels were incubated for 40 hours in reaction buffer and stained with Coomassie Brilliant Blue.
We next examined the effects of CD9 on the invading ability of tumor cells. There are some reports that forced expression of CD9 in tumor cells reduced their motility, resulting in reduction of metastatic ability.36,39,40 We thus estimated the invading ability of HT1080/mock, HT1080/CD9, and HT1080/CD44 cells using a Matrigel invasion assay (Figure 2C) and gelatin zymography (Figure 2D). Consistent with the previous reports, CD9 expression slightly, but significantly, reduced HT1080 cell invasion (Figure 2C). However, ectopic expression of CD9 did not affect the activity of matrix metalloproteinase (MMP)–2 and MMP-9 (Figure 2D). CD9 probably reduced motility rather than invasion of HT1080 cells. CD44 expression also reduced HT1080 cell invasion to the Matrigel without affecting MMP-2 and MMP-9 activities. Interestingly, CD44 expression exhibited little effect on metastasis in vivo (Figure 1B). These results suggest that attenuation of motility might not be the reason for in vivo metastasis suppression by CD9 in HT1080 cells.
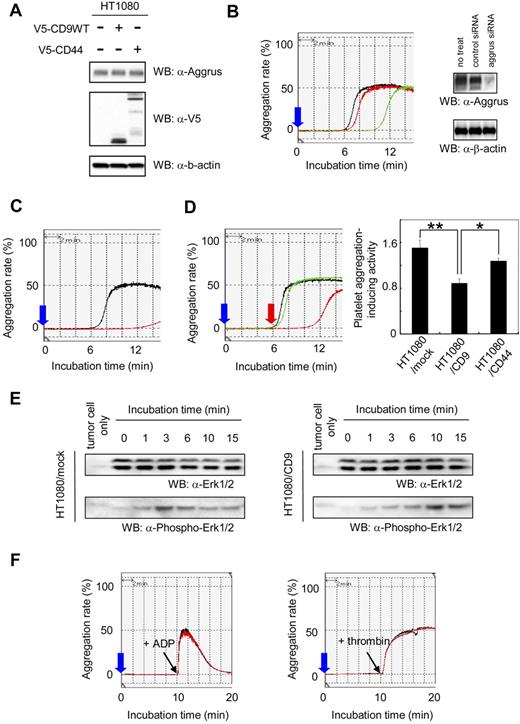
We hypothesized that CD9 affects the state of tumor cells circulating in the bloodstream (eg, protection and embolization of tumor cells in microvasculature). One possible target of CD9 was tumor cell-induced platelet aggregation. The aggregation was associated with large tumor-platelet aggregate formation, protection of tumor cells from immunologic assault, and embolization in the microvasculature.32,33 We previously identified Aggrus as a platelet aggregation-inducing factor,26 and discovered that Aggrus promoted pulmonary metastasis in vivo.27 Immunoblot analysis showed that HT1080 cells endogenously expressed Aggrus. Ectopic expression of CD9 or CD44 did not affect the expression level of endogenous Aggrus in HT1080 cells (Figure 3A). To estimate the activity and dependency of Aggrus on HT1080-induced platelet aggregation, a platelet aggregation assay was used with HT1080 cells that had been transfected with aggrus-specific siRNA (Figure 3B) or cells that had been preincubated with a neutralizing antibody to Aggrus (Figure 3C). Platelet aggregation induced by HT1080 cells was attenuated by both aggrus gene silencing and the neutralizing-antibody treatment. We also examined lung retention and experimental metastasis of HT1080 cells in vivo, in the presence or absence of the neutralizing antibody. We found that the antibody treatment reduced lung retention and pulmonary metastasis of HT1080 cells (data not shown). These results indicate that Aggrus is associated with platelet aggregation, lung retention, and in vivo metastatic potency of HT1080 cells.
Effect of CD9 expression on Aggrus-induced platelet aggregation. (A) HT1080 cells that had been stably transfected with expression vector encoding nothing, V5-tagged CD9, or V5-tagged CD44 were lysed and immunoblotted with antibodies to Aggrus, V5 tag, or β-actin. (B) HT1080 cells that had been transfected with nothing (black line), control siRNA (red line), or aggrus-specific siRNA (green line) were incubated with mouse PRP. Then, light transmittance was monitored for 15 minutes. The blue arrow indicates when cells are added. Aggrus expression level in each sample was estimated by Western blot analysis with an antibody to Aggrus or β-actin (right panels). (C) HT1080 cells were preincubated with control rat IgG (black line) or rat anti-Aggrus antibody (red line). Then, the reactions were incubated with mouse PRP. Light transmittance was monitored for 15 minutes. (D) Stably transfected polyclonal HT1080/mock (black line), HT1080/CD9 (red line), and HT1080/CD44 (green line) were incubated with mouse platelet-rich plasma. Then, light transmittance was monitored for 15 minutes (left panel). The blue arrow indicates when cells are added; red arrow, when platelet aggregation begins. Platelet aggregation–inducing activity was calculated using the following formula: platelet aggregation-inducing activity = 1 / A, where A is the time from reaction-starting point to maximum value point. Each point represents a mean plus or minus SD of triplicate experiments. *P < .05; **P < .005 using the Student t test. (E) Stably transfected polyclonal HT1080/mock and HT1080/CD9 cells were added to platelets, and reactions were terminated by the addition of 2 × lysis buffer after indicated times. The lysates were electrophoresed and immunoblotted with an antibody to Erk1/2 and phospho-Erk1/2. (F) CHO cells were transiently transfected with a CD9-expressing vector (CHO/CD9; red line) or an empty vector (CHO/mock; black line). Mouse PRP was preincubated with CHO/mock or CHO/CD9 for 10 minutes. Then, 10 μL of ADP (40 μM in PBS; left panel) or thrombin (50 U/mL in PBS; right panel) was added to the reactions.
Effect of CD9 expression on Aggrus-induced platelet aggregation. (A) HT1080 cells that had been stably transfected with expression vector encoding nothing, V5-tagged CD9, or V5-tagged CD44 were lysed and immunoblotted with antibodies to Aggrus, V5 tag, or β-actin. (B) HT1080 cells that had been transfected with nothing (black line), control siRNA (red line), or aggrus-specific siRNA (green line) were incubated with mouse PRP. Then, light transmittance was monitored for 15 minutes. The blue arrow indicates when cells are added. Aggrus expression level in each sample was estimated by Western blot analysis with an antibody to Aggrus or β-actin (right panels). (C) HT1080 cells were preincubated with control rat IgG (black line) or rat anti-Aggrus antibody (red line). Then, the reactions were incubated with mouse PRP. Light transmittance was monitored for 15 minutes. (D) Stably transfected polyclonal HT1080/mock (black line), HT1080/CD9 (red line), and HT1080/CD44 (green line) were incubated with mouse platelet-rich plasma. Then, light transmittance was monitored for 15 minutes (left panel). The blue arrow indicates when cells are added; red arrow, when platelet aggregation begins. Platelet aggregation–inducing activity was calculated using the following formula: platelet aggregation-inducing activity = 1 / A, where A is the time from reaction-starting point to maximum value point. Each point represents a mean plus or minus SD of triplicate experiments. *P < .05; **P < .005 using the Student t test. (E) Stably transfected polyclonal HT1080/mock and HT1080/CD9 cells were added to platelets, and reactions were terminated by the addition of 2 × lysis buffer after indicated times. The lysates were electrophoresed and immunoblotted with an antibody to Erk1/2 and phospho-Erk1/2. (F) CHO cells were transiently transfected with a CD9-expressing vector (CHO/CD9; red line) or an empty vector (CHO/mock; black line). Mouse PRP was preincubated with CHO/mock or CHO/CD9 for 10 minutes. Then, 10 μL of ADP (40 μM in PBS; left panel) or thrombin (50 U/mL in PBS; right panel) was added to the reactions.
Next, we checked the platelet aggregation-inducing activity of HT1080/mock, HT1080/CD9, and HT1080/CD44 cells. The activity was calculated using the following formula: platelet aggregation-inducing activity = 1/A, where A is the time from reaction-starting point to maximum value point. As shown in Figure 3D, ectopic expression of CD9, but not CD44, down-regulated the platelet aggregation-inducing activity in HT1080 cells (Figure 3D; **P < .005; *P < .05). Characteristics of this inhibition were prolonged lag phase and minimal effect on either the slope of the aggregation curve or maximal aggregation. Therefore, we analyzed intraplatelet phosphorylation of Erk1/2, which was used an indicator of platelet activation.41,42 The expression level of Erk1/2 in HT1080 cells was very low compared with that of platelets (Figure 3E; tumor cell only). As shown in Figure 3E, addition of HT1080/mock cells rapidly increased the phospho-Erk1/2 level. However, addition of HT1080/CD9 cells slowly increased the phospho-Erk1/2 level. The delay of intraplatelet phosphorylation of Erk1/2 might be associated with the prolonged lag phase after HT1080/CD9 addition (Figure 3D). Importantly, CD9 expression seemed not to affect the maximal signal intensity of phospho-Erk1/2 level, which was correlated with no alteration of maximum platelet aggregation (Figure 3D). To exclude the possibility that CD9 acts directly on platelets to inhibit their activation, we prepared CHO cells that had been transiently transfected with a vector encoding nothing (CHO/mock) or CD9 (CHO/CD9). After incubating cells with mouse platelet-rich plasma for 10 minutes, ADP and thrombin were added. ADP and thrombin are the most common platelet aggregation inducer of tumor cells. The ADP- or thrombin-induced platelet aggregation was not affected by pretreatment of the platelets with CD9 (Figure 3F). These results suggest that CD9 suppresses platelet aggregation not by directly affecting platelet activity but by interfering with Aggrus function.
CD9 formed a complex via transmembrane domains 1 and 2 and colocalized with Aggrus
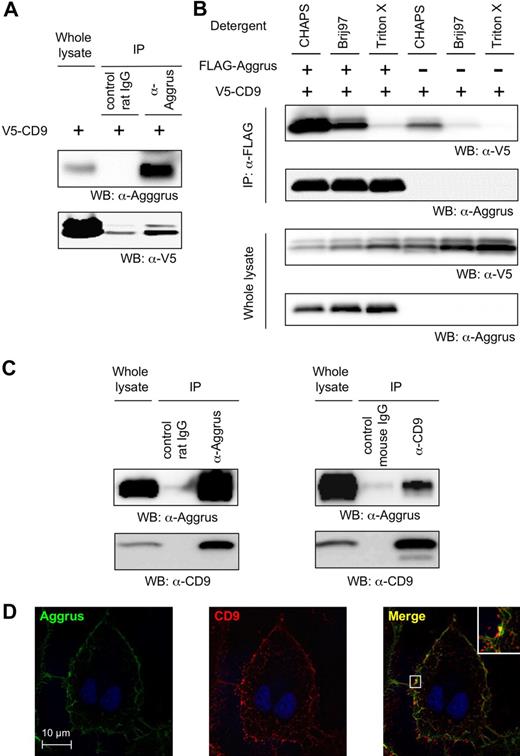
Next, we examined the direct interaction between CD9 and Aggrus. When endogenous Aggrus was immunoprecipitated with an anti-Aggrus antibody, CD9 was coimmunoprecipitated (Figure 4A). To determine the strength of interaction between CD9 and Aggrus, we transfected CHO cells with FLAG-tagged Aggrus and V5-tagged CD9. Then, cells were lysed with buffer containing 1% CHAPS, Brij 97, or Triton X. Interaction between CD9 and Aggrus was observed in the buffer containing 1% CHAPS or Brij 97 (Figure 4B). The results indicate that the interaction was a level 2 interaction of tetraspanins and was likely to be significant for tetraspanin function.35 We screened and found that a human glioblastoma cell line, LN319, enodogenously expressed both CD9 and Aggrus on its surface. When the endogenous CD9 protein expressed on LN319 cells was immunoprecipitated with an antibody to CD9 in the buffer containing 1% CHAPS, endogenous Aggrus was coimmunoprecipitated and vice versa (Figure 4C). An immunostaining experiment showed that CD9 was localized in a dotlike pattern on the LN319 cell membrane, suggesting its localization in tetraspanin-enriched microdomains (tetraspanin web; Figure 4D). Aggrus was colocalized on the overall cell membrane, but in specific regions, strong colocalization of CD9 and Aggrus could be detected (Figure 4D right panel, inset). These results suggest that CD9 and Aggrus interacted and colocalized on specific membrane regions.
Interaction between CD9 and Aggrus. (A) HT1080 cells were stably transfected with V5-tagged, CD9-encoding vector (HT1080/CD9). Then, endogenous Aggrus was immunoprecipitated with an anti-Aggrus antibody in buffer containing 1% CHAPS. Exogenous CD9 was coimmunoprecipitated with the anti-Aggrus antibody. (B) CHO cells were transfected with expression plasmids encoding FLAG-tagged Aggrus and V5-tagged CD9. Then, the cells were solubilized with buffer containing 1% CHAPS, Brij 97, or Triton X. The FLAG-tagged Aggrus was immunoprecipitated with an anti-FLAG agarose. The immunoprecipitated proteins were electrophoresed and immunoblotted with an antibody to V5 tag or Aggrus. (C) LN319 cells were solubilized with buffer containing 1% CHAPS. Then, endogenous proteins were immunoprecipitated with control IgG or antibody to CD9 or Aggrus. The immunoprecipitants were electrophoresed and immunoblotted with an antibody to Aggrus or CD9. (D) LN319 cells were immunostained with antibodies to CD9 and Aggrus. Aggrus was visualized by incubation with Alexa Fluor 488–conjugated goat anti–rat IgG (left panel). CD9 was visualized by incubating with Alexa Fluor 633–conjugated goat anti–mouse IgG (middle panel). Merged image is shown in right panel.
Interaction between CD9 and Aggrus. (A) HT1080 cells were stably transfected with V5-tagged, CD9-encoding vector (HT1080/CD9). Then, endogenous Aggrus was immunoprecipitated with an anti-Aggrus antibody in buffer containing 1% CHAPS. Exogenous CD9 was coimmunoprecipitated with the anti-Aggrus antibody. (B) CHO cells were transfected with expression plasmids encoding FLAG-tagged Aggrus and V5-tagged CD9. Then, the cells were solubilized with buffer containing 1% CHAPS, Brij 97, or Triton X. The FLAG-tagged Aggrus was immunoprecipitated with an anti-FLAG agarose. The immunoprecipitated proteins were electrophoresed and immunoblotted with an antibody to V5 tag or Aggrus. (C) LN319 cells were solubilized with buffer containing 1% CHAPS. Then, endogenous proteins were immunoprecipitated with control IgG or antibody to CD9 or Aggrus. The immunoprecipitants were electrophoresed and immunoblotted with an antibody to Aggrus or CD9. (D) LN319 cells were immunostained with antibodies to CD9 and Aggrus. Aggrus was visualized by incubation with Alexa Fluor 488–conjugated goat anti–rat IgG (left panel). CD9 was visualized by incubating with Alexa Fluor 633–conjugated goat anti–mouse IgG (middle panel). Merged image is shown in right panel.
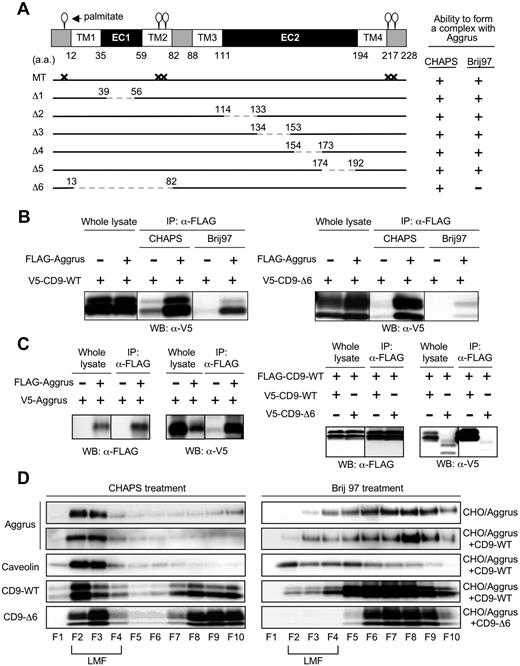
To identify the binding site in CD9, we prepared several CD9 point and deletion mutants. We generated a deletion mutant lacking the shorter extracellular loop (EC1) (Δ1: 39-56 deletion) or lacking the longer extracellular loop (EC2) (Δ2: 114-133 deletion; Δ3: 134-153 deletion; Δ4: 154-173 deletion; and Δ5: 174-192 deletion; Figure 5A). We also established a deletion mutant lacking TM1, EC1, and TM2 (Δ6: 13-82 deletion). Because palmitoylation supports assembly and function of tetraspanins, we generated a mutant that was unable to be palmitoylated (MT: C9S, C78S, C79S, C218S, and C219S; Figure 5A). Immunoprecipitation analysis showed that all the generated mutants, except CD9-Δ6, interacted with Aggrus under both CHAPS and Brij 97 treatment (data not shown), just as CD9-WT did (Figure 5B left panels). CD9-Δ6 interacted with Aggrus in the presence of CHAPS but not Brij 97 (Figure 5B right panels). CD9 TM1 and TM2 have been reported to be important in intra- and intermolecular interactions, and the interactions are fundamental in forming primary units within the tetraspanin web.43 In addition, the 3 other tetraspanins we tested could form complexes with Aggrus (data not shown). These results suggest that Aggrus constantly localizes and interacts with CD9 in the tetraspanin web.
Identification of CD9 mutant forming a weak complex with Aggrus and the tetraspanin web. (A) Schematic representation of the generated CD9 mutants: shorter extracellular loop (EC1)–deletion mutant (Δ1: 39-56 deletion); longer extracellular loop (EC2)–deletion mutants (Δ2: 114-133 deletion; Δ3: 134-153 deletion; Δ4: 154-173 deletion; and Δ5: 174-192 deletion); TM1, EC1, and TM2 deletion mutant (Δ6: 13-82 deletion); and palmitoylation-deficient point mutant (MT: C9S, C78S, C79S, C218S, and C219S). (B) CHO cells were transiently transfected with V5-tagged CD9-WT (left panels) or CD9-Δ6 (right panels) together with or without FLAG-tagged Aggrus. After solubilization with 1% CHAPS or Brij 97, the FLAG-tagged Aggrus was immunoprecipitated with an anti-FLAG agarose. The immunoprecipitants were analyzed by Western blot with an antibody to V5 tag. (C) CHO cells were transfected with plasmid encoding nothing or FLAG-Aggrus together with V5-Aggrus (left panels). In some experiments, CHO cells were transfected with plasmids encoding V5-CD9-WT or V5-CD9-Δ6 together with FLAG-CD9-WT (right panels). These cells were solubilized with lysis buffer containing 1% CHAPS. After immunoprecipitation with an anti-FLAG agarose, the immunoprecipitants were analyzed by Western blot with antibodies to V5 or FLAG. (D) CHO cells were transiently transfected with FLAG-Aggrus expressing plasmid together with plasmids encoding nothing (CHO/Aggrus), V5-CD9-WT (CHO/Aggrus+CD9-WT), or V5-CD9-Δ6 (CHO/Aggrus+CD9-Δ6). Then, cells were solubilized in lysis buffer containing 1% CHAPS or Brij 97. Each cell lysate was centrifuged in sucrose gradients and fractioned. Then, each fraction was electrophoresed and analyzed by Western blot with antibodies to Aggrus, caveolin, FLAG tag, or V5 tag. LMFs were concentrated in fraction numbers 2 through 4 (F2-F4).
Identification of CD9 mutant forming a weak complex with Aggrus and the tetraspanin web. (A) Schematic representation of the generated CD9 mutants: shorter extracellular loop (EC1)–deletion mutant (Δ1: 39-56 deletion); longer extracellular loop (EC2)–deletion mutants (Δ2: 114-133 deletion; Δ3: 134-153 deletion; Δ4: 154-173 deletion; and Δ5: 174-192 deletion); TM1, EC1, and TM2 deletion mutant (Δ6: 13-82 deletion); and palmitoylation-deficient point mutant (MT: C9S, C78S, C79S, C218S, and C219S). (B) CHO cells were transiently transfected with V5-tagged CD9-WT (left panels) or CD9-Δ6 (right panels) together with or without FLAG-tagged Aggrus. After solubilization with 1% CHAPS or Brij 97, the FLAG-tagged Aggrus was immunoprecipitated with an anti-FLAG agarose. The immunoprecipitants were analyzed by Western blot with an antibody to V5 tag. (C) CHO cells were transfected with plasmid encoding nothing or FLAG-Aggrus together with V5-Aggrus (left panels). In some experiments, CHO cells were transfected with plasmids encoding V5-CD9-WT or V5-CD9-Δ6 together with FLAG-CD9-WT (right panels). These cells were solubilized with lysis buffer containing 1% CHAPS. After immunoprecipitation with an anti-FLAG agarose, the immunoprecipitants were analyzed by Western blot with antibodies to V5 or FLAG. (D) CHO cells were transiently transfected with FLAG-Aggrus expressing plasmid together with plasmids encoding nothing (CHO/Aggrus), V5-CD9-WT (CHO/Aggrus+CD9-WT), or V5-CD9-Δ6 (CHO/Aggrus+CD9-Δ6). Then, cells were solubilized in lysis buffer containing 1% CHAPS or Brij 97. Each cell lysate was centrifuged in sucrose gradients and fractioned. Then, each fraction was electrophoresed and analyzed by Western blot with antibodies to Aggrus, caveolin, FLAG tag, or V5 tag. LMFs were concentrated in fraction numbers 2 through 4 (F2-F4).
We investigated localization of Aggrus, CD9-WT, and CD9-Δ6 in the tetraspanin web using 2 different criteria. First, the tetraspanin web is consisted of many kinds of molecules, so molecules within tetraspanin web are likely to have a homophilic interaction. Second, molecules within the tetraspanin web are expected to localize in LMFs in sucrose gradients because the ratio of lipid to protein in the tetraspanin web is higher than in surrounding parts. To prove a homophilic interaction, we prepared CHO cells expressing FLAG-tagged Aggrus together with V5-tagged Aggrus. We also generated CHO cells expressing FLAG-tagged CD9-WT together with V5-tagged CD9-WT or V5-tagged CD9-Δ6. Immunoprecipitation after immunoblot analysis revealed the homophilic interaction of Aggrus and CD9-WT (Figure 5C). In contrast, CD9-Δ6 could not bind to CD9-WT. Next, to examine localization in LMF, we solubilized CHO/Aggrus, CHO/Aggrus+CD9-WT, and CHO/Aggrus+CD9-Δ6 cells in lysis buffer containing 1% CHAPS or Brij 97. Lysates were centrifuged in sucrose gradients and were fractioned. Because caveolin is well known to appear in lipid-enriched microdomains, we used it as a positive control for LMFs. We found that Aggrus, CD9-WT and CD9-Δ6 were localized in LMF (fraction numbers 2-4) under CHAPS treatment (Figure 5D). Under Brij 97 treatment, Aggrus, CD9-WT, and CD9-Δ6 all shifted to higher-density membrane fractions; however, it was most complete for CD9-Δ6. CD9 expression did not affect Aggrus distribution to LMFs under both CHAPS and Brij 97 treatment. These results suggest that Aggrus constantly localized and interacted with CD9 in the tetraspanin web. Because CD9-Δ6 could not strongly interact with the tetraspanin web, CD9-Δ6 could not form a complex with Aggrus.
CD9 TM1 and TM2 involvement in Aggrus-induced platelet aggregation and metastasis formation
To clearly show the direct link between CD9-Aggrus interaction and Aggrus inactivation, we prepared CHO/mock or CHO/Aggrus cells that had been transfected with plasmids encoding nothing (CHO/mock+mock or CHO/Aggrus+mock), CD9-WT (CHO/Aggrus+CD9-WT), CD9-Δ6 (CHO/Aggrus+CD9-Δ6), or CD44 (CHO/Aggrus+CD44). The expression of CD9 and CD44 did not affect the expression level of Aggrus in CHO/Aggrus cells (Figure 6A top right panel). Consistent with the data on HT1080 cells (Figure 3D), overexpression of CD9-WT attenuated the platelet aggregation-inducing activity of Aggrus (CHO/Aggrus+CD9-WT; Figure 6A). In contrast to CD9-WT, CD9-Δ6 exhibited minimum inhibitory effect on Aggrus function (Figure 6A). We also examined pulmonary metastasis of the transfectants and confirmed that the number of lung surface tumor nodules in mice injected with CHO/Aggrus+CD9-WT cells was significantly lower than that in mice injected with CHO/Aggrus+mock cells (Figure 6B; *P < .05). Although the number of CHO/Aggrus + CD9-Δ6 was slightly reduced compared with the number of CHO/Aggrus+mock, the extent of reduction was not significant (Figure 6B; NS indicates not significant).
Effect of CD9-Δ6 expression on platelet aggregation and metastasis induced by forced expression of Aggrus. (A) CHO/mock and CHO/Aggrus cells were transiently transfected with vectors encoding nothing (mock), CD9-WT, CD9-Δ6, or CD44. The transfected polyclonal CHO/mock+mock (pink line), CHO/Aggrus+mock (black line), CHO/Aggrus+CD9-WT (red line), CHO/Aggrus+CD9-Δ6 (green line), and CHO/Aggrus+CD44 (blue line) cells were incubated with mouse PRP. The light transmittance was monitored for 15 minutes. Blue arrow indicates time when cells are added, and red arrow indicates time when platelet aggregation begins (top left panel). Platelet aggregation-inducing activity was calculated using the following formula: volume = W2 × L / 2, where W is the short diameter and L is the long diameter. Data are means plus or minus SD of triplicate experiments (bottom panel). *P < .05; NS indicates not significant by Student t test. Expression level of each protein was confirmed by Western blot with antibodies to Aggrus, V5 tag, or β-actin (top right panels). (B) CHO/mock+mock, CHO/Aggrus+mock, CHO/Aggrus+CD9-WT, CHO/Aggrus+CD9-Δ6, and CHO/Aggrus+CD44 cells were injected intravenously into BALB/c-nu/nu mice (n = 6). At 28 days after inoculation, the number of lung surface tumor nodules was counted. Representative pictures of the lungs (left panels) and detailed numbers of the lung metastasis nodules (right panel) are shown. Data are means plus or minus SD. *P < .05; NS indicates not significant by Student t test.
Effect of CD9-Δ6 expression on platelet aggregation and metastasis induced by forced expression of Aggrus. (A) CHO/mock and CHO/Aggrus cells were transiently transfected with vectors encoding nothing (mock), CD9-WT, CD9-Δ6, or CD44. The transfected polyclonal CHO/mock+mock (pink line), CHO/Aggrus+mock (black line), CHO/Aggrus+CD9-WT (red line), CHO/Aggrus+CD9-Δ6 (green line), and CHO/Aggrus+CD44 (blue line) cells were incubated with mouse PRP. The light transmittance was monitored for 15 minutes. Blue arrow indicates time when cells are added, and red arrow indicates time when platelet aggregation begins (top left panel). Platelet aggregation-inducing activity was calculated using the following formula: volume = W2 × L / 2, where W is the short diameter and L is the long diameter. Data are means plus or minus SD of triplicate experiments (bottom panel). *P < .05; NS indicates not significant by Student t test. Expression level of each protein was confirmed by Western blot with antibodies to Aggrus, V5 tag, or β-actin (top right panels). (B) CHO/mock+mock, CHO/Aggrus+mock, CHO/Aggrus+CD9-WT, CHO/Aggrus+CD9-Δ6, and CHO/Aggrus+CD44 cells were injected intravenously into BALB/c-nu/nu mice (n = 6). At 28 days after inoculation, the number of lung surface tumor nodules was counted. Representative pictures of the lungs (left panels) and detailed numbers of the lung metastasis nodules (right panel) are shown. Data are means plus or minus SD. *P < .05; NS indicates not significant by Student t test.
These results indicate that platelet aggregation and pulmonary metastasis induced by Aggrus could be attenuated by interaction with CD9 on the tetraspanin web.
Discussion
Some would argue that treatments directed against metastasis are too late because cells have already escaped from the primary tumor. However, there has been considerable improvement in survival after adjuvant radiation and chemotherapy designed to eliminate disseminated cells after surgical removal of the primary tumor. To establish treatments against metastasis, identifying and understanding the specific genes that control metastasis is necessary. Generally, a metastasis suppressor is distinguished from a tumor suppressor and is defined as a gene that prevents ectopic dissemination without blocking tumor formation. More than 20 metastasis suppressors have been identified, but their precise mechanisms of action are largely unknown. Recently, a tetraspanin family member, CD82/KAI1, believed to be a metastasis suppressor, has been shown to interact with a protein (DARC [Duffy antigen receptor for chemokines]) on endothelial cells and to induce senescence of tumor cells,44 indicating a role in preventing ectopic dissemination. Another tetraspanin member, CD9, is believed to suppress migration signals by forming membrane complexes with integrin receptors,4,5,45 and has been identified as a metastasis suppressor.13,36-38 However, it has not been clearly or concretely elucidated how the motility change acts, or which metastatic step is affected. We confirmed herein that CD9 expression reduced motility of HT1080 cells (Figure 2C). Tumor cell motility might not be directly associated with in vivo metastatic ability because CD44 expression also reduced motility of HT1080 cells, without affecting in vivo metastatic potential (Figure 1B). We also found that CHO cells did not exhibit an invasive ability in vitro, even though the cells were transfected with Aggrus or CD9 (data not shown), while Aggrus-transfected CHO cells formed many lung metastatic foci in vivo (Figure 6B).
One key to success in metastasis formation is tumor survival in the bloodstream. Tumor cell–induced platelet aggregation is believed to form the large tumor-platelet aggregate, leading to tumor cell protection from host immunity and cell embolization in the microvasculature.32,33 The metastatic potential is not simply an inherent trait of cancer cells, but it is substantially modified by tumor microenvironment. Platelets release a number of soluble factors (eg, PDGF, TGF-β, ADP, fibronectin) after aggregation, which may make the microenvironment suitable for metastasis formation.46-48 Considering these features, tumor cell–induced platelet aggregation promotes ectopic dissemination without affecting primary tumor formation. Conversely, this suggests that inhibition of platelet aggregation may slow down the rate of tumor progression and metastasis.
In this study, we showed that CD9 expression reduced lung retention of tumor cells 4 hours after inoculation (Figure 1C). This means that CD9 expression affected the extremely early steps of the metastatic process (ie, circulation and arresting in microvasculature). We confirmed that CD9 expression reduced platelet-aggregating activity of HT1080 and CHO cells (Figures 3D and 6A). Because HT1080-mediated platelet aggregation occurred after 6 minutes of contact between HT1080 cells and platelets (Figure 3), tumor cells in the lung after 5 minutes of HT1080 inoculation might be temporary entrapment before platelet aggregation–induced stable lung retention. That might be the reason why no difference in lung retention after 5 minutes of tumor inoculation could be found among 3 samples (Figure 1C). It is well known that tumor cells induce platelet aggregation mainly by the synthesis of thrombin and ADP. However, platelet activation induced by thrombin and ADP was not suppressed by CD9 (Figure 3F). These results suggest that CD9 suppressed platelet aggregation by interfering with other factors involved in platelet function. Aggrus-inducing platelet aggregation is observed in the presence of specific thrombin and ADP inhibitors,49 and Aggrus can activate platelets washed free from other plasma components26 through activating intraplatelet signaling cascade.28 We have shown here that reducing platelet aggregation and metastasis by CD9 expression required interaction between CD9 and Aggrus via TM1 and TM2 (Figure 6A,B). Because CD9 lacking TM1 and TM2 could not strongly interact with the tetraspanin web (Figure 5C,D), CD9-Δ6 could not inhibit Aggrus function on the tetraspanin web. We confirmed that CD9 expression delayed intraplatelet phosphorylation without affecting maximal signal intensity (Figure 3E). There is a correlation between the delayed ERK phosphorylation and the increased lag time in the aggregation studies when platelets were incubated with CD9 expressing HT1080 cells, although we cannot determine whether this is causal. In the formation of metastasis in vivo, the delay of platelet aggregation would be sufficient in metastasis suppression because tumor cells in circulation are destroyed very rapidly by sheer force or the host immune system.32 In addition, we examined whether CD9 expression affected Aggrus binding to CLEC-2, which is one of the counterparts of Aggrus on platelets.28,29 CD9 expression did not affect the Aggrus binding to CLEC-2 (data not shown). This result suggested that CD9 expression might inhibit some platelet-aggregating pathways after Aggrus binding to CLEC-2 (eg, CLEC-2 multimerization). Cross-linking of CLEC-2 by anti–CLEC-2 antibody can induce platelet aggregation in the absence of specific ligands.30 Thus, CLEC-2 multimerization was reported to be associated with CLEC-2–mediated platelet aggregation. CD9 might interfere with CLEC-2 aggregation and suppress platelet aggregation. Thus, we need to examine its effect on CLEC-2 multimerization in the future.
Therapeutic use of platelet inhibitors against metastasis has been challenged by many investigators, and an anticancer effect has often been suggested.50-52 However, applying these results to effective anticancer therapies has not been established because the state of the platelets of a patient with cancer is extremely fluid and elusive. Patients with cancer can show thrombocytosis or thrombocytopenia, depending on their circumstances. High platelet counts, or thrombocytosis, are often seen in patients with cancer. It is thought that tumors secrete substances to stimulate platelet production.53 Furthermore, in patients with advanced cancer, the expression of adhesion molecules on platelets is increased, indicating the platelet activation.54 In fact, concomitant venous thromboembolism (VTE) is well recognized in cancer. An antiplatelet approach is effective not only for metastasis but also for hypercoagulability, such as in VTE. On the other hand, patients with cancer undergoing treatment with cytotoxic agents often suffer from bleeding due to platelet toxicity, or thrombocytopenia. Because platelets play an important role in maintaining normal hemostasis even in the absence of acute injury, patients with cancer suffering from thrombocytopenia regularly receive platelet transfusions. It is possible that this life-saving therapy simultaneously facilitates metastasis. Effective platelet-modulating therapies must exhibit specificity for the pathologic tumor cell–platelet interaction without affecting normal platelet function. In this study, suppression of platelet aggregation was confirmed as an entirely new function of CD9 with regard to metastasis suppression. This suppression depended on Aggrus expression on the tumor cell surface (Figure 3). These tumor cell–platelet–specific mechanisms must be more clearly elucidated in order to determine the feasibility of antiplatelet therapy.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Drs N. Sugimura and N. Onda (Olympus) for their technical support.
This study was supported in part by special grants from Ministry of Education, Culture, Sports, Science and Technology, Japan 17016012 and 18390020 (to T.T. and N.F., respectively); a grant from Takeda Science Foundation, Japan (to N.F.); and a grant from the Cell Science Research Foundation, Japan (to N.F.).
Authorship
Contribution: Y.N. and N.F. designed the research; Y.N. performed the research; M.N., Y.K., K.M., H.A., and T.T. contributed new analytical tools; and Y.N. and N.F. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Naoya Fujita, Division of Experimental Chemotherapy, Cancer Chemotherapy Center, Japanese Foundation for Cancer Research, 3-10-6, Ariake, Koto-ku, Tokyo 135-8550, Japan; e-mail: naoya.fujita@jfcr.or.jp.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal