Abstract
Aurora-A kinase (Aur-A) is a member of the serine/threonine kinase family that regulates the cell division process, and has recently been implicated in tumorigenesis. In this study, we identified an antigenic 9–amino-acid epitope (Aur-A207-215: YLILEYAPL) derived from Aur-A capable of generating leukemia-reactive cytotoxic T lymphocytes (CTLs) in the context of HLA-A*0201. The synthetic peptide of this epitope appeared to be capable of binding to HLA-A*2402 as well as HLA-A*0201 molecules. Leukemia cell lines and freshly isolated leukemia cells, particularly chronic myelogenous leukemia (CML) cells, appeared to express Aur-A abundantly. Aur-A–specific CTLs were able to lyse human leukemia cell lines and freshly isolated leukemia cells, but not normal cells, in an HLA-A*0201–restricted manner. Importantly, Aur-A–specific CTLs were able to lyse CD34+ CML progenitor cells but did not show any cytotoxicity against normal CD34+ hematopoietic stem cells. The tetramer assay revealed that the Aur-A207-215 epitope–specific CTL precursors are present in peripheral blood of HLA-A*0201–positive and HLA-A*2402–positive patients with leukemia, but not in healthy individuals. Our results indicate that cellular immunotherapy targeting Aur-A is a promising strategy for treatment of leukemia.
Introduction
Cellular immunotherapy for malignancies targeting various tumor-associated antigens has been developed.1,2 Recently, some attractive target antigens recognized by leukemia-reactive cytotoxic T lymphocytes (CTLs), such as WT1 and PR1, have been discovered and phase 1/2 clinical studies of cancer immunotherapy targeting these antigens have been conducted; however, the clinical response against hematologic malignancies remains unsatisfactory.3,4 To establish effective cancer immunotherapy, identification of target antigens that are recognized efficiently by tumor-specific CTLs is necessary. Antigens that can serve as ideal targets recognizable by tumor-specific CTLs need to have several essential characteristics. First, their expression should be limited to, or abundant in, tumor cells rather than normal cells. Second, the antigens should be efficiently processed in tumor cells and expressed on the cell surface in context with common HLA molecules. Third, target antigens should play an important role in tumorigenesis and/or progression of malignancies, because their expression is essential for tumor survival.
Aurora-A kinase (Aur-A) is a member of the serine/threonine kinase family, and the Aur-A gene is located at chromosome 20q13, a region frequently amplified in breast cancer.5 Aur-A is mainly expressed in the G2/M phase of the cell cycle and regulates mitotic cell division in normal cells.6-8 Among normal tissues, Aur-A is expressed exclusively in testis, but in various kinds of cancer it is aberrantly overexpressed, and associated with poor prognosis.9-14 Overexpression of Aur-A determined by amplification of Aur-A mRNA has also been widely observed in hematologic malignancies.15-19 Aur-A overexpression has been linked with centrosome amplification, aneuploidy, and chromosome instability.20,21 Furthermore, ectopic overexpression of Aur-A efficiently transforms immortalized rodent fibroblasts.9,20 These data strongly suggest that Aur-A is one of the fundamental cancer-associated genes and a potential target for cancer treatment. In addition, previous reports have demonstrated that silencing of the gene encoding Aur-A in cancer cells results in inhibition of their growth and enhancement of the cytotoxic effect of anticancer agents.22 Therefore the development of small molecules with an Aur-A–inhibitory function may make it possible to reduce or block the oncogenic activity of Aur-A. On the basis of this concept, clinical studies using Aur-A inhibitors for cancer treatment are now under way; however, their clinical efficacy is still unknown.23-27 The biologic characteristics of Aur-A mentioned above suggest that it is an ideal target for tumor-specific CTLs, and that cancer immunotherapy targeting Aur-A could be feasible. In this study, therefore, we attempted to verify the feasibility of cellular immunotherapy for leukemia targeting Aur-A.
Methods
Synthetic peptides
Candidate peptides derived from Aur-A with high binding affinity for the HLA-A*0201 or HLA-A*2402 molecule were predicted algorithmically by the BIMAS program (http://www-bimas.cit.nih.gov/molbio/hla_bind/). On the basis of these data, peptides with favorable binding affinity for the HLA-A*0201 or HLA-A*2402 molecule were selected and synthesized (Thermo Electron; Greiner Bio-One, Tokyo, Japan). Amino acid sequences of the peptides used in this study are listed in Table 1. All the peptides were synthesized with a purity exceeding 80%.
Binding affinities of synthetic peptides
| HLA . | Position . | Length, mer . | Sequence . | Score . | Fluorescence index . |
|---|---|---|---|---|---|
| A*0201 | Aur-A271-279 | 9 | KIADFGWSV | 3911 | 0.93 |
| A*0201 | Aur-A63-71 | 9 | KLVSSHKPV | 243 | 0.23 |
| A*0201 | Aur-A207-215 | 9 | YLILEYAPL | 147 | 1.47 |
| A*0201 | WT17-15 | 9 | DLNALLPAV | 12 | 0.06 |
| A*0201 | CMVpp65495-503 | 9 | NLVPMVATV | 160 | 1.71 |
| A*2402 | Aur-A207-215 | 9 | YLILEYAPL | 6 | 0.99 |
| A*2402 | WT17-15 | 9 | DLNALLPAV | 0.18 | 0.02 |
| A*2402 | WT1235-243Y | 9 | CYTWNQMNL | 200 | 4.5 |
| HLA . | Position . | Length, mer . | Sequence . | Score . | Fluorescence index . |
|---|---|---|---|---|---|
| A*0201 | Aur-A271-279 | 9 | KIADFGWSV | 3911 | 0.93 |
| A*0201 | Aur-A63-71 | 9 | KLVSSHKPV | 243 | 0.23 |
| A*0201 | Aur-A207-215 | 9 | YLILEYAPL | 147 | 1.47 |
| A*0201 | WT17-15 | 9 | DLNALLPAV | 12 | 0.06 |
| A*0201 | CMVpp65495-503 | 9 | NLVPMVATV | 160 | 1.71 |
| A*2402 | Aur-A207-215 | 9 | YLILEYAPL | 6 | 0.99 |
| A*2402 | WT17-15 | 9 | DLNALLPAV | 0.18 | 0.02 |
| A*2402 | WT1235-243Y | 9 | CYTWNQMNL | 200 | 4.5 |
The binding affinities of synthetic peptides for HLA molecules were predicted by computer algorithms available on the National Institutes of Health BIMAS website (http://www-bimas.cit.nih.gov/molbio/hla_bind). The binding affinities of synthetic peptides for HLA molecules were evaluated by MHC stabilization assay as detailed in “HLA peptide–binding assay.”
HLA peptide–binding assay
Binding affinity of peptides for the HLA-A*0201 or HLA-A*2402 molecule was assessed by an HLA-A*0201 or HLA-A*2402 stabilization assay as described previously.28,29 Briefly, the HLA-A*0201–positive cell line (T2) or the HLA-A*2402 gene–transfected T2 cell line (T2-A24) was plated in 24-well plates at 106 cells per well and incubated overnight with the candidate peptides at a concentration of 10 μM in serum-free RPMI 1640 medium. The T2 and T2-A24 cells were washed twice with phosphate-buffered saline (PBS), and then incubated with fluorescein isothiocyanate (FITC)–conjugated anti–HLA-A2 or HLA-A24 monoclonal antibody (MoAb; One Lambda, Canoga Park, CA) at 4°C for 20 minutes. The cells were washed and suspended in 1 mL PBS and analyzed using a flow cytometer (FACSCalibur; Becton Dickinson, San Jose, CA). Measurement of mean fluorescence intensity and analysis of data were done with CellQuest Software (Becton Dickinson). The fluorescence index (FI) was calculated as FI = (sample mean − background mean) / background mean.
Cell lines, freshly isolated leukemia cells, and normal cells
Approval for this study was obtained from the institutional review board of Ehime University Hospital. Written informed consent was obtained from all patients, healthy volunteers, and parents of cord blood donors in accordance with the Declaration of Helsinki.
B-lymphoblastoid cell lines (B-LCLs) were established by transformation of peripheral blood B lymphocytes with Epstein-Barr virus. LCLs, T2, T2-A24, and leukemia cell lines were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS). The HLA-A*0201 gene–transfected C1R cell line (C1R-A*0201; kindly provided by Dr A. John Barrett, National Heart, Lung, and Blood Institute [NHLBI], Bethesda, MD) was cultured in RPMI 1640 medium supplemented with 10% FCS and 2 mM l-glutamine. Peripheral blood mononuclear cells (PBMCs) and bone marrow mononuclear cells (BMMCs) from leukemia patients and healthy volunteers, and cord blood mononuclear cells (CBMCs) from healthy donors were isolated and stored in liquid nitrogen until use. All leukemia samples contained more than 95% leukemia cells. CD34+ cells from BMMCs and CBMCs were isolated using CD34+ cell-isolating immunomagnetic beads (MACS beads; Miltenyi Biotec, Auburn, CA). In some experiments, BMMCs and CBMCs were stained with FITC-conjugated anti-CD34 MoAb and phycoerythrin (PE)–conjugated anti-CD38 MoAb, and CD34+CD38high cells and CD34+CD38low cells were sorted with an EPICS ALTRA cell sorter (Beckman-Coulter, Fullerton, CA).
Generation of Aur-A peptide–specific CTL lines
Aur-A peptide–specific CTLs were generated as described previously.30 Briefly, monocytes (CD14+ mononuclear cells) were isolated from PBMCs of HLA-A*0201–positive individuals using CD14+ cell-isolating MACS beads. Monocytes were cultured in RPMI 1640 medium supplemented with 10% FCS, 75 ng/mL recombinant human granulocyte-macrophage colony-stimulating factor, 10 ng/mL recombinant human interleukin 4 (IL-4; R&D Systems, Minneapolis, MN), and 100 U/mL recombinant human tumor necrosis factor-α (Dainippon Pharmaceutical, Osaka, Japan) to generate mature dendritic cells (DCs). CD8+ T lymphocytes isolated from PBMCs using CD8+ cell-isolating MACS beads were plated in 96-well round-bottomed plates at 105 cells per well and stimulated with 104 autologous DCs pulsed with synthetic peptide derived from Aur-A at a concentration of 10 μM. The cells were cultured in RPMI 1640 medium supplemented with 10% human AB serum. After 7 days, the cells were restimulated with 104 autologous DCs pulsed with Aur-A peptide, and 10 U/mL IL-2 (Boehringer Mannheim, Mannheim, Germany) was added 4 days later. After culturing for a further 3 days (day 15 of culture), the cells were stimulated with 105 autologous PBMCs treated with mitomycin C (MMC; Kyowa Hakko, Tokyo, Japan) pulsed with Aur-A peptide. Thereafter, the cells were restimulated weekly by MMC-treated autologous PBMCs pulsed with Aur-A peptide. The Aur-A peptide–specific cytotoxic activity of growing cells was examined by standard 51Cr-release assay.
Cytotoxicity assays
The standard 51Cr-release assays were performed as described previously.31 Briefly, 10451Cr-labeled (Na251CrO4; New England Nuclear, Boston, MA) target cells and various numbers of effector cells in 200 μL RPMI 1640 medium supplemented with 10% FCS were seeded into 96-well round-bottom plates. The target cells were incubated with or without synthetic peptide for 2 hours before adding the effector cells. To assess the HLA class I restriction of cytotoxicity, target cells were incubated with an anti–HLA class I framework MoAb (w6/32; ATCC, Manassas, VA) or an anti–HLA-DR MoAb (L243; ATCC) at an optimal concentration (10 μg/mL) for 1 hour before adding the effector cells. Aur-A peptide specificity of cytotoxicity was examined by cold target inhibition assay as follows. 51Cr-labeled target cells (hot targets) were mixed with various numbers of 51Cr-unlabeled Aur-A peptide–loaded HLA-A*0201–positive LCLs or with 51Cr-unlabeled Aur-A peptide–loaded HLA-A*0201–negative LCLs (cold targets). After incubation with the effector cells for 5 hours, 100 μL supernatant was collected from each well. The percentage of specific lysis was calculated as: (experimental release cpm − spontaneous release cpm) / (maximal release cpm − spontaneous release cpm) × 100 (%).
Quantitative analysis of Aur-A mRNA expression
Total RNA was extracted from each sample with an RNeasy Mini Kit (QIAGEN, Hilden, Germany) in accordance with the manufacturer's instructions. Quantitative real-time polymerase chain reaction (QRT-PCR) of Aur-A mRNA (Hs00269212_m1) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA (4326317E) as an internal control was performed using the TaqMan Gene Expression assay (Applied Biosystems, Foster City, CA) in accordance with the manufacturer's instructions using an ABI Prism 7700 Sequence Detection System (Applied Biosystems). The expression level of Aur-A mRNA was corrected by reference to that of GAPDH mRNA, and the relative amount of Aur-A mRNA in each sample was calculated by the comparative ΔCt method.
Western blotting of Aur-A protein
Western blotting was performed as follows. Briefly, 106 cells were lysed in lysis buffer (25 mM HEPES, pH 7.5, 1% NP-40, 50 mM NaCl, 5 mM EDTA) with a protease inhibitor cocktail (Roche Diagnostics, Basel, Switzerland), and then incubated on ice, sonicated, frozen, and thawed. After centrifugation at 12 000g for 15 minutes at 4°C, the supernatant was collected as the lysate. After addition of sodium dodecyl sulfate (SDS) buffer, the total cell lysates were subjected to 10% SDS–polyacrylamide gel electrophoresis (PAGE), and blotted onto nitrocellulose membranes. The blots were reacted with anti–Aur-A mouse MoAb (Abcam, Cambridge, United Kingdom) followed by incubation with horseradish peroxidase–conjugated anti–mouse IgG antibody (GE Healthcare, Little Chalfont, United Kingdom). The probed proteins were visualized using an enhanced chemiluminescence system (GE Healthcare). The blotted membranes were also examined with anti–β-actin mouse MoAb (Sigma-Aldrich, St Louis, MO) to confirm that samples of equal volume had been loaded.
Detection of Aur-A207-215–specific CTL precursors in leukemia patients and healthy individuals by tetramer assays and enzyme-linked immunospot assays
HLA-A*0201/Aur-A207-215 peptide and HLA-A*2402/Aur-A207-215 peptide tetramers were produced as described previously.32,33 Briefly, recombinant HLA-A*0201 or HLA-A*2402 and the β2-microglobulin molecule were generated by the gene-transfer method. Expression of the HLA heavy chain was limited to the extracellular domain, and the C terminus of the domain was modified by addition of a substrate sequence for the biotinylating enzyme BirA. Monomeric HLA-peptide complexes were folded in vitro by adding the HLA protein to β2-microglobulin in the presence of Aur-A207-215 (YLILEYAPL), HIV-1 p17 Gag77-85 (SLYNTVATL), or HIV-1 Env584-592 (RYLRDQQLL) peptide. After gel purification, the HLA complex was biotinylated using recombinant BirA enzyme (Avidity, Denver, CO), and HLA-peptide tetramers were made by mixing the biotinylated HLA with PE-labeled streptavidin (Molecular Probes, Eugene, OR) at a molar ratio of 4:1.
PBMCs from HLA-A*0201– or HLA-A*2402–positive leukemia patients and healthy individuals were seeded in 24-well plates at 1.5 × 106 per well in the presence of the Aur-A207-215 peptide at a concentration of 10 μM in RPMI 1640 medium supplemented with 10% human AB serum and 10 U/mL IL-2. After culturing for 14 days, Aur-A207-215–specific CTL frequencies in cultured cells were examined by tetramer staining. Cultured PBMCs were stained with FITC-conjugated anti-CD8 MoAb and the tetramer at a concentration of 20 μg/mL at 4°C for 20 minutes. After washing twice, stained cells were analyzed using a FACSCalibur and Cell Quest Software.
Enzyme-linked immunospot (ELISPOT) assays were carried out as described previously.33 Briefly, 96-well flat-bottom MultiScreen-HA plates with a nitrocellulose base (Millipore, Bedford, MA) were coated with 10 μg/mL anti–interferon-γ (IFN-γ) MoAb (R&D Systems) and incubated overnight at 4°C. After being washed with PBS, the plates were blocked with the assay medium for 1 hour at 37°C. T2-A24 cells (5.0 × 104/well) were pulsed with Aur-A207-215 peptide at a concentration of 10 μM or with PBS alone, and incubated in RPMI 1640 medium supplemented with 10% FCS for 1 hour at 37°C. Then, the responder cells generated were seeded into each well to mix with the target peptide–loaded T2-A24 cells, and the plates were incubated in a 5% CO2 incubator at 37°C for 20 hours. After incubation, plates were washed vigorously with PBS containing 0.1% Tween 20. A polyclonal rabbit anti–IFN-γ antibody (Endgen, Woburn, MA) was added to each well and the plates was left for 90 minutes at room temperature, followed by exposure to peroxidase-conjugated goat anti–rabbit IgG (Zymed, San Francisco, CA) for an additional 90 minutes. To reveal IFN-γ–specific spots, 100 μL 0.1 M sodium acetate buffer (pH 5.0) containing 3-amino-9-ethylcarbazole (Sigma-Aldrich) and 0.015% H2O2 were added to each well. After 40 minutes, the color reaction was interrupted by washing with water, and the plates were dried. Diffuse large spots were counted under a dissecting microscope (Figure S1A, available on the Blood website; see the Supplemental Materials link at the top of the online article).
Results
Binding activities of Aur-A peptides for HLA-A*0201 and HLA-A*2402 molecules
The BIMAS-predicted binding scores and results of the binding assay for the HLA-A*0201 and HLA-A*2402 molecules with the 3 candidate Aur-A peptides and the positive and negative control peptides are summarized in Table 1. Among the 3 candidate Aur-A peptides, Aur-A207-215 showed high binding affinity for HLA-A*0201 in comparison with the others. Interestingly, Aur-A207-215 peptide appeared to be capable of binding to HLA-A*2402 as well as HLA-A*0201. These data suggest that Aur-A207-215 peptide can elicit Aur-A–specific CTLs.
Establishment of an Aur-A207-215 peptide–specific CTL line
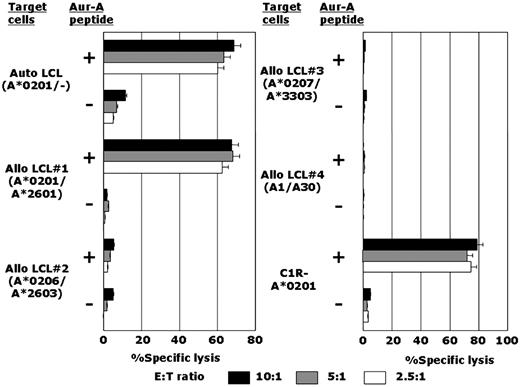
By repeated stimulation of CD8+ T lymphocytes with Aur-A peptide–loaded autologous DCs, as detailed in “Methods,” an Aur-A207-215 peptide–specific CTL line, designated AUR-1, was established from an HLA-A*0201–positive individual. It was possible to generate Aur-A207-215 peptide–specific CTLs from 2 other HLA-A*0201–positive individuals; however, long-term maintenance of these CTL lines was unsuccessful. Therefore, detailed studies of the functional characteristics of Aur-A–specific CTLs were performed using AUR-1. Establishment of Aur-A63-71–specific or Aur-A271-279–specific stable CTL lines was unsuccessful. As shown in Figure 1, AUR-1 showed strong cytotoxicity against Aur-A207-215 peptide–loaded autologous and allogeneic HLA-A*0201–positive LCLs but not Aur-A207-215 peptide–unloaded HLA-A*0201–positive LCLs. AUR-1 did not show any cytotoxicity against Aur-A207-215 peptide–loaded HLA-A*0201–negative allogeneic LCLs. Autologous LCLs loaded with other HLA-A*0201–binding peptides were not lysed by AUR-1 (data not shown). To confirm HLA-A*0201 restriction of Aur-A207-215 peptide–specific cytotoxicity mediated by AUR-1, cytotoxic activity against the HLA-A*0201 gene–transfectant cell line C1R-A*0201 was examined. AUR-1 was cytotoxic to C1R-A*0201 cells only in the presence of Aur-A207-215 peptide, and this cytotoxicity was significantly attenuated by anti–HLA class I MoAb but not by anti–HLA-DR MoAb (data not shown). These data indicate that Aur-A207-215–specific cytotoxicity of AUR-1 is restricted by HLA-A*0201.
Establishment of an HLA-A*0201–restricted and Aur-A207-215 peptide–specific CTL line, AUR-1. The cytotoxicity of the CTL line designated AUR-1 against various LCLs and HLA-A*0201 gene–transfected cells (C1R-A*0201), which were loaded or unloaded with Aur-A207-215 peptide, was determined by 4-hour 51Cr-release assays at effector-to-target (E:T) ratios of 10:1, 5:1, and 2.5:1.
Establishment of an HLA-A*0201–restricted and Aur-A207-215 peptide–specific CTL line, AUR-1. The cytotoxicity of the CTL line designated AUR-1 against various LCLs and HLA-A*0201 gene–transfected cells (C1R-A*0201), which were loaded or unloaded with Aur-A207-215 peptide, was determined by 4-hour 51Cr-release assays at effector-to-target (E:T) ratios of 10:1, 5:1, and 2.5:1.
Aur-A207-215–specific and HLA-A*0201–restricted lysis of Aur-A–expressing leukemia cell lines by AUR-1
Aur-A mRNA expression levels in leukemia cell lines and PBMCs of healthy people as a control were assessed by the QRT-PCR method. The amount of Aur-A mRNA in each cell line relative to that in the chronic myelogenous leukemia (CML) cell line K562 was calculated. Similarly, Aur-A protein expression levels in leukemia cell lines and normal PBMCs were examined by Western blotting. As shown in Figure 2A, Aur-A appeared to be expressed abundantly in all the leukemia cell lines examined, including acute myelogenous leukemia (AML) and CML cell lines. In contrast, expression of Aur-A in normal PBMCs was undetectable.
Expression of Aur-A in leukemia cell lines and the cytotoxicity of AUR-1 against leukemia cell lines. (A) Expression of Aur-A mRNA and protein in leukemia cell lines and normal PBMCs. Expression levels of Aur-A mRNA in the cells were determined by QRT-PCR as detailed in “Methods.” The level of Aur-A mRNA expression in the K562 leukemia cell line, which strongly expresses Aur-A, is shown as 1.0 and the expression levels in the cells were calculated relative to this value. Aur-A protein expression was examined by Western blotting using anti–Aur-A antibody and anti–β-actin antibody as the control. (B) Cytotoxicity of the Aur-A207-215–specific CTL line AUR-1 against leukemia cell lines. The cytotoxicity of AUR-1 to HLA-A*0201–positive and HLA-A*0201–negative leukemia cell lines was determined by 4-hour 51Cr-release assays at E/T ratios of 10:1, 5:1, and 2.5:1. (C) HLA class I restriction of cytotoxicity mediated by AUR-1 against leukemia cells. The cytotoxicity of AUR-1 against leukemia cell lines (GANMO-1 and CMK11-5) was determined by 4-hour 51Cr-release assays at an E/T ratio of 2.5:1 in the presence or absence of anti–HLA class I MoAb or anti–HLA-DR MoAb. (D) Cold target inhibition assays. 51Cr-labeled GANMO-1 cells (5 × 103 cells) were mixed with various numbers of 51Cr-unlabeled Aur-A207-215 peptide–loaded autologous LCL cells (○) or with 51Cr-unlabeled Aur-A207-215 peptide–loaded HLA-A*0201–negative allogeneic LCL cells (●). The cytotoxicity of AUR-1 to the mixture of 51Cr-labeled and unlabeled target cells was determined by 4-hour 51Cr-release assays at an effector-to-51Cr-labeled target cell ratio of 10:1.
Expression of Aur-A in leukemia cell lines and the cytotoxicity of AUR-1 against leukemia cell lines. (A) Expression of Aur-A mRNA and protein in leukemia cell lines and normal PBMCs. Expression levels of Aur-A mRNA in the cells were determined by QRT-PCR as detailed in “Methods.” The level of Aur-A mRNA expression in the K562 leukemia cell line, which strongly expresses Aur-A, is shown as 1.0 and the expression levels in the cells were calculated relative to this value. Aur-A protein expression was examined by Western blotting using anti–Aur-A antibody and anti–β-actin antibody as the control. (B) Cytotoxicity of the Aur-A207-215–specific CTL line AUR-1 against leukemia cell lines. The cytotoxicity of AUR-1 to HLA-A*0201–positive and HLA-A*0201–negative leukemia cell lines was determined by 4-hour 51Cr-release assays at E/T ratios of 10:1, 5:1, and 2.5:1. (C) HLA class I restriction of cytotoxicity mediated by AUR-1 against leukemia cells. The cytotoxicity of AUR-1 against leukemia cell lines (GANMO-1 and CMK11-5) was determined by 4-hour 51Cr-release assays at an E/T ratio of 2.5:1 in the presence or absence of anti–HLA class I MoAb or anti–HLA-DR MoAb. (D) Cold target inhibition assays. 51Cr-labeled GANMO-1 cells (5 × 103 cells) were mixed with various numbers of 51Cr-unlabeled Aur-A207-215 peptide–loaded autologous LCL cells (○) or with 51Cr-unlabeled Aur-A207-215 peptide–loaded HLA-A*0201–negative allogeneic LCL cells (●). The cytotoxicity of AUR-1 to the mixture of 51Cr-labeled and unlabeled target cells was determined by 4-hour 51Cr-release assays at an effector-to-51Cr-labeled target cell ratio of 10:1.
AUR-1 exerted cytotoxicity against the HLA-A*0201–positive leukemia cell lines GANMO-1 and CMK11-5, but not against the HLA-A*0201–negative cell lines MEG01, KAZZ, OUN-1, and K562 (Figure 2B). As shown in Figure 2C, cytotoxicity against leukemia cell lines mediated by AUR-1 was inhibited by addition of anti–HLA class I framework MoAb but not anti–HLA-DR MoAb. The cold target inhibition assay showed that the cytotoxicity of AUR-1 against the HLA-A*0201–positive leukemia cell line was significantly abrogated by adding 51Cr-unlabeled Aur-A207-215 peptide–loaded autologous LCL, but not HLA-A*0201–negative allogeneic LCL, indicating that the cytotoxicity of AUR-1 against leukemia cells is Aur-A specific (Figure 2D). These results show that AUR-1 can exert cytotoxicity against leukemia cell lines in an HLA-A*0201–restricted manner through recognition of the Aur-A207-215 epitope that is naturally processed from Aur-A protein in leukemia cells and presented on the cell surface in the context of HLA class I molecules.
Freshly isolated leukemia cells, but not normal PBMCs or normal mitotic cells, express Aur-A abundantly and are lysed by AUR-1
Next, we examined whether Aur-A–specific CTLs can discriminate freshly isolated leukemia cells from normal cells and whether AUR-1 can lyse freshly isolated leukemia cells as well as leukemia cell lines. As shown in Figure 3A, Aur-A appeared to be overexpressed in a wide spectrum of leukemia, including acute lymphoblastic leukemia (ALL), AML, and CML, as reported previously.17-19 Among the various kinds of leukemia, CML cells express a very high level of Aur-A mRNA. In contrast, expression levels of Aur-A mRNA in normal PBMCs and phytohemagglutinin (PHA)–stimulated peripheral blood T lymphocytes (normal mitotic cells) were extremely low in comparison with those in freshly isolated leukemia cells.
Expression of Aur-A in freshly isolated leukemia cells and the cytotoxicity of AUR-1 against freshly isolated leukemia cells. (A) Expression of Aur-A mRNA in freshly isolated leukemia cells, normal PBMCs, and PHA-stimulated T lymphocytes. Expression levels of Aur-A mRNA in freshly isolated leukemia cells and normal cells were determined using samples obtained from 7 patients with ALL, 17 patients with AML, 10 patients with CML in chronic phase, 4 healthy individuals, and PHA-stimulated T lymphoblasts obtained from 3 healthy individuals. To prepare PHA-stimulated T lymphoblasts, PBMCs were cultured in RPMI 1640 medium supplemented with 10% FCS and PHA at an appropriate concentration for 4 days. The level of Aur-A mRNA in normal PBMCs is shown as 1.0 and the expression levels in samples were calculated relative to this value. (B) Cytotoxicity of AUR-1 against freshly isolated leukemia cells and normal cells. The cytotoxicity of AUR-1 against HLA-A*0201–positive and HLA-A*0201–negative freshly isolated leukemia cells, HLA-A*0201–positive normal PBMCs, and HLA-A*0201–positive normal PHA-stimulated T lymphoblasts was determined by 51Cr-release assays at E:T ratios of 20:1, 10:1, and 5:1. Expression levels of Aur-A mRNA in samples are also shown.
Expression of Aur-A in freshly isolated leukemia cells and the cytotoxicity of AUR-1 against freshly isolated leukemia cells. (A) Expression of Aur-A mRNA in freshly isolated leukemia cells, normal PBMCs, and PHA-stimulated T lymphocytes. Expression levels of Aur-A mRNA in freshly isolated leukemia cells and normal cells were determined using samples obtained from 7 patients with ALL, 17 patients with AML, 10 patients with CML in chronic phase, 4 healthy individuals, and PHA-stimulated T lymphoblasts obtained from 3 healthy individuals. To prepare PHA-stimulated T lymphoblasts, PBMCs were cultured in RPMI 1640 medium supplemented with 10% FCS and PHA at an appropriate concentration for 4 days. The level of Aur-A mRNA in normal PBMCs is shown as 1.0 and the expression levels in samples were calculated relative to this value. (B) Cytotoxicity of AUR-1 against freshly isolated leukemia cells and normal cells. The cytotoxicity of AUR-1 against HLA-A*0201–positive and HLA-A*0201–negative freshly isolated leukemia cells, HLA-A*0201–positive normal PBMCs, and HLA-A*0201–positive normal PHA-stimulated T lymphoblasts was determined by 51Cr-release assays at E:T ratios of 20:1, 10:1, and 5:1. Expression levels of Aur-A mRNA in samples are also shown.
The cytotoxicity of AUR-1 against freshly isolated leukemia cells was examined by standard 51Cr-release assay. Because the frequency of HLA-A*0201 in the Japanese population is less than 10%, only 3 HLA-A*0201–positive leukemia samples were available. As expected, all HLA-A*0201–positive freshly isolated leukemia cells were lysed by AUR-1; however, HLA-A*0201–negative freshly isolated leukemia cells, HLA-A*0201–positive normal PBMCs, and HLA-A*0201–positive PHA lymphoblasts were resistant to AUR-1–mediated cytotoxicity (Figure 3B). Taken together, Aur-A–specific CTLs appeared to be capable of discriminating leukemia cells from normal cells in an HLA-restricted manner.
CD34+ leukemia progenitor cells, but not CD34+ normal hematopoietic progenitor cells, express Aur-A abundantly and are susceptible to AUR-1–mediated cytotoxicity
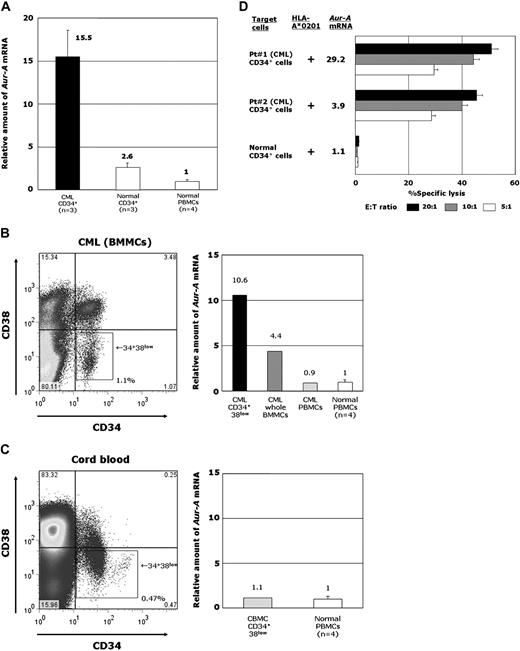
Because it is important to determine whether Aur-A–specific CTLs can specifically recognize and lyse leukemia progenitors, we further examined Aur-A expression and susceptibility to AUR-1–mediated cytotoxicity of CD34+ fractions in leukemia cells and normal hematopoietic cells. The CD34+ cells were sorted from BMMCs of patients with CML and CBMCs, and their Aur-A mRNA expression was examined by QRT-PCR. As shown in Figure 4A, the expression levels of Aur-A mRNA in CML CD34+ progenitor cells appeared to be significantly higher than in normal CD34+ hematopoietic progenitor cells. Because it is suggested that leukemia stem cells and normal hematopoietic stem cells are present in the CD34+CD38low fraction, we further examined Aur-A expression in CD34+CD38low cells of CML BMMCs and CBMCs. Consequently, it appeared that Aur-A mRNA was abundantly expressed in the CD34+CD38low fraction of CML (Figure 4B); however, the expression level of Aur-A mRNA in the CD34+CD38low fraction of normal hematopoietic progenitors was significantly low (Figure 4C).
Expression of Aur-A in CD34+CD38low fractions of CML cells and normal hematopoietic progenitor cells, and cytotoxicity of AUR-1 against CD34+ CML cells and CD34+ normal hematopoietic stem cells. (A) Expression levels of Aur-A mRNA in CD34+ cells isolated from BMMCs of patients with CML, CD34+ cells isolated from normal BMMCs and CBMCs, and normal PBMCs. Expression levels of Aur-A mRNA in leukemic CD34+ cells, normal hematopoietic stem cells, and normal PBMCs were determined using 3 samples of CML BMMCs, 1 sample of normal BMMCs, 2 samples of CBMCs, and 4 samples of normal PBMCs. The level of Aur-A mRNA in normal PBMCs is shown as 1.0 and the expression levels in samples were calculated relative to this value. (B) Representative data of Aur-A mRNA expression in the CD34+CD38low fraction of BMMCs, whole BMMCs, and PBMCs isolated from a patient with CML in chronic phase and PBMCs isolated from 4 healthy individuals. The CD34+CD38low cells were collected using a cell sorter. (C) Representative data of Aur-A mRNA expression in the CD34+CD38low fraction of CBMCs isolated from a normal donor and PBMCs isolated from 4 healthy individuals. The CD34+CD38low cells were collected using a cell sorter. (D) Cytotoxicity of AUR-1 against CD34+ leukemia progenitor cells and normal CD34+ hematopoietic progenitor cells. The cytotoxicity of AUR-1 against CD34+ leukemia cells isolated from 2 HLA-A*0201–positive patients with CML and normal CD34+ hematopoietic progenitor cells isolated from an HLA-A*0201–positive cord blood donor was determined by 51Cr-release assays at E/T ratios of 20:1, 10:1, and 5:1. Expression levels of Aur-A mRNA in samples are also shown.
Expression of Aur-A in CD34+CD38low fractions of CML cells and normal hematopoietic progenitor cells, and cytotoxicity of AUR-1 against CD34+ CML cells and CD34+ normal hematopoietic stem cells. (A) Expression levels of Aur-A mRNA in CD34+ cells isolated from BMMCs of patients with CML, CD34+ cells isolated from normal BMMCs and CBMCs, and normal PBMCs. Expression levels of Aur-A mRNA in leukemic CD34+ cells, normal hematopoietic stem cells, and normal PBMCs were determined using 3 samples of CML BMMCs, 1 sample of normal BMMCs, 2 samples of CBMCs, and 4 samples of normal PBMCs. The level of Aur-A mRNA in normal PBMCs is shown as 1.0 and the expression levels in samples were calculated relative to this value. (B) Representative data of Aur-A mRNA expression in the CD34+CD38low fraction of BMMCs, whole BMMCs, and PBMCs isolated from a patient with CML in chronic phase and PBMCs isolated from 4 healthy individuals. The CD34+CD38low cells were collected using a cell sorter. (C) Representative data of Aur-A mRNA expression in the CD34+CD38low fraction of CBMCs isolated from a normal donor and PBMCs isolated from 4 healthy individuals. The CD34+CD38low cells were collected using a cell sorter. (D) Cytotoxicity of AUR-1 against CD34+ leukemia progenitor cells and normal CD34+ hematopoietic progenitor cells. The cytotoxicity of AUR-1 against CD34+ leukemia cells isolated from 2 HLA-A*0201–positive patients with CML and normal CD34+ hematopoietic progenitor cells isolated from an HLA-A*0201–positive cord blood donor was determined by 51Cr-release assays at E/T ratios of 20:1, 10:1, and 5:1. Expression levels of Aur-A mRNA in samples are also shown.
Since a sufficient number of CD34+CD38low cells could not be obtained, whole CD34+ cells were used as target cells for cytotoxicity assays (Figure 4D). As expected, AUR-1 exerted strong cytotoxicity against CD34+ cells isolated from CML BMMCs of 2 patients. In contrast, AUR-1 did not show any cytotoxicity against normal CD34+ hematopoietic progenitor cells. These data strongly suggest that Aur-A–specific CTLs can discriminate leukemia progenitor cells from normal hematopoietic stem cells and selectively inhibit the growth of leukemia stem cells, and that immunotherapy targeting Aur-A is effective and safe.
Presence of Aur-A207-215–specific CTL precursors in peripheral blood of leukemia patients
When considering the feasibility of cellular immunotherapy for leukemia targeting Aur-A, it seems important to clarify whether Aur-A–specific CTL precursors are present in patients with leukemia. Aur-A207-215–specific CTL precursors in HLA-A*0201–positive patients with leukemia including AML in complete remission (CR) after allogeneic hematopoietic stem cell transplantation, ALL in CR after chemotherapy, and CML in the chronic phase before imatinib therapy, and 8 healthy subjects were analyzed by tetramer assay. Because Aur-A207-215 peptide can bind to HLA-A*2402 as well as HLA-A*0201, we also examined Aur-A207-215–specific CTL precursors in 2 HLA-A*2402–positive patients with CML in chronic phase after therapy with interferon or imatinib and 2 healthy individuals. Since we were unable to detect Aur-A–specific CTL precursors when freshly isolated lymphocytes were used for assays, PBMCs were stimulated with Aur-A207-215 peptide and then analyzed. Representative data of tetramer assays for HLA-A*0201–positive and HLA-A*2402–positive patients with leukemia are shown in Figure 5A. The frequencies of Aur-A207-215–specific CTL precursors in HLA-A*0201–positive and HLA-A*2402–positive patients with leukemia and healthy individuals are summarized in Figure 5B. Consequently, Aur-A207-215–specific CTL precursors were apparently detected in both HLA-A*0201–positive and HLA-A*2402–positive patients with leukemia. The frequency of Aur-A207-215–specific CTL precursors in leukemia patients appeared to be significantly higher than that in healthy individuals (0.25% ± 0.1% for leukemia patients, and 0.05% ± 0.03% for healthy individuals; P < .001). These data strongly suggest that Aur-A–specific CTL precursors are primed in patients with leukemia, and that vaccination with Aur-A peptide may efficiently induce an Aur-A–specific immune response in leukemia patients. To determine whether Aur-A207-215–specific CTL precursors detected by tetramer assays are indeed functional, we performed tetramer assays and ELISPOT assays using the same samples simultaneously, and determined the correlation between the 2 sets of data. PBMCs isolated from 7 HLA-A*0201– or HLA-A*2402–positive individuals were used for tetramer assays and ELISPOT assays. Consequently, the frequencies of Aur-A207-215 peptide–specific CTL precursors detected by these 2 different assay systems appeared to be closely correlated (r = 0.817; Figure S1B). These data strongly suggest that Aur-A tetramer–positive cells certainly have a functional response to stimulation with Aur-A.
Detection of Aur-A207-215–specific CTL precursors in patients with leukemia. (A) Representative data of the tetramer assay for Aur-A207-215–specific CTL precursors. PBMCs isolated from HLA-A*0201–positive and HLA-A*2402–positive patients with CML in chronic phase were stimulated with Aur-A207-215 peptide and then stained with HLA-A*0201/Aur-A207-215 tetramer and HLA-A*2402/Aur-A207-215 tetramer, respectively. HLA-A*0201/HIV-1 p17 Gag77-85 (SLYNTVATL) tetramer and HLA-A*2402/HIV-1 Env584-592 (RYLRDQQLL) tetramer were used as negative controls. (B) Summary of tetramer assays for Aur-A207-215–specific CTL precursors. PBMCs isolated from 3 HLA-A*0201–positive patients with leukemia (a patient with AML in complete remission after allogeneic stem cell transplantation, a patient with ALL in complete remission after chemotherapy, and a patient with untreated CML in chronic phase; ■), 2 HLA-A*2402–positive patients with leukemia (2 patients with CML in chronic phase after therapy with interferon or imatinib; ▴), 8 HLA-A*0201–positive healthy individuals (□), and 2 HLA-A*2402–positive healthy individuals (▵) were stained with HLA-A*0201/Aur-A207-215 or HLA-A*2402/Aur-A207-215 tetramer. The frequency of Aur-A207-215–specific CTL precursors in the patients with leukemia was significantly higher than that in healthy individuals (Student t test; P < .001).
Detection of Aur-A207-215–specific CTL precursors in patients with leukemia. (A) Representative data of the tetramer assay for Aur-A207-215–specific CTL precursors. PBMCs isolated from HLA-A*0201–positive and HLA-A*2402–positive patients with CML in chronic phase were stimulated with Aur-A207-215 peptide and then stained with HLA-A*0201/Aur-A207-215 tetramer and HLA-A*2402/Aur-A207-215 tetramer, respectively. HLA-A*0201/HIV-1 p17 Gag77-85 (SLYNTVATL) tetramer and HLA-A*2402/HIV-1 Env584-592 (RYLRDQQLL) tetramer were used as negative controls. (B) Summary of tetramer assays for Aur-A207-215–specific CTL precursors. PBMCs isolated from 3 HLA-A*0201–positive patients with leukemia (a patient with AML in complete remission after allogeneic stem cell transplantation, a patient with ALL in complete remission after chemotherapy, and a patient with untreated CML in chronic phase; ■), 2 HLA-A*2402–positive patients with leukemia (2 patients with CML in chronic phase after therapy with interferon or imatinib; ▴), 8 HLA-A*0201–positive healthy individuals (□), and 2 HLA-A*2402–positive healthy individuals (▵) were stained with HLA-A*0201/Aur-A207-215 or HLA-A*2402/Aur-A207-215 tetramer. The frequency of Aur-A207-215–specific CTL precursors in the patients with leukemia was significantly higher than that in healthy individuals (Student t test; P < .001).
Discussion
In the present study, we demonstrated that Aur-A is an ideal target antigen of cellular immunotherapy for leukemia, based on the following findings. First, Aur-A is broadly overexpressed in various types of leukemia but not in normal tissues except for testis, which is negative for HLA expression. Second, an Aur-A–derived peptide, Aur-A207-215, can bind to HLA-A*0201 and HLA*2402 molecules and elicit Aur-A–specific CTLs. Third, Aur-A is efficiently processed in leukemia cells, and leukemia cell lines and freshly isolated leukemia cells, but not normal cells, are lysed by Aur-A–specific CTLs in an HLA class I–restricted manner. Fourth, Aur-A–specific CTL precursors are certainly present in the peripheral blood of patients with leukemia.
One of the important characteristics of proteins that could be used as ideal tumor-associated antigens for cancer immunotherapy is an essential role in tumorigenesis and/or tumor progression. Aur-A is localized mainly at spindle poles and the mitotic spindle during mitosis, where it regulates the functions of centrosomes, spindles, and kinetochores required for proper mitotic progression. Recent studies have revealed that Aur-A is frequently overexpressed in various cancer cells, indicating its involvement in tumorigenesis.9-14 Overexpression of Aur-A contributes to genetic instability and tumorigenesis by disrupting the proper assembly of the mitotic checkpoint complex at the level of the Cdc20-BubR1 interaction.34 Its overexpression also causes resistance to apoptosis induced by taxol in human cancer cell lines.35,36 Moreover, Aur-A is a key regulatory component of the p53 pathway, as its overexpression leads to increased p53 degradation, thus facilitating oncogenic transformation.37 In addition, Aur-A expression in tumors is often associated with poor histologic differentiation and poor prognosis.12-14 These characteristics indicate that Aur-A is an ideal target antigen for cancer immunotherapy. Although Aur-A is also expressed in normal cells during mitosis, its expression level in normal tissue is quite low; therefore, normal mitotic cells are resistant to Aur-A–specific CTL-mediated cytotoxicity, as shown in the present study.
As reported previously,17-19 the present study demonstrated that Aur-A is overexpressed widely in various types of leukemia including AML, ALL, and CML. Among the leukemias, CML cells appeared to express a large amount of Aur-A. It was also found that Aur-A is abundantly expressed in the CD34+CD38low fraction of CML cells. Previous gene expression profiling analysis has also shown that mitogen-activated protein kinases, which activate mitotic kinases including Aurora kinases, are overexpressed in CD34+ progenitor cells in CML.38 In contrast to overexpression of Aur-A in the CD34+CD38low fraction of CML cells, the expression level of Aur-A in the CD34+CD38low fraction of normal hematopoietic progenitors appeared to be markedly lower than that in leukemic cells. We therefore addressed the question of whether Aur-A–specific CTLs can lyse leukemic progenitors. Because a sufficient number of CD34+CD38low cells could not be obtained, CD34+ cells were used as target cells. Consequently, in parallel with the expression levels of Aur-A, CD34+ CML cells but not CD34+ normal hematopoietic progenitor cells were efficiently lysed by Aur-A–specific CTLs. Although the detailed characteristics of leukemic stem cells are still obscure, they are considered to be present in the CD34+CD38low fraction.39-41 Taken together, targeting of Aur-A may be effective for eradicating leukemic stem cells.
Another interesting finding of this study was that Aur-A207-215 peptide is able to bind to HLA-A*2402 as well as to HLA-A*0201. Although AUR-1 could not recognize the complex of Aur-A207-215 peptide and HLA-A*0206 or HLA-A*0207, this peptide can bind to the HLA-A*0206 molecule (data not shown; written communication from Dr K. Udaka, Kochi University, Nangoku, Japan, August 5, 2007). Binding of a single peptide to both HLA-A*0201 and HLA-A*2402 has also been reported previously for a WT1-derived peptide (WT1235-243; CMTWNQMNL),30,42 which is now used as a cancer peptide vaccine. Since CTLs recognize a tumor-associated epitope in the context of HLA class I molecules, identification of a peptide that can bind to common HLA types is essential for development of a universal cancer peptide vaccine. Because HLA-A*2402 is the most common HLA type in the Japanese population, Aur-A207-215 is a promiscuous peptide and therefore likely useful for development of a cancer vaccine for Asian as well as white patients.
To date, 3 Aurora kinases, Aur-A, Aur-B, and Aur-C, have been identified in mammals. The Aurora kinases show different subcellular localization patterns and perform distinct tasks during cell division.43 These molecules show a similar domain organization: a N-terminal domain of 39-129 residues, a protein kinase domain, and a short C-terminal domain of 15-20 residues. The N-terminal domain of Aurora kinases shows low sequence conservation, and this determines selectivity during protein-protein interactions.44 In contrast, the catalytic domain is more highly conserved. Importantly, Aur-A207-215 is located in the catalytic domain and the conserved residues of Aur-A, Aur-B, and Aur-C (Figure S2). Interestingly, the Aur-B149-157, Aur-B151-159, and Aur-C83-91 peptides, which are derived from the catalytic domain of Aur-B and Aur-C, can bind to HLA-A*0201 and HLA*2402 molecules (Figure S3), suggesting that these residues could be a universal target epitope for cancer immunotherapy.
In summary, we have demonstrated for the first time that Aur-A is a potentially ideal target of cellular immunotherapy for leukemia. When considering the evidence that Aur-A is overexpressed widely in various kinds of cancer, Aur-A–targeting cancer immunotherapy may be universally applicable. On the basis of our present data, we are now planning a clinical trial of Aur-A peptide vaccination for cancer patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful for the skilled technical assistance of Ms Junko Mizumoto and Dr Kenji Kameda, Ehime University (Toon, Japan). We thank Dr A. John Barrett, NHLBI/NIH (Bethesda, MD), for providing the C1R-A*0201 cell line. We also thank Dr Hiroo Saji, HLA Laboratory, Japan, for HLA typing.
This work was supported in part by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan; a Grant-in-Aid for Cancer Research (no. 19-14) from the Ministry of Health, Labor and Welfare; and from the Uehara Memorial Foundation.
Authorship
Contribution: T.O. and H.F. designed and performed the research and wrote the paper; K.S., T.A., Y.Y., and T.H. discussed and interpreted the experimental results and provided clinical materials; K.K. made and supplied the tetramer; and M.Y. de-signed the research, wrote and edited the paper, and provided financial support.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Masaki Yasukawa, Department of Bioregulatory Medicine, Ehime University Graduate School of Medicine, Toon, Ehime 791-0295, Japan; e-mail: yasukawa@m.ehime-u.ac.jp.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal