Abstract
Hematopoietic progenitor cells derived from human embryonic stem cells (hESCs) develop into diverse mature hematopoietic lineages, including lymphocytes. Whereas functional natural killer (NK) cells can be efficiently generated in vitro from hESC-derived CD34+ cells, studies of T- and B-cell development from hESCs have been much more limited. Here, we demonstrate that despite expressing functional Notch-1, CD34+ cells from hESCs did not derive T cells when cocultured with OP9 cells expressing Delta-like 1, or in fetal thymus organ culture. hESC-derived CD34+ cells also did not produce B cells in vitro. In contrast, CD34+ cells isolated from UCB routinely generated T and B cells when cultured in the same conditions. Notably, both undifferentiated hESCs, and sorted hESC-derived populations with hematopoietic developmental potential exhibited constitutive expression of ID family genes and of transcriptional targets of stem cell factor–induced signaling. These pathways both inhibit T-cell development and promote NK-cell development. Together, these results demonstrate fundamental differences between hESC-derived hematopoietic progenitors and analogous primary human cells. Therefore, hESCs can be more readily supported to differentiate into certain cell types than others, findings that have important implications for derivation of defined lineage-committed populations from hESCs.
Introduction
Human embryonic stem cells (hESCs) provide an important model system to define the mechanisms that mediate cellular development. hESC-derived hematopoietic progenitor cells efficiently produce erythroid, myeloid, and lymphoid lineage cells in vitro.1-4 We previously defined an in vitro culture system to generate natural killer (NK) cells from hESCs.5 hESC-derived NK cells express surface receptors characteristic of primary NK cells, kill tumor target cells, and produce interferon-γ when stimulated with cytokines. These results suggest that hESC-derived progenitors may also readily commit to the T-cell lineage in vitro, since T and NK lymphocytes are developmentally closely related.6,7
One study has used an in vivo model to examine the T-cell potential of hESCs.8 Galic et al injected hESC-derived hematopoietic progenitor cells into human thymus/fetal liver (Thy/Liv) grafts in severe combined immunodeficient-human (SCID-hu) mice. This study demonstrated T-cell development after 3 to 5 weeks in vivo, although in a less efficient manner than what has been observed with hematopoietic progenitor cells from human fetal liver (FL), bone marrow (BM), or umbilical cord blood (UCB)9-11 evaluated in SCID-hu mice. Although useful, SCID-hu mice are not optimal to evaluate development of specific phenotypic cell populations over time, and the effects of specific molecular signaling pathways are difficult to quantify via this SCID-hu system. Therefore, in vitro models of lymphocyte development are needed, although despite the considerable interest in hematopoietic development of hESCs, in vitro studies have not provided significant evidence of functional T and B lymphocyte maturation of hESC-derived hematopoietic progenitors. Although one study identified a small percentage of CD19+ B cells and expression of CD3 gene transcripts, no CD4+ or CD8+ phenotypic cells were characterized.1 Another study also demonstrated development of a limited number of CD19+ cells derived from hESCs, although again there were no more specific studies of this population and no evidence of T-cell development.12
Here, we cocultured hESC-derived hematopoietic progenitor cells with OP9 stromal cells that ectopically express the Notch ligand Delta-like 1 (DL1). The OP9-DL1 system has been used very effectively to analyze the T-lineage potential of mouse bone marrow, FL, and mouse embryonic stem cell (mESC)–derived hematopoietic progenitors, as well as of human bone marrow and UCB cells.13-16 Signaling induced by DL1, but not by other Notch ligands, is crucial for T-cell lineage commitment.17 In the absence of Notch-1 in vivo, B cells completely replace T cells in the thymus, whereas transgenic expression of a constitutively active form of Notch-1 induces ectopic T-cell development in the BM.18-20 As an alternative system, we also used the fetal thymic organ culture (FTOC) system to analyze T-cell commitment of hESC-derived hematopoietic cells.
Surprisingly, the results described here demonstrate a complete absence of T or B cells observed in vitro from hESC-derived hematopoietic progenitors. In contrast, UCB-derived progenitor cells exhibited effective T- and B-cell development in both systems. The lack of T- or B-cell development by hESCs corresponded to clear differences in expression between hESC-derived and UCB-isolated hematopoietic progenitor cells of ID family genes and SLUG, a primary downstream target of stem cell factor (SCF) signaling. Complementary studies from other models show both ID gene expression and SCF signaling promote NK-cell development and expansion while inhibiting T- and B-cell development.21-24 Therefore, these findings have important implications for in vitro studies of lymphoid development from hESCs.
Methods
Cell culture
The hESC line H9 (Wicell, Madison, WI) was maintained as undifferentiated cells as previously described.25,26 hESC karyotype was routinely monitored and found to be normal. S17 mouse stromal cells (a generous gift of Dr K. Dorshkind, University of California Los Angeles), OP9-DL1, OP9-GFP, and AFT024 were used as previously described.5,14
hESC differentiation and enrichment of hematopoietic progenitor populations
hESCs were cocultured with S17 stromal cells as previously described.2,27 Total cells were harvested after 14 to 17 days of differentiation. Single-cell suspensions were prepared by successive treatment with collagenase type IV (Invitrogen Life Technologies, Carlsbad, CA) and 0.05% trypsin/EDTA (Mediatech, Manassas, VA) containing 2% chicken serum (Sigma-Aldrich, St Louis, MO). Hematopoietic progenitor populations were enriched to 70% to 90% purity from hESC- and UCB-derived single-cell suspensions using EasySep selection kits (StemCell Technologies, Vancouver, BC). Hematopoietic populations were also sorted using a FACSAria (BD Biosciences, San Jose, CA), with similar results.
Flow cytometry
Single-cell suspensions were analyzed by staining with antibodies specific for CD34, CD45, CD31, CD4, CD8, CD7, CD1a, CD56, and Notch-1 (BD Pharmingen, San Jose, CA). Live cells were distinguished from dead cells with propidium iodide or 7-amino actinomycin D. Data were collected on a FACSCalibur flow cytometer (BD Biosciences) and analysis performed using FlowJo software (TreeStar, Ashland, OR).
In vitro generation of T and B cells
hESC- and UCB-derived populations were plated onto preconfluent layers of OP9-DL1 and OP9-GFP stromal cells in 24-well plates (Nunc, Rochester, NY). Cells were cultured as previously described.13,15 NK-cell development was induced by transferring hematopoietic populations onto irradiated AFT024 stromal cells, as previously described.5 B-cell development was induced by transferring hematopoietic populations onto MS-5 stromal cell layers as previously described.28
Fetal thymus organ culture
Hematopoietic progenitor cells (1000-10 000) were incubated in 30 μL T-cell culture medium in microtiter plates (Nunc) with thymic lobes, irradiated with 20 Gy, from 14.5-day postcoitus Balb/c mouse embryos obtained from timed matings. After 2 days, lobes were transferred to transwell plates (Costar, Corning, NY) and cultured for 12 days in T-cell culture medium. Lobes were then pushed through a cell strainer to produce a single-cell suspension. These cells were analyzed by flow cytometry for surface expression of human CD4 and CD8. This study received Institutional Animal Care and Use Committee (IACUC) approval for the use of mice.
RT-PCR and Q-RT-PCR
For quantitative polymerase chain reaction (Q-PCR) and reverse-transcription (RT)–PCR, total RNA was prepared from enriched populations using an RNeasy kit (Qiagen, Valencia, CA). The RNA was treated with Turbo DNAse (Ambion, Austin, TX), and complementary DNA was prepared using Superscript III reverse transcriptase (Invitrogen Life Technologies). RT-PCR was performed with the resulting cDNA using the primers in Table 1.
RT-PCR primers used
| Primer name . | Direction . | Application . | Sequence, 5′-3′ . |
|---|---|---|---|
| Actin | Sense | RT-PCR | ATCTGGCACCACACCTTCTACAATGAGCTGCG |
| Actin | Antisense | RT-PCR | CGTCATACTCCTGCTTGCTGATCCACATCTGC |
| CD122 | Sense | RT-PCR | CCCAGTTCACATGCTTCTAC |
| CD122 | Antisense | RT-PCR | GACAACTTGGAGGGAGATG |
| E2A | Sense | RT-PCR | CGAGGAGAACACGTCAGCGG |
| E2A | Antisense | RT-PCR | GCTCGGCCTTCTGCTCTG |
| ID1 | Sense | RT-PCR | GCTCATCGACTACATCAGGG |
| ID1 | Antisense | RT-PCR | CACAGAGCACGTAATTCCTC |
| ID2 | Sense | RT-PCR | TCAGCCTGCATCACCAGAGA |
| ID2 | Antisense | RT-PCR | CCATTCAACTTGTCCTCC |
| ID3 | Sense | RT-PCR | AGGGAAGGGCCCGGCAGCTG |
| ID3 | Antisense | RT-PCR | TTCCGGCAGGAGAGGTTCCC |
| ID4 | Sense | RT-PCR | ATGGGATGAGGAAATGCTTG |
| ID4 | Antisense | RT-PCR | TGGAGGAAGGAAAGCAGAAA |
| IL-7R | Sense | RT-PCR | GTCAACATCACCAATCTGG |
| IL-7R | Antisense | RT-PCR | GTAAGATAGGATCCATCTCC |
| Notch-1 | Sense | RT-PCR | CACTGTGGGCGGGTCC |
| Notch-1 | Antisense | RT-PCR | GTTGTATTGGTTCGGCACCAT |
| RAG-1 | Sense | RT-PCR | GAGCAAGGTACCTCAGCCAG |
| RAG-1 | Antisense | RT-PCR | AACAATGGCTGAGTTGGGAC |
| SCF | Sense | RT-PCR | CTACGAGATATGGTAATACAA |
| SCF | Antisense | RT-PCR | ATCGCTACTGCTGTCATTC |
| Slug | Sense | RT-PCR | AAAAAGCCAAACTACAGCGAAC |
| Slug | Antisense | RT-PCR | GAGGTGTCAGATGGAGGAGGG |
| Actin | Sense | Q-RT-PCR | TACCTCATGAAGATCCTCA |
| Actin | Antisense | Q-RT-PCR | TTCGTGGATGCCACAGGAC |
| ID2 | Sense | Q-RT-PCR | TCAGCCTGCATCACCAGAGA |
| ID2 | Antisense | Q-RT-PCR | CCATTCAACTTGTCCTCC |
| ID3 | Sense | Q-RT-PCR | TCGGAACGCAGTCTGGCCATC |
| ID3 | Antisense | Q-RT-PCR | CTCGGCTGTCTGGATGGGAAG |
| Primer name . | Direction . | Application . | Sequence, 5′-3′ . |
|---|---|---|---|
| Actin | Sense | RT-PCR | ATCTGGCACCACACCTTCTACAATGAGCTGCG |
| Actin | Antisense | RT-PCR | CGTCATACTCCTGCTTGCTGATCCACATCTGC |
| CD122 | Sense | RT-PCR | CCCAGTTCACATGCTTCTAC |
| CD122 | Antisense | RT-PCR | GACAACTTGGAGGGAGATG |
| E2A | Sense | RT-PCR | CGAGGAGAACACGTCAGCGG |
| E2A | Antisense | RT-PCR | GCTCGGCCTTCTGCTCTG |
| ID1 | Sense | RT-PCR | GCTCATCGACTACATCAGGG |
| ID1 | Antisense | RT-PCR | CACAGAGCACGTAATTCCTC |
| ID2 | Sense | RT-PCR | TCAGCCTGCATCACCAGAGA |
| ID2 | Antisense | RT-PCR | CCATTCAACTTGTCCTCC |
| ID3 | Sense | RT-PCR | AGGGAAGGGCCCGGCAGCTG |
| ID3 | Antisense | RT-PCR | TTCCGGCAGGAGAGGTTCCC |
| ID4 | Sense | RT-PCR | ATGGGATGAGGAAATGCTTG |
| ID4 | Antisense | RT-PCR | TGGAGGAAGGAAAGCAGAAA |
| IL-7R | Sense | RT-PCR | GTCAACATCACCAATCTGG |
| IL-7R | Antisense | RT-PCR | GTAAGATAGGATCCATCTCC |
| Notch-1 | Sense | RT-PCR | CACTGTGGGCGGGTCC |
| Notch-1 | Antisense | RT-PCR | GTTGTATTGGTTCGGCACCAT |
| RAG-1 | Sense | RT-PCR | GAGCAAGGTACCTCAGCCAG |
| RAG-1 | Antisense | RT-PCR | AACAATGGCTGAGTTGGGAC |
| SCF | Sense | RT-PCR | CTACGAGATATGGTAATACAA |
| SCF | Antisense | RT-PCR | ATCGCTACTGCTGTCATTC |
| Slug | Sense | RT-PCR | AAAAAGCCAAACTACAGCGAAC |
| Slug | Antisense | RT-PCR | GAGGTGTCAGATGGAGGAGGG |
| Actin | Sense | Q-RT-PCR | TACCTCATGAAGATCCTCA |
| Actin | Antisense | Q-RT-PCR | TTCGTGGATGCCACAGGAC |
| ID2 | Sense | Q-RT-PCR | TCAGCCTGCATCACCAGAGA |
| ID2 | Antisense | Q-RT-PCR | CCATTCAACTTGTCCTCC |
| ID3 | Sense | Q-RT-PCR | TCGGAACGCAGTCTGGCCATC |
| ID3 | Antisense | Q-RT-PCR | CTCGGCTGTCTGGATGGGAAG |
Results
Inhibited T- and B-cell development from hESC-derived cells
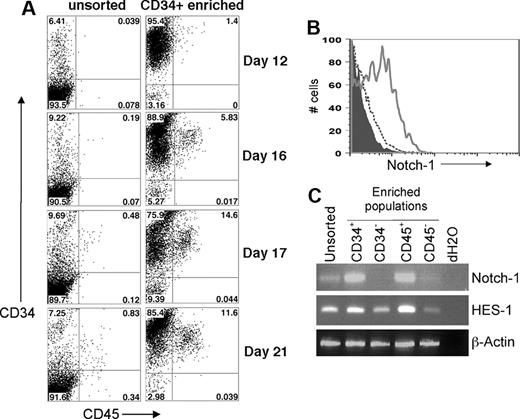
hESCs allowed to differentiate for one week by coculture with murine stromal cell lines, such as S17, M210, and OP9, in medium containing either fetal bovine serum (FBS) or defined cytokines routinely differentiate into a significant population of CD34+ cells.1,2,29-32 Many of the early CD34+ cells coexpress CD31, and after 14 to 21 days a population of CD34+ CD45+ cells, which we have previously reported, develops (Figure 1A).27,29,33 This CD34+ CD45+ population can be further enriched by sorting the human CD34+ and CD45+ cells (Figure 1A).5 We have previously shown that purifying CD34+ CD31+ or CD34+ CD45+ cells results in greater enrichment of hematopoietic activity than sorting for CD34+ cells alone.2,27,29,31 Purified CD34− cells derived from hESCs do not exhibit any detectable hematopoietic activity.
Development of hESC-derived hematopoietic progenitor cells with expression of functional Notch-1. (A) Time course of hematopoietic progenitor cell production by hESCs. Coculture of undifferentiated hESCs with S17 stromal cells for the indicated number of days was followed by flow cytometric analysis of surface expression of CD34 and CD45. These results demonstrate surface antigen expression both on the unsorted cell population and the enriched CD34+ cell population. (B) hESC-derived cells were harvested after 14 days of culture on S17 stromal cells and analyzed by flow cytometry for expression of CD34 and Notch-1. The data were electronically gated to distinguish CD34+ cells (solid line) from CD34− nonhematopoietic cells (dashed line). Isotype control staining is also indicated (filled histogram). (C) After 20 days of culture on S17 cells, total RNA was prepared from unsorted hESC/S17 cells or from the indicated populations of CD34+, CD34−, CD45+, or CD45− cells. cDNA was prepared by reverse transcription, and expression of Notch-1, HES-1, and Actin was analyzed by PCR.
Development of hESC-derived hematopoietic progenitor cells with expression of functional Notch-1. (A) Time course of hematopoietic progenitor cell production by hESCs. Coculture of undifferentiated hESCs with S17 stromal cells for the indicated number of days was followed by flow cytometric analysis of surface expression of CD34 and CD45. These results demonstrate surface antigen expression both on the unsorted cell population and the enriched CD34+ cell population. (B) hESC-derived cells were harvested after 14 days of culture on S17 stromal cells and analyzed by flow cytometry for expression of CD34 and Notch-1. The data were electronically gated to distinguish CD34+ cells (solid line) from CD34− nonhematopoietic cells (dashed line). Isotype control staining is also indicated (filled histogram). (C) After 20 days of culture on S17 cells, total RNA was prepared from unsorted hESC/S17 cells or from the indicated populations of CD34+, CD34−, CD45+, or CD45− cells. cDNA was prepared by reverse transcription, and expression of Notch-1, HES-1, and Actin was analyzed by PCR.
To determine whether hematopoietic cells are responsive to signals required to induce T-cell lineage commitment and differentiation, we analyzed hESC-derived hematopoietic progenitors for surface expression of Notch-1 (Figure 1B). hESC-derived CD34+ cells, but not CD34− cells, expressed Notch-1. The level of expression found on hESC-derived CD34+ cells was consistent with previous reports of Notch-1 expression on other hematopoietic populations.34,35 We also examined expression of NOTCH-1 and one of its primary transcriptional targets, HES-136 by RT-PCR (Figure 1C). We observed clear expression of NOTCH-1 in CD34+ and CD45+ cells, compared with little or no expression in the CD34− and CD45− populations (Figure 1C). Although HES-1 expression was observed in all sorted cell populations, HES-1 expression was greater in the CD34+ and CD45+ populations (Figure 1C). These results indicate that hESC-derived hematopoietic progenitors are receptive to Notch-1–induced signaling.
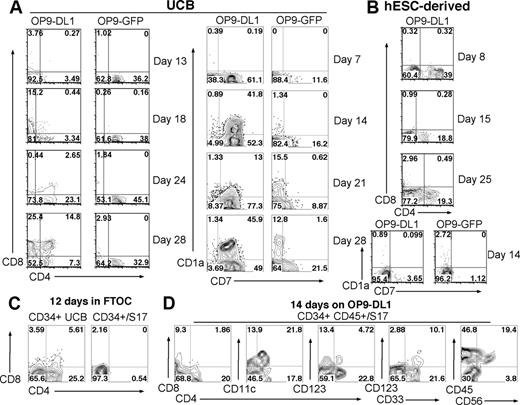
When cocultured on OP9-DL1, CD34+ UCB cells followed a clear developmental progression: first expressing CD7, followed by coexpression of CD7 and CD1a, then becoming CD4+ CD8+ double-positive (DP) T-lineage–committed cells (Figure 2A). Importantly, these CD34+ UCB cells consist of hematopoietic precursor/progenitor cells rather than precommitted lymphoid populations, all of which are completely removed by positive selection of CD34+ cells and/or depletion of lineage marker (Lin)–expressing cells. As expected, CD34+ UCB cells cultured with OP9-GFP cells (which do not express DL1) expressed either CD7 or CD1a (but not both) and did not commit to the T-cell lineage (Figure 2A). A direct comparison between UCB- and hESC-derived cells on day 14 shows that hESC-derived CD34+ cells generated few CD7+ or CD1a+ cells, and no cells expressing both CD7 and CD1a when cocultured with either OP9-DL1 or OP9-GFP stromal cells (Figure 2B). In addition, hESC-derived CD34+ cells produced a population of CD4+ CD8− cells, but never generated any CD4+ CD8+ T cells. These hESC-derived progenitor populations failed to proliferate or survive in OP9 coculture. By 28 days, there were few remaining hESC-derived cells left in the cultures. This is in contrast to NK-promoting cultures on AFT024 stroma, in which hESC-derived progenitors proliferated extensively.5
Development of T cells in vitro from UCB- but not from hESC-derived hematopoietic progenitor cells. (A) CD34+ UCB cells or (B) CD34+ hESC-derived cells were cocultured with OP9-DL1 or OP9-GFP stromal cells. Cells were harvested at the indicated time points and analyzed by flow cytometry for surface expression of CD4, CD8, CD7, and CD1a. (C) FTOC culture of UCB- and hESC-derived hematopoietic progenitors. CD34+ cells from UCB and hESC/S17 sources were cultured for 12 days in mouse fetal thymic lobes. Cultured cells were analyzed by flow cytometry for expression of CD4 and CD8 using antibodies specific for human proteins. (D) CD34+ CD45+ hESC-derived progenitors were cocultured with OP9-DL1 for 14 days and analyzed by flow cytometry for expression of CD4, CD8, CD11c, CD123, CD33, CD45, and CD56.
Development of T cells in vitro from UCB- but not from hESC-derived hematopoietic progenitor cells. (A) CD34+ UCB cells or (B) CD34+ hESC-derived cells were cocultured with OP9-DL1 or OP9-GFP stromal cells. Cells were harvested at the indicated time points and analyzed by flow cytometry for surface expression of CD4, CD8, CD7, and CD1a. (C) FTOC culture of UCB- and hESC-derived hematopoietic progenitors. CD34+ cells from UCB and hESC/S17 sources were cultured for 12 days in mouse fetal thymic lobes. Cultured cells were analyzed by flow cytometry for expression of CD4 and CD8 using antibodies specific for human proteins. (D) CD34+ CD45+ hESC-derived progenitors were cocultured with OP9-DL1 for 14 days and analyzed by flow cytometry for expression of CD4, CD8, CD11c, CD123, CD33, CD45, and CD56.
As a second method to promote T-cell development in vitro, we also compared the T-lineage potential of UCB- and hESC-derived progenitors using the FTOC system.37 Again, CD34+ UCB cells produced clear populations of DP, CD4+ CD8− and CD4− CD8+ single-positive (SP) T cells, whereas hESC-derived CD34+ cells were not able to generate any T-lineage populations in FTOC (Figure 2C), supporting the results observed for OP9-DL1 stromal cell coculture.
Because the CD34+ population of hESC-derived cells is still heterogeneous, we next evaluated the T-cell potential of CD34+ CD45+ hESC-derived cells, which are more enriched in hematopoietic progenitor activity than single-sorted CD34+ cells.29 Again, these CD34+ CD45+ progenitors produced CD4+ CD8− cells but no significant CD4+ CD8+ cells after 14 days in OP9-DL1 stromal coculture (Figure 2D), the same result as obtained from single-sorted CD34+ hESC-derived progenitors (Figure 2B). Further characterization of the CD4+ CD8− cells that develop from CD34+CD45+ hESC-derived cells demonstrated approximately 50% were CD11c+ and approximately 20% were CD123lo, of which almost all were CD33+ (Figure 2D). None of these hESC-derived hematopoietic progenitors ever produced CD3ϵ+ cells in OP9-DL1 coculture (data not shown). The phenotype of the CD4+ CD8− hESC-derived cells is consistent with myeloid dendritic cells.38 Although CD34+ UCB-derived progenitors also produced a population of CD4+ CD123+ BDCA-2+ plasmacytoid dendritic cells when cultured on OP9 stromal cells, a similar population of CD4+ CD123+ BDCA-2+ plasmacytoid dendritic cells was never generated from hESC-derived progenitors (data not shown).
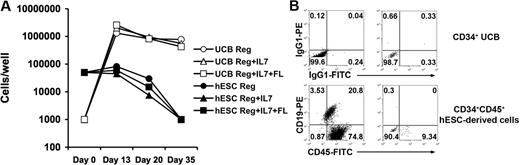
Next, we compared the ability of hESC- and UCB-derived hematopoietic progenitors to produce B cells in vitro. CD19+CD45+ cells can be derived from CD34+ UCB cells cocultured with MS5 cells in media containing defined cytokine combinations (either SCF + G-CSF alone, SCF + G-CSF + IL-7, or SCF + G-CSF + IL-7 + Flt3L; Figure 3).39 In contrast, hESC-derived CD34+ progenitors did not proliferate or derive CD19+ CD45+ cells upon coculture with MS5 cells using any of the cytokine combinations (Figure 3A). Similar results finding lack of B-cell development were also seen, starting with fewer hESC-derived progenitor cells (10 000 or 1000 cells per well), the same numbers of starting cells per well found effective for the UCB progenitor cells. Therefore, this lack of B-cell development is not due to a nonspecific inhibitory effect of elevated numbers of the hESC-derived cells cultured under these defined conditions.
B-cell–promoting culture of UCB- and hESC-derived hematopoietic progenitors. (A) CD34+ UCB cells (○, ▵, □) proliferate in B-cell promoting culture but CD34+ hESC-derived cells (●, ▴, ■) do not. Hematopoietic progenitors were seeded into coculture on MS-5 stromal cells in RPMI 1640 containing 10% FBS, 1% P/S, and 10 ng/mL each of SCF and G-CSF (Reg: ○,●), or additionally supplemented with 10 ng/mL IL-7 (Reg + IL-7: ▵, ▴), or with 10 ng/mL IL-7 and Flt3L (Reg + IL-7 + Flt3L: □, ■). Hematopoietic cells were harvested after 13, 20, or 35 days and live cells counted to determine proliferation. (B) CD34+ UCB cells form B cells in MS-5 culture but CD34+ hESCs do not. Cells cultured for 35 days on MS-5 cells plus SCF and G-CSF were collected and analyzed for expression of human CD45 and CD19 by flow cytometry. Similar phenotypic results were seen with the other cytokine conditions in panel A.
B-cell–promoting culture of UCB- and hESC-derived hematopoietic progenitors. (A) CD34+ UCB cells (○, ▵, □) proliferate in B-cell promoting culture but CD34+ hESC-derived cells (●, ▴, ■) do not. Hematopoietic progenitors were seeded into coculture on MS-5 stromal cells in RPMI 1640 containing 10% FBS, 1% P/S, and 10 ng/mL each of SCF and G-CSF (Reg: ○,●), or additionally supplemented with 10 ng/mL IL-7 (Reg + IL-7: ▵, ▴), or with 10 ng/mL IL-7 and Flt3L (Reg + IL-7 + Flt3L: □, ■). Hematopoietic cells were harvested after 13, 20, or 35 days and live cells counted to determine proliferation. (B) CD34+ UCB cells form B cells in MS-5 culture but CD34+ hESCs do not. Cells cultured for 35 days on MS-5 cells plus SCF and G-CSF were collected and analyzed for expression of human CD45 and CD19 by flow cytometry. Similar phenotypic results were seen with the other cytokine conditions in panel A.
Evaluation of ID expression in UCB- and hESC-derived cells
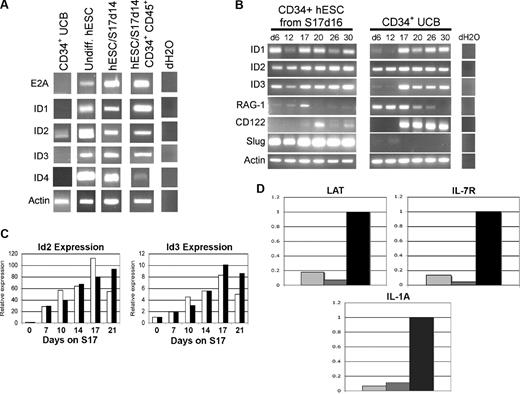
To identify specific cellular and genetic mechanisms that may account for the inability of hESC-derived hematopoietic progenitor cells to derive T cells in vitro, we compared hESC- and UCB-derived cells for expression of genes known to regulate T-, B-, and NK-cell development. Initial studies analyzed hESC- and UCB-derived cells for expression of ID family genes ID1, ID2, ID3, and ID4 (Figure 4). Both ID2 and ID3 promote NK-cell development and repress T- and B-cell development in both humans and mice by inhibiting E-protein family of basic helix-loop-helix (bHLH) transcription factors such as E2A.21-23,40-43 Both undifferentiated hESCs and hESC-derived hematopoietic progenitor cells constitutively expressed ID1, ID2, and ID3, whereas ID4 was observed more prominently in undifferentiated and unsorted hESC-derived cells than in CD34+ CD45+ sorted cells (Figure 4A). hESCs also constitutively expressed E2A, indicating that ID genes would be able to inhibit E-protein–mediated transcription during hESC differentiation (Figure 4A). In contrast, uncultured CD34+ UCB cells expressed low levels of ID2, but did not express ID1, ID3, or ID4 (Figure 4A). CD34+ UCB cells also did not express E2A, suggesting less potential activity of ID2 to effect T-cell development in these cells.
hESC-derived hematopoietic cells constitutively express transcription factors that promote NK-cell development from lymphoid progenitor cells. (A) UCB CD34+ cells, undifferentiated hESCs, and indicated differentiated hESC-derived cell populations were analyzed for expression of E2A, ID1, ID2, ID3, ID4, and ACTIN by RT-PCR. hESC/S17 represents hESC allowed to differentiate on S17 stromal cells for 14 days and analyzed either as an unsorted population, or sorted for CD34+CD45+, CD34+ Flk1−, or CD34+ Flk1+ cells as indicated. (B) Gene expression in hESC- and UCB-derived hematopoietic progenitor cells during NK differentiation. CD34+ hESC/S17 cells at 16 days of differentiation (left panels) and CD34+ UCB (center panels) cells were cultured in NK supporting conditions for indicated numbers of days, then analyzed at the indicated days for expression of ID1, ID2, ID3, RAG-1, CD122, SLUG, and ACTIN genes by RT-PCR. (C) Q-RT-PCR of ID2 and ID3 expression from hESCs cocultured with S17 stromal cells in standard differentiation conditions (□, RPMI + 15% FBS) or with serum-free medium containing BMP-4 (■, StemPro + BMP-4). Cells were harvested after the indicated number of days and analyzed by Q-RT-PCR for ID2, ID3, and ACTIN expression. cDNA content was normalized according to actin levels, and expression of ID2 and ID3 was calculated relative to day-0 expression levels. Data from 1 of 2 replicate studies are shown. (D) Q-RT-PCR analysis of E2A-responsive genes LAT, IL7Rα, and IL1a in hESC/S17 cells at 16 days of differentiation ( ), sorted hESC-derived CD34+CD45+ cells (
), sorted hESC-derived CD34+CD45+ cells ( ), and CD34+ cells from UCB (■). Results are presented as relative gene expression normalized to UCB CD34+ cells.
), and CD34+ cells from UCB (■). Results are presented as relative gene expression normalized to UCB CD34+ cells.
hESC-derived hematopoietic cells constitutively express transcription factors that promote NK-cell development from lymphoid progenitor cells. (A) UCB CD34+ cells, undifferentiated hESCs, and indicated differentiated hESC-derived cell populations were analyzed for expression of E2A, ID1, ID2, ID3, ID4, and ACTIN by RT-PCR. hESC/S17 represents hESC allowed to differentiate on S17 stromal cells for 14 days and analyzed either as an unsorted population, or sorted for CD34+CD45+, CD34+ Flk1−, or CD34+ Flk1+ cells as indicated. (B) Gene expression in hESC- and UCB-derived hematopoietic progenitor cells during NK differentiation. CD34+ hESC/S17 cells at 16 days of differentiation (left panels) and CD34+ UCB (center panels) cells were cultured in NK supporting conditions for indicated numbers of days, then analyzed at the indicated days for expression of ID1, ID2, ID3, RAG-1, CD122, SLUG, and ACTIN genes by RT-PCR. (C) Q-RT-PCR of ID2 and ID3 expression from hESCs cocultured with S17 stromal cells in standard differentiation conditions (□, RPMI + 15% FBS) or with serum-free medium containing BMP-4 (■, StemPro + BMP-4). Cells were harvested after the indicated number of days and analyzed by Q-RT-PCR for ID2, ID3, and ACTIN expression. cDNA content was normalized according to actin levels, and expression of ID2 and ID3 was calculated relative to day-0 expression levels. Data from 1 of 2 replicate studies are shown. (D) Q-RT-PCR analysis of E2A-responsive genes LAT, IL7Rα, and IL1a in hESC/S17 cells at 16 days of differentiation ( ), sorted hESC-derived CD34+CD45+ cells (
), sorted hESC-derived CD34+CD45+ cells ( ), and CD34+ cells from UCB (■). Results are presented as relative gene expression normalized to UCB CD34+ cells.
), and CD34+ cells from UCB (■). Results are presented as relative gene expression normalized to UCB CD34+ cells.
Next, to further evaluate the role of these genes during lymphocyte development in vitro, we analyzed changes in gene expression in hESC- and UCB-derived hematopoietic progenitors during NK cell–promoting culture. Both hESC- and UCB-derived cells initially expressed RAG-1 (Figure 4B), required for development of T and B lymphocytes, but not NK cells.44 As expected, RAG-1 expression declined and became undetectable in both the hESC- and UCB-derived cells at time points after NK-cell lineage commitment.5 hESC-derived hematopoietic progenitors maintained ID gene expression at all time points analyzed (Figure 4B). In contrast, CD34+ UCB cells began to express ID1 and ID3 only after 2 weeks in NK cell–promoting culture (Figure 4B).5,45 ID1 and ID3 expression in UCB cells began concurrently with expression of CD122, a marker of commitment to the NK-cell lineage (Figure 4B). ID2 was expressed in both hESC- and UCB-derived hematopoietic progenitors, but did not prevent UCB cells from undergoing T-cell development (Figure 2A). This difference may reflect the relatively greater contributions that ID3 makes to the T-cell versus NK-cell lineage developmental pathway in humans compared with mice.21,46
To determine the relative induction of ID gene expression in hESCs by our FBS-containing differentiation medium, we compared it with defined serum-free medium supplemented with BMP-4, a known inducer of ID gene expression.29,32,47 Culture in the presence of either FBS or supplemental BMP-4 increased expression of both ID2 and ID3 compared with undifferentiated hESCs (Figure 4C). These data show that our hESC differentiation methods produce increases in ID gene expression that match those observed in the presence of BMP-4. This increasing expression may subsequently affect the lymphoid lineage differentiation of hESC-derived hematopoietic progenitors. In addition, we tested expression of 3 E2A-responsive genes in the hESC and UCB populations and found that expression of the IL-7 receptor α chain (IL7R), interleukin 1a (IL1a), and LAT are all much more highly expressed in the UCB-CD34+ cells than the bulk population of unsorted hESC-derived cells, or the sorted hESC-derived CD34+ CD45+ cells (Figure 4D).
SCF-mediated signaling in hESC- and UCB-derived cells
We also examined specific cell signaling pathways that may affect lymphocyte development. SCF signaling is essential for NK-cell development48 and inhibits progression from the DN to the DP stage during T-cell development.24 Interestingly, hESC- and UCB-derived progenitors showed a clear difference in their expression of SLUG, a primary downstream target of SCF signaling.49 hESC-derived cells expressed SLUG at all observed time points, whereas UCB cells never expressed SLUG (Figure 4B). Therefore, SCF signaling appears to be more active in hESC- than in UCB-derived hematopoietic progenitors, and may alter T- versus NK-cell developmental potential.
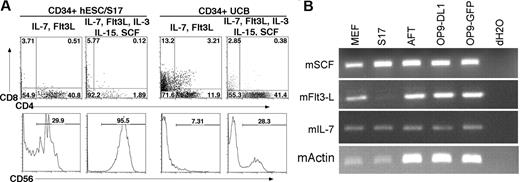
Finally, to better define the role of cytokines in these lymphoid developmental systems, we next evaluated development of T and NK cells in media supplemented only with the cytokines used during OP9-DL1 culture, interleukin-7 (IL-7) and Flt3L, or with addition of IL-3, IL-15, and SCF, used to promote NK-cell development. In cultures with IL-7 and Flt3L, hESC-derived CD34+ cells produced 28% CD56+ NK cells on OP9-DL1 cells. However, when IL-3, IL-15, and SCF were also added, CD56+ CD45+ NK cells accounted for greater than 95% of the cells derived (Figure 5A). IL-3, IL-15, and SCF also increased the proportion of NK cells in cultures of CD34+ UCB cells on OP9-DL1, from 6.56% to 27.8%, whereas CD4+ CD8+ T-lineage cells declined from 3.2% to 0.38%. This decline in DP T cells was accompanied by an increased prevalence of CD4+ CD8− cells, from 11.9% to 41.4% (Figure 5A). We also found that MEF, S17, AFT024, OP9-DL1, and OP9-GFP stromal cell lines all express SCF, Flt3L, and IL-7, with the exception of Flt3L in S17 cells, which may affect the development of cocultured hESC-derived cells (Figure 5B).
The effect of cytokines on lymphoid lineage differentiation in vitro. (A) Addition of SCF, IL-3, and IL-15 promotes increased NK-cell differentiation. CD34+ hESC/S17 (left panels) and CD34+ UCB (right panels) cells were cocultured with OP9-DL1 stromal cells for 30 days. Cultures were supplemented with exogenous IL-7 and Flt3L (T-cell conditions) or with IL-7, Flt3L, SCF, IL-3, and IL-15 (NK-cell conditions). Cells were harvested and analyzed by flow cytometry for expression of CD4, CD8, and CD56. (B) To determine other sources of cytokines that could stimulate hESC-derived cells, the indicated mouse stromal cell lines were analyzed by RT-PCR for expression of the cytokines Scf, Flt3L, IL-7, and β-actin.
The effect of cytokines on lymphoid lineage differentiation in vitro. (A) Addition of SCF, IL-3, and IL-15 promotes increased NK-cell differentiation. CD34+ hESC/S17 (left panels) and CD34+ UCB (right panels) cells were cocultured with OP9-DL1 stromal cells for 30 days. Cultures were supplemented with exogenous IL-7 and Flt3L (T-cell conditions) or with IL-7, Flt3L, SCF, IL-3, and IL-15 (NK-cell conditions). Cells were harvested and analyzed by flow cytometry for expression of CD4, CD8, and CD56. (B) To determine other sources of cytokines that could stimulate hESC-derived cells, the indicated mouse stromal cell lines were analyzed by RT-PCR for expression of the cytokines Scf, Flt3L, IL-7, and β-actin.
Discussion
In vitro culture methods are invaluable to define specific cellular and genetic mechanisms that mediate lymphocyte development. Whereas one previous study indicated that hESCs can produce T lymphocytes in vivo using the SCID-hu model,8 we surprisingly find that hESC-derived hematopoietic progenitors that can be routinely supported to derive NK cells are not able to undergo effective T-cell development in vitro. We are able to use these in vitro culture methods to demonstrate T-cell development is blocked at a very early developmental stage, before the progenitors could coexpress CD7 and CD1a, a requisite stage prior to becoming DP T cells.50 hESC-derived CD34+ cells, when cultured with OP9-DL1 stromal cells, produced a prominent population of CD4+ CD8− cells but no CD4+CD8+ cells. These CD4+ CD8− cells consisted primarily of myeloid dendritic cells, based on their coexpression of CD11c, CD4, CD123, and CD33.38 CD34+ UCB cells cocultured with OP9-GFP produced a similar population of CD4+CD8− cells, although these UCB-derived cells were likely plasmacytoid dendritic cells, as they also expressed BDCA-2 in addition to CD4, but not CD11c (data not shown).51 Despite expression of Notch-1, CD34+ hESC-derived cells were not able to commit to the T-cell lineage in the presence of DL1. hESC-derived CD34+ cells also failed to produce CD19+ B cells under in vitro conditions that lead to clear B-cell development from UCB CD34+ cells (Figure 3). Higher starting numbers of hESC-derived progenitors did not inhibit B-cell development. CD19+ cells were not observed in any experiment when the same number of hESC- and UCB-derived progenitor cells were used to initiate the B-cell culture assay (data not shown). These results demonstrate that lymphoid development potential of CD34+ hESC-derived cells strongly favors the NK-cell lineage at the expense of both T and B cells.
These studies demonstrate at least 2 possible mechanisms may account for the inability of hESC-derived hematopoietic progenitor cells to develop into T cells. First, expression of ID genes likely presents a transcriptional block to T- and B-cell commitment (Figure 4). Whereas CD34+ UCB cells exhibited high levels of ID1 and ID3 expression only after they had been cultured in NK cell–promoting culture for more than 2 weeks, CD34+ hESC-derived cell populations had high levels of ID gene expression at all stages of culture from the undifferentiated state to hematopoietic progenitors to fully committed NK cells. Treatment with BMP-4, known to activate ID gene expression, resulted in an increase in ID2 and ID3 expression similar to hESCs treated with serum. These findings suggest that hESCs may be prevented from becoming T or B cells due to constitutive expression of ID1, ID2, and ID3, which inhibits the activity of E-proteins required for T- and B-cell development.42,52 This hypothesis is further supported by finding reduced expression of E2A-responsive genes (IL7R, IL1, and LAT) in the hESC-derived populations compared with the UCB-CD34+ cells. Therefore, the increased ID expression in the hESC-derived cells may inhibit expression of these genes important for T-cell development.
We also found that DL1 is extensively expressed both on mouse embryonic fibroblasts (MEFs) as well as hESCs prior to initiation of their hematopoietic differentiation (data not shown), and others have also recently shown expression of Notch and Notch ligands on hESCs.53 This expression may contribute to the constitutive ID gene expression observed in undifferentiated hESCs, since ID genes are known downstream targets of Notch-1 signaling.53,54 These results suggest a means by which hESC- and UCB-derived hematopoietic progenitors could have differing ID gene expression profiles, leading to distinct lymphoid lineage developmental potential. In addition, expression of DL1 by hESCs during maintenance and differentiation may inhibit the ability of these cells to respond to DL1 due to cis-mediated inhibition of Notch signaling.55
Cytokine-mediated signaling is a second mechanism that may affect hESC-derived lymphoid cell development. T-cell development in OP9-DL1 culture is enhanced by addition of exogenous human IL-7 and Flt3L. We found that UCB cells could be diverted away from T-cell development and toward the NK-cell lineage despite the presence of DL1 by additional supplementation with IL-3, IL-15, and SCF. These conditions lead to an increase in non–T-lineage CD4+ CD8− cells. This indicates that the influence of cytokine signaling can supersede the effects of DL1 signaling through Notch-1 in stromal cell cocultures.24 Cytokines produced by the stromal cells may influence the transcriptional profile of hESCs. Although most stromal cells used in coculture with hESCs are derived from mice, many murine cytokines are able to signal through their cognate human receptors. An important example is SCF, a cytokine that has profound effects on T- versus NK-cell lineage choice.56,57 All the murine stromal cells used in these studies expressed Scf, indicating that hESC developmental potential could be affected by cross-species cytokine signaling at all stages of culture. This possibility is supported by other studies that demonstrate SCF effectively promotes NK-cell development from human CD34+ cells58 and that exogenous SCF markedly inhibits T-cell differentiation of mouse bone marrow cells in OP9-DL1 cultures.24 It is also supported by our finding that hESC-derived cells, but not UCB cells, express high levels of the transcription factor SLUG, a downstream target of SCF/c-kit signaling.49 Although SCF/c-kit signaling has other downstream targets in addition to SLUG,59,60 our results indicate a clear signaling difference between hESC- and UCB-derived progenitors. This difference may be due to the distinctly different environments, in vitro and in vivo, in which these 2 populations are generated. We propose that ectopic SCF/c-kit signaling in vitro may contribute to the block in T-cell development of hESC-derived hematopoietic progenitors.
These studies provide key insight into where hESC-derived cell populations fit into a specific stage of human embryogenesis. hESC-derived hematopoietic cells likely reflect an embryonic or fetal developmental stage at which hematolymphoid progenitor cells form only NK cells and lack T- and B-cell potential. Indeed, Peault and colleagues examined CD34+ hematopoietic cells from human yolk sac at approximately 3 to 4 weeks of development.61 Interestingly, in vitro culture found these cells were capable only of NK-cell development, and not T- and B-cell production. Erythroid cells derived from hESCs display a similar fetal stage of hematopoietic development.4,62,63 For example, hESC-derived erythroid colonies express mainly embryonic and fetal globin proteins (ϵ- and γ-globin) without a switch to adult (β) globin expression.62 Similarly, studies of pancreatic and cardiomyocyte development from hESCs also suggests these cells are more closely related to fetal cells then adult tissue.64,65 The findings presented here describe mechanisms that may play important roles in at least one of the divergences between embryonic and adult hematopoietic development. Elucidating these differences is of high importance to fully understand how to improve the utility of hESCs both as a widely applicable model system of cellular development and as a source for promising new cellular therapies.
Finally, our results highlight an important issue that applies to the derivation of virtually all cell types from hESCs, and not just lymphocytes. The methods used to obtain hematopoietic progenitors from hESCs and from UCB (or other postnatal tissue) present a clear difference between these populations. In vitro culture and differentiation of hESCs leads to complex conditions and results in a heterogeneous mixture of progenitor cells that are stimulated by stromal cells and multiple soluble proteins that can effect hESC lineage differentiation and development. Therefore, these studies highlight the critical need to consider the effects that in vitro culture has on the potential of hESCs to produce specific cell lineages.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Rebecca Marcus, Jon Linehan, Julie Morris, Matt Painschab, Shawn Mahmud, and Geneve Awong for technical assistance and collaboration on this project.
These studies were supported by National Institutes of Health (Bethesda, MD) R01 HL77923 (D.S.K.), a University of Minnesota Grant-in-Aid Award (D.S.K.), and a University of Minnesota thesis dissertation award (P.S.W.). J.C.Z.-P. is supported by a Canada Research Chair in Developmental Immunology, and by the National Cancer Institute of Canada (Toronto, ON).
National Institutes of Health
Authorship
Contribution: C.H.M. and P.S.W. designed and performed experiments and wrote the paper; Z.N. designed and performed experiments; J.C.Z.-P. provided essential reagents, assisted with experimental design, and edited the paper; D.S.K. designed experiments, and wrote and edited the paper; and all authors read and approved the final paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dan S. Kaufman, 2001 6th Street SE, Minneapolis, MN 55455-3007; e-mail: kaufm020@umn.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal