Abstract
Mast cells (MCs) play critical roles in allergy and inflammation, yet their development remains controversial due to limitations posed by traditional animal models. The zebrafish provides a highly efficient system for studying vertebrate hematopoiesis. We have identified zebrafish MCs in the gill and intestine, which resemble their mammalian counterparts both structurally and functionally. Carboxypeptidase A5 (cpa5), a MC-specific enzyme, is expressed in zebrafish blood cells beginning at 24 hours post fertilization (hpf). At 28 hpf, colocalization is observed with pu.1, mpo, l-plastin, and lysozyme C, but not fms or cepbα, identifying these early MCs as a distinct myeloid population arising from a common granulocyte/monocyte progenitor. Morpholino “knock-down” studies demonstrate that transcription factors gata-2 and pu.1, but not gata-1 or fog-1, are necessary for early MC development. These studies validate the zebrafish as an in vivo tool for studying MC ontogeny and function with future capacity for modeling human MC diseases.
Introduction
Mast cells (MCs) play central roles in allergic and inflammatory reactions.1,2 Stimulation of cell-surface receptors, such as C-KIT and the high-affinity IgE receptor,1,2 results in the release of mediators from cytoplasmic granules, including tryptase and histamine.2 MC number and function are regulated by their development, proliferation, migration, and survival.1 Barriers to understanding these processes include accessibility and imaging limitations posed by traditional animal models. The zebrafish has proven itself to be a robust and highly conserved model for studying vertebrate hematopoiesis.3 Here, we provide the first evidence that the zebrafish possesses MC equivalents that share structural and functional characteristics with their mammalian counterparts. Furthermore, we demonstrate the utility of the zebrafish as an in vivo tool in dissecting the contribution of transcription factors to MC development.
Methods
Zebrafish were maintained, bred, and developmentally staged according to Westerfield.4 Use of zebrafish in this study was approved by the Dalhousie University Animal Care Committee. Zebrafish gills and intestine were fixed in 10% neutral buffered formalin, and standard staining procedures were applied (Figure 1A-F). Immunohistochemistry was facilitated by antigen retrieval (Figure 1I,J). For electron microscopy, tissues were fixed overnight in 2% glutaraldehyde in 0.1 M caccodylate and postfixed in 1% osmium tetroxide. Thin sections (90 nm) were stained in 25% uranyl acetate in methanol and lead citrate.
Bromophenol blue and 10 μg compound 48/80 or saline were injected intraperitoneally, and blood was extracted by cardiac puncture after 2.5 minutes. Tryptase activity was measured in plasma spectrophotometrically at 415 nm by the release of p-nitroanilide from N-benzoyl-DL-arginine-p-nitroanilide (BAPNA), a tryptase substrate.
Digoxogenin- or fluorescein isothiocyanate (FITC)–labeled antisense mRNA probes for zebrafish carboxypeptidase A5 (cpa5), α-globin, cebpα, pu.1, myeloperoxidase (mpo), l-plastin, lysozyme C, fms, gata-2, and gata-1 were synthesized according to the published literature.5-8 Whole-mount single or double mRNA in situ hybridization (ISH) assays were adapted from standard protocol.5 Images were taken on a Leica MZ16F with a Leica DFC 490 camera (5× objective; Leica, Wetzlar, Germany). cpa5-FITC–labeled Fast Red–stained 28 hours post fertilization (hpf) and 7 days post fertilization (dpf) embryos were dissociated using Blendzyme 3 (Roche Applied Sciences, Indianapolis, IN), and a strained cell suspension was centrifuged for 10 minutes at 4500g and resuspended in 400 μL of 0.9× phosphate buffered saline (PBS)/5% fetal bovine serum (FBS) for fluorescence-activated cell sorting (FACS) and cytospin.
Sections (5 μm) of intestinal tissue were deparaffinized with xylene and rehydrated with graded alcohols. ISH (whole mount protocol) was performed adding 400 μg/mL levamisole after NBT-BCIP staining (methyl green counterstain).
gata-1, gata-2, and friend of gata-1 (fog-1) morpholinos and controls were purchased from Genetools (Philomath, OR); pu.1 morpholino and control9 were kindly provided by Dr Jennifer Rhodes (Dana-Farber Cancer Institute, Boston, MA). Morpholinos were diluted to a working concentration (gata-1, 1.0 mM; gata-2, 1.0 mM; fog-1, 0.8 mM; and pu.1, 0.5 mM) with 1% phenol red; 1 nL was injected into zebrafish embryos at the 1- to 4-cell stage.
Results and discussion
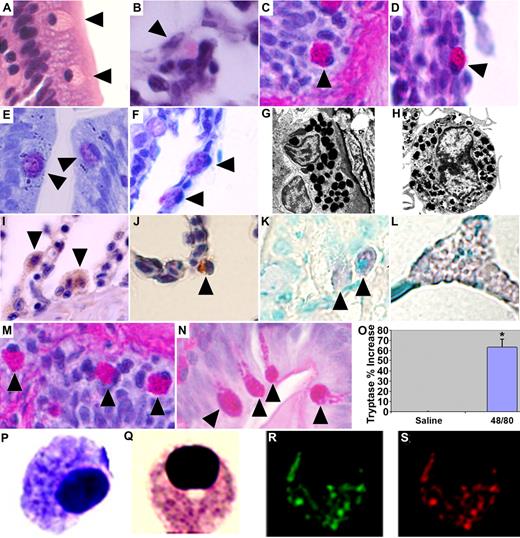
MCs were identified in gill and intestinal sections of adult wild-type zebrafish—anatomic equivalents to well-established sites where MCs reside as part of the innate immune system in mammals.2 These cells contain an ovoid eccentric nucleus and prominent eosinophilic granules on hematoxylin and eosin (H&E) staining, which stained positively with periodic acid-Schiff (PAS), a feature shared with the eosinophil/basophil population previously identified among the hematopoietic elements of the zebrafish kidney.5 Mammalian MCs and basophils similarly share some common structural features.10 Granules appeared metachromatic after toluidine blue staining, a pathognomonic characteristic of mammalian MCs11 (Figure 1A-F). Electron microscopy demonstrated an eccentrically placed nucleus and dense homogenous granules closely approximating the appearance of murine MCs (Figure 1G,H). Immunohistochemistry demonstrated a positive reaction to polyclonal anti-human C-KIT and monoclonal anti-human MC tryptase (Figure 1I,J). Intraperitoneal injection of compound 48/80, an agent shown to induce MC degranulation in both mammals and other teleost fish,12,13 resulted in increased numbers of activated degranulating intestinal MCs and a significant elevation in plasma tryptase levels compared with saline-injected controls (Figure 1M-O). Tryptase release is a reliable reflection of MC burden or reaction severity,14 suggesting that the zebrafish can serve as a robust in vivo system for evaluating vertebrate MC responses.
Zebrafish MCs structurally and functionally resemble their mammalian counterparts. H&E staining. (A) Intestine. (B) Gill. Periodic acid-Schiff (PAS). (C) Intestine. (D) Gill. Toluidine blue. (E) Intestine. (F) Gill. MCs in each panel indicated by ▶. Transmission electron microscope (Phillips 300; FEI, Hillsboro, OR) images of (G) a zebrafish intestinal MC (20 000× magnification) and (H) a mouse bone marrow–derived MC (26 000× magnification). Zebrafish MCs demonstrate a positive reaction to (I) a polyclonal antibody raised against human CD117 (C-KIT) antigen (Dako Cytomation, Carpinteria, CA) and (J) a monoclonal anti–human MC tryptase antibody (gills; Dako Cytomation). Biotinylated Universal Linker (Dako Cytomation) secondary antibody and 3,3′-diaminobenzidine for chromogenic detection (hematoxylin counterstain; 100× objective). RNA ISH using digoxigenin-labeled RNA antisense probe to zebrafish cpa5 demonstrates positive staining in (K) intestinal MCs and (L) pancreas; MCs indicated by ▶. Adult zebrafish injected intraperitoneally with 10 μg of compound 48/80 demonstrate (N) MC degranulation compared with (M) saline-injected controls. PAS staining, 100× objective; MCs in each panel indicated by ▶. (O) Increased plasma tryptase levels compared with saline-injected control fish. Presented as means plus SEM of 3 experiments with 4 to 6 fish per group; *P < .05 (t test). Cytospin of FACS-sorted, FITC-labeled, Fast Red–stained cpa5+ cells isolated from zebrafish embryos at 7 dpf demonstrate morphology consistent with MCs. (P) Toluidine blue. (Q) Wright-Giemsa. (R) Green channel (FITC). (S) Red channel (Fast Red; also Figure S2). (A-F, I-N) Images obtained at 100×/1.3 NA oil-immersion objective with a Nikon Eclipse E600, Nikon DXM 1200 camera, and ACT-1 software (all Nikon, Tokyo, Japan). Panels assembled using Adobe Photoshop CS3 Extended version 10.0 (Adobe Systems, San Jose, CA). (P-S) Images obtained at 100×/1.4 NA oil immersion objective with a Zeiss Axioplan 2 (Zeiss, Jena, Germany) and Axiocam HRc digital camera, Axiovision 4.6 with multi-channel fluorescence module (Diagnostic Instruments, Sterling Heights, MI).
Zebrafish MCs structurally and functionally resemble their mammalian counterparts. H&E staining. (A) Intestine. (B) Gill. Periodic acid-Schiff (PAS). (C) Intestine. (D) Gill. Toluidine blue. (E) Intestine. (F) Gill. MCs in each panel indicated by ▶. Transmission electron microscope (Phillips 300; FEI, Hillsboro, OR) images of (G) a zebrafish intestinal MC (20 000× magnification) and (H) a mouse bone marrow–derived MC (26 000× magnification). Zebrafish MCs demonstrate a positive reaction to (I) a polyclonal antibody raised against human CD117 (C-KIT) antigen (Dako Cytomation, Carpinteria, CA) and (J) a monoclonal anti–human MC tryptase antibody (gills; Dako Cytomation). Biotinylated Universal Linker (Dako Cytomation) secondary antibody and 3,3′-diaminobenzidine for chromogenic detection (hematoxylin counterstain; 100× objective). RNA ISH using digoxigenin-labeled RNA antisense probe to zebrafish cpa5 demonstrates positive staining in (K) intestinal MCs and (L) pancreas; MCs indicated by ▶. Adult zebrafish injected intraperitoneally with 10 μg of compound 48/80 demonstrate (N) MC degranulation compared with (M) saline-injected controls. PAS staining, 100× objective; MCs in each panel indicated by ▶. (O) Increased plasma tryptase levels compared with saline-injected control fish. Presented as means plus SEM of 3 experiments with 4 to 6 fish per group; *P < .05 (t test). Cytospin of FACS-sorted, FITC-labeled, Fast Red–stained cpa5+ cells isolated from zebrafish embryos at 7 dpf demonstrate morphology consistent with MCs. (P) Toluidine blue. (Q) Wright-Giemsa. (R) Green channel (FITC). (S) Red channel (Fast Red; also Figure S2). (A-F, I-N) Images obtained at 100×/1.3 NA oil-immersion objective with a Nikon Eclipse E600, Nikon DXM 1200 camera, and ACT-1 software (all Nikon, Tokyo, Japan). Panels assembled using Adobe Photoshop CS3 Extended version 10.0 (Adobe Systems, San Jose, CA). (P-S) Images obtained at 100×/1.4 NA oil immersion objective with a Zeiss Axioplan 2 (Zeiss, Jena, Germany) and Axiocam HRc digital camera, Axiovision 4.6 with multi-channel fluorescence module (Diagnostic Instruments, Sterling Heights, MI).
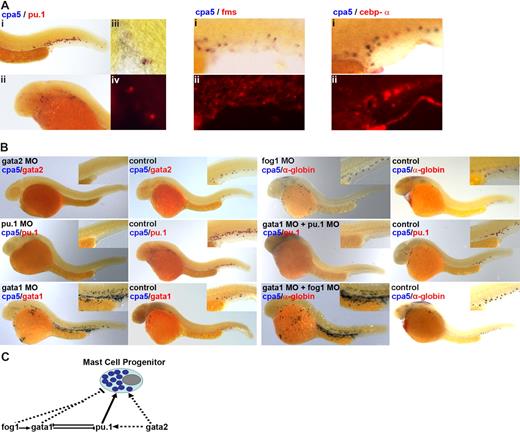
We identified zebrafish carboxypeptidase A5 (cpa5), a protein that shares 64% identity with human CPA1 expressed in exocrine pancreas and 38% identity with CPA3 found in human MCs. Zebrafish cpa5 pancreatic expression has been previously identified,15 and embryonic blood cell expression was demonstrated in a large-scale ISH screen.16 In adults, cpa5 expression was restricted to morphologically identified MCs and pancreatic tissue (Figure 1K,L). In embryos, cpa5 expression was restricted to hematopoietic cells present in the anterior lateral paraxial mesoderm and in smaller numbers around the intermediate cell mass (sites of embryonic hematopoiesis3,6 ), beginning at 24 hpf. cpa5-expressing cells reached a peak by 28 hpf, where they congregated at both sites of embryonic hematopoiesis and in circulation over the yolk sac, persisting through 7 days post-fertilization (dpf). By 72 hpf, cpa5 expression could also be observed in the pancreas (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Other zebrafish homologs of mammalian carboxypeptidases, including cpa1 and cpb1, were not expressed in zebrafish blood cells (data not shown). FACS analysis and cytospin of cpa5-FITC–labeled Fast Red–stained cells revealed a predominance of cells with a morphologic appearance in keeping with mammalian MCs17 (Figures 1P-S and S2). cpa5 expression colocalized in a proportion of embryonic myeloid cells expressing the early myeloid transcription factor, pu.1 as well as mpo,5,18 l-plastin,6 and lysozyme-C.19 These latter 3 genes were previously characterized as granulocyte (mpo) or monocyte (l-plastin, lysozyme C) specific, but more recent zebrafish data have implicated a more panmyeloid expression profile.8,20,21 Interestingly, no colocalization was observed between cpa5 and fms, a gene exclusively expressed on monocytes,8 or for cpa5 and cepb-α, a transcription factor required for neutrophil and basophil cell fate22 (Figures 2A and S3). These data establish cpa-5–expressing cells as a unique myeloid subpopulation arising from a cell with both granulocyte and monocyte potential, in keeping with the model of mastopoiesis posed by Arinobu et al.22 This model contends that MCs and basophils arise from a common granulocyte/monocyte progenitor, with cepb-α functioning as the transcriptional switch determining cell fate.
cpa5 identifies zebrafish MC progenitors. (A) Double whole-mount ISH using a digoxigenin-labeled RNA antisense probe to zebrafish cpa5 (blue) and FITC-labeled RNA anti-sense probe (red) to pu.1, mpo, l-plastin, and lysozyme C (see Figure S3) demonstrate coexpression of cpa5 in a proportion of cells: (i) tail, (ii) head/yolk sac (5× objective). Evidence of coexpression is shown by colocalization observed in higher-magnification images of selected cells: (iii) brightfield, (iv) fluorescence (10× objective). No colocalization is observed for fms and cebp-α: (i) brightfield, (ii) fluorescence (8× objective). (B) gata-2 and pu.1 are both required for the development of early MCs as evidenced by the absence of cpa5 expression in gata-2 and pu.1 morphants, whereas gata-1 morphants paradoxically demonstrate increased numbers of cpa5+ cells. Fog-1 is dispensable for early MC development as evidenced by wild-type cpa5 expression in fog-1 morphants. Compound gata-1/pu.1 morphants demonstrate an absence of cpa5 expression, whereas compound gata-1/fog-1 morphants show a dramatic increase in numbers of cpa5+ cells. Lateral views, anterior left and dorsal at the top (28 hpf, 5× objective). Inset boxes demonstrate a higher-magnification view of the tail and the region around the intermediate cell mass. (C) Proposed model of transcription factor interactions required for early MC development. Established interactions are represented by —, and potential interactions by …. All images obtained with Leica application suite version 2.4.OR (Leica Microsystems, Heerbrugg, Switzerland); figure panels assembled using Adobe Photoshop CS3 Extended version 10.0 (Adobe Systems).
cpa5 identifies zebrafish MC progenitors. (A) Double whole-mount ISH using a digoxigenin-labeled RNA antisense probe to zebrafish cpa5 (blue) and FITC-labeled RNA anti-sense probe (red) to pu.1, mpo, l-plastin, and lysozyme C (see Figure S3) demonstrate coexpression of cpa5 in a proportion of cells: (i) tail, (ii) head/yolk sac (5× objective). Evidence of coexpression is shown by colocalization observed in higher-magnification images of selected cells: (iii) brightfield, (iv) fluorescence (10× objective). No colocalization is observed for fms and cebp-α: (i) brightfield, (ii) fluorescence (8× objective). (B) gata-2 and pu.1 are both required for the development of early MCs as evidenced by the absence of cpa5 expression in gata-2 and pu.1 morphants, whereas gata-1 morphants paradoxically demonstrate increased numbers of cpa5+ cells. Fog-1 is dispensable for early MC development as evidenced by wild-type cpa5 expression in fog-1 morphants. Compound gata-1/pu.1 morphants demonstrate an absence of cpa5 expression, whereas compound gata-1/fog-1 morphants show a dramatic increase in numbers of cpa5+ cells. Lateral views, anterior left and dorsal at the top (28 hpf, 5× objective). Inset boxes demonstrate a higher-magnification view of the tail and the region around the intermediate cell mass. (C) Proposed model of transcription factor interactions required for early MC development. Established interactions are represented by —, and potential interactions by …. All images obtained with Leica application suite version 2.4.OR (Leica Microsystems, Heerbrugg, Switzerland); figure panels assembled using Adobe Photoshop CS3 Extended version 10.0 (Adobe Systems).
We used a morpholino-based strategy to interrogate the roles of several transcription factors in MC development. We demonstrated that gata-2 and pu.1 are both required for early MC development in zebrafish, as gata-2 and pu.1 morpholino-injected morphants demonstrate severely decreased to absent cpa5 expression. gata-1 morphants paradoxically demonstrate abundant cpa5+ cells, likely due to unopposed pu.1 expression.9 This supposition was supported by the absence of cpa5 expression in compound gata-1/pu.1 morphants. Alternatively, cpa5+ MC progenitors may accumulate at the expense of mature MCs that require gata-1 expression, as observed in Gata-1low mice.23 Fog-1 has recently been suggested to antagonize MC development.24,25 Zebrafish fog-1 morphants maintained cpa5 expression, confirming fog-1 is dispensable for early MC development. Interestingly, a large expansion of cpa5+ cells was seen when gata-1 was simultaneously knocked down, suggesting the permissive effect of fog-1 inhibition on MC progenitor development may be enhanced in the absence of gata-1 (Figure 2B,C).
With the discovery of zebrafish MC counterparts, we have contributed to the establishment of a complete myeloid cell repertoire in this species and demonstrated that the developmental and technical opportunities afforded by the zebrafish for studying other lineages can be applied to MC biology. Continuation of these efforts has the potential for significantly expanding our understanding of vertebrate mastopoiesis and MC function. Moreover, these studies set the stage for harnessing the transgenic capabilities of the zebrafish to model inflammatory and malignant human MC diseases, with the future capacity for high-throughput inhibitor screening.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Patricia Colp for assistance with immunohistochemistry, Marlene Henry for assistance with electron microscopy, and Sarah Bugden for assistance with FACS analysis. We thank Alan Cantor, John Kanki, and Jean Marshall for their critical review of the manuscript; Jennifer Rhodes and Leonard Zon for helpful discussion; and Jocelyn Jaques for administrative assistance.
J.N.B. is supported by a Dalhousie University Clinical Scholar Award and a Canadian Institutes of Health Research–Nova Scotia Health Research Foundation Regional Partnership Award.
Authorship
Contribution: J.T.D. and E.M.T. performed research and analyzed data; J.S. and S.D. performed research; R.B.F. contributed pathology expertise and provided reagents; B.H.P. contributed reagents and analyzed data; T.J.L. designed research and analyzed data; and J.N.B. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jason N. Berman, Division of Pediatric Hematology/Oncology, Departments of Pediatrics and Microbiology/Immunology, Dalhousie University, IWK Health Centre, PO Box 9700, 5850/5980 University Avenue, Halifax, Nova Scotia B3K 6R8 Canada; e-mail: jason.berman@iwk.nshealth.ca.