In this issue of Blood, Rimsza and colleagues present an embarrassment of riches. They report the use of a multiplex quantitative nuclease protection assay to measure, in FFPET, prognostic genes for DLBCL in patients treated with rituximab. Their approach overcomes 3 significant problems in the field, namely the difficulty of working with FFPET, the requirement for a simple method for clinical measurement of prognostic genes, and the need for a prognostic gene signature applicable to patients treated in the rituximab era.
Microarray gene expression profiling has resulted in an exponential increase in the understanding of cancer. It has also identified a plethora of putative novel diagnostic and prognostic biomarkers. These are increasingly important as traditional markers, either clinical or cellular, are beginning to lose discriminatory power, especially in the emerging era of personalized medicine. However, almost all diagnostic material is in the form of formalin-fixed paraffin embedded tissue (FFPET), in which gene expression profiling is compromised by RNA degradation and cross-linking, and this has hindered translation of microarray-identified biomarkers to clinical practice.1 Several groups have sought to overcome this, either by use of modified RNA extraction and labeling for microarray analysis,2 by measurement of candidate prognostic genes by real-time PCR3 or by bead-based approaches, such as the DASL platform.4 These studies demonstrate the feasibility of gene expression profiling in FFPET but admit reduced sensitivity compared with fresh/frozen tissue, though Malumbres and colleagues reported successful measurement of a 6-gene model in FFPET in diffuse large B-cell lymphoma (DLBCL) patients treated with R-CHOP.5 Additionally, while real-time PCR is possible in FFPET, there remains a need for multiplex measurement of several genes, particularly as the power of prognostic gene signatures is often based on more than one gene. Furthermore, identification of specific prognostic groups often suggests new drug targets, increasing the importance of FFPET-based studies capable of extending the range and depth of clinical material for translational study.
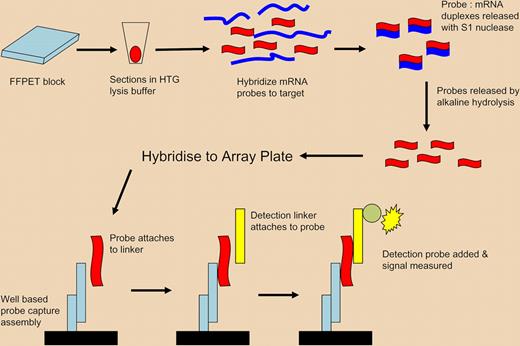
Diagram of ArrayPlate assay. Proprietary lysis buffer (HTG) is used to lyse and permeabilize FFPET tissue sections. Specific nuclease protection probes are added, which hybridize to all mRNA, either soluble or still cross-linked. S1 nuclease is added to remove nonspecific RNA, leaving only target mRNA/probe duplexes. The hybridized-specific probes are released by hydrolysis and the isolated probes are transferred to an ArrayPlate well for detection using linkers, detection probes, and a chemiluminescent substrate. The method enables expression profiling of multiple mRNAs without RNA extraction by the use of short probes capable of hybridisation to both soluble and insoluble/cross-linked mRNA of importance in FFPET. Adapted by permission from Macmillan Publishers. Roberts RA, Sabalos CM, LeBlanc ML, et al. Quantitative nuclease protection assay in paraffin-embedded tissue replicates prognostic microarray gene expression in diffuse large-B-cell lymphoma. Lab Invest. 2007;87:979-997.
Diagram of ArrayPlate assay. Proprietary lysis buffer (HTG) is used to lyse and permeabilize FFPET tissue sections. Specific nuclease protection probes are added, which hybridize to all mRNA, either soluble or still cross-linked. S1 nuclease is added to remove nonspecific RNA, leaving only target mRNA/probe duplexes. The hybridized-specific probes are released by hydrolysis and the isolated probes are transferred to an ArrayPlate well for detection using linkers, detection probes, and a chemiluminescent substrate. The method enables expression profiling of multiple mRNAs without RNA extraction by the use of short probes capable of hybridisation to both soluble and insoluble/cross-linked mRNA of importance in FFPET. Adapted by permission from Macmillan Publishers. Roberts RA, Sabalos CM, LeBlanc ML, et al. Quantitative nuclease protection assay in paraffin-embedded tissue replicates prognostic microarray gene expression in diffuse large-B-cell lymphoma. Lab Invest. 2007;87:979-997.
The biology and behavior of DLBCL is highly heterogeneous. Several gene expression profiling studies have provided greater prediction of disease behavior, separating DLBCL into at least 2 distinct molecular subtypes: a favorable germinal center B-cell type (76% 5-year survival) and an unfavorable activated B-cell type (16% 5-year survival).6 Rituximab has dramatically altered outcome in DLBCL and its introduction has been contemporaneous with these studies. Therefore, their results must be tempered by their analysis of patients treated in the pre-rituximab era, with some reports of loss of prognostic distinction between germinal center (GC) and non-GC types in patients treated with rituximab.7 Monti et al addressed this problem by unsupervised analysis of samples of DLBCL from untreated patients to determine an a priori biological signature predictive of new therapeutic targets,8 demonstrating the importance of identifying a prognostic gene signature in patients treated with rituximab.
The paper by Rimsza et al uses an elegant novel methodology to solve, at a single stroke, the multiple problems outlined above. The ArrayPlate method (High Throughput Genomics (HTG), Tuscon, AZ) enables gene expression measurement without the need for RNA isolation, reverse transcription, or amplification, each of which are technically difficult in FFPET and in combination can lead to confounding results, while the use of short 50-mer probes enables hybridization, despite RNA fragmentation, usually to fragments approximately 200bp long. It is intriguing that the majority of the prognostic genes were unchanged between the pre-rituximab and rituximab treatment groups, in contrast to some other studies, but this study investigated a wide range of genes from several other studies and this may have increased the possibility of identifying a common set of genes, while the ArrayPlate method captures all mRNA, whether soluble or insoluble/cross-linked, and avoids a RNA extraction step, further minimizing possible processing losses. It follows that the “key biological aspects of DLBCL were not affected by addition” of rituximab, refocusing on the pathobiology underlying differential outcomes in DLBCL. From this, Rimsza et al investigate the contribution of 2 genes, HLA-DR and MYC, representing the immune response and proliferation, in outcome, demonstrating that aggressive DLBCL was characterized by low levels of HLA-DR and high levels of MYC, and identifying loss of immune surveillance and high proliferation as the key biological features in aggressive B-cell lymphoma. It is interesting biologically, and important clini-cally, that this key feature holds whether rituximab is used or not.
In conclusion, Rimza et al advertise their method as having “the potential to change the practice of pathology.” A masterful use of the understatement!
Conflict-of-interest disclosure: The author declares no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal