Abstract
Lymphatic vessel growth and activation, mediated by vascular endothelial growth factor (VEGF)–C and/or VEGF-A, have important roles in metastasis and in chronic inflammation. We aimed to comprehensively identify downstream molecular targets induced by VEGF-A or VEGF-C in lymphatic endothelium by analyzing the time-series transcriptional profile of treated human dermal lymphatic endothelial cells (LECs). We identified a number of genes, many not previously known to be involved in lymphangiogenesis, that were characterized either as early response genes, transiently induced genes, or progressively induced genes. Endothelial-specific molecule-1 (ESM-1) was one of the genes that were most potently induced by both VEGF-A and VEGF-C. Whereas ESM-1 induction by VEGF-A was mainly dependent on activation of VEGFR-2, VEGF-C–mediated induction depended on the activity of both VEGFR-2 and VEGFR-3. Incubation of LECs with ESM-1 increased the stimulatory effects of both VEGF-A and VEGF-C on LEC proliferation and migration, whereas ESM-1 alone had no effect. Importantly, VEGF-A (or VEGF-C) induction of LEC proliferation and migration were significantly inhibited by siRNA-mediated silencing of ESM-1 in vitro and in vivo. These studies reveal ESM-1 as a novel mediator of lymphangiogenesis and as a potential target for the inhibition of pathologic lymphatic vessel activation.
Introduction
The lymphatic vascular system has an important role in the maintenance of tissue fluid homeostasis, in the afferent phase of the immune response and in acute and chronic inflammation.1–3 Recent studies have revealed that lymphatic vessels also have an active role in the metastasis of malignant tumor cells to regional lymph nodes.4–6 In particular, tumors can induce lymphangiogenesis via release of the lymphangiogenic growth factors vascular endothelial growth factor (VEGF)–C or VEGF-D, leading to increased rates of metastasis to the draining sentinel lymph nodes and beyond.4–6 Indeed, studies have revealed that tumor-induced lymphangiogenesis is the most significant prognostic indicator for the occurrence of regional lymph node metastasis in malignant melanomas of the skin.7 More recently, it has been found that tumors can also induce lymphangiogenesis within their draining lymph nodes, even before they metastasize,8,9 and that induction of lymph node lymphangiogenesis promotes the further metastasis to distant lymph nodes and to organs.8 Thus, tumor-induced lymphatic growth and activation represents a novel potential target for treating or preventing advanced cancer
During the last few years, several mediators of lymphangiogenesis have been identified. Hepatocyte growth factor (HGF; also known as scatter factor) induces proliferation, migration, and tube formation of lymphatic endothelial cells (LECs) and also increases lymphangiogenesis in vivo.10 In addition, fibroblast growth factor (FGF)–2 promotes lymphatic vessel growth in the mouse cornea11,12 and also stimulates proliferation and migration of LECs by binding to the receptor FGFR-3, which is up-regulated by the transcription factor Prox1 in lymphatic endothelium.13 Other growth factors with effects on the lymphatic vasculature include platelet-derived growth factor-BB, insulin-like growth factors 1 and 2,14,15 angiopoietin-1,16,17 and adrenomedullin.18 Despite the growing number of novel potential lymphangiogenic factors, there is strong evidence that growth factors of the VEGF family represent the most important lymphangiogenic stimuli in the majority of human and experimental cancers.
VEGF-C promotes lymphangiogenesis by activating VEGF receptor (VEGFR)–2 and VEGFR-3 on LECs.19 VEGF-C–deficient mice fail to develop a functional lymphatic system,20 and transgenic expression of soluble VEGFR-3 results in pronounced lymphedema.19 Recently, VEGF-A was identified as a strong lymphangiogenic mediator. Adenoviral delivery of murine VEGF-A164 to the skin of mice strongly promotes lymphatic vessel growth, and transgenic mice that overexpress murine VEGF-A164, specifically in the skin, show increased lymphangiogenesis during wound healing and inflammation.3,21–23 Importantly, when VEGF-A transgenic mice were subjected to a standard chemically induced multistep skin carcinogenesis regimen, there was increased proliferation of VEGFR-2–expressing tumor-associated lymphatic vessels, leading to an increased incidence of lymph node metastasis.21 The relative importance of direct VEGF-A–induced signaling via VEGFR-2 versus the potential induction of a paracrine stimulatory loop via up-regulation of VEGF-C expression by LECs has remained unclear. Moreover, in contrast to the detailed investigation of the effects of VEGF-A on the blood vasculature,24 the downstream targets of VEGF-A (as well as of VEGF-C) in the lymphatic vasculature have remained unknown.
In this study, we aimed to comprehensively identify downstream molecular targets induced by VEGF-A or VEGF-C in lymphatic endothelium. To this end, we treated human dermal microvascular LECs with VEGF-A or VEGF-C for up to 24 hours and then used gene microarray technology to perform time-series transcriptional profiling. We identified a number of genes, many not previously known to be involved in lymphangiogenesis, that were characterized as early response genes, transiently induced genes, or progressively induced genes. Endothelial-specific molecule-1 (ESM-1) was one of the genes that were most potently induced by both VEGF-A and VEGF-C. Whereas ESM-1 induction by VEGF-A was mainly dependent on activation of VEGFR-2, VEGF-C–mediated induction depended on the activity of both VEGFR-2 and VEGFR-3. We found that incubation of LECs with ESM-1 increased the stimulating effects of both VEGF-A and VEGF-C on LEC proliferation and migration, whereas incubation with ESM-1 alone had no effect. Importantly, VEGF-A– (or VEGF-C)–induced induction of LEC proliferation and migration was significantly inhibited by siRNA-mediated silencing of ESM-1 in vitro and in vivo. Together, these studies reveal that ESM-1 is a novel mediator of lymphangiogenesis and a potential target for the inhibition of VEGF-A– or VEGF-C–induced pathologic lymphatic vessel growth and activation.
Methods
Cells
Primary human dermal microvascular LECs were isolated from neonatal human foreskin by immunomagnetic purification as previously described.25 The lineage-specific differentiation was confirmed by real-time reverse transcription–polymerase chain reaction (RT-PCR) for the lymphatic vascular markers Prox1, LYVE-1, and podoplanin, and for the blood vascular endothelial markers VEGFR-1 and VEGF-C, as well as by immunostaining for CD31, LYVE-1, and Prox1, as described.25 LECs were seeded onto fibronectin-coated culture dishes (10 μg/mL; BD Biosciences, Bedford, MA) and were cultured in endothelial cell basal medium (EBM; Cambrex Bio Science, Walkersville, MD) supplemented with 20% fetal bovine serum (FBS; Invitrogen, Grand Island, NY), 2 mM l-glutamine, antibiotic-antimycotic solution, 10 μg/mL hydrocortisone and N6,2′-O-dibutyryl-adenosine 3′,5′-cyclic monophosphate (25 μg/mL; all from Fluka, Buchs, Switzerland). Cells were used at passages 7 or 8.
Microarray analyses
Primary LECs were starved of serum overnight in EBM supplemented with 0.2% bovine serum albumin (BSA). Cells were treated or not for 1 hour, 4 hours, 8 hours, or 24 hours with recombinant human VEGF-A165 (R&D Systems, Minneapolis, MN; 20 ng/mL) or mature human VEGF-C (R&D Systems; 500 ng/mL). Total cellular RNA was isolated using the Trizol reagent (Invitrogen, Carlsbad, CA) and was extracted with chloroform, precipitated with isopropanol, washed with 70% ethanol, and dissolved in DNase-free/RNase-free distilled water. The concentration of RNA was measured using a NanoDrop ND-1000 spectrophotometer (Witec AG, Littau, Switzerland), and RNA quality was assessed using a 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA).
Digoxigenin-UTP–labeled cRNA was generated and amplified from 500 ng of total RNA using the NanoAmp RT-IVT Labeling Kit (Applied Biosystems, Foster City, CA) following the manufacturer's protocol, and was hybridized to Applied Biosystems Human Genome Survey Microarrays V2.0. Chemiluminescence detection, image acquisition, and analysis were performed using the Chemiluminescence Detection Kit (Applied Biosystems) and the Applied Biosystems 1700 Chemiluminescent Microarray Analyzer following the manufacturer's protocol. A total of 3 biological replicates were generated for each treatment condition (VEGF-A and VEGF-C) and for each time point (0 hours, 1 hour, 4 hours, 8 hours, and 24 hours). Microarray data are accessible at http://www.ncbi.nlm.nih.gov/geo/ (GSE11228).
Microarray data analysis
Raw data were normalized using variance stabilization and normalization (VSN), a model derived from the variance-versus–mean dependence for microarray intensity data,26 available from R/Bioconductor.27 In a second step, probes which had a signal-to-noise ratio (S/N ratio) of 3 or greater, flag (error) value of 5000 or less in at least 2 of the 3 replicates for each time point were further subjected to statistical analyses. Differentially expressed genes were identified using the multivariate Empirical Bayes (EB) analysis (R package: time course R/Bioconductor). The multivariate-EB procedure focuses on moderating the denominator of the multivariate t statistics, and ranks genes according to the moderated statistic to reduce the number of false positives and false negatives resulting from very small or very large replicate variances or covariances.28 The Short Time Series Expression Miner (STEM)29 was used to identify early response, transiently up-regulated, progressively induced, and down-regulated clusters. STEM implements a clustering method that depends on a set of distinct and representative short temporal expression profiles, and each probe in the dataset is assigned to a profile with the closest match. The expected number of probes assigned to each profile was estimated by permutation and the statistically significantly overexpressed (P < .05) profiles are then identified. The preprocessed datasets of 3 independent experiments were imported into STEM. Experimental profiles with a minimal correlation of 0.7 with the predetermined model profiles were then clustered together. Pathway analyses were performed using the PANTHER (protein analysis through evolutionary relationships) protein classification system, which classifies proteins into families/subfamilies, molecular functions, biological processes, and biological pathways.30 Pathways that were overrepresented in the group of genes up-regulated after VEGF-A or VEGF-C treatment were identified, and the statistical significance of the overrepresentation was calculated by a random overlapping, using binomial tests to determine P values, with all of the genes represented on the Applied Biosystems Human Genome Survey Microarray serving as the reference list.31
siRNA transfection, receptor blocking experiments, and qRT-PCR
siRNA transfection was performed using the Basic Nucleofector Kit for primary mammalian endothelial cells (Amaxa Biosystems, Cologne, Germany) according to the manufacturer's protocol. The following siRNAs against ESM-1 were used: 5′-GGUUUGUAAAAGAAGAAUCtt-3′, 5′-GGUGUCAGCCUUCUAAUGGtt-3′, and 5′-GCUGCAUAAGCUGUUAGGUtt-3′, as well as control siRNA (silencer negative control no. 1 siRNA; Ambion, Huntingdon, United Kingdom). Antibodies against the extracellular domain of human VEGFR-2 (1121b)32 and of human VEGFR-3 (hF4-3C5),33 as well as rat negative control IgG, were kindly provided by Bronek Pytowski (Imclone Systems, New York, NY). The expression of ESM-1 mRNA was quantified by TaqMan real-timeRT-PCR using the AB 7900 HT fast real-time PCR system (Applied Biosystems). The probes and primers for ESM-1 were predesigned (Hs00199831 m1; Applied Biosystems). Each reaction was normalized for the expression of β-actin (forward primer, 5′-TCACCGAGCGCGGCT-3′; reverse primer, 5′-TAATGTCACGCACGATTTCCC-3′; probe, 5′-JOE-CAGCTTCACCACCACGGCCGAG-TAMRA-3′).
Immunoblotting
LECs were treated with 0 or 20 ng/mL human VEGF-A165 for 24 or 48 hours and were homogenized in lysis buffer, as previously described.10 Supernatants were concentrated using a Centricon Ultracel YM-10 membrane filter device with a nominal molecular weight limit of 10 000 (Millipore, Bedford, MA). The protein concentrations were determined using the NanoOrange Protein Quantitation Kit (Molecular Probes, Eugene, OR). The lysates (100 μg total protein each) and supernatants (50 μg) were subjected to SDS–polyacrylamide gel electrophoresis, using NuPAGE 10% BT gels, 1.0 mm, 12 well and NuPAGE MES SDS running buffer (Invitrogen). The proteins were transferred onto a Trans-Blot Transfer Medium pure nitrocellulose membrane (BioRad, Hercules, CA) and then immunoblotted with the human anti–ESM-1 goat polyclonal antibody (0.2 μg/mL; R&D Systems). Blocking was performed with 5% nonfat dry milk in 0.1% Tween20 (Sigma-Aldrich, St Louis, MO) in phosphate-buffered saline (PBS). Specific binding was detected by the ECL Plus Western Blotting Detection System (GE Healthcare, Little Chalfont, United Kingdom). Equal loading was confirmed using an antibody against β-actin (Sigma-Aldrich).
Cell proliferation and migration assays
LECs (2 × 103) were seeded onto fibronectin-coated 96-well plates. Quintruplicate wells were treated with different concentrations of recombinant human ESM-1 (0.01 ng/mL to 2500 ng/mL; R&D Systems) and with 20 ng/mL VEGF-A or 100 ng/mL VEGF-C. After 72 hours, cells were incubated with 4-methylumbelliferyl heptanoate (MUH; Sigma-Aldrich). The fluorescence intensity, proportional to the number of viable cells, was measured using a Spectra Max GEMINI EM fluorescence reader (Bucher Biotec AG, Basel, Switzerland). For endothelial cell migration assays, control- or ESM-1 siRNA–transfected LECs were grown to 100% confluency and starved of serum overnight. The following day, a cell-free wound zone was created by scraping the monolayer with a sterile pipette tip. The cells were washed with PBS, and then the medium was changed to EBM/0.1% BSA containing PBS, VEGF-A (20 ng/mL), ESM-1 (1 μg/mL), or a combination of VEGF-A and ESM-1. The cells were incubated in 5% CO2 at 37°C for 48 hours. Representative images were taken at 5 × magnification directly after wounding and then again 48 hours later using an AxioCam MRm camera attached to an Axiovert 200M microscope (Carl Zeiss AG; Feldbach, Switzerland). Computer-assisted morphometric wound area analyses were performed using the IP-LAB software (Scanalytics, Fairfax, VA). All experiments were performed 3 times. Statistical analyses were performed using the unpaired Student t test.
Matrigel assay, immunofluorescence stainings, and morphometric analyses
Lymphangiogenesis was evaluated in vivo using a matrigel plug assay, as described previously.10,34,35 Friend virus B (FVB) wild-type mice (female, 6 to 8 weeks old) were anesthetized and given subcutaneous injections into the lower flank skin with 100 μL of Matrigel (BD Biosciences) containing either human VEGF-A165 (500 ng/mL) and Silencer Negative control siRNA (10 μg/mL; catalog no. 4635; Ambion, Austin, TX) or VEGF-A165 and murine ESM-1 siRNA (10 μg/mL; Ambion, catalog no. 16804; n = 5 per group). After 7 days, skin samples were embedded in optimal cutting temperature compound (OCT; Sakura Finetek, Torrance, CA). Immunofluorescence analyses were performed on 8-μm cryostat sections, as described,3,36 using a rabbit polyclonal antibody against mouse LYVE-1 (Upstate Biotechnology, Charlottesville, VA) and a monoclonal rat antibody against mouse CD31 (BD Biosciences Pharmingen, San Diego, CA). For the detection of ESM-1 in the skin of VEGF-A transgenic mice (female, 8 weeks old),3,37 murine ESM-1 antibody (R&D Systems) was used. Corresponding secondary antibodies were labeled with AlexaFluor488 or AlexaFluor594 (Molecular Probes). Nuclei were counterstained with 20 μg/mL Hoechst 33342 (Molecular Probes). Sections were examined by an Axioskop2 microscope (Carl Zeiss AG), and images were captured at 20× magnification with an AxioCam MRm digital camera and the following objectives: Plan-APOCHROMAT 10×/0.45 MA and Plan-NEOPLAN 20×/0.5 NA (both from Zeiss). Image acquisition in the individual fluorescent channels was accomplished using Axio Vision4.4 software (Zeiss). Adobe Photoshop CS2 (Adobe Systems, San Jose, CA) was used to adjust image brightness and for image overlay. Computer-assisted morphometric vessel analyses of representative LYVE-1 and CD31 double-stained sections were performed using the IP-LAB software as described.3 A total of 3 individual fields per section were examined and the number of vessels per square millimeter, the average vessel size, and the average tissue area covered by vessels were determined. Statistical analysis was performed using the unpaired Student t test.
Results
Microarray analysis reveals novel mediators of VEGF-A– and VEGF-C–induced effects on LECs
Both VEGF-A and VEGF-C promote lymphangiogenesis in vivo and increase LEC proliferation and migration in vitro.3,8,21,22,38,39 To identify genes involved in lymphangiogenesis that are mediated by these factors, we incubated human dermal microvascular LECs with either VEGF-A or VEGF-C for 0, 1, 4, 8 and 24 hours in triplicate, followed by gene microarray analyses using the chemiluminescence-based Applied Biosystems Human Genome Microarrays platform. The lineage-specific differentiation of the LECs used for these studies was confirmed by quantitative TaqMan RT-PCR in comparison with matched blood vascular endothelial cells (BECs). LECs expressed higher levels of Prox1 (80.2-fold), LYVE-1 (288.3-fold), and podoplanin (55.2-fold) than BECs, and BECs expressed higher levels of VEGF-C (13.1-fold) and VEGFR-1 (21.7-fold). These results were in agreement with previous publications, in which Prox1 expression was up-regulated in LECs by 4.6-fold to 74.1-fold, LYVE-1 expression by 3.7-fold to 90.7-fold, and podoplanin expression by 16-fold to 141.7-fold; VEGF-C expression was higher in BECs by 3.7-fold to 42.2-fold; and VEGFR-1 expression was higher by 3.5-fold to 8.9-fold.25,40–43
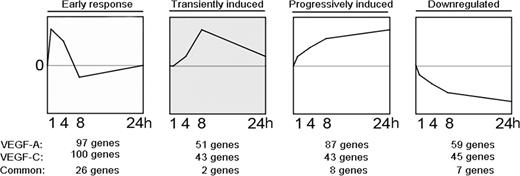
We investigated the genes modulated by VEGF-A and/or VEGF-C by applying multivariate EB statistics, which ranks genes on the basis of their sequential expression over time and the reproducibility at each time point.44–47 We next performed STEM analysis29 to determine which significantly modulated genes cluster together based on their temporal regulation pattern (Figure 1). For the VEGF-A–treated LECs, we identified 97 genes clustering into the early-response genes group (peak at time point 1 hour), 51 genes into the transiently induced genes (peak between 4 and 8 hours), 87 genes into the progressively induced genes group (progressive increase of expression over time), and 59 genes into the down-regulated genes group (progressive decrease over time). For the VEGF-C–treated LECs, 100 genes clustered into the early-response genes group, 43 into the transiently induced genes group, and 43 into the progressively induced genes group; 45 were down-regulated (Figure 1). The early response gene cluster revealed the most overlapping genes induced by both VEGF-A and VEGF-C (n = 26) compared with the other temporal clusters, and included known early response genes such as EGR1, EGR2, and EGR3 (Table 1). As previously described for blood vascular endothelium,48–51 DSCR1 was one of the most strongly induced VEGF-A early response genes (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Among the progressively induced genes, we identified several genes that have been previously reported to be involved in the mediation of lymphangiogenesis, including VEGF-C and angiopoietin-2,39,52 along with asp-like, microcephaly associated (ASPM), TTK protein kinase (TTK), and kinesin family member 14 (KIF14; Table 1). Fatty acid binding protein 3 (FABP3), SHC SH2-domain binding domain 1 (SHCBP1), and cell division cycle 2 (CDC2) were highly up-regulated by both VEGF-A and VEGF-C in LECs. The list of significantly modulated genes is provided in Table S1.
Microarray time course analysis of LECs treated with VEGF-A or VEGF-C reveals 4 major temporally regulated gene clusters. Transcription profiling and STEM analysis of LECs stimulated with VEGF-A or VEGF-C, respectively, for 1, 4, 8, or 24 hours revealed 97 and 100 genes specifically up-regulated at 1 hour (early response genes), 51 and 43 genes up-regulated transiently (transiently induced), 87 and 43 genes up-regulated progressively over time (progressively induced) and 59 and 45 genes down-regulated over time (down-regulated).
Microarray time course analysis of LECs treated with VEGF-A or VEGF-C reveals 4 major temporally regulated gene clusters. Transcription profiling and STEM analysis of LECs stimulated with VEGF-A or VEGF-C, respectively, for 1, 4, 8, or 24 hours revealed 97 and 100 genes specifically up-regulated at 1 hour (early response genes), 51 and 43 genes up-regulated transiently (transiently induced), 87 and 43 genes up-regulated progressively over time (progressively induced) and 59 and 45 genes down-regulated over time (down-regulated).
Top 10 genes induced by VEGF-A or VEGF-C
| . | Early response genes . | Transiently induced genes . | Progressively induced genes . | Down-regulated genes . | ||||
|---|---|---|---|---|---|---|---|---|
| VEGFA . | VEGFC . | VEGFA . | VEGFC . | VEGFA . | VEGFC . | VEGFA . | VEGFC . | |
| 1 | EGR3 | EGR3 | PLAUR | LOC388466 | ESM1 | ESM1 | CA4 | FGFR1 |
| 2 | NR4A2 | EGR2 | FLT1 | FLJ46358 | ANGPT2 | FABP3 | BRUNOL5 | LTB |
| 3 | EGR2 | NR4A1 | SHC4 | TIGD1 | FABP3 | ASPM | TGFA | SCARF2 |
| 4 | NR4A1 | F3 | LRP8 | CAMK2B | DGKD | ST8SIA4 | MAN1C1 | TMEM100 |
| 5 | F3 | EGR1 | NPAS2 | HLA-DOB | ASPM | LOC338579 | LTB | TMC8 |
| 6 | EGR1 | FOS | ITPKA | CDC14A | ST8SIA4 | VEGFC | LFNG | C1orf61 |
| 7 | FOS | ATF3 | ATAD3B | ETV1 | CXCR4 | NP | TMEM100 | C2orf23 |
| 8 | ATF3 | PTGS2 | RNU3IP2 | GABBR1 | CDC45L | C20orf128 | SLC2A12 | LAMA2 |
| 9 | PTGS2 | BHLHB2 | NOL5A | LY6H | KIF2C | ITGB1BP2 | SDC2 | WASF2 |
| 10 | FOSB | DUSP5 | C1orf21 | KIAA1913 | SHCBP1 | GALNT8 | SPATA12 | KCNQ1 |
| . | Early response genes . | Transiently induced genes . | Progressively induced genes . | Down-regulated genes . | ||||
|---|---|---|---|---|---|---|---|---|
| VEGFA . | VEGFC . | VEGFA . | VEGFC . | VEGFA . | VEGFC . | VEGFA . | VEGFC . | |
| 1 | EGR3 | EGR3 | PLAUR | LOC388466 | ESM1 | ESM1 | CA4 | FGFR1 |
| 2 | NR4A2 | EGR2 | FLT1 | FLJ46358 | ANGPT2 | FABP3 | BRUNOL5 | LTB |
| 3 | EGR2 | NR4A1 | SHC4 | TIGD1 | FABP3 | ASPM | TGFA | SCARF2 |
| 4 | NR4A1 | F3 | LRP8 | CAMK2B | DGKD | ST8SIA4 | MAN1C1 | TMEM100 |
| 5 | F3 | EGR1 | NPAS2 | HLA-DOB | ASPM | LOC338579 | LTB | TMC8 |
| 6 | EGR1 | FOS | ITPKA | CDC14A | ST8SIA4 | VEGFC | LFNG | C1orf61 |
| 7 | FOS | ATF3 | ATAD3B | ETV1 | CXCR4 | NP | TMEM100 | C2orf23 |
| 8 | ATF3 | PTGS2 | RNU3IP2 | GABBR1 | CDC45L | C20orf128 | SLC2A12 | LAMA2 |
| 9 | PTGS2 | BHLHB2 | NOL5A | LY6H | KIF2C | ITGB1BP2 | SDC2 | WASF2 |
| 10 | FOSB | DUSP5 | C1orf21 | KIAA1913 | SHCBP1 | GALNT8 | SPATA12 | KCNQ1 |
We next applied the PANTHER annotation and classification analysis to identify biologic pathways with time-specific regulation by VEGF-A. Among the 14 molecular functions significantly overrepresented after 1 hour of VEGF-A treatment were transcription factors and signaling molecules (P < .001; Table 2 left columns). Within the transiently and progressively induced gene clusters, genes encoding cytokine receptors and growth factors were significantly overrepresented. The most significantly overrepresented molecular functions after 24 hours included cytoskeletal and microtubule binding proteins (P < .001). According to their biologic process annotations, genes induced after 1 hour were significantly involved in transcription, cell proliferation, and cell-cycle control (Table 2; right columns). Among the overrepresented pathways after 24 hours of VEGF-A treatment were protein modification and phosphorylation. The results for VEGF-C–treated LECs are provided in Table S2.
Pathway classification analysis of VEGF-A–induced genes in LECs
| Molecular function . | 1 h . | 4 h . | 8 h . | 24 h . | Biological process . | 1 h . | 4 h . | 8 h . | 24 h . |
|---|---|---|---|---|---|---|---|---|---|
| Cytokine receptor | ++ | ++ | ++ | ++ | Cell proliferation and differentiation | +++ | + | + | + |
| Phosphorylase | ++ | ++ | ++ | ++ | Developmental processes | +++ | + | + | + |
| Growth factor | + | +++ | + | + | Nucleoside, nucleotide, and nucleic acid metabolism | +++ | + | + | + |
| Kinase modulator | + | + | + | + | Intracellular signaling cascade | ++ | ++ | + | + |
| Kinase inhibitor | + | + | ++ | − | MAPKKK cascade | +++ | ++ | +++ | − |
| Phosphatase | ++ | + | +++ | − | Immunity and defense | ++ | ++ | ++ | − |
| Basic helix-loop-helix transcription factor | + | + | + | − | Receptor protein tyrosine kinase signaling pathway | + | + | ++ | − |
| Signaling molecule | +++ | +++ | + | − | JNK cascade | + | + | + | − |
| Carbohydrate phosphatase | ++ | +++ | +++ | − | Signal transduction | +++ | +++ | + | − |
| Kinase | + | + | − | +++ | Protein phosphorylation | +++ | + | − | +++ |
| Select regulatory molecule | + | − | + | + | Ligand-mediated signaling | +++ | +++ | − | − |
| Protein phosphatase | + | − | ++ | − | Cell communication | +++ | ++ | − | − |
| Transcription factor | +++ | − | − | − | Cell cycle | + | − | + | +++ |
| Metalloprotease | + | − | − | − | Cell-cycle control | +++ | − | + | +++ |
| Defense/immunity protein | − | + | + | − | Protein modification | + | − | − | ++ |
| RNA-binding protein | − | + | + | − | Inhibition of apoptosis | ++ | − | − | + |
| Replication origin binding protein | − | − | + | ++ | Cell-surface receptor–mediated signal transduction | ++ | − | − | − |
| Transaminase | − | − | ++ | + | mRNA transcription | +++ | − | − | − |
| Transferase | − | − | + | − | Cell adhesion–mediated signaling | + | − | − | − |
| Microtubule binding motor protein | − | − | − | +++ | Neurogenesis | + | − | − | − |
| Microtubule family cytoskeletal protein | − | − | − | +++ | Complement-mediated immunity | − | + | + | + |
| Cytoskeletal protein | − | − | − | +++ | Angiogenesis | − | + | + | + |
| Protein kinase | − | − | − | ++ | Amino acid biosynthesis | − | − | + | ++ |
| Actin binding motor protein | − | − | − | + | Mitosis | − | − | − | +++ |
| DNA topoisomerase | − | − | − | + | Chromosome segregation | − | − | − | +++ |
| DNA strand-pairing protein | − | − | − | + | Cytokinesis | − | − | − | +++ |
| DNA helicase | − | − | − | + | DNA replication | − | − | − | ++ |
| Molecular function . | 1 h . | 4 h . | 8 h . | 24 h . | Biological process . | 1 h . | 4 h . | 8 h . | 24 h . |
|---|---|---|---|---|---|---|---|---|---|
| Cytokine receptor | ++ | ++ | ++ | ++ | Cell proliferation and differentiation | +++ | + | + | + |
| Phosphorylase | ++ | ++ | ++ | ++ | Developmental processes | +++ | + | + | + |
| Growth factor | + | +++ | + | + | Nucleoside, nucleotide, and nucleic acid metabolism | +++ | + | + | + |
| Kinase modulator | + | + | + | + | Intracellular signaling cascade | ++ | ++ | + | + |
| Kinase inhibitor | + | + | ++ | − | MAPKKK cascade | +++ | ++ | +++ | − |
| Phosphatase | ++ | + | +++ | − | Immunity and defense | ++ | ++ | ++ | − |
| Basic helix-loop-helix transcription factor | + | + | + | − | Receptor protein tyrosine kinase signaling pathway | + | + | ++ | − |
| Signaling molecule | +++ | +++ | + | − | JNK cascade | + | + | + | − |
| Carbohydrate phosphatase | ++ | +++ | +++ | − | Signal transduction | +++ | +++ | + | − |
| Kinase | + | + | − | +++ | Protein phosphorylation | +++ | + | − | +++ |
| Select regulatory molecule | + | − | + | + | Ligand-mediated signaling | +++ | +++ | − | − |
| Protein phosphatase | + | − | ++ | − | Cell communication | +++ | ++ | − | − |
| Transcription factor | +++ | − | − | − | Cell cycle | + | − | + | +++ |
| Metalloprotease | + | − | − | − | Cell-cycle control | +++ | − | + | +++ |
| Defense/immunity protein | − | + | + | − | Protein modification | + | − | − | ++ |
| RNA-binding protein | − | + | + | − | Inhibition of apoptosis | ++ | − | − | + |
| Replication origin binding protein | − | − | + | ++ | Cell-surface receptor–mediated signal transduction | ++ | − | − | − |
| Transaminase | − | − | ++ | + | mRNA transcription | +++ | − | − | − |
| Transferase | − | − | + | − | Cell adhesion–mediated signaling | + | − | − | − |
| Microtubule binding motor protein | − | − | − | +++ | Neurogenesis | + | − | − | − |
| Microtubule family cytoskeletal protein | − | − | − | +++ | Complement-mediated immunity | − | + | + | + |
| Cytoskeletal protein | − | − | − | +++ | Angiogenesis | − | + | + | + |
| Protein kinase | − | − | − | ++ | Amino acid biosynthesis | − | − | + | ++ |
| Actin binding motor protein | − | − | − | + | Mitosis | − | − | − | +++ |
| DNA topoisomerase | − | − | − | + | Chromosome segregation | − | − | − | +++ |
| DNA strand-pairing protein | − | − | − | + | Cytokinesis | − | − | − | +++ |
| DNA helicase | − | − | − | + | DNA replication | − | − | − | ++ |
+ indicates P < .05; ++, P < .01; +++, P < .001; and −, not statistically significant.
ESM-1 expression is potently induced in LECs by VEGF-A and VEGF-C
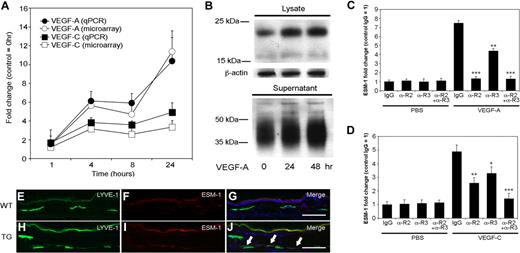
In the progressively induced gene cluster, ESM-1 was one of the most potently up-regulated genes after VEGF-A and VEGF-C treatment. Quantitative TaqMan real-time RT-PCR analyses revealed a more than 10-fold induction of ESM-1 mRNA expression following 24 hours of VEGF-A treatment, and a more than 4-fold induction following 24 hours of treatment with VEGF-C (Figure 2A), confirming the microarray results. Immunoblot analyses confirmed that ESM-1 protein expression also strongly increased in both LEC lysates and supernatants following 24 and 48 hours of VEGF-A treatment, compared with the control LECs (Figure 2B). The larger molecular weight of ESM-1 protein in supernatants, and its appearance as a smear on the gel, is due to the extensive glycosylation of the secreted molecule, as previously shown in human umbilical vein endothelial cells.53
Expression of ESM-1 in LECs is induced by VEGF-A and VEGF-C via VEGFR-2 and VEGFR-3. (A) Quantitative RT-PCR (qPCR) analysis revealed that compared with untreated controls, LECs expressed levels of ESM-1 mRNA that were more than 10-fold higher after 24 hours of stimulation with VEGF-A (●), and more than 4-fold higher after 24 hours of stimulation with VEGF-C (■); these findings confirmed those of the microarray expression results (○, □) (B) Immunoblot analysis confirmed that LECs stimulated with VEGF-A for 24 or 48 hours expressed much higher levels of ESM-1 in the cell lysates and in cell supernatants compared with untreated controls (0 hours). β-actin levels were used as gel loading controls. (C) Treatment of LECs with VEGF-A compared with PBS strongly induced the expression of ESM-1 in the presence of control antibody (IgG). ESM-1 induction was completely inhibited by a VEGFR-2–blocking antibody (αR2). Incubation with a VEGFR-3–blocking antibody (αR3) partially reduced the induction of ESM-1 by VEGF-A, and a combination of both blocking antibodies (αR2 + αR3) inhibited the VEGF-A–mediated ESM-1 induction. (D) Compared with PBS, VEGF-C also induced the expression of ESM-1. Incubation with either an αR2 or αR3 only partially blocked the ESM-1 induction by VEGF-C; ESM-1 induction by VEGF-C was completely prevented by a combination of both receptors. ***P < .001; **P < .005; *P < .01. Error bars represent standard deviation. The LYVE-1+ lymphatic vessels in the skin of wild-type mice did not express ESM-1 (E-G), whereas the LYVE-1+ lymphatic vessels of VEGF-A transgenic mice (H-J) did express ESM-1 (panel J; arrows). Scale bars equal 100 μm.
Expression of ESM-1 in LECs is induced by VEGF-A and VEGF-C via VEGFR-2 and VEGFR-3. (A) Quantitative RT-PCR (qPCR) analysis revealed that compared with untreated controls, LECs expressed levels of ESM-1 mRNA that were more than 10-fold higher after 24 hours of stimulation with VEGF-A (●), and more than 4-fold higher after 24 hours of stimulation with VEGF-C (■); these findings confirmed those of the microarray expression results (○, □) (B) Immunoblot analysis confirmed that LECs stimulated with VEGF-A for 24 or 48 hours expressed much higher levels of ESM-1 in the cell lysates and in cell supernatants compared with untreated controls (0 hours). β-actin levels were used as gel loading controls. (C) Treatment of LECs with VEGF-A compared with PBS strongly induced the expression of ESM-1 in the presence of control antibody (IgG). ESM-1 induction was completely inhibited by a VEGFR-2–blocking antibody (αR2). Incubation with a VEGFR-3–blocking antibody (αR3) partially reduced the induction of ESM-1 by VEGF-A, and a combination of both blocking antibodies (αR2 + αR3) inhibited the VEGF-A–mediated ESM-1 induction. (D) Compared with PBS, VEGF-C also induced the expression of ESM-1. Incubation with either an αR2 or αR3 only partially blocked the ESM-1 induction by VEGF-C; ESM-1 induction by VEGF-C was completely prevented by a combination of both receptors. ***P < .001; **P < .005; *P < .01. Error bars represent standard deviation. The LYVE-1+ lymphatic vessels in the skin of wild-type mice did not express ESM-1 (E-G), whereas the LYVE-1+ lymphatic vessels of VEGF-A transgenic mice (H-J) did express ESM-1 (panel J; arrows). Scale bars equal 100 μm.
Although VEGFR-2 is the only known receptor for VEGF-A on LECs, some of the observed VEGF-A effects might have been mediated by an indirect pathway involving up-regulation of VEGF-C, which might lead to VEGFR-3 activation. Moreover, the mature form of human VEGF-C used for the experiments can bind to both VEGFR-2 and VEGFR-3. We therefore investigated the relative contribution of VEGFR-2 and VEGFR-3 toward the induction of ESM-1 by VEGF-A and VEGF-C using blocking antibodies specific for VEGFR-2 or VEGFR-3. Treatment of LECs with VEGF-A strongly induced the expression of ESM-1 in the presence of a control IgG, whereas ESM-1 induction was completely inhibited by a VEGFR-2–blocking antibody (P < .001; Figure 2C). Incubation with a VEGFR-3–blocking antibody partially reduced the induction of ESM-1 by VEGF-A (P < .005), and a combination of both blocking antibodies inhibited the VEGF-A–mediated ESM-1 induction (P < .001; Figure 2C). VEGF-C also induced the expression of ESM-1 in the presence of control IgG, though less potently than VEGF-A (Figure 2D). Incubation with either anti–VEGFR-2 or anti–VEGFR-3 only partially blocked the ESM-1 induction by VEGF-C (P < .005 and P < .01, respectively; Figure 2D). Combined blockade of VEGFR-2 and VEGFR-3 completely prevented VEGF-C–mediated induction of ESM-1 expression (P < .001).
To investigate whether ESM-1 expression by lymphatic endothelium is up-regulated by VEGF-A in vivo, we performed differential immunofluorescence analyses of skin samples obtained from VEGF-A transgenic mice to analyze levels of ESM-1 and the lymphatic-specific hyaluronan receptor LYVE-1. VEGF-A transgenic mice express the murine VEGF-A164 under control of the epidermis-specific keratin 14 promoter, leading to increased VEGF-A levels within the skin.3,37 Although ESM-1 was not detected in lymphatic vessels in the skin of wild-type mice (Figure 2E-G), a subset of LYVE-1+ lymphatic vessels in the skin of VEGF-A transgenic mice expressed ESM-1 (Figure 2H-J).
ESM-1 promotes LEC proliferation and migration induced by VEGF-A and VEGF-C
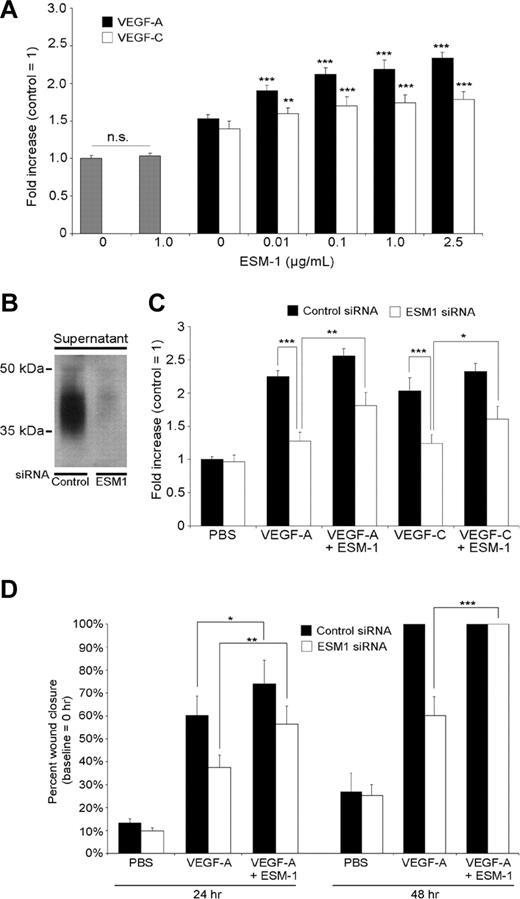
Because VEGF-A and VEGF-C each promoted ESM-1 expression by LECs, we investigated whether ESM-1 might modulate the effects of these growth factors on LEC function. Incubation of LECs with ESM-1 (0.1 ng/mL to 1 μg/mL) did not affect LEC proliferation (Figure 3A; data not shown). However, the addition of ESM-1 to VEGF-A or VEGF-C significantly increased the stimulatory effects of each growth factor on LEC proliferation, with a minimal effective concentration of 0.01 μg/mL (Figure 3A). Addition of 1 μg/mL of ESM-1 resulted in a 43% increase of VEGF-A–induced LEC proliferation (P < .001) and in a 22% increase of VEGF-C–induced LEC proliferation (P < .001). We investigated whether silencing of ESM-1 expression by siRNAs might affect the proliferative effects of VEGF-A and VEGF-C on LECs. Transfection of LECs with ESM-1 siRNAs reduced the ESM-1 protein levels compared with control siRNA–transfected LECs (Figure 3B). When ESM-1 siRNA–transfected LECs were treated with 30 ng/mL VEGF-A or 200 ng/mL VEGF-C, the proliferation-inducing effect of each growth factor was potently suppressed, compared with control siRNA–transfected LECs (P < .005; Figure 3C). Addition of human recombinant ESM-1 protein to ESM-1 siRNA–transfected LECs partially restored the level of proliferation induction by VEGF-A and VEGF-C (P < .005 and P < .05, respectively; Figure 3C).
ESM-1 promotes LEC proliferation and migration induced by VEGF-A and VEGF-C. (A) Addition of ESM-1 together with VEGF-A (20 ng/mL) or VEGF-C (100 ng/mL) significantly increased the stimulatory effects of both growth factors on LEC proliferation with a minimal effective concentration of 0.01 μg/mL, whereas treatment with ESM-1 alone had no effect. (B) Transfection of LECs with ESM-1 siRNAs reduced ESM-1 protein levels (right lane) compared with control siRNA–transfected LEC (left lane). (C) The proliferation-inducing effects of 30 ng/mL VEGF-A and of 200 ng/mL VEGF-C were suppressed in ESM-1 siRNA–transfected LECs but not in control siRNA–transfected LECs. Addition of 1 μg/mL of ESM-1 promoted LEC proliferation (+14% for VEGF-A, P < .001; +14% for VEGF-C, P = .006). Addition of 1 μg/mL ESM-1 protein to ESM-1 siRNA–transfected LECs partially restored the induction of proliferation by VEGF-A and VEGF-C. (D) Monolayer wound healing assay: ESM-1 (1 μg/mL) promoted the promigratory effect of VEGF-A in a monolayer wound assay after 24 hours (+14% increase in cell migration; P = .012), whereas LECs transfected with ESM-1 siRNA showed a significantly reduced migratory response to VEGF-A compared with control (siRNA-transfected) LECs. At the 48-hour time point, both VEGF-A–treated cultures and cultures treated with VEGF-A plus ESM-1 already showed a 100% wound closure. Addition of 1 μg/mL ESM-1 protein to ESM-1 siRNA–transfected LECs significantly restored the effect of VEGF-A on LEC migration after 24 and 48 hours. *P < .05; **P < .005; ***P < .001. Error bars represent standard deviation.
ESM-1 promotes LEC proliferation and migration induced by VEGF-A and VEGF-C. (A) Addition of ESM-1 together with VEGF-A (20 ng/mL) or VEGF-C (100 ng/mL) significantly increased the stimulatory effects of both growth factors on LEC proliferation with a minimal effective concentration of 0.01 μg/mL, whereas treatment with ESM-1 alone had no effect. (B) Transfection of LECs with ESM-1 siRNAs reduced ESM-1 protein levels (right lane) compared with control siRNA–transfected LEC (left lane). (C) The proliferation-inducing effects of 30 ng/mL VEGF-A and of 200 ng/mL VEGF-C were suppressed in ESM-1 siRNA–transfected LECs but not in control siRNA–transfected LECs. Addition of 1 μg/mL of ESM-1 promoted LEC proliferation (+14% for VEGF-A, P < .001; +14% for VEGF-C, P = .006). Addition of 1 μg/mL ESM-1 protein to ESM-1 siRNA–transfected LECs partially restored the induction of proliferation by VEGF-A and VEGF-C. (D) Monolayer wound healing assay: ESM-1 (1 μg/mL) promoted the promigratory effect of VEGF-A in a monolayer wound assay after 24 hours (+14% increase in cell migration; P = .012), whereas LECs transfected with ESM-1 siRNA showed a significantly reduced migratory response to VEGF-A compared with control (siRNA-transfected) LECs. At the 48-hour time point, both VEGF-A–treated cultures and cultures treated with VEGF-A plus ESM-1 already showed a 100% wound closure. Addition of 1 μg/mL ESM-1 protein to ESM-1 siRNA–transfected LECs significantly restored the effect of VEGF-A on LEC migration after 24 and 48 hours. *P < .05; **P < .005; ***P < .001. Error bars represent standard deviation.
We next investigated whether ESM-1 might also modulate the effects of VEGF-A or VEGF-C on LEC migration. Using a standard monolayer wound assay in vitro, we found that 1 μg/mL ESM-1 promoted the migration-enhancing effect of VEGF-A (+14%; P = .012) compared with only VEGF-A (Figure 3D). LECs transfected with ESM-1 siRNA showed a significantly reduced migratory response to VEGF-A (P < .005) compared with control siRNA-transfected LECs. At the 48-hour time point, both VEGF-A–treated cultures and cultures treated with VEGF-A plus ESM-1 already showed a 100% wound closure. Addition of recombinant ESM-1 protein to ESM-1 siRNA–transfected LECs restored the stimulatory effect of VEGF-A on LEC migration at 24 and 48 hours (Figure 3D). Comparable results were seen when LECs were treated with VEGF-C (data not shown).
ESM-1 promotes lymphatic vessel activation by VEGF-A in vivo
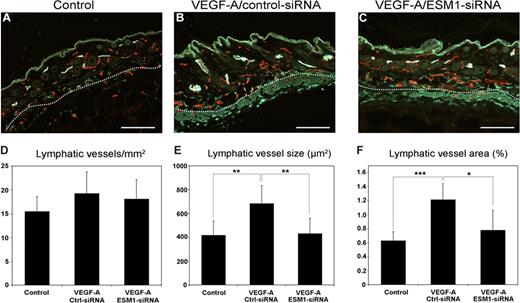
Because ESM-1 expression by LECs was strongly up-regulated by VEGF-A in vitro, and was also up-regulated on lymphatic vessels in the skin of VEGF-A transgenic mice, we investigated whether ESM-1 might also promote the effects of VEGF-A on lymphatic endothelium in vivo. To this end, we used an established Matrigel implantation assay in FVB wild-type mice. Matrigels containing PBS or VEGF-A, together with either ESM-1 siRNA or control siRNA, were subcutaneously injected into mice. After 7 days, tissue samples were obtained and frozen sections were subjected to differential immunofluorescence analyses for the lymphatic marker LYVE-1 and the panendothelial marker CD31. In the skin surrounding Matrigel implants containing VEGF-A and control siRNA, LYVE-1+ lymphatic vessels were enlarged (Figure 4B) compared with Matrigel implants that contained only PBS (Figure 4A). In contrast, lymphatic vessels showed a normal morphology and were not enlarged in the skin surrounding Matrigel implants containing VEGF-A and ESM-1 siRNA (Figure 4C). Computer-assisted morphometric analyses of LYVE-1/CD31-stained sections demonstrated that the density of lymphatic vessels was comparable among all 3 groups (Figure 4D). However, the mean size of the lymphatic vessels was significantly larger in the skin surrounding the Matrigel implants containing VEGF-A and control siRNA, compared with Matrigels containing PBS alone (P < .005; Figure 4E). Importantly, lymphatic vessels in the skin surrounding implants containing VEGF-A and ESM-1 siRNA were of a comparable size to those found surrounding PBS-containing implants and were significantly smaller than those surrounding VEGF-A/control siRNA implants (P < .005; Figure 4E). Similarly, the average tissue area covered by lymphatic vessels was significantly increased surrounding VEGF-A/control siRNA implants, compared with PBS-containing (P < .001) or VEGF-A/ESM-1 siRNA–containing implants (P < .05; Figure 4F).
Targeting ESM-1 by siRNA inhibits VEGF-A effects on lymphatic vessels in vivo. Differential immunostains for CD31 (red) and LYVE-1 (green) reveal that, compared with Matrigels that contain PBS (control) (A), LYVE-1+ lymphatic vessels (green) surrounding Matrigels containing VEGF-A and control siRNA were enlarged (B). Lymphatic vessels were not enlarged surrounding Matrigels containing VEGF-A and ESM-1 siRNA (C). Scale bars equal 100 μm. Computer-assisted morphometric analyses of LYVE-1/CD31–stained sections showed comparable density of lymphatic vessels in all groups (D). The mean size of lymphatic vessels was significantly larger surrounding Matrigels containing VEGF-A and control siRNA (Ctrl-siRNA) than Matrigels containing PBS (control) (E) or VEGF-A and ESM-1 siRNA (ESM1-siRNA). The mean tissue area covered by lymphatic vessels was significantly increased surrounding VEGF-A/Ctrl-siRNA implants compared with PBS-containing (control) or VEGF-A/ESM1-siRNA–containing implants (F). *P < .05; **P < .005; ***P < .001.
Targeting ESM-1 by siRNA inhibits VEGF-A effects on lymphatic vessels in vivo. Differential immunostains for CD31 (red) and LYVE-1 (green) reveal that, compared with Matrigels that contain PBS (control) (A), LYVE-1+ lymphatic vessels (green) surrounding Matrigels containing VEGF-A and control siRNA were enlarged (B). Lymphatic vessels were not enlarged surrounding Matrigels containing VEGF-A and ESM-1 siRNA (C). Scale bars equal 100 μm. Computer-assisted morphometric analyses of LYVE-1/CD31–stained sections showed comparable density of lymphatic vessels in all groups (D). The mean size of lymphatic vessels was significantly larger surrounding Matrigels containing VEGF-A and control siRNA (Ctrl-siRNA) than Matrigels containing PBS (control) (E) or VEGF-A and ESM-1 siRNA (ESM1-siRNA). The mean tissue area covered by lymphatic vessels was significantly increased surrounding VEGF-A/Ctrl-siRNA implants compared with PBS-containing (control) or VEGF-A/ESM1-siRNA–containing implants (F). *P < .05; **P < .005; ***P < .001.
Discussion
To comprehensively identify downstream molecular targets induced by VEGF-A or VEGF-C in lymphatic endothelium, we performed transcriptional profiling of LECs exposed to each growth factor for up to 24 hours. This is the first study into the comprehensive downstream mediators of VEGF-A in the lymphatic endothelium.
Microarray time course studies provide the ability to monitor the temporal behavior of biologic processes through the sequential measurement of the expression of tens of thousands of genes.54–57 However, given the large number of genes evaluated in these experiments, and the often rather small number of replicates, the variances are usually poorly estimated. To analyze gene expression of triplicate samples at 5 different time points, we applied the multivariate EB approach to inference.44,45 This is a model-based strategy for introducing moderation into the analysis that has been reported to generate the least number of false positives and false negatives.26 Using STEM analysis,29 we identified distinct temporal clusters of genes modulated by VEGF-A and VEGF-C. Within the early response cluster—with a peak of expression after 1 hour—we observed a predominance of transcription factors and signaling molecules. Several of these, such as the early growth response genes EGR1, EGR2, and EGFR3, have been previously described as early VEGF-A target genes in blood vascular endothelium.58–60 Our finding that Down syndrome critical region protein-1 (DSCR1) was one of the most potently induced early response targets in LECs is in agreement with previous results in cultured blood vascular endothelial cells.48–51 DSCR1 belongs to a family of proteins that interact with the phosphatase calcineurin A and has been shown to repress several inflammatory genes in human endothelial cells by inhibiting nuclear translocation of nuclear factor of activated T cells (NFAT), a transcription factor that is regulated by calcineurin A.48,61 It is of interest that the overlap among VEGF-A and VEGF-C targets was highest in the early response cluster compared with the transiently induced and the progressively induced gene clusters. Among the cluster of genes with progressively increasing expression after VEGF-A and VEGF-C treatment were angioipoietin-2, which has been previously shown to be indispensable for the normal develop-ment of the lymphatic vasculature in mice,62 and VEGF-C, indicating the possible induction of an autocrine growth pathway via VEGFR-3 activation.
Importantly, we found ESM-1 to be one of the most potently induced genes by both VEGF-A and VEGF-C in cultured LECs; ESM-1 was induced at both the mRNA and the protein levels. ESM-1 expression was also detectable on lymphatic vessels in the skin of transgenic mice with chronically elevated levels of VEGF-A but not in the skin of wild-type mice, indicating that ESM-1 is also a target of VEGF-A in vivo. ESM-1 is a dermatan sulfate proteoglycan secreted by endothelial cells that has been suggested to have a role in the regulation of cell adhesion in inflammatory disorders and in tumor progression.63 Whereas the precise function of ESM-1 is still unclear, it has been proposed to inhibit the interaction between intercellular adhesion molecule-1 (ICAM-1) and the integrin LFA-1 on lymphocytes and monocytes.64 Recently, an increased level of ESM-1 mRNA expression was reported to represent one of the most significant molecular elements of a poor prognosis signature in several types of tumor types, including lung cancer.65 Moreover, overexpression of ESM-1 in human embryonic kidney 293 cells promoted tumor growth in a mouse xenotransplantation model.66
Our current study revealed that ESM-1 promoted the mitogenic and promigratory activity of both VEGF-A and VEGF-C on cultured LECs, whereas addition of ESM-1 alone did not affect LEC functions in vitro, similar to blood vascular endothelium.67 Moreover, siRNA-mediating ESM-1 silencing inhibited the activation of LECs by VEGF-A and VEGF-C in vitro and by VEGF-A in vivo. Together, the results indicate that VEGF-A/VEGF-C–mediated induction of ESM-1 represents an autocrine, positive feedback loop to further promote the stimulatory effects of both growth factors on lymphatic endothelium. ESM-1 has been previously shown to bind to HGF and to increase the HGF-mediated proliferation of human embryonic kidney cells in a manner similar to that of heparin; the single dermatan sulfate chain of ESM-1, covalently attached to serine 137, appears to be required for this effect, since the nonglycanated form of ESM-1 did not promote the effects of HGF.63 Thus, it will be of interest to investigate whether ESM-1 also binds to VEGF-A and VEGF-C to increase their interaction with VEGFR-2 and/or VEGFR-3 on LECs.
The relative contribution of direct activation of VEGFR-2 versus possible indirect effects on VEGFR-3, via induction of its ligand VEGF-C, toward the lymphangiogenic effects of VEGF-A have remained unclear.9 In our study, we found that inhibition of VEGFR-2 with a blocking antibody completely abrogated the VEGF-A–mediated induction of ESM-1 in LECs, clearly indicating that VEGFR-2 is essential for mediating VEGF-A effects. However, specific blockade of VEGFR-3 resulted in a partial inhibition of the ESM-1 induction by VEGF-A. Together with the observed induction of VEGF-C expression after VEGF-A treatment, these findings indicate that VEGF-A might mediate its lymphangiogenic effects, in part, via activation of an autocrine loop in LECs, which leads to VEGFR-3 activation by VEGF-C. It remains unclear whether or not VEGF-A might also exert effects on the formation of heterodimers of VEGFR-2 and VEGFR-3, which might then affect receptor tyrosine phosphorylation.68 However, the effects of VEGF-A on LEC gene expression were, in general, stronger than those of VEGF-C, and the VEGF-A–mediated induction of ESM-1 was only partially blocked by the anti–VEGFR-3 blocking antibody. Our study also demonstrates that both VEGFR-2 and VEGFR-3 are required for the full activity of VEGF-C on LEC gene expression; inhibition of each receptor alone only partially inhibited the induction of ESM-1 by VEGF-C, whereas combined blockade completely abrogated the VEGF-C effect. These findings are in agreement with results demonstrating that the mature form of the VEGF-C protein, which was used for this study, efficiently binds to and activates both receptors.1 Overall, our studies reveal ESM-1 as a novel mediator of lymphangiogenesis and as a potential target for the inhibition of VEGF-A– or VEGF-C–induced pathologic lymphatic vessel growth and activation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Institutes of Health grants CA69184 and CA86410, Swiss National Fund grant 3100A0-108207, Austrian Science Foundation grant S9408-B11, Cancer League Zurich, and Commission of the European Communities grant LSHC-CT-2005-518178 (M.D.).
National Institutes of Health
Authorship
Contribution: J.S. designed and performed research, analyzed the data, and wrote the paper; R.H. performed research and analyzed data; and M.D. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michael Detmar, Institute of Pharmaceutical Sciences, Swiss Federal Institute of Technology, ETH Zurich, Wolfgang-Pauli-Str 10, HCI H303, CH-8093, Zurich, Switzerland; e-mail: michael.detmar@pharma.ethz.ch.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal