Abstract
The transcription factor GATA1 coordinates timely activation and repression of megakaryocyte gene expression. Loss of GATA1 function results in excessive megakaryocyte proliferation and disordered terminal platelet maturation, leading to thrombocytopenia and leukemia in patients. The mechanisms by which GATA1 does this are unclear. We have used in vivo biotinylated GATA1 to isolate megakaryocyte GATA1-partner proteins. Here, several independent approaches show that GATA1 interacts with several proteins in the megakaryocyte cell line L8057 and in primary megakaryocytes. They include FOG1, the NURD complex, the pentameric complex containing SCL/TAL-1, the zinc-finger regulators GFI1B and ZFP143, and the corepressor ETO2. Knockdown of ETO2 expression promotes megakaryocyte differentiation and enhances expression of select genes expressed in terminal megakaryocyte maturation, eg, platelet factor 4 (Pf4). ETO2-dependent direct repression of the Pf4 proximal promoter is mediated by GATA-binding sites and an E-Box motif. Consistent with this, endogenous ETO2, GATA1, and the SCL pentameric complex all specifically bind the promoter in vivo. Finally, as ETO2 expression is restricted to immature megakaryocytes, these data suggest that ETO2 directly represses inappropriate early expression of a subset of terminally expressed megakaryocyte genes by binding to GATA1 and SCL.
Introduction
The evolutionarily conserved family of C4 zinc-finger GATA transcription factors coordinate differentiation and proliferation, in a cell-context specific manner, to ensure that appropriate numbers of terminally mature cells arise from stem/progenitor cells through development and adult life.1 In human hemopoiesis, these processes generate approximately 1010 new blood cells daily. Moreover, the number and mix of cells produced can be altered rapidly as required. When these processes go awry, diseases such as leukemia can occur. Currently, there is an incomplete understanding of how hemopoietic transcription factors cooperate with other nuclear regulators to coordinate terminal maturation of lineage-restricted precursors leading to cell-cycle exit
GATA1, the founding member of the GATA family, has critical functions during myelopoiesis. Gain-of-function experiments show that GATA1 specifies erythroid cells and megakaryocytes at the expense of granulocytes/macrophages in fish2,3 and mice.4,5 In erythroid cells and megakaryocytes, sustained GATA1 expression is required for terminal maturation. Germ line GATA1 deletion results in arrested terminal erythroid and megakaryocyte maturation.6-8 In red cells, this causes apoptosis of proerythroblasts, whereas in megakaryocytes it leads to marked immature megakaryoblast hyperproliferation and abnormal platelet production. Germ line GATA1 mutations in patients results in thrombocytopenia, platelet maturation defects, anemia, β-thalassemia, porphyria, and neutropenia.9 Moreover, acquired GATA1 mutations are a required event in the megakaryocyte hyperproliferative disorders of transient abnormal myelopoiesis and acute megakaryoblastic leukemia in neonates and children with Down syndrome.10
In erythrocytes and megakaryocytes, GATA1 both activates red cell-11 and megakaryocyte-specific7,8,12,13 gene expression and represses genes associated with proliferation (Gata2, c-myc, Nab2, and Kit11,13-15 ), and alternate cell fate (Prg2, Eosinophil Major Basic Protein16 ) by mechanisms that are incompletely understood.
GATA1 needs to bind DNA at either single (A/T)GATA(A/G) or GATC sites17 or complex inverted or repeated double (A/T)GATA(A/G) sites and (A/T)GATA(A/G):GAT motifs18 in red cells19 and megakaryocytes.20 GATA1 transcriptional activity is also modulated by interaction with proteins in transcriptionally active multiprotein complexes. These have been best characterized in red cells. Here, GATA1 exists in a complex with the multitype zinc finger protein FOG1.16,21,22 Elegant studies in mice23 and of human patients24,25 have confirmed the importance of this interaction in erythroid maturation. There are probably to be at least 2 GATA-FOG1 complexes: one that activates and another that represses expression and includes the MeCP1/NURD repressor complex, which directly interacts with FOG1.16,22
In erythroid cells, GATA1 is also part of a transcriptional activating pentameric complex, which includes SCL/TAL-1 (hereafter called SCL), E2A, Ldb1, and LMO2,26 which occupies regulatory sequences in the Gata1,27,28 α-globin,29 and glycophorin A30 genes. This complex has been isolated biochemically from erythroid cells.16,31 When this pentameric complex also contains ETO232 (a corepressor protein implicated in human leukemogenesis33 ), bound to E2A,34 the complex is repressive.31,35,36 In red cells, GATA1 is also bound to the transcription factor Gfi1b,16 the chromatin remodeling complex ACH/WCRF,16,37 the ETS transcription factor Fli-138 and, in the hemopoietic cell line, K562, to the basic helix-loop-helix protein HERP2 (hairy-enhancer-of split related factor).39 Finally, GATA1 also binds itself to form GATA1 multimers.40 Several proteins have also been shown to immunoprecipitate with GATA1 when overexpressed in heterologous cells or by GST pull-down assays. Examples include RUNX1,41 EKLF,42,43 and SP1.43
In megakaryocytes, the nature of GATA1 containing complexes is less clear. GATA1 is bound by FOG112,21 and the pentameric complex.31 Beyond this, it is unclear whether other erythroid GATA1-containing complexes exist in megakaryocytes (and whether they serve the same function) and/or whether megakaryocyte-specific GATA1 complexes occur. Here, we describe megakaryocyte GATA1-containing protein complexes to help further our understanding of the mechanism of GATA1 action during megakaryopoiesis.
Methods
Institutional Review Board approval for this study was obtained from the United Kingdom Government Home Office.
Constructs
The Escherichia coli BirA biotin ligase gene44 was inserted as a polymerase chain reaction (PCR) fragment downstream of the human EF1α promoter in a vector with a puromycin resistance gene (pEF1α-BirA). Wild-type biotin tagged44 murine Gata1 cDNA was subcloned downstream of the EF1α promoter in a vector with a neomycin resistance gene (pEF1α-bioGATA1).
Cell lines and transfections
L8057 cells (murine megakaryoblastic cell line) were cultured and differentiated as described45 and transfected as described.28 The cell line was derived from an irradiated C3H/He mouse. Stable clones expressing high levels of BirA mRNA were selected by Northern blotting and subsequently transfected with pEF1α-bioGATA1. Nuclear extracts were prepared as described.46
Streptavidin pull-down
Pull-downs were performed with 1 mg or 120 mg nuclear extract as described previously.44 Briefly, paramagnetic streptavidin beads were blocked by washing 3 times in binding buffer (139 mm KCl, 12 mM NaCl, 0.8 mM MgCl2, 20 mM Tris-HCl, pH 7.9, 0.5% NP-40, 0.2 mM ethylenediaminetetraacetic acid, 20% glycerol, 1 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride, protease inhibitor cocktail [Roche Diagnostics, Indianapolis, IN]) supplemented with 200 ng/μL purified ovalbumin; 20 μL beads was used per 1 mg nuclear extract. Binding was done in binding buffer at 4°C overnight on a rocking platform, followed by 6 washes in washing buffer (200 mM KCl, 12 mM NaCl. 0.8 mM MgCl2, 20 mM Tris, pH 7.9, 0.5% NP-40, 0.2 mM ethylenediaminetetraacetic acid, 20% glycerol, 1 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail [Roche Diagnostics]). Bound material was eluted by boiling for 10 minutes in Laemmli protein sample loading buffer.
Mass spectrometry
Proteins eluted from the beads after binding of the BirA or BirA/biotag GATA1 nuclear extracts were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis on a 4% to 12% polyacrylamide gel (Invitrogen, Carlsbad, CA) and stained with Colloidal Coomassie Blue (Invitrogen). Each lane was cut out and divided into 25 gel plugs. Trypsinization and mass spectrometry analysis of these gel slices was performed by high performance liquid chromatography-Q-time of flight (HPLC-Q-TOF) at Birmingham University Mass Spectrometer Facility (Birmingham, United Kingdom). For the peptide search, the following parameters were used: fixed modifications, carbamidomethyl; variable modifications, oxidation; peptide charge 2+ and 3+ and the mass spectrometry data were analyzed using MASCOT software (Matrix Science, London, United Kingdom) for peptide assignment. Peptide matches were confirmed by BLAST search.
Gel filtration
Protein complexes were fractionated on Superose 6 10/30 column (AKTA FPLC; GE Healthcare, Little Chalfont, United Kingdom), precipitated with 100% trichloroacetic acid, and analyzed by Western blotting.
Western blotting
Western blot analysis was performed as previously described.47 Primary and secondary antibodies used are described in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Signal intensities were measured with a Typhoon Imager (GE Healthcare).
Coimmunoprecipitation and sequential immunoprecipitations
Coimmunoprecipitation and sequential immuoprecipitations (IPs) were performed as described previously.16,31 For IP with anti-GATA1 N6 antibody (Santa Cruz Biotechnology, Santa Cruz, CA), a rabbit antirat bridging antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) bound to beads was used.
Immunofluorescence and colocalization
L8057 cells and primary megakaryocytes were fixed on coverslips in 4% paraformaldehyde and permeabilized in 0.1% Triton-phosphate-buffered saline. Antibody dilutions were described previously31 ; in addition, GATA1 (1/100 dilution) FOG1 (1/50 dilution), GFI1B (1/100 dilution), and ZFP143 (1/400 dilution) were used. Secondary antibodies were Cy2-conjugated goat antirat, Cy3-conjugated donkey antirabbit, Cy3-conjugated donkey antigoat, and Cy3-conjugated donkey antimouse (all from Jackson ImmunoResearch Laboratories). Nucleic acids were counterstained with Cy5-conjugated ToPro-3 (Invitrogen). Images were captured with a fluorescent microscope (Olympus BX51) and Macprobe 4.3 software. Fluorescence intensities were measured with Metamorph software (Molecular Devices, Sunnyvale, CA) as previously described.48 Colocalization was measured as competition between pairs of antibodies. Briefly, for each pair of antibodies, 3 slides were immunolabeled: 1 with antibody A alone, another with B alone, and a third with A and B. After completion of the immunolabeling procedure, the intensities for A and B in 3 slides were measured. If competition exists, the intensity of A and/or B in the combined slide should be reduced relative to the slides stained with the antibodies alone. Further explanation of the conceptual basis of the technique is set out in Figure S1.
Purification of murine primary megakaryocytes
Details are supplied in Document S1.
Luciferase reporter assays
Twenty-four hours after being plated in 12-well plates, 293T cells were transfected with Lipofectamine 2000 reagent, according to the manufacturer's instructions (Invitrogen). The full-length rat Pf4 promoter (kind gift from K. Ravid) was subcloned into the pGL4.10 luciferase reporter plasmid (Promega, Southampton, United Kingdom). Deletion constructs and mutants were generated by PCR and verified by sequencing. PCR primer sequences are available on request. Expression plasmids have been described previously.31 At 48 hours after transfection, cells were lysed and luciferase and β-galactosidase (to monitor transfection efficiency) activities were measured using standard procedures. Each transfection was performed in triplicate, and data presented are from 3 to 5 independent experiments.
Lentiviral packaging and infection
shRNA oligonucleotides were expressed from the pTRIP-ΔU3 lentivirus expressing an EF1α-GFP reporter cassette (kind gift from P. H. Romeo). Lentiviral supernatants were made and infections undertaken as described.31,35 At 96 hours after infection, GFP-positive cells were sorted on a MoFlo FACsorter (DakoCytomation; Dako North America, Carpinteria, CA). Sorted cells were used immediately or returned to culture. Nuclear extract and total RNA was isolated 4 days after sort (when > 95% of cells were GFP-positive) for Western blot and Taqman real-time reverse-transcribed (RT)–PCR analysis.
Real-time RT-PCR
RNA was isolated from 105 cells using an RNeasy Micro kit (Qiagen, Hilden, Germany) and treated with DNase I. cDNA synthesis and real-time PCR were performed as previously described.49 cDNA from L8057 cells was used to estimate the range of linearity of each combination of primers and probe. Taqman RT-PCR primer and probe sequences are available on request. Gene expression ratios were calculated relative to the cycle threshold (Ct) value for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) according to the following equation: relative ratio = 2(Ct Gapdh − Ct primer/probe).
Chromatin immunoprecipitation
Cross-linked chromatin was prepared from 107 L8057 cells per antibody as described,28,50 except for chromatin immunoprecipitation (ChIP) with anti-ETO2 antibody where 4 × 107 cells were cross-linked with 2 mM ethylene glycol-bis succinimidylsuccinate (Pierce Chemical, Cramlington, United Kingdom; 20 minutes at room temperature), followed by 1% formaldehyde (10 minutes at room temperature). Sonication was performed for 6 × 30 seconds at 50% amplitude. Antibodies used are described in the Document S1. For each antibody, at least 3 independent chromatin preparations were immunoprecipitated. Taqman PCR primers and probes were designed with Primer Express 2.0 software and are available on request (Applied Biosystems, Foster City, CA). Duplicate PCR reactions on each immunoprecipitated template were performed on a Sequence Detection 7000 thermocycler (Applied Biosystems). Quantitation of the enrichment at a particular test point with every antibody (including IgG) was initially normalized with respect to input and then with respect to a negative control gene locus Gapdh as described before.28,50
Results
In vivo biotinylation of GATA1 to identify GATA1-interacting proteins
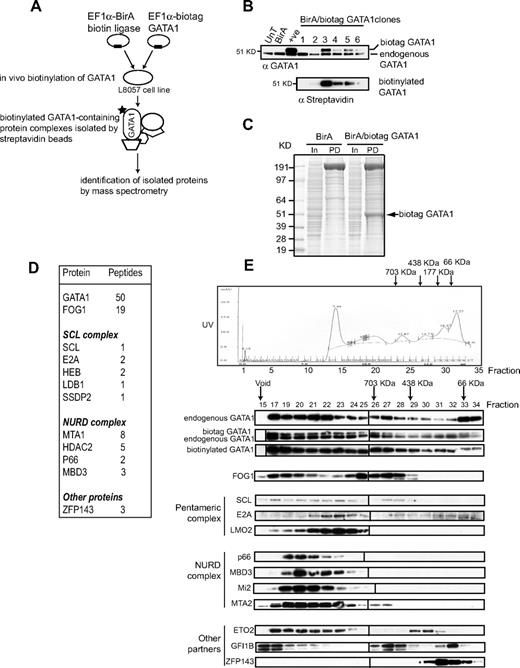
To identify GATA1-interacting proteins in megakaryocytes, we used a 1-step purification method44 that allows isolation of biotinylated GATA1-containing protein complexes by streptavidin beads (Figure 1A). The immature megakaryoblastic cell line L8057 was first stably transfected with a biotin ligase (BirA) expression plasmid. Clones expressing high levels of biotin ligase RNA (data not shown) were then transfected with a second plasmid expressing murine GATA1 with an N-terminal 23-amino acid tag (biotag GATA1) that allows for in vivo biotinylation. Previous studies have shown that this tag does not alter GATA1 function.16
Isolation of GATA1-containing complexes in L8057 megakaryocyte cells. (A) Scheme of in vivo protein biotinylation and purification by streptavidin beads. (B) Nuclear extracts from wild-type (UnT), BirA-expressing (BirA), biotag-GATA1–expressing (+ ve), and 6 independent L8057 clones expressing biotag GATA1 and BirA biotin ligase were tested by Western blot with anti-GATA1 antibody (top panel). The top band corresponds to the slower migrating biotag GATA1; bottom band, endogenous GATA1. The blot was probed with antistreptavidin-HRP antibody, which confirms biotinylation of GATA1 in extracts from clones 3, 4, 5, and 6. (C) Crude nuclear extracts from cells transfected with either BirA alone (BirA) or BirA/biotag GATA1 (BirA/biotag GATA1) were incubated with streptavidin-coated beads. Precipitated proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (PD lane) and stained with Coomassie blue. Approximately 20 μg crude extract was loaded as input (In lane).  indicates biotinylated GATA1 (biotag GATA1), as determined by mass spectrometry. (D) Table of proteins and the number of peptides precipitated by streptavidin beads and identified by mass spectrometry. (E) Gel filtration analysis (top). An example of fractionation of crude nuclear extracts from L8057 cells transfected with BirA/biotag GATA1 on a Superose 6 column. Similar results were obtained from wild-type nuclear extracts.
indicates biotinylated GATA1 (biotag GATA1), as determined by mass spectrometry. (D) Table of proteins and the number of peptides precipitated by streptavidin beads and identified by mass spectrometry. (E) Gel filtration analysis (top). An example of fractionation of crude nuclear extracts from L8057 cells transfected with BirA/biotag GATA1 on a Superose 6 column. Similar results were obtained from wild-type nuclear extracts.  indicates position where protein molecular markers elute. The UV profile indicates that proteins elute in a broad fractionation profile. Fractions were taken from the Superose 6 column, precipitated, and analyzed by Western blotting for GATA1 and several potential protein partners (bottom panels). The antibody used is indicated on the lefthand side of the panel. Note that endogenous GATA1 and biotag-GATA1 have a similar elution profile. Vertical line(s) have been inserted to indicate a repositioned gel lane.
indicates position where protein molecular markers elute. The UV profile indicates that proteins elute in a broad fractionation profile. Fractions were taken from the Superose 6 column, precipitated, and analyzed by Western blotting for GATA1 and several potential protein partners (bottom panels). The antibody used is indicated on the lefthand side of the panel. Note that endogenous GATA1 and biotag-GATA1 have a similar elution profile. Vertical line(s) have been inserted to indicate a repositioned gel lane.
Isolation of GATA1-containing complexes in L8057 megakaryocyte cells. (A) Scheme of in vivo protein biotinylation and purification by streptavidin beads. (B) Nuclear extracts from wild-type (UnT), BirA-expressing (BirA), biotag-GATA1–expressing (+ ve), and 6 independent L8057 clones expressing biotag GATA1 and BirA biotin ligase were tested by Western blot with anti-GATA1 antibody (top panel). The top band corresponds to the slower migrating biotag GATA1; bottom band, endogenous GATA1. The blot was probed with antistreptavidin-HRP antibody, which confirms biotinylation of GATA1 in extracts from clones 3, 4, 5, and 6. (C) Crude nuclear extracts from cells transfected with either BirA alone (BirA) or BirA/biotag GATA1 (BirA/biotag GATA1) were incubated with streptavidin-coated beads. Precipitated proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (PD lane) and stained with Coomassie blue. Approximately 20 μg crude extract was loaded as input (In lane).  indicates biotinylated GATA1 (biotag GATA1), as determined by mass spectrometry. (D) Table of proteins and the number of peptides precipitated by streptavidin beads and identified by mass spectrometry. (E) Gel filtration analysis (top). An example of fractionation of crude nuclear extracts from L8057 cells transfected with BirA/biotag GATA1 on a Superose 6 column. Similar results were obtained from wild-type nuclear extracts.
indicates biotinylated GATA1 (biotag GATA1), as determined by mass spectrometry. (D) Table of proteins and the number of peptides precipitated by streptavidin beads and identified by mass spectrometry. (E) Gel filtration analysis (top). An example of fractionation of crude nuclear extracts from L8057 cells transfected with BirA/biotag GATA1 on a Superose 6 column. Similar results were obtained from wild-type nuclear extracts.  indicates position where protein molecular markers elute. The UV profile indicates that proteins elute in a broad fractionation profile. Fractions were taken from the Superose 6 column, precipitated, and analyzed by Western blotting for GATA1 and several potential protein partners (bottom panels). The antibody used is indicated on the lefthand side of the panel. Note that endogenous GATA1 and biotag-GATA1 have a similar elution profile. Vertical line(s) have been inserted to indicate a repositioned gel lane.
indicates position where protein molecular markers elute. The UV profile indicates that proteins elute in a broad fractionation profile. Fractions were taken from the Superose 6 column, precipitated, and analyzed by Western blotting for GATA1 and several potential protein partners (bottom panels). The antibody used is indicated on the lefthand side of the panel. Note that endogenous GATA1 and biotag-GATA1 have a similar elution profile. Vertical line(s) have been inserted to indicate a repositioned gel lane.
As shown in Figure 1B (top panel), Western blot analysis shows that 4 L8057 clones (clones 3-6) stably express both endogenous GATA1 and a slightly higher-molecular-weight GATA1 protein that is biotinylated (bottom panel, the same blot is reprobed with streptavidin-horseradish peroxidase conjugate). Clone 5 was used for subsequent experiments as it expresses biotag GATA1 at a lower level than endogenous GATA1. Therefore, in these cells, the overall level of GATA1 protein is less than 2-fold greater than in untransfected L8057 cells. This reduces, but does not eliminate, the chance that the stoichiometry and nature of GATA1-containing complexes are perturbed.
Nuclear extracts from L8057 clones expressing BirA alone and BirA/biotag GATA1 were incubated with streptavidin beads, and the precipitated proteins separated by gel electrophoresis. As expected, many cellular proteins are naturally biotinylated (Figure 1C BirA Pull Down [PD] lane), but protein sequence analysis identified proteins that were specifically detected only in BirA/biotag GATA1, but not BirA, expressing cells. The most prominent of these is biotag GATA1 (Figure 1C BirA/biotag GATA1 PD lane, Figure 1D, and data not shown). Sequence analysis confirmed that peptides corresponding to known partners of GATA1, FOG1, members of the SCL, and NURD complexes were precipitated in cells expressing BirA/biotag GATA1 (Figure 1D). In addition, the zinc finger protein ZFP143/Staf51-53 (the human homolog is known as ZNF14354,55 ) was also identified as a putative GATA1 interacting protein for the first time.
GATA1 is part of multiple distinct complexes
We next asked whether the predicted and newly identified potential GATA1-interacting partner proteins eluted with GATA1 in high molecular weight protein complexes. Gel filtration profile of nuclear extracts from the BirA/biotag GATA1-expressing L8057 cells shows that proteins elute from more than 703 kDa through less than 66 kDa (Figure 1E). The top 3 Western blot panels of Figure 1E show that endogenous GATA1 is detected throughout the elution profile and that this is mirrored by biotag GATA1. This suggests that endogenous and biotag GATA1 are present in complexes of similar molecular weight. In contrast, GATA1-interacting proteins have distinct and more restricted elution profiles. FOG1 is not detected below fraction 29 (∼ 438 kDa). Higher-molecular-weight FOG1-containing fractions 15 to 24 coelute with NURD complex members with the suggestion that lower molecular weight FOG1-containing complexes may not contain the full NURD complex. Although there is overlap with elution of the NURD member containing complexes, members of the pentameric complex have a distinct elution profile and are most prominent in fractions 21 to 25. Finally, we also studied the elution profile of other suspected and known GATA1 partner proteins. Examples here include ETO2,31,35,36 GFI1B,16 and ZFP143 (data presented here). ETO2 (fractions 17-23 and 29-31) and GFI1B (fractions 15-17, 26-28, and 32,33) both elute in distinct, widely separated regions, suggesting that these proteins may participate in multiple distinct complexes. Finally, ZFP143 (66 kDa) elutes in a tight peak lower-molecular-weight fractions (fractions 30-33), suggesting that some of it may be complexed in monomeric form with low- molecular-weight proteins.
Validation of interaction between GATA1 and potential protein partners
Streptavidin pull-down.
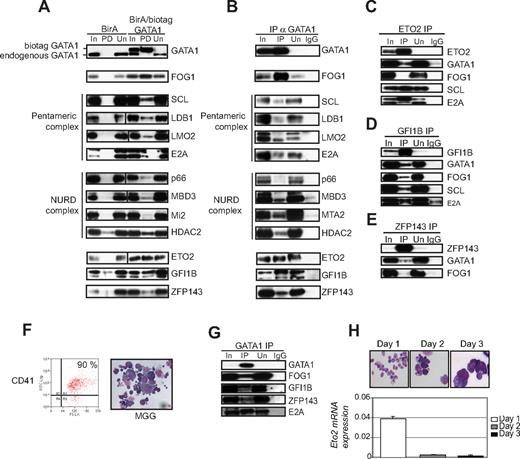
Next, we sought to validate interactions between GATA1 and its potential partners. First, we looked, by Western blot analysis to determine whether GATA1-interacting proteins were precipitated after incubation with streptavidin specifically from nuclear extracts from cells expressing BirA/biotag GATA1 (Figure 2A, lane marked PD). Known GATA1 partners FOG1 and members of the pentameric complex (SCL, LDB1, LMO2, E2A) were detected in the precipitate. In addition, we confirmed that p66, MBD3, Mi2, and HDAC2 (members of the NURD complex) and GFI1B16 physically interact with GATA1 in megakaryocytic cells. However, the results with HDAC2 and GFI1B in this experiment have to be interpreted with caution as they can be detected in the precipitate with extracts from BirA only expressing cells (see PD lane for BirA control pull-downs). We also documented an interaction between GATA1 and ETO2 and GATA1 and ZFP143 by streptavidin pull-down. All potential interacting proteins were specifically pulled down by streptavidin in cells expressing BirA/biotag GATA1 but not cells expressing only BirA. These data confirm that in vivo biotinylation of GATA1 in our experiments can identify bona fide GATA1 partners.
Validation of interaction between GATA1 and partner proteins in L8057 cells and primary megakaryocytes. (A) Nuclear extracts from BirA biotin ligase expressing (BirA) and BirA and biotag GATA1 expressing (BirA/biotag GATA1) cells were studied. In indicates input lane (20 μg crude nuclear extract); PD, pull-down lane (nuclear extracts precipitated with streptavidin beads); and Un, unbound supernatant lane (proteins not bound to streptavidin). The antibodies used in the Western blot analysis are indicated on the right of the panels. Vertical line(s) have been inserted to indicate a repositioned gel lane. Biotag GATA1 is pulled down from BirA/bioGATA1-transfected cells but is absent from BirA-only cells (top panel). The positions of endogenous (bottom band) and biotag GATA1 (top band) are indicated. (B) Nuclear extracts from untransfected L8057 cells were immunoprecipitated (IP) with αGATA1 antibody. In indicates 20 μg crude nuclear extract; IP, proteins immunoprecipitated by αGATA1 antibody; Un, unbound proteins left in the supernatant after immunoprecipitation; and IgG, control was immunoprecipitation performed with the corresponding normal IgG. Antibodies used in Western blot analysis are indicated on the right of the panels. FOG1, all members of the pentameric complex, several members of the NuRD complex, as well as ETO2, GFI1B, and ZFP143 all coimmunoprecipitate with GATA1. (C-E) Reverse coimmunoprecipitation experiments. L8057 cell nuclear extracts were immunoprecipitated with antibodies against ETO2 (C), GFI1B (D), or ZFP143 (E). Lanes are marked as in panel A. Antibodies used in Western blot analysis are marked on the right of the panels. (F) Mouse bone marrow cells were cultured for 3 days with thrombopoietin. Percentage of CD41 expressing primary megakaryocytes was assessed by fluorescence-activated cell sorter analysis (left). May-Grunwald-Giemsa staining (right) shows that the morphology of the cells used for immunoprecipitation experiments were a mixture of immature and mature megakaryocytes. (G) Nuclear extracts prepared from primary megakaryocytes shown in panel F were immunoprecipitated with αGATA1 antibody. Coimmunoprecipitated proteins were detected by Western blot analysis. Antibodies used in the Western blot analysis are indicated on the right of the panels. (H) Analysis of Eto2 mRNA expression during megakaryocyte maturation. Mouse bone marrow cells were cultured with thrombopoietin for 3 days. CD41 expressing cells were isolated on each day of culture. May-Grunwald-Giemsa staining shows the morphology of the cells isolated on each day (top panel). All panels were photographed at 40× magnification. These cells were used to extract RNA and make cDNA. Eto2 mRNA levels were quantitated relative to GAPDH levels by Taqman real-time RT-PCR, at each time point (bottom panel).
Validation of interaction between GATA1 and partner proteins in L8057 cells and primary megakaryocytes. (A) Nuclear extracts from BirA biotin ligase expressing (BirA) and BirA and biotag GATA1 expressing (BirA/biotag GATA1) cells were studied. In indicates input lane (20 μg crude nuclear extract); PD, pull-down lane (nuclear extracts precipitated with streptavidin beads); and Un, unbound supernatant lane (proteins not bound to streptavidin). The antibodies used in the Western blot analysis are indicated on the right of the panels. Vertical line(s) have been inserted to indicate a repositioned gel lane. Biotag GATA1 is pulled down from BirA/bioGATA1-transfected cells but is absent from BirA-only cells (top panel). The positions of endogenous (bottom band) and biotag GATA1 (top band) are indicated. (B) Nuclear extracts from untransfected L8057 cells were immunoprecipitated (IP) with αGATA1 antibody. In indicates 20 μg crude nuclear extract; IP, proteins immunoprecipitated by αGATA1 antibody; Un, unbound proteins left in the supernatant after immunoprecipitation; and IgG, control was immunoprecipitation performed with the corresponding normal IgG. Antibodies used in Western blot analysis are indicated on the right of the panels. FOG1, all members of the pentameric complex, several members of the NuRD complex, as well as ETO2, GFI1B, and ZFP143 all coimmunoprecipitate with GATA1. (C-E) Reverse coimmunoprecipitation experiments. L8057 cell nuclear extracts were immunoprecipitated with antibodies against ETO2 (C), GFI1B (D), or ZFP143 (E). Lanes are marked as in panel A. Antibodies used in Western blot analysis are marked on the right of the panels. (F) Mouse bone marrow cells were cultured for 3 days with thrombopoietin. Percentage of CD41 expressing primary megakaryocytes was assessed by fluorescence-activated cell sorter analysis (left). May-Grunwald-Giemsa staining (right) shows that the morphology of the cells used for immunoprecipitation experiments were a mixture of immature and mature megakaryocytes. (G) Nuclear extracts prepared from primary megakaryocytes shown in panel F were immunoprecipitated with αGATA1 antibody. Coimmunoprecipitated proteins were detected by Western blot analysis. Antibodies used in the Western blot analysis are indicated on the right of the panels. (H) Analysis of Eto2 mRNA expression during megakaryocyte maturation. Mouse bone marrow cells were cultured with thrombopoietin for 3 days. CD41 expressing cells were isolated on each day of culture. May-Grunwald-Giemsa staining shows the morphology of the cells isolated on each day (top panel). All panels were photographed at 40× magnification. These cells were used to extract RNA and make cDNA. Eto2 mRNA levels were quantitated relative to GAPDH levels by Taqman real-time RT-PCR, at each time point (bottom panel).
Coimmunoprecipitation.
Next, physical interactions between endogenous proteins were tested by coimmunoprecipitation in untransfected L8057 megakaryoblastic cells (Figure 2B). As positive controls, we confirmed the interaction between GATA1 and FOG1, and GATA1 with all 4 other members of the pentameric complex (SCL, E2A, LDB1, and LMO2). We note that there are 2 bands in the input and unbound lanes when the blot is detected with the anti-LMO2 antibody. This may represent alternatively spliced LMO2 proteins or proteins with different posttranslational modifications. Only the lower molecular weight protein species interacts with GATA1 (Figure 2A,B). For the first time, we showed that GATA1 also coimmunoprecipitated with members of the NURD complex (MBD3, MTA2, and HDAC2) in megakaryocytic cells. Interaction was also detected between GATA1 and ETO2, GFI1B, and ZFP143. To confirm interaction between endogenous proteins, reverse coimmunoprecipitation (Figure 2C-E) showed that ETO2 bound to GATA1, SCL, and E2A but not FOG1. In contrast, GFI1B bound FOG1 as well as GATA1, SCL, and E2A. ZFP143 bound only a small proportion of GATA1, even less of FOG1 but not SCL and E2A (data not shown). Identical results were obtained when the coimmunoprecipitation and reverse immunoprecipitation experiments were repeated with DNase I and RNase treated nuclear extracts, to exclude interactions mediated by either nonspecific or specific binding to nucleic acids (data not shown).
To determine whether the interactions between GATA1, FOG1, ETO2, GFI1B, and ZFP143 were present in primary megakaryocytes, we isolated megakaryocytes (Figure 2F) from bone marrow, harvested from 5-fluorouracil–treated mice that had been cultured for 3 days with thrombopoietin. The 3-day culture allowed expansion of megakaryocyte numbers (to get enough for coimmunoprecipitation experiment) and megakaryocyte maturation. The purity of the megakaryocytes was approximately 90% as assessed by CD41 expression by fluorescence-activated cell sorter analysis and by cell morphology (Figure 2F). In nuclear extracts from primary megakaryocytes, immunoprecipitated GATA1 interacted with FOG1, GFI1B, ZFP143, and E2A (Figure 2G).
However, in contrast to L8057 immature megakaryoblastic cells, we could not detect an interaction between ETO2 and GATA1 in mature primary megakaryocytes (data not shown). One explanation for this could be that, like during erythroid differentiation,31 Eto2 mRNA expression decreases during megakaryocyte maturation. To determine whether this was the case, we quantitated Eto2 mRNA expression from RNA prepared from CD41-expressing bone marrow cells harvested from mice treated with 5-fluorouracil and cultured for 1, 2, and 3 days with thrombopoietin (Figure 2H). Day 1 cultures primarily consist of CD41+ immature megakaryoblasts, and as the cells mature through day 2 to day 3, they acquire morphologic features of more mature megakaryocytes (Figure 2H) and express higher levels of the megakaryocyte genes Glycoprotein IBα, Glycoprotein VI, and Glycoprotein IX (data not shown). Eto2 mRNA was expressed at least a log higher in mRNA extracted from day 1 cells compared with that extracted from days 2 and 3 cells, where it was barely detected. Thus, absence of detectable GATA1-ETO2 interaction by coimmunoprecipitation in day 3 megakaryocytes may be resulting from low ETO2 protein expression in this cell population. However, we were able to confirm that ETO2 and GATA1 did interact in primary megakaryoblasts by immunofluorescence (see “Colocalization”).
Colocalization.
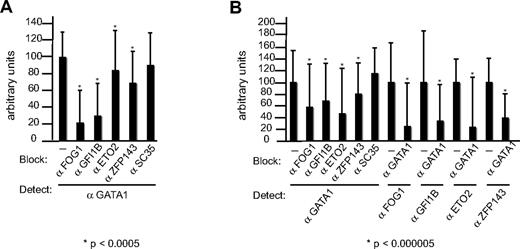
To further document this association, we performed colocalization experiments. L8057 cells (Figure 3A) and primary immature (day 1; Figure 3B) and mature megakaryocytes (day 3, data not shown) were fixed and immunolabeled with αGATA1 and αFOG1, αGFI1B, αZFP143, αETO2, and control antibodies to the splicing factor SC35. Staining showed that all proteins were primarily located in the nucleus (compared with 4,6 diamidino-2-phenylindole staining, data not shown). To quantitate the degree of colocalization and exclude that overlap of staining patterns was not adventitious, we used a high-resolution approach that takes advantage of the ability of an antibody to block access of another antibody to its antigen.48,56 In contrast to conventional immunofluorescence analysis indicating that 2 targets lie within 200 nm, this approach reveals targets that lie within a few nanometers and therefore are very probably to interact with each other. In the absence of blocking antibodies, the intensity of fluorescence detected in nuclei on incubation with αGATA1 antibodies was arbitrarily set to 100 (Figure 3A,B). In L8057 cells (Figure 3A) and primary immature megakaryocytes (Figure 3B), the intensity of the signal emitted from αGATA1 antibodies (the detecting antibody) was significantly reduced on coincubation with αFOG1, αGFI1B, αZFP143, and αETO2 (blocking antibodies) but not with control antibodies to SC35. In the case of ETO2, the decreased access is less obvious. However, the difference is still significant, as so many events are counted. Reverse blocking experiments performed in primary immature megakaryocytes (Figure 3B) showed that access of αFOG1, αGFI1B αZFP143, and αETO2 (detecting antibodies) was obviously reduced by αGATA1 antibody. In contrast, αGATA1 antibody did not reduce access to antibodies to SC35 (data not shown). Similar data for colocalization were obtained for mature megakaryocytes, except that the ETO2 staining was not detected in mature megakaryocytes.
GATA1 colocalizes with partner proteins in L8057 cells and primary megakaryocytes. Degree of colocalization (shown on the y-axis) is revealed by antibody blocking in L8057 (A) and primary megakaryocyte nuclei (B). αFOG1, -GFI1B, -ETO2, and -ZFP143 antibodies, but not αSC35 antibody, block access to αGATA1 antibody (A,B). Conversely, αGATA1 antibody blocks access of αFOG1, -GFI1B, -ETO2, and -ZFP143 antibodies, to their respective epitopes (B).
GATA1 colocalizes with partner proteins in L8057 cells and primary megakaryocytes. Degree of colocalization (shown on the y-axis) is revealed by antibody blocking in L8057 (A) and primary megakaryocyte nuclei (B). αFOG1, -GFI1B, -ETO2, and -ZFP143 antibodies, but not αSC35 antibody, block access to αGATA1 antibody (A,B). Conversely, αGATA1 antibody blocks access of αFOG1, -GFI1B, -ETO2, and -ZFP143 antibodies, to their respective epitopes (B).
In conclusion, the physical interactions between GATA1 and known (FOG1, members of the pentameric and NURD complexes) and predicted (GFI1B and ETO2) partners were validated by 3 independent methods (pull-down, immunoprecipitation, and colocalization) in megakaryocytic cells. In addition, for the first time, we demonstrate that GATA1 interacts with ZFP143. It is important to note that these experiments do not distinguish whether the interactions between these proteins and GATA1 is direct or indirect.
Nature of GATA1-containing complexes
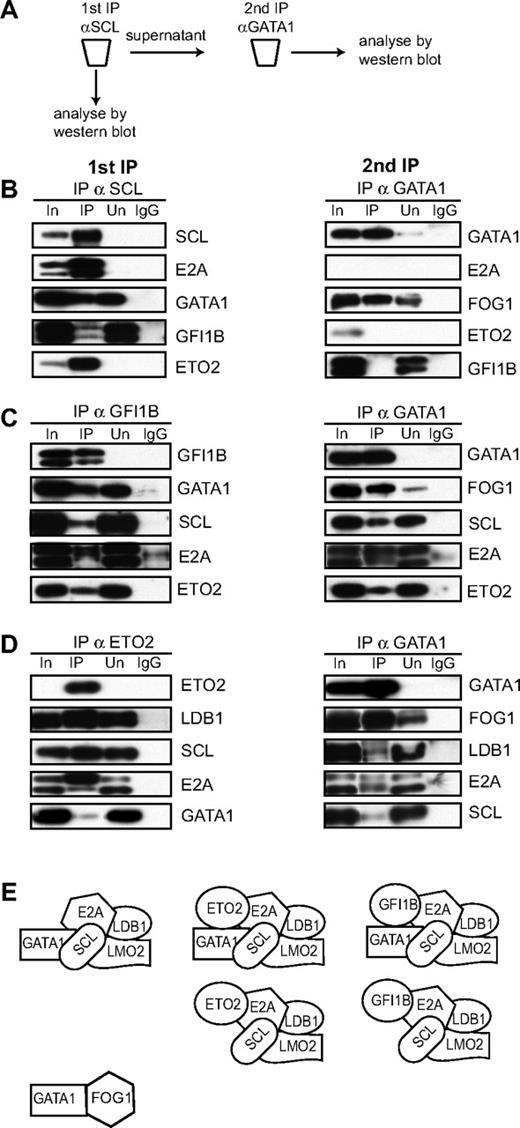
To gain a better idea of the nature of the GATA1-containing protein complexes, we performed sequential immunodepletion experiments of L8057 nuclear extracts. The experimental scheme is shown in Figure 4A. L8057 nuclear extracts were first immunodepleted with αSCL antibody (Figure 4B top left panel). The immunoprecipitate efficiently depletes SCL and its heterodimeric partner E2A as none is detected in the supernatant. SCL also interacts with GATA1, GFI1B, and ETO2, which are present in the immunoprecipitate. These 3 proteins also exist in complexes not containing SCL, as each protein is also detected either in the unbound fraction or in the input (supernatant) for the sequential IP with αGATA antibody. When the supernatant from the SCL immunoprecipitate is then immunoprecipitated with αGATA1 antibody (Figure 4B top right panel), GATA1 is detected in the input and immunoprecipitated lanes, as expected. The immunoprecipitation is efficient as very little GATA1 is detected in the unbound supernatant. GATA1 that is not complexed to SCL is now detected with FOG1 but not E2A, ETO2, or GFI1B. These data suggest that the GATA1-FOG1 complex exists independently of GATA1 in complexes with SCL, E2A, ETO2, and GFI1B. It also suggests, but does not prove, that GATA1 only exists in a complex with E2A, ETO2, and GFI1B in the presence of SCL. In part, the note of caution reflects the data in Figure 1E where ETO2 and GFI1B elute without SCL but with GATA1 (after fraction 29, ie, < 400 kDa), although clearly coelution of GATA1 with ETO2 and GFI1B in these low MW fractions does not mean that GATA1, ETO2, and/or GFI1B physically interact.
Characterization of GATA1-containing protein complexes in L8057 cells. (A) Scheme of the sequential immunoprecipitation experiments. (B-D) L8057 cell crude nuclear extracts were first immuno-depleted with αSCL (B), αGFI1B (C), or αETO2 (D) antibodies (left panels). The lanes are named as in Figure 2. Coimmunoprecipitated proteins were analyzed by Western blot using antibodies indicated on the right of the panels. A second immunoprecipitation was performed on the depleted supernatant (right panels) using αGATA1 antibodies. Coimmunoprecipitated proteins were analyzed by Western blot using antibodies marked on the right of the panels. (E) Models of the possible composition of protein complexes from L8057 cells derived from experiments in this figure.
Characterization of GATA1-containing protein complexes in L8057 cells. (A) Scheme of the sequential immunoprecipitation experiments. (B-D) L8057 cell crude nuclear extracts were first immuno-depleted with αSCL (B), αGFI1B (C), or αETO2 (D) antibodies (left panels). The lanes are named as in Figure 2. Coimmunoprecipitated proteins were analyzed by Western blot using antibodies indicated on the right of the panels. A second immunoprecipitation was performed on the depleted supernatant (right panels) using αGATA1 antibodies. Coimmunoprecipitated proteins were analyzed by Western blot using antibodies marked on the right of the panels. (E) Models of the possible composition of protein complexes from L8057 cells derived from experiments in this figure.
When L8057 nuclear extracts are first immunoprecipitated with αGFI1B antibodies (Figure 4C left panel), the depletion is efficient (no GFI1B is detected in the unbound fraction) and GFI1B interacts with GATA1, SCL, E2A, and ETO2. When the supernatant from the immunodepletion with αGFI1B is immunoprecipitated with αGATA1 antibody (Figure 4C right panel), all of GATA1 is efficiently precipitated. This remainder of GATA1 interacts with FOG1, SCL, E2A, and ETO2.
In the last immunodepletion experiment, L8057 nuclear extracts were depleted with αETO2 antibody first (Figure 4D left panel). The depletion was very efficient, with no ETO2 detected in the unbound fraction. In the immunoprecipitate, SCL, E2A, LDB1, and a small amount of GATA1 are detected. When the supernatant is immunoprecipitated with αGATA1 antibody, all of the remaining GATA1 is immunoprecipitated and detected with FOG1, SCL, E2A, and LDB1.
At a first approximation, the sum of the sequential immunodepletions suggests that GATA1 exists in multiple complexes (Figure 4E). Some of GATA1 is bound to FOG1 (Figure 4B right panel); some of GATA1 is bound to SCL (which is often complexed to E2A). SCL/E2A also exist in multiple complexes, some with GATA1 and some without GATA1. In some complexes, GATA1, SCL, and E2A may be in a complex with GFI1B and ETO2 (Figure 4B left panel; 4C left panel), with ETO2 but not GFI1B (Figure 4C right panel) and finally without either GFI1B or ETO2 (Figure 4D right panel). SCL, E2A, and ETO2 are also in a complex without GATA1 and GFI1B (as these proteins are detected in the supernatant, Figure 4C right panel). There are at least 2 caveats to the interpretation of these findings. First, the interactions are dependent on the conditions used in the immunoprecipitation and may not reflect what occurs in the nucleus. Second, these experiments do not identify which protein interactions are direct and which are indirect. Finally, these experiments do not establish all the components of the complexes.
Reduction of ETO2 protein levels augments megakaryocyte differentiation
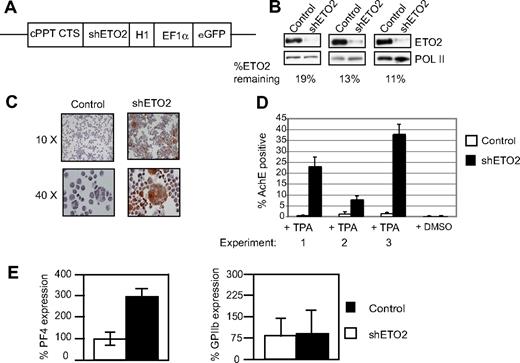
We then focused on understanding the function of ETO2 and its interaction with GATA1 during megakaryocyte maturation, as this had not previously been studied. In 3 independent experiments, pools of L8057 cells were infected with a lentivirus expressing a shRNA directed against either Eto2 mRNA or a control scrambled sequence (Figure 5A). In these experiments, ETO2 protein levels were reduced to between 11% and 19% of normal levels compared with L8057 cells infected with a control shRNA (Figure 5B). These infected pools were induced to undergo megakaryocyte differentiation by culture with the phorbol ester 12-0 tetradecanoylphorbol-13-acetate (TPA).45 The extent of differentiation was scored morphologically by counting the number of cells that express the enzyme acetylcholine esterase, a marker of mature megakaryocytes (AchE+ cells; Figure 5C). Compared with infection with lentivirus-expressing control sequence, infection with shRNA directed against ETO2 resulted in an 8- to 20-fold increase in AchE+ TPA-cultured L8057 cells (Figure 5D), and this was coupled with a 2- to 3-fold increase in AchE mRNA levels (data not shown). Expression of 2 other megakaryocyte-specific genes, Pf4 and GPIX, but not GPIIB, was also increased (Figure 5E and data not shown). In conclusion, these data suggest that ETO2 restrains megakaryocyte differentiation by either directly or indirectly repressing megakaryocyte-specific gene expression.
Knock down of ETO2 in L8057 cells. (A) Diagram of lentiviral construct used to express Eto2-directed shRNA (shETO2). cPPT indicates central polypurine tract; CTS, central termination sequence; and H1, H1 promoter. EF1α promoter and the eGFP reporter gene are shown. (B) Knock down of ETO2 protein expression (top panels) was assessed in L8057 cells infected with either control shRNA (Control, lefthand lane) or shRNA directed against ETO2 (shETO2, righthand lane) by Western blot analysis in 3 independent experiments. The blots were probed with anti-POL II antibody (bottom panels) to control for protein loading. Percentage knock down of normalized ETO2 protein expression is shown. (C,D) L8057 cells expressing a control shRNA or ETO2 shRNA were induced to undergo megakaryocytic differentiation with TPA. Cells were stained for acetylcholine esterase (AchE) activity after 4 days of induction (C). Images were acquired using an Olympus BX60 microscope with a QImaging camera (Surrey, BC) using an Olympus lens at 10×/0.5 and 40×/0.5 numeric aperture objectives. Openlab version 3 software (Improvision, Coventry, United Kingdom) was used for image acquisition, and images were exported into Adobe Photoshop version CS2 (Adobe Systems, San Jose, CA) for processing. Note the increased number of brown staining (AchE-positive) cells that are larger in size, indicative of greater megakaryocyte maturation, when cells are infected with the ETO2 shRNA. The percentage of AchE-positive cells counted in 3 independent experiments (D). Cells treated with DMSO served as a control for TPA induction. (E) Pf4 (left) and GPIIb (right) mRNA levels were quantitated relative to GAPDH levels by Taqman real-time RT-PCR in L8057 cells infected with virus expressing either control shRNA (▭) or shETO2 ( ). Data shown in panels D and E are the average of 3 independent experiments, and error bars represent 2 SD.
). Data shown in panels D and E are the average of 3 independent experiments, and error bars represent 2 SD.
Knock down of ETO2 in L8057 cells. (A) Diagram of lentiviral construct used to express Eto2-directed shRNA (shETO2). cPPT indicates central polypurine tract; CTS, central termination sequence; and H1, H1 promoter. EF1α promoter and the eGFP reporter gene are shown. (B) Knock down of ETO2 protein expression (top panels) was assessed in L8057 cells infected with either control shRNA (Control, lefthand lane) or shRNA directed against ETO2 (shETO2, righthand lane) by Western blot analysis in 3 independent experiments. The blots were probed with anti-POL II antibody (bottom panels) to control for protein loading. Percentage knock down of normalized ETO2 protein expression is shown. (C,D) L8057 cells expressing a control shRNA or ETO2 shRNA were induced to undergo megakaryocytic differentiation with TPA. Cells were stained for acetylcholine esterase (AchE) activity after 4 days of induction (C). Images were acquired using an Olympus BX60 microscope with a QImaging camera (Surrey, BC) using an Olympus lens at 10×/0.5 and 40×/0.5 numeric aperture objectives. Openlab version 3 software (Improvision, Coventry, United Kingdom) was used for image acquisition, and images were exported into Adobe Photoshop version CS2 (Adobe Systems, San Jose, CA) for processing. Note the increased number of brown staining (AchE-positive) cells that are larger in size, indicative of greater megakaryocyte maturation, when cells are infected with the ETO2 shRNA. The percentage of AchE-positive cells counted in 3 independent experiments (D). Cells treated with DMSO served as a control for TPA induction. (E) Pf4 (left) and GPIIb (right) mRNA levels were quantitated relative to GAPDH levels by Taqman real-time RT-PCR in L8057 cells infected with virus expressing either control shRNA (▭) or shETO2 ( ). Data shown in panels D and E are the average of 3 independent experiments, and error bars represent 2 SD.
). Data shown in panels D and E are the average of 3 independent experiments, and error bars represent 2 SD.
ETO2 represses the Pf4 promoter by recruitment via GATA and E-box sites
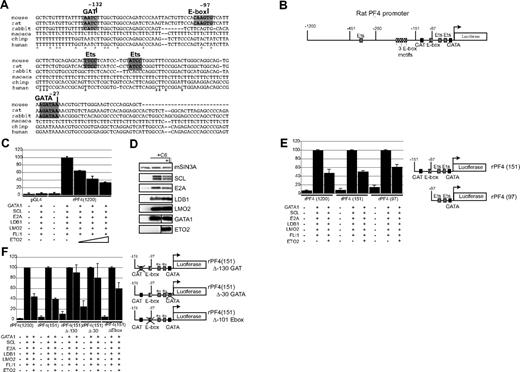
To investigate the mechanism of ETO2-mediated repression of megakaryocyte gene expression, we studied how ETO2 repressed Pf4 expression. Analysis of the Pf4 gene promoter reveals a GATA, GAT, E-box, and 2 ETS sequence motifs (Figure 6A,B) within the proximal 1200 base pairs (bps). We tested the transcriptional activity of the rat Pf4 gene promoter fragment attached to a Luciferase reporter gene in 293T fibroblasts. Maximal luciferase gene expression was detected when 293T cells were cotransfected with expression plasmids encoding for all the members of the pentameric complex (GATA1, SCL, E2A, LDB1, LMO2) and FLI1 (Figure 6C). We added FLI1 protein to the assay as it has been shown to cooperate with GATA1 in activating megakaryocyte gene expression.12 However, when increasing amounts of ETO2 expression plasmid were cotransfected, luciferase expression was incrementally reduced by 70%. Western blot analysis showed that all the expression plasmids were expressed (Figure 6D). These data suggest that ETO2 may directly repress the Pf4 promoter.
ETO2 directly represses the Pf4 promoter in transactivation assays via GATA, GAT, and E-box sites. (A) Comparison of the nucleotide sequence of the 5′ upstream region of the Pf4 gene from 6 species. Sequences highlighted in bold or by a box indicate a GAT, E-box, ETS, and GATA motifs. *Conserved nucleotides. Coordinates in nucleotides are with respect to the transcriptional start site. (B) Schematic representation of the luciferase gene reporter construct (pGL4.10) under the control of the enhancer/promoter regions of the rat Pf4 gene. Coordinates in nucleotides are with respect to the Pf4 gene transcriptional start site. Sequence motifs for transcription factors are marked. ↱shows the position of the transcriptional start site. The luciferase gene is drawn as an open box. (C) 293T fibroblasts were transiently transfected with either a promoterless luciferase gene (pGL4.10) or a luciferase gene regulated by 1200 nucleotides 5′ of the transcriptional start site of the rat Pf4 gene (rPF4 1200). Cells were also transfected with expression vectors expressing the indicated transcription factors. The triangle represents transfection with increasing amounts of ETO2 expression vector (50, 150, and 300 ng). Expression of luciferase in cells transfected with rPF4 1200 with expression vectors for all 6 transcriptional regulators (C6: GATA1, SCL, E2A, LDB1, LMO2, and FLI1) was set to 100%. (D) Western blot analysis confirms expression of transfected proteins in 293T cells in the absence (+ C6) or presence (+ C6 + E) of the ETO2 expression plasmid. Expression of mSIN3A protein serves as a loading control. (E) Expression of luciferase protein in 293T cells transfected with either full-length rPF4 1200 or truncated forms of the rat Pf4 promoter (rPF4 151 and rPF4 97) with C6 factors in the presence or absence of 150 ng ETO2 expressing plasmid. (F) Transcriptional effect of point mutations of the GAT motif at −130 and GATA site at −30 and E-box motif at −102 (relative to the transcriptional start site) in the construct rPF4 151 on luciferase gene expression were tested in 293T cells in the presence of the transcription factors indicated at the bottom of the diagram. In panels C, E, and F, the results are the mean plus or minus 2 SD of 3 to 5 independent experiments.
ETO2 directly represses the Pf4 promoter in transactivation assays via GATA, GAT, and E-box sites. (A) Comparison of the nucleotide sequence of the 5′ upstream region of the Pf4 gene from 6 species. Sequences highlighted in bold or by a box indicate a GAT, E-box, ETS, and GATA motifs. *Conserved nucleotides. Coordinates in nucleotides are with respect to the transcriptional start site. (B) Schematic representation of the luciferase gene reporter construct (pGL4.10) under the control of the enhancer/promoter regions of the rat Pf4 gene. Coordinates in nucleotides are with respect to the Pf4 gene transcriptional start site. Sequence motifs for transcription factors are marked. ↱shows the position of the transcriptional start site. The luciferase gene is drawn as an open box. (C) 293T fibroblasts were transiently transfected with either a promoterless luciferase gene (pGL4.10) or a luciferase gene regulated by 1200 nucleotides 5′ of the transcriptional start site of the rat Pf4 gene (rPF4 1200). Cells were also transfected with expression vectors expressing the indicated transcription factors. The triangle represents transfection with increasing amounts of ETO2 expression vector (50, 150, and 300 ng). Expression of luciferase in cells transfected with rPF4 1200 with expression vectors for all 6 transcriptional regulators (C6: GATA1, SCL, E2A, LDB1, LMO2, and FLI1) was set to 100%. (D) Western blot analysis confirms expression of transfected proteins in 293T cells in the absence (+ C6) or presence (+ C6 + E) of the ETO2 expression plasmid. Expression of mSIN3A protein serves as a loading control. (E) Expression of luciferase protein in 293T cells transfected with either full-length rPF4 1200 or truncated forms of the rat Pf4 promoter (rPF4 151 and rPF4 97) with C6 factors in the presence or absence of 150 ng ETO2 expressing plasmid. (F) Transcriptional effect of point mutations of the GAT motif at −130 and GATA site at −30 and E-box motif at −102 (relative to the transcriptional start site) in the construct rPF4 151 on luciferase gene expression were tested in 293T cells in the presence of the transcription factors indicated at the bottom of the diagram. In panels C, E, and F, the results are the mean plus or minus 2 SD of 3 to 5 independent experiments.
To localize the DNA sequences mediating ETO2 repressive activity at the rat Pf4 promoter, deletional constructs of the Pf4 promoter were tested (Figure 6E). Equivalent repression of Pf4 gene expression was seen with either 151 bps or 1200 bps of promoter sequence. A 97-bps Pf4 promoter also mediated ETO2 repression, but possibly less efficiently.
To localize specific DNA binding sites for ETO2-mediated repression, we mutated the GAT, GATA, and E-box binding sites within the 151 bps promoter (Figure 6F). Mutation of either the GAT or GATA site (130 bps and 30 bps upstream of the transcriptional start site, respectively) abrogated significant ETO2-mediated repression. Mutation of the E-box reduced the ETO2-mediated repression, but possibly to a lesser extent. The combination of these data argues that ETO2 requires the GAT, GATA sites, and to a lesser extent the E-box motif to mediate maximal repression of the Pf4 promoter. This suggests that ETO2 may need to bind both to GATA1 (bound to the GAT and GATA sites; for further discussion of GATA/GAT sites, see “Discussion”) and possibly E-box binding proteins, to be recruited to the Pf4 promoter.
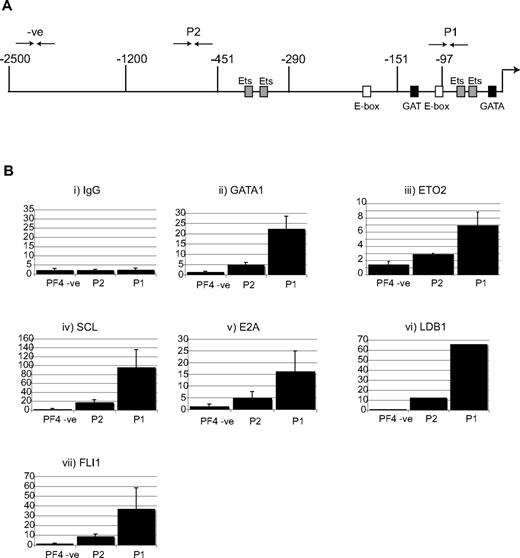
This was confirmed by chromatin immunoprecipitation experiments at the endogenous Pf4 promoter in L8057 cells (Figure 7). There was highly significant enrichment of sequences approximately 100 bps upstream of the transcriptional start site when chromatin is immunoprecipitated not only with αGATA1 and αETO2 antibodies but also antibodies directed against members of the pentameric complex SCL, E2A, LDB1, and finally αFLI1 antibodies. In conclusion, biochemical (Figure 5), mutational (Figure 6F), and ChIP data (Figure 7) support a model where ETO2 interacts with GATA1 and the pentameric SCL/E2A complex (SCL/E2A/GATA1/LDB1) and is recruited to GAT, GATA, and E-box sites at the Pf4 promoter to repress Pf4 transcription, presumably early in megakaryocyte differentiation when ETO2 is maximally expressed (Figure 2H).
Recruitment of the pentameric complex and ETO2 to the Pf4 promoter in L8057 cells. (A) Schematic representation of the endogenous mouse PF4 locus. Coordinates in nucleotides are with respect to the transcriptional start site. Sequence motifs for transcription factors are marked. The ↱shows the position of the transcriptional start site. The 3 end-to-end arrows above the locus (-ve, P2, and P1) represent the location of the genomic sequences amplified by Taqman real-time PCR in the chromatin immunoprecipitation assay (ChIP) in panel B. (B) ETO2 and the pentameric complex co-occupy the Pf4 locus in L8057 cells. ChIP was performed using chromatin isolated from L8057 cells and antibodies directed against (i) GATA1, (ii) ETO2, (iii) SCL, (iv) E2A, (v) LDB1, and (vi) FLI1. Immunoprecipitated material was analyzed by Taqman real-time RT-PCR. The y-axis represents the fold enrichment normalized with respect to IgG and a control locus, the Gapdh promoter, at specific sites in the Pf4 promoter region (PF4-ve, P2, and P1).
Recruitment of the pentameric complex and ETO2 to the Pf4 promoter in L8057 cells. (A) Schematic representation of the endogenous mouse PF4 locus. Coordinates in nucleotides are with respect to the transcriptional start site. Sequence motifs for transcription factors are marked. The ↱shows the position of the transcriptional start site. The 3 end-to-end arrows above the locus (-ve, P2, and P1) represent the location of the genomic sequences amplified by Taqman real-time PCR in the chromatin immunoprecipitation assay (ChIP) in panel B. (B) ETO2 and the pentameric complex co-occupy the Pf4 locus in L8057 cells. ChIP was performed using chromatin isolated from L8057 cells and antibodies directed against (i) GATA1, (ii) ETO2, (iii) SCL, (iv) E2A, (v) LDB1, and (vi) FLI1. Immunoprecipitated material was analyzed by Taqman real-time RT-PCR. The y-axis represents the fold enrichment normalized with respect to IgG and a control locus, the Gapdh promoter, at specific sites in the Pf4 promoter region (PF4-ve, P2, and P1).
Discussion
GATA1 is present in multiple distinct complexes
Four independent approaches (interaction with biotinylated GATA1, coimmunoprecipitation, reverse coimmunoprecipitation, and colocalization) showed that megakaryocyte GATA1 is bound to a known megakaryocyte partner, FOG1,21,23 and also other proteins. Some of these have been detected as GATA1 partners in red cells: the NURD complex (presumably binding via FOG116,22 ), the pentameric SCL/E-protein/LMO2/Ldb1 complex (presumably interacting via LMO257 ), ETO2 (that interacts directly via E proteins34 ), and GFI1B. Finally, binding of biotinylated and endogenous GATA1 to the zinc finger protein ZFP143 is an interaction that has not previously been reported. As mentioned before, the data do not define if these protein interactions with GATA1 are direct or indirect.
Gel filtration and immunodepletion studies show GATA1 is present in distinct complexes. Whereas GATA1 elutes throughout the elution profile, FOG1 elutes in 2 broad peaks. In one peak, it elutes with members of the NURD complex, whereas the other peak corresponds to lower molecular weight complexes, and here, GATA1 and FOG1 are probably to be in a complex with the widely expressed zinc finger protein ZBP89.46
In contrast, the SCL/E2A/Ldb1/LMO2/GATA1 pentameric complex elutes at a different position from the 2 FOG1 peaks, supporting our immunodepletion data, and results in red cells,16 that GATA1-FOG1 and GATA1-pentameric complexes are distinct. The SCL immunodepletion studies suggest that, in L8057 cells, interaction with ETO2 and GFI1B is mediated via SCL and its partners. However, only a minority of GFI1B coelutes with SCL on gel filtration and similarly only a small fraction of GFI1B immunoprecipitates with GATA1 in megakaryocytes and MEL cells.16 Moreover, a recent study shows that in vivo biotinylated and endogenous GFI1B binds principally to the corepressor CoREST, the histone demethylase LSD1, and HDAC1 and 2.58 ZFP143, a 66-kDa protein, elutes between 66 and approximately 400 kDa, away from the FOG1, and pentameric complexes, suggesting that GATA1-ZFP143 interaction may be in yet another distinct complex(es).
ZFP143, a previously unrecognized GATA1-interacting protein
ZFP143, a 7 C2-H2 zinc finger protein, is a widely expressed DNA binding regulator that is conserved from human to zebrafish. It activates both POLII and POLIII regulated promoters.51,54,55 Bioinformatic analyses coupled with large-scale ChIP studies suggest that human ZNF143 could bind up to approximately 2500 evolutionarily conserved ZNF143 binding sites, at approximately 2000 gene promoters, many of which are associated with CpG islands.55 It will be informative to determine how many of these promoters are co-occupied and coregulated by GATA1. The functional importance of the ZFP143-GATA1 interaction is unclear. Our preliminary data show that in vitro differentiation of ES cells heterozygous for a loss-of-function ZFP143 allele fail to generate embryoid bodies, suggesting it is required early in development. This also precluded us from studying hemopoiesis from these in vitro-differentiated ES cells.
ETO2-GATA1 and megakaryocyte differentiation
The ETO protein family is conserved through evolution from human through to Drosophila (Nervy protein). In human and mouse, there are 3 ETO genes. In human, the first 2, MTG8 (encoding ETO protein) and MTG16 (encoded ETO2 protein), are located at the translocation breakpoints associated with acute myeloid leukemia.33,59 The third gene, MTGR1, encodes MTG-related-1 protein. All 3 contain 4 conserved nervy homology domains that function as protein-protein interaction modules. ETO proteins can repress transcription by distinct mechanisms: promoting repressive chromatin marks, by binding class I HDACs (HDAC1, 2, and 3), and the corepressor N-CoR60-62 and erasing activating chromatin, by displacing the HAT p300/CBP from E-proteins.34 In L8057 cells experiments, we detected an interaction between GATA1 and HDAC2 but it is unclear whether this is mediated by the NURD complex or by ETO2.
As primary megakaryocytes differentiate, Eto2 mRNA levels decline, by approximately 10-fold, to almost undetectable. Moreover, the GATA1-ETO2 protein interaction is only detectable in early immature primary megakaryoblasts and L8057 cells. This is reminiscent of erythroid cells where ETO2 mRNA and proteins levels decline during erythropoiesis and result in loss of the SCL-ETO2 interaction.31,35 Functional studies demonstrated that ETO2 protein knockdown enhances megakaryocyte maturation of phorbol ester-induced L8057 cells. Taken together, this suggests that ETO2 may prevent premature megakaryocyte differentiation in early megakaryoblasts.
The mechanism by which ETO2 restrains megakaryocyte differentiation is probably to be, at least in part, direct transcriptional repression of select terminal megakaryocyte genes, such as Platelet factor 4, Acetyl cholinesterase, and GPIX. At these genes, one mechanism to recruit ETO2 to cis-elements is probably to be physical interaction with E-proteins34 bound to E-box motifs. What then are the roles of the GATA and GAT sites? Closely located GAT and GATA motifs, at a number of GATA1 regulated promoters, are high affinity GATA1 binding sites.18 Furthermore, functionally important hemopoietic cis-elements (including those in the Gata1 locus27,63 ) include combinations of E-box motifs and GATA sites (including GAT sites) that bind versions of the pentameric SCL/GATA complex. Thus, at the Pf4 promoter, our mutagenesis, ChIP and coimmunoprecipitation data support the hypothesis that GATA1 bound to GATA and GAT sites and SCL and E2A bound to the E-box motif interact with LMO2, Ldb1 and ETO2 (at a minimum) allowing for ETO-2-mediated repression.
In conclusion, our observations suggest that ETO2 works with GATA1 and E-proteins to help coordinate timing of terminal megakaryocyte gene expression and thus terminal maturation. This is reminiscent of the proposed role ETO2 plays in end-stage erythropoiesis,31,35 highlighting similarities, at a molecular level, of terminal maturation of these 2 closely related lineages.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors acknowledge use of the preparative nuclear extract protocol (Alan Cantor), the ETO2 siRNA vector (Nicolas Goardon and Paul Henri Romeo, Cochin, France), the pCMVΔ8.9 and pCMV-VSV-G plasmids (Didier Trono, Whitehead), ZNF143 antibody (Gary Kunkel, Texas A&M University), LDB1 and LMO2 antibodies (Stuart Orkin, Harvard University), and rat Pf4 promoter fragment (Katya Ravid, Boston University).
I.H. was funded by an MRC PhD studentship and the Oxford Partnership Comprehensive Biomedical Research Center with funding from the Department of Health's National Institute for Health Research Biomedical Research Centres funding scheme.
Authorship
Contribution: I.H., F.I., C.P., and P.V. designed the experiments; I.H., J.D., and F.I. performed the experiments; P.V. and C.P. supervised the work; I.H., J.D., J.S., F.I., C.P., and P.V. analyzed the results; I.H. and P.V. created the figures; and all authors contributed to the writing of the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Paresh Vyas, MRC Molecular Haematology Unit and Department of Haematology, Weatherall Institute of Molecular Medicine, John Radcliffe Hospital, Oxford OX3 9DU, United Kingdom; e-mail: paresh.vyas@imm.ox.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal