B-cell lymphoma is the most common immune system malignancy. TCL1 transgenic mice (TCL1-tg), in which TCL1 is ectopically expressed in mature lymphocytes, develop multiple B- and T-cell leukemia and lymphoma subtypes, supporting an oncogenic role for TCL1 that probably involves AKT and MAPK-ERK signaling pathway augmentation. Additional, largely unknown genetic and epigenetic alterations cooperate with TCL1 during lymphoma progression. We examined DNA methylation patterns in TCL1-tg B-cell tumors to discover tumor-associated epigenetic changes, and identified hypermethylation of sprouty2 (Spry2). Sprouty proteins are context-dependent negative or positive regulators of MAPK-ERK pathway signaling, but their role(s) in B-cell physiology or pathology are unknown. Here we show that repression of Spry2 expression in TCL1-tg mouse and human B-cell lymphomas and cell lines is associated with dense DNA hypermethylation and was reversed by inhibition of DNA methylation. Spry2 expression was induced in normal splenic B cells by CD40/B-cell receptor costimulation and regulated a negative feedback loop that repressed MAPK-ERK signaling and decreased B-cell viability. Conversely, loss of Spry2 function hyperactivated MAPK-ERK signaling and caused increased B-cell proliferation. Combined, these results implicate epigenetic silencing of Spry2 expression in B lymphoma progression and suggest it as a companion lesion to ectopic TCL1 expression in enhancing MAPK-ERK pathway signaling.
Introduction
Hyperactivation of the RAS-RAF-MEK-ERK signaling pathway (MAPK-ERK pathway) promotes a variety of cancers, including hematologic malignancies, by enhancing cell proliferation and cell survival and by affecting differentiation.1,2 The MAPK-ERK pathway can be constitutively activated by tumor type–specific gain-of-function mutations in (or overexpression of) upstream pathway members, such as growth factor receptors, RAS, and RAF, and by loss-of-function mutations in (or repression of) negative pathway regulators, such as NF1, SPROUTY, or SPRED proteins.3
Ectopic expression of T-cell leukemia 1 (TCL1), a gene that encodes a 14-kDa protein that augments AKT pathway signaling in lymphocytes, is common in mature B-cell leukemias and lymphomas.4,–6 A causative role for ectopic TCL1 expression in lymphocyte transformation is supported by the observation that a spectrum of T- and B-cell tumors form in TCL1 transgenic (TCL1-tg) mice, in which the human TCL1 gene is ectopically expressed under the control of an Eμ-B29 promoter in mature lymphocytes.7 The mechanism(s) by which ectopic TCL1 expression promotes T and B lymphocyte transformation is not fully understood. In T cells, TCL1 augments AKT and MAPK-ERK signaling with each pathway independently contributing to increase T-cell proliferation in TCL1-tg mice.8 There is also evidence that the tumorigenic activity of TCL1 involves more than augmentation of AKT signaling because it has been shown that increasing AKT activity in B cells to levels higher than are detected in TCL1-tg B cells by deleting Pten, a negative regulator of AKT, fails to cause lymphocyte transformation.9 Moreover, it is not known what effect TCL1 has on MAPK-ERK signaling in B cells. The observation that in humans and TCL1-tg mice there is a long latency before B- and T-cell tumors form after ectopic expression of TCL1 is consistent with the conclusion that additional changes beyond TCL1-mediated augmentation of AKT signaling are needed to transform lymphocytes.
Because TCL1 expression is more frequently dysregulated in B-cell compared with T-cell tumors, we have focused on its role in B-cell malignancies. B-cell lymphomas from TCL1-tg mice display aneuploidy and chromosomal translocations. In many of these lymphomas, trisomy 15 and its associated c-MYC overexpression results in a lesion that histologically and molecularly resembles human Burkitt lymphoma (BL).10,11 In addition to genetic alterations, TCL1-tg B-cell tumors also display reproducible genome-wide patterns of aberrant DNA hypermethylation, as revealed by restriction landmark genome scanning (RLGS) of more than 2000 genetic loci.12,13 DNA hypermethylation promotes chromatin compaction and gene silencing, with consistent repression of specific tumor suppressor genes associated with multiple types of cancer, including leukemias and lymphomas.14,,–17 Our recent RLGS survey of TCL1-tg B-cell lymphomas detected aberrant DNA hypermethylation in many candidate tumor suppressor genes, including Epha7, Foxd3, STK39, and sprouty2 (Spry2), which is the subject of this study.12 Importantly, tumor-associated genetic or epigenetic alterations may or may not contribute to tumor causation or progression, which must be tested.
Spry2 is 1 of 4 mammalian homologs of the Drosophila sprouty gene,18 which encode 32- to 34-kDa proteins that share a highly conserved carboxyl-terminal cysteine-rich sprouty domain. They function as negative feedback regulators of signaling via multiple receptor tyrosine kinase (RTK) pathways, including the fibroblast growth factor and epidermal growth factor (EGF) receptor pathways. Sequence divergence between the amino-termini of the different sprouty family members is responsible for distinct protein-protein interactions, which are thought to result in functional differences between sprouty proteins.3,19 In the normal mouse embryo, the different sprouty genes are coexpressed in specific cell populations before mid-gestation, but at later stages their expression domains become more distinct.20 Typically, sprouty proteins antagonize growth factor–mediated cell proliferation, cell migration, and differentiation by modulating RTK signaling and suppressing the MAPK-ERK signaling pathway.3,19,21
Alternatively, sprouty can increase cell proliferation by enhancing MAPK-ERK signaling in naive T cells after T-cell receptor (TCR) engagement or after EGF receptor (EGFR) activation in multiple nonlymphoid cell types.19,22,,–25 Precisely how sprouty proteins modulate RTK signaling is not yet clear, and several mechanisms for pathway inhibition or activation have been proposed, including antagonism of MAPK-ERK pathway signaling at the level of RAS activation, RAF activation, or upstream of RAS by sequestering the adaptor, GRB2.3,19,21 SPRY2 also can augment EGF-dependent MAPK-ERK signaling by sequestering a negative pathway regulator, CBL (Cas-BR-M murine ectopic retroviral transforming sequence homolog), which ubiquitinates the EGFR, causing its degradation.23,24
As important repressors of RTK signaling, sprouty proteins are increasingly being recognized for their roles in opposing pro-growth signaling responses that could inhibit malignant transformation or progression.26 Sprouty genes are repressed or silenced in cancers of the prostate,27,28 lung,29 liver,30 and breast,31 where they are thought to function mainly as tumor suppressors by antagonizing the MAPK-ERK pathway. Consistent with this role, the expression of sprouty family members is induced through a negative feedback mechanism that reduces MAPK-ERK signaling and causes oncogene-induced senescence, providing a potential mechanism for opposing malignant transformation or progression.32 However, SPRY2 also has a recently discovered role in tumor progression by enhancing levels of EGFR and up-regulating EGF-mediated MAPK-ERK pathway signaling after sarcomatous transformation.33 This result highlights a potentially complex role for sprouty proteins in cancer from distinct activities as either tumor suppressors or tumor promoters.
As yet, there is little evidence that sprouty family members play a role in B-cell lymphomas, except for one recent study showing that SPRY2 repression worsens patient outcome and its expression inhibits tumor growth in diffuse large B-cell lymphoma (DLBCL).34 Here, we show that Spry2 expression is induced by CD40 and B-cell receptor (BCR) costimulation through an ERK-dependent negative feedback loop that dampens physiologic MAPK-ERK pathway signaling, including TCL1-augmented signaling. Spry2 expression is also repressed or silenced by dense DNA hypermethylation in mouse TCL1-tg and human B-cell lymphomas as well as mouse and human B lymphoma cell lines. Loss of Spry2 function increases ERK-dependent mature B-cell proliferation and leads to increased numbers of B cells in vivo, providing mechanistic insight and support for a tumor suppressor role for SPRY2 in B-cell lymphoma progression.
Methods
Biologic materials
TCL1-tg and Spry2-null mice have been described.7,35 TCL1-tg B-cell lymphomas were obtained from mouse spleens and classified with Mouse Models of Human Cancer Consortium criteria.36 Mice studies were performed with University of California–Los Angeles Animal Research Committee approval. Tumor cells comprised 74% to 95% of the total cells from affected TCL1-tg spleens by flow cytometry (data not shown). B cells were isolated to more than 95% purity from pooled spleens of 3- to 4-month-old C57BL/6 mice by AUTOMACS magnetic sorting (Miltenyi Biotec, Auburn, CA) using anti-phycoerythrin (PE) microbeads (Miltenyi Biotec) with anti–CD4-PE, anti–CD8-PE, anti–Gr1-PE antibodies (Abs; all from BD Biosciences PharMingen, San Diego, CA), as previously described.10 Human tissue was obtained from the University of California–Los Angeles Tissue Procurement Core Laboratory with Institutional Review Board approval. Human B cells were isolated to more than 95% purity from tonsils by Ficoll-Paque Premium (GE Healthcare, Little Chalfont, United Kingdom) separation and magnetic bead sorting with anti-PE microbeads and anti-CD19-PE Ab (BD Biosciences PharMingen). Nalm-6 human pre-B cells were provided by Dr D. Rawlings (University of Washington, Seattle, WA), 3-1 mouse pre-B cells were provided by Dr R. Wall (University of California–Los Angeles),37 and all other B-cell lines were obtained from ATCC (Manassas, VA). Mouse B-cell tumor lines were from pre-B (3-1) immature B (Wehi), and mature B-cell (A20, Bal117) stages of development. Human B-cell tumor lines were from pre-B (Nalm-6) and mature B-cell (2F7, Bjab, BL41, P3HR1, Raji, Ramos, and BCBL1) stages of development. Cell lines and primary B cells were maintained at 5% CO2 in RPMI 1640 with 10% fetal bovine serum, l-glutamine, and penicillin/streptomycin. Nalm-6 cells stably expressing SPRY2, CA-MEK1 (constitutively active MEK1, MEK1D218,D222), or an empty control vector were generated using spin transfection with retroviruses expressing MSCV-SPRY2-IRES-PURO, MSCV-CA-MEK1-IRES-PURO, or MSCV-IRES-PURO as previously described.38 CA-MEK1 was cloned from the Addgene plasmid 15 268, pBabe-Puro-MEK-DD.39 5-aza-2′-deoxycytidine (5-aza-dC) alone or in combination with trichostatin A (TSA) treatment of human and mouse B-cell lines was performed as previously described.12
Nucleic acid isolation and quantitative polymerase chain reaction
Genomic DNA (gDNA) and total RNA were isolated using Trizol reagent (Invitrogen, Carlsbad, CA). NotI RLGS DNA methylation profiles were generated as previously described.12 cDNA was reverse-transcribed from 2 μg RNA using random hexamers and the Superscript First Strand Synthesis System (Invitrogen). SYBR green real-time quantitative polymerase chain reaction (PCR) was performed with an Applied Biosystems 7700 sequencer (Foster City, CA) as described.12 Mouse and human samples were analyzed for 36b4 (Rplp0) or GAPDH expression, respectively, as normalization controls. Primer sequences are available on request.
Genomic bisulfite sequencing and methylation-specific PCR
Genomic bisulfite sequencing (GBS) was performed as described.12 In brief, sodium bisulfite–treated gDNA was PCR amplified (primers available on request), PCR products were cloned into the pCR2.1 TOPO vector (Invitrogen), and individual clones were sequenced. Methylation-specific PCR (MSP) was performed as previously described.12 Primer sets amplified a 229-bp or 221-bp product located at the same genomic region of the mouse Spry2 promoter (forward primers from −311 to −291 and reverse primers from −313 to −285) and a 209-bp product located at the same genomic region of the human SPRY2 promoter (forward primers from −425 to −401 and reverse primers from −241 to −217).
Western blot
Proteins were resolved by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, and incubated overnight with anti-SPRY1, anti-SPRY2, anti-TCL1, anti-P44/42 MAPK (9102; Cell Signaling Technology, Danvers, MA), anti–phospho-P44/p42 MAPK (9101; Cell Signaling Technology), or anti-ACTIN (A2066; Sigma-Aldrich, St Louis, MO) Abs, followed by a 1-hour incubation with horseradish peroxidase–linked secondary Ab (Jackson ImmunoResearch Laboratories, West Grove, PA).
Induction of SPRY2 protein expression
Isolated mouse splenic B cells were stimulated with 1.5 μg/mL anti-CD40 Ab (553722; BD Biosciences PharMingen) for 24 hours, followed by 10 μg/mL anti-IgM Ab (115-006-020; Jackson ImmunoResearch Laboratories) for up to 24 hours as indicated. Nalm-6 cells were serum-starved overnight and then stimulated with 10 μg/mL anti-IgM Ab (109-005-129; Jackson ImmunoResearch Laboratories) for the indicated times. Nalm-6 and mouse B cells were incubated for 30 minutes with either 10 μg/mL of the MEK1/2 inhibitor U0126 (9903; Cell Signaling Technology) or dimethyl sulfoxide carrier just before stimulation.
Flow cytometry
Spleen cells from Spry2-null and wild-type (WT) mice at 7 to 8 weeks of age (sex-matched littermates) were stained with anti–B220-PE (RA3-6B2; BD Biosciences PharMingen), anti-CD69–fluorescein isothiocyanate (FITC; HI.2F3; BD Biosciences PharMingen), and anti-CD25–APC (PC61; eBioscience, San Diego, CA) Abs before and after stimulation with anti-CD40 and anti-IgM Abs, as described in “Induction of SPRY2 protein expression,” followed by flow cytometry. Apoptosis was determined with annexin V–FITC and propidium iodide staining and flow cytometry. Cells were analyzed on FACSCalibur or LSR (BD Biosciences, San Jose, CA) using FlowJo software (TreeStar, Ashland, OR).
Cell proliferation and viability assays
Proliferation of B cells isolated from Spry2-null and WT, sex-matched littermate spleens was assessed in triplicate assays of 2 × 105 cells/well using anti-CD40 plus anti-IgM stimulation, with or without U0126, as described in “Induction of SPRY2 protein expression.” After 44 hours, cells were pulsed with 0.5 μCi [3H]-thymidine for 16 hours and then harvested onto glass fiber filters, and counts per minute were determined. Viability of Nalm-6 cells seeded in triplicate at 104 cells/well in 96-well plates was assessed by MTT assays as described by the manufacturer (ATCC). In brief, the tetrazolium MTT reagent (3-[4, 5-dimethylthiazolyl-2]-2, 5-diphenyltetrazolium bromide) was added to each well for 2 hours at the indicated time, and then each sample was lysed overnight using the MTT detergent. Viability was determined by measuring the absorbance at 570 nm for each sample.
Statistical analysis
Statistical significance was calculated with the 2-tailed Student t test, and all P values less than .05 were considered significant.
Results
DNA hypermethylation and repression of Spry2 expression in TCL1-tg B-cell lymphomas
RLGS profiles of genome-wide DNA methylation patterns were generated previously from 11 TCL1-tg B-cell tumors, 6 pooled premalignant TCL1-tg spleens, and 3 WT spleens. Tumors with more than 50% loss of intensity at specific spots were scored as methylated for that locus, whereas tumors with less than 50% loss of spot intensity were scored as unmethylated/partially methylated, providing a more stringent scoring criteria than established.14,40 RLGS profile comparisons revealed spot loss; therefore, advanced DNA methylation, of a methylation-sensitive Not1 site within the Spry2 promoter in 3 of 6 Burkitt-like lymphoma (BLL1-6, formerly called LBL1-6), 1 of 3 diffuse large B-cell lymphoma (DLBCL1-3), and 1 of 2 marginal zone lymphoma (MZL1,2) samples (Figure 1A).12 One tumor, BLL1, scored as unmethylated/partially methylated for Spry2, whereas 2 progeny tumors, BLL1a and BLL1b, established by transfer of BLL1 into 2 different individual syngeneic mice, scored as methylated for Spry2. In general, the overall methylation pattern at a given locus was stable, with the majority (estimated at > 99%) of 1741 RLGS loci scoring the same between parent and progeny tumors. One explanation for the difference in methylation at the Spry2 locus between parent and progeny is that there was a small subpopulation of Spry2 methylated cells in the parent tumor that was below the level of RLGS detection, and it was those few cells from which the progeny tumors developed. Alternatively, de novo Spry2 methylation in a progeny TCL1-tg BLL may have occurred over time.
Spry2 is DNA-hypermethylated, and its expression is repressed in TCL1-tg B-cell lymphomas. (A) Spry2 RLGS locus (arrow) is shown for WT mouse spleen and 3 TCL1-tg B- cell tumors. Comparisons to the WT spot intensity scored BLL3 as unmethylated/partially methylated (> 50% of WT) and BLL1a and MZL1 as DNA hypermethylated (< 50% of WT) for the surveyed NotI site.14,40 (B) GBS of a CpG-rich region located between −728 and −484 in the mouse Spry2 promoter. Each row represents the sequence of an individual clone of WT spleen, MZL2, or BLL1a cells. ○, represents unmethylated CpG sites; ●, methylated sites; squares, 2 CpG loci within the Not1 site. (C) Spry2 expression in WT or TCL1-tg B-cell lymphomas was determined by quantitative PCR. Average Spry2 expression for each group (WT, hypermethylated, and unmethylated/partially methylated lymphomas) is shown by a dashed line. Spry2 expression was normalized to 36b4 (Rplp0) expression. DNA methylation status for WT or TCL1-tg B-cell lymphomas was also assessed by MSP. The top and bottom bands correspond to PCR products amplified by primers specific for methylated (M) or unmethylated (U) CpG sites after bisulfite conversion.
Spry2 is DNA-hypermethylated, and its expression is repressed in TCL1-tg B-cell lymphomas. (A) Spry2 RLGS locus (arrow) is shown for WT mouse spleen and 3 TCL1-tg B- cell tumors. Comparisons to the WT spot intensity scored BLL3 as unmethylated/partially methylated (> 50% of WT) and BLL1a and MZL1 as DNA hypermethylated (< 50% of WT) for the surveyed NotI site.14,40 (B) GBS of a CpG-rich region located between −728 and −484 in the mouse Spry2 promoter. Each row represents the sequence of an individual clone of WT spleen, MZL2, or BLL1a cells. ○, represents unmethylated CpG sites; ●, methylated sites; squares, 2 CpG loci within the Not1 site. (C) Spry2 expression in WT or TCL1-tg B-cell lymphomas was determined by quantitative PCR. Average Spry2 expression for each group (WT, hypermethylated, and unmethylated/partially methylated lymphomas) is shown by a dashed line. Spry2 expression was normalized to 36b4 (Rplp0) expression. DNA methylation status for WT or TCL1-tg B-cell lymphomas was also assessed by MSP. The top and bottom bands correspond to PCR products amplified by primers specific for methylated (M) or unmethylated (U) CpG sites after bisulfite conversion.
Here, the methylation status of Spry2 promoter in tumors with available gDNA was examined in greater detail. Consistent with what was previously reported, more than 50% spot loss at the Spry2 RLGS locus (ie, dense hypermethylation) was detected in tumors BLL1a, BLL2, and MZL1, compared with no or partial methylation in BLL3, MZL2, DLBCL1, or DLBCL2. Likewise, no spot loss was observed in nonmalignant TCL1-tg or WT splenic RLGS profiles (Figure 1A). GBS of a 244-bp cytosine-guanine dinucleotide (CpG)–rich region that contained 35 CpG sites within the Spry2 promoter was performed to assess the extent of DNA methylation and validate the RLGS data (Figure 1B). Results showed 2.2 CpG sites per clone (6%) were cytosine-methylated in WT spleen cells, 2.3 CpG sites per clone (7%) were methylated in nonmalignant TCL1-tg spleen cells (data not shown), and 6.3 CpG sites (18%) were methylated in MZL2, scored as an unmethylated/partially methylated TCL1-tg tumor by RLGS. In contrast, an average of 16.6 CpG sites per Spry2 promoter clone (47%) were DNA methylated in BLL1a, including at least 1 of 2 tandem CpG dinucleotides present in the surveyed Not1 site that was detected by RLGS in 7 of 10 sequenced GBS clones (Figure 1B). MSP also confirmed the DNA methylation detected by RLGS (Figure 1C) in these tumors and in an additional 9 TCL1-tg B-cell lymphomas (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). The data indicate that progressive DNA methylation of the Spry2 promoter occurs from essentially no methylation in WT spleen cells to partial or dense methylation in TCL1-tg B-cell lymphomas.
To determine whether the extent of DNA methylation of the Spry2 promoter was associated with gene silencing, steady-state Spry2 expression was analyzed by quantitative PCR for each sample. Compared with the average Spry2 expression in nonmalignant spleen cells, tumors with Spry2 DNA methylation showed an average 8.3-fold reduction in Spry2 expression, whereas tumors with no or partial methylation showed an average 2.9-fold increase in Spry2 expression, which may be a suppressive response to increased MAPK-ERK signaling (Figures 1C, S1).
Although TCL1-tg B-cell tumors were more than 74% B cells by flow cytometry, heterologous cell types including T cells were present, which could express Spry222 and affect interpretations of GBS and gene expression data. To further validate the primary sample results, the promoter methylation status and expression of Spry2 in mouse B-cell tumor cell lines were determined (Figure 2A,B). GBS of the Spry2 promoter showed very dense DNA methylation, with an average of 94% of CpG sites methylated per clone in Wehi, Bal 117, and A20 cells, including almost complete methylation of the 2 CpG sites within the RLGS-surveyed Not1 site. In contrast, the 3-1 pre–B-cell line was essentially devoid of methylation with only 0.3% of CpG sites methylated (Figure 2A). MSP validated these GBS data (Figure 2B). A comparison by quantitative PCR showed that Spry2 expression was more than 1000-fold higher in 3-1 cells than in Wehi cells, which have characteristics of immature B cells, or in Bal 117 and A20 cells, which have characteristics of mature B cells (Figure 2B). Thus, there was a perfect correlation between the methylation status of sequences in the Spry2 promoter region and Spry2 expression. Further supporting epigenetic silencing of Spry2, treatment of Wehi, Bal 117, and A20 cell lines with the DNA methyltransferase inhibitor 5-aza-dC alone or in combination with the histone deacetylase inhibitor TSA reactivated Spry2 expression in a dose-dependent manner (Figure 2C). In contrast, treatment of unmethylated 3-1 cells with 5-aza-dC alone or in combination with TSA did not augment Spry2 expression. Collectively, these results demonstrate repression of Spry2 that depended on the extent of DNA methylation in both murine lymphoma cell lines and B-cell tumors of a TCL1-tg mouse model of human lymphoma.
Spry2 is DNA hypermethylated, and its expression is repressed in mouse B-cell lymphoma lines but can be reactivated by treatment with a demethylating agent. (A) GBS of a CpG-rich region located between −728 and −484 in the Spry2 promoter in mouse B-cell lines 3-1, Wehi, Bal 117, and A20. Each row represents the sequence of an individual clone. ○ represents unmethylated CpG sites; ●, methylated sites; and squares, 2 CpG loci within the Not1 site. (B) Spry2 expression was assayed by quantitative PCR in 3-1, Wehi, Bal 117, and A20 mouse B-cell lines. N.D. indicates not detected. Spry2 expression is normalized to 36b4 (Rplp0) expression. DNA methylation status for each tumor cell line was also assessed by MSP. The top and bottom bands correspond to PCR products amplified by primers specific for methylated (M) or unmethylated (U) CpG sites after bisulfite conversion. (C) Spry2 expression assayed in triplicate without and with 5-aza-dC and/or 100 nm TSA treatment.
Spry2 is DNA hypermethylated, and its expression is repressed in mouse B-cell lymphoma lines but can be reactivated by treatment with a demethylating agent. (A) GBS of a CpG-rich region located between −728 and −484 in the Spry2 promoter in mouse B-cell lines 3-1, Wehi, Bal 117, and A20. Each row represents the sequence of an individual clone. ○ represents unmethylated CpG sites; ●, methylated sites; and squares, 2 CpG loci within the Not1 site. (B) Spry2 expression was assayed by quantitative PCR in 3-1, Wehi, Bal 117, and A20 mouse B-cell lines. N.D. indicates not detected. Spry2 expression is normalized to 36b4 (Rplp0) expression. DNA methylation status for each tumor cell line was also assessed by MSP. The top and bottom bands correspond to PCR products amplified by primers specific for methylated (M) or unmethylated (U) CpG sites after bisulfite conversion. (C) Spry2 expression assayed in triplicate without and with 5-aza-dC and/or 100 nm TSA treatment.
DNA hypermethylation and repression of SPRY2 expression in human B-cell lymphomas
To assess the potential relevance for SPRY2 methylation in human B-cell lymphomas, MSP of patient samples showed that 5 of 7 DLBCLs and 1 of 1 BL contained DNA methylation of the SPRY2 promoter (Figure 3A). SPRY2 promoter methylation was associated with repressed average SPRY2 expression by quantitative PCR in 4 of 6 lymphoma samples (Figure 3A). Interestingly, 2 unmethylated/partially methylated samples showed increased SPRY2 expression compared with sorted B cells, again suggesting a suppressive response to increased MAPK-ERK signaling (Figures 1C, S1). GBS of a 188-bp CpG-rich region that contained 34 CpG sites within the SPRY2 promoter was performed on 2 methylated and 1 unmethylated/partially methylated patient tumors to assess the extent of DNA methylation and validate the MSP data. Results showed less than 1 CpG site methylated per clone in unmethylated/partially methylated DLBCL7, 29.4 CpG sites per clone (86%) methylated in DLBCL5, and 16.5 CpG sites (48%) methylated in BL1, validating the MSP results (Figure 3B). Interestingly, almost all of the unmethylated CpG sites in DLBCL5 and BL1 were contained in completely unmethylated clones. This indicates DNA methylation heterogeneity at the SPRY2 locus that may originate from the tumor cells themselves or from heterogeneous elements in the tumor microenvironment, such as reacting lymphocytes, endothelial cells, macrophages, and fibroblasts.
DNA methylation of the SPRY2 promoter is associated with reduced SPRY2 expression in patient lymphoma samples. (A) SPRY2 expression normalized to GAPDH expression was assessed by quantitative PCR in 7 DLBCLs and 1 BL, and the DNA methylation status for each tumor was assessed by MSP. The top and bottom bands correspond to PCR products amplified by primers specific for methylated (M) or unmethylated (U) CpG sites after bisulfite conversion. Estimated percentage of tumor cells in each sample is shown in parentheses. (B) GBS of a CpG-rich region located between −163 and +25 in the SPRY2 promoter in DLBCL5, DLBCL7, and BL1 patient samples. Each row represents the sequence of an individual clone. ○ represents unmethylated CpG sites; ●, methylated CpG sites.
DNA methylation of the SPRY2 promoter is associated with reduced SPRY2 expression in patient lymphoma samples. (A) SPRY2 expression normalized to GAPDH expression was assessed by quantitative PCR in 7 DLBCLs and 1 BL, and the DNA methylation status for each tumor was assessed by MSP. The top and bottom bands correspond to PCR products amplified by primers specific for methylated (M) or unmethylated (U) CpG sites after bisulfite conversion. Estimated percentage of tumor cells in each sample is shown in parentheses. (B) GBS of a CpG-rich region located between −163 and +25 in the SPRY2 promoter in DLBCL5, DLBCL7, and BL1 patient samples. Each row represents the sequence of an individual clone. ○ represents unmethylated CpG sites; ●, methylated CpG sites.
The DNA methylation and expression status of human SPRY2 was determined in sorted tonsil B cells, Epstein-Barr virus (EBV)–immortalized peripheral B lymphocytes (PBLs), and B-cell tumor lines (Figure 4A). MSP demonstrated SPRY2 promoter DNA methylation in 2F7, P3HR1, BL41, Ramos, Bjab, and BCBL1 mature B-cell tumor lines and unmethylated/partially methylated SPRY2 promoters in sorted tonsil B cells, PBLs, the Nalm-6 pre–B-cell tumor line, and the Raji mature B-cell tumor line. Consistent with results from the mouse B-cell tumor lines (Figure 2A,B), SPRY2 promoter methylation was associated with more than 100-fold repressed SPRY2 expression by quantitative PCR analysis compared with SPRY2 expression in mature B cells sorted from tonsil (Figure 4A).
SPRY2 is DNA hypermethylated, and its expression is repressed but can be reactivated by treatment with a demethylating agent in human B-cell lymphoma lines. (A) SPRY2 expression normalized to GAPDH expression was assessed by quantitative PCR in human tonsil B cells (B-cell sort), EBV-immortalized PBL, and a panel of human B-cell lymphoma lines. N.D. indicates not detected. DNA methylation status for sorted B cells, PBLs, and each tumor cell line was assessed by MSP. The top and bottom bands correspond to PCR products amplified by primers specific for methylated (M) or unmethylated (U) CpG sites after bisulfite conversion. (B) GBS of a CpG-rich region located between −163 and +25 in the SPRY2 promoter in human tonsil B cells (B-cell sort) and human B-cell lines Nalm-6, 2F7, P3HR1, and BL41. Each row represents the sequence of an individual clone. ○ represents unmethylated CpG sites; ●, methylated CpG sites. (C) SPRY2 expression normalized to GAPDH expression was assayed in triplicate by quantitative PCR without and with 5-aza-dC and/or 100 nm TSA treatment.
SPRY2 is DNA hypermethylated, and its expression is repressed but can be reactivated by treatment with a demethylating agent in human B-cell lymphoma lines. (A) SPRY2 expression normalized to GAPDH expression was assessed by quantitative PCR in human tonsil B cells (B-cell sort), EBV-immortalized PBL, and a panel of human B-cell lymphoma lines. N.D. indicates not detected. DNA methylation status for sorted B cells, PBLs, and each tumor cell line was assessed by MSP. The top and bottom bands correspond to PCR products amplified by primers specific for methylated (M) or unmethylated (U) CpG sites after bisulfite conversion. (B) GBS of a CpG-rich region located between −163 and +25 in the SPRY2 promoter in human tonsil B cells (B-cell sort) and human B-cell lines Nalm-6, 2F7, P3HR1, and BL41. Each row represents the sequence of an individual clone. ○ represents unmethylated CpG sites; ●, methylated CpG sites. (C) SPRY2 expression normalized to GAPDH expression was assayed in triplicate by quantitative PCR without and with 5-aza-dC and/or 100 nm TSA treatment.
GBS of the SPRY2 promoter was performed on sorted tonsil B cells and 4 tumor cell lines to confirm and quantify the MSP results (Figure 4B). Methylation of approximately 90% of CpG sites within an 188-bp CpG-rich region of the proximal SPRY2 promoter was detected in all 3 BL cell lines (2F7, P3HR1, and BL41), whereas less than 1% of the CpG sites were methylated in the SPRY2 promoter of Nalm-6 cells, and no methylated CpG sites were detected in sorted mature B cells. SPRY2 expression was induced in a dose-dependent manner in 2F7, P3HR1, and BL41 BL cells with 5-aza-dC with or without TSA treatment (Figure 4C), further supporting a role for epigenetic control of SPRY2 expression.
A direct role for dense DNA hypermethylation in repressing SPRY2 expression was examined using increasing-length SPRY2 promoter-luciferase reporter constructs41 that were unmethylated or hypermethylated in vitro at CG dinucleotides with SssI methyltransferase and transfected into human embryonic kidney (HEK) 293T cells (Figure S2A). Dense CpG methylation of all 3 reporter constructs resulted in almost complete suppression of SPRY2 promoter activity, with luciferase activity reduced 50- to 200-fold compared with unmethylated reporter constructs (Figure S2B). Combined, these data strongly link dense DNA methylation at the SPRY2 promoter with repressed SPRY2 expression in mouse and human mature B-cell tumor lines and primary B-cell lymphomas.
Induction of Spry2 expression by CD40 and B-cell receptor costimulation is ERK1/2-dependent
Sprouty gene expression is induced by RTK signaling as part of a feedback control mechanism to modulate signaling of the ERK pathway.13,29 However, induction of the expression of a specific sprouty family member is context-specific.3 For example, in CD4+ T cells, Spry1 is the family member that is induced by TCR stimulation and regulates a negative feedback loop that suppresses ERK activity.22 As yet, it is not known which, if any, family member responds to B cell–specific signaling. However, given that SPRY2 is expressed in human B cells (Figures 3A, 4A) and is repressed in mouse and human B-cell tumors (Figures 1,Figure 2,Figure 3–4, S2), SPRY2 seemed a logical choice to examine as a potential regulator of B-cell ERK pathway signaling. The combination of CD40 and BCR signaling stimulates B-cell proliferation and survival, resulting in a B-cell expansion that forms a blast population of cells with germinal center (GC) B-cell features.42,43 Therefore, SPRY2 protein expression was determined by Western blot analysis in mouse splenic B cells stimulated ex vivo with anti-CD40 Ab for 24 hours followed by BCR costimulation (Figure 5A). SPRY2 protein expression began to increase at approximately 10 to 12 hours of CD40/BCR costimulation, with up to a 3.4-fold increase by 24 hours, as determined by densitometry. SPRY1 protein expression was also evaluated because it is induced in T cells after TCR stimulation; however, SPRY1 protein was not detected with or without CD40/BCR costimulation. Thus, SPRY1 is not functionally redundant with SPRY2 in B cells (Figure 5B).
Induction of SPRY2 protein expression by CD40/BCR costimulation in B cells is ERK1/2 and TCL1 dependent. (A) SPRY2 protein expression in sorted WT B cells (pooled from 3 mice) stimulated with anti-CD40 Ab for 24 hours followed by costimulation with anti-IgM Ab for the indicated times. Freshly isolated splenic B cells before CD40 or BCR stimulations are shown in lane 1. The number beneath each lane shows the SPRY2 protein level normalized to the ACTIN level, as determined by densitometry. (B) SPRY1 and SPRY2 protein expression in WT B cells. Freshly isolated splenic B cells (pooled from 2 mice) before (−) or after stimulation (+) with anti-CD40 for 24 hours, followed by anti-IgM for 24 hours. Human embryonic kidney (HEK) 293T cells overexpressing SPRY1 or SPRY2 were used as positive controls (data not shown). (C) SPRY2 protein expression in WT B cells pooled from 2 mice costimulated as in panel B without (−) or with (+) U0126, a MEK1/2 inhibitor. (D) CD19-sorted human tonsil B cells were stimulated for the indicated times with phorbol 12-myristate 13-acetate and ionomycin, followed by Western blot. (E) Nalm-6 cells were infected with a retrovirus expressing a constitutively active form of MEK1 (CA-MEK1) or an empty vector control, followed by stable cell line establishment and Western blot. (F) ERK1/2 phosphorylation time course in WT (wt) and nonmalignant TCL1-tg (tg) splenic B cells costimulated by CD40/BCR without (−) or with (+) U0126. (G) SPRY2 protein expression in nonmalignant TCL1-tg (tg) or WT (wt) splenic B cells costimulated for 18 hours as in panel A. B cells were pooled from 2 TCL1-tg or 2 WT mouse spleens. The number beneath each lane shows the SPRY2 protein level normalized to the ACTIN level, as determined by densitometry. The mean and SD from this and a repeat experiment are plotted (right), normalized to the expression level for WT uninduced B cells. Data are representative of 2 (C-E), 3 (A,B,F), or 4 (G) independent experiments.
Induction of SPRY2 protein expression by CD40/BCR costimulation in B cells is ERK1/2 and TCL1 dependent. (A) SPRY2 protein expression in sorted WT B cells (pooled from 3 mice) stimulated with anti-CD40 Ab for 24 hours followed by costimulation with anti-IgM Ab for the indicated times. Freshly isolated splenic B cells before CD40 or BCR stimulations are shown in lane 1. The number beneath each lane shows the SPRY2 protein level normalized to the ACTIN level, as determined by densitometry. (B) SPRY1 and SPRY2 protein expression in WT B cells. Freshly isolated splenic B cells (pooled from 2 mice) before (−) or after stimulation (+) with anti-CD40 for 24 hours, followed by anti-IgM for 24 hours. Human embryonic kidney (HEK) 293T cells overexpressing SPRY1 or SPRY2 were used as positive controls (data not shown). (C) SPRY2 protein expression in WT B cells pooled from 2 mice costimulated as in panel B without (−) or with (+) U0126, a MEK1/2 inhibitor. (D) CD19-sorted human tonsil B cells were stimulated for the indicated times with phorbol 12-myristate 13-acetate and ionomycin, followed by Western blot. (E) Nalm-6 cells were infected with a retrovirus expressing a constitutively active form of MEK1 (CA-MEK1) or an empty vector control, followed by stable cell line establishment and Western blot. (F) ERK1/2 phosphorylation time course in WT (wt) and nonmalignant TCL1-tg (tg) splenic B cells costimulated by CD40/BCR without (−) or with (+) U0126. (G) SPRY2 protein expression in nonmalignant TCL1-tg (tg) or WT (wt) splenic B cells costimulated for 18 hours as in panel A. B cells were pooled from 2 TCL1-tg or 2 WT mouse spleens. The number beneath each lane shows the SPRY2 protein level normalized to the ACTIN level, as determined by densitometry. The mean and SD from this and a repeat experiment are plotted (right), normalized to the expression level for WT uninduced B cells. Data are representative of 2 (C-E), 3 (A,B,F), or 4 (G) independent experiments.
FGF2-mediated induction of SPRY2 expression in fibroblasts requires ERK1/2 activation.44 To determine whether induction of SPRY2 protein expression in CD40/BCR costimulated B cells likewise requires ERK1/2 activity, mouse splenic B cells were preincubated with the MEK1/2-specific inhibitor, U0126, followed by CD40/BCR costimulation. U0126 blocked CD40/BCR-mediated induction of SPRY2 (Figure 5C). Furthermore, costimulation with phorbol 12-myristate 13-acetate plus ionomycin, which is known to robustly activate ERK1/2,45 induced SPRY2 expression in CD19-sorted tonsil B cells (Figure 5D). Overexpression of a constitutively active MEK1 (CA-MEK1 or MEK1D218,D222), which activates the MAPK-ERK1/2 pathway,46 also induced the robust expression of SPRY2 in Nalm-6 cells (Figure 5E). The combined blocking, activation, and overexpression data indicate that MAPK-ERK pathway activation is required to induce SPRY2 expression in B cells.
Because ERK1/2 signaling activity is increased by TCL1 in mature peripheral T cells,8 the effect on ERK1/2 activity of ectopic TCL1 expression that occurs in TCL1-tg B cells was determined. Nonmalignant splenic B cells from TCL1-tg mice showed increased ERK1/2 phosphorylation compared with WT control spleen B cells after CD40/BCR costimulation, and this activity was completely blocked by preincubation with the MEK1/2 inhibitor, U0126 (Figure 5F). Moreover, SPRY2 protein expression was determined to be 2-fold increased in nonmalignant TCL1-tg mouse B cells than in WT B cells after CD40/BCR costimulation (Figure 5G), suggesting that TCL1 augmentation of the ERK pathway enhances SPRY2 protein expression. Consistent with this conclusion, Spry2 expression was elevated relative to the level in WT spleen cells in those TCL1-tg and human B-cell lymphomas in which there was little to no methylation of the Spry2 promoter (Figures 1C, 3A).
SPRY2 overexpression reduces ERK1/2 activation and induces B-cell apoptosis
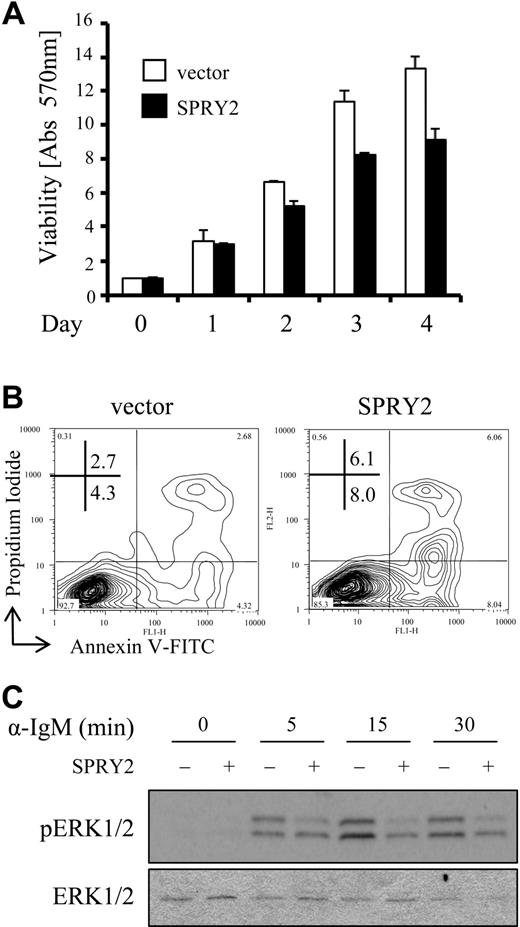
An increase in expression of sprouty family members causes a decrease in cell migration and proliferation, as well as senescence in nonlymphoid cell types,3,32 suggesting a potential inhibitory function for SPRY2 protein in B cells. To determine whether overexpression of SPRY2 has a similar effect on B cells, Nalm-6 cells were infected with a SPRY2-expressing retrovirus and the effects on ERK1/2 activation, cell viability, and cell proliferation were examined. Nalm-6 cells were chosen because they displayed abundant TCL1 expression,4 demonstrated ERK1/2-dependent induction of SPRY2 protein with BCR engagement (Figure S3A), and were amenable to infection with a SPRY2-expressing retrovirus, which was not tolerated by other B-cell lines (data not shown). Stably transduced Nalm-6 cells expressed 10-fold more SPRY2 protein than control-infected Nalm-6 cells by densitometry (Figure S3B) and displayed decreased viability by MTT assay compared with control vector–only cells (Figure 6A). No change in the cell- cycle profile between SPRY2 overexpressing and control cells was detected (data not shown), although a 2- to 3-fold increase in apoptosis was identified by annexin V–FITC and propidium iodide staining (Figure 6B). Reduced cell viability was paralleled by a reduction in BCR-stimulated phosphorylation of ERK1/2 (Figure 6C), suggesting a potential mechanism whereby a decrease in ERK pathway signaling leads to an increase in cell death.47 Incubation with U0126 resulted in increased apoptosis of a similar magnitude to that observed with retrovirus-mediated overexpression of SPRY2 in Nalm-6 cells (Figure S3C). These data support the hypothesis that SPRY2-mediated reduction in ERK1/2 activity causes a decrease in Nalm-6 cell viability.
SPRY2 overexpression decreases B-cell viability and reduces ERK1/2 signaling activity. (A) Nalm-6 cells infected with control (vector) or SPRY2-expressing retroviruses (SPRY2) were evaluated for viability by MTT assay over a 4-day time course. The data are from triplicate samples normalized to day 0 absorbance at 570 nm, which is set to 1.0. (B) Nalm-6 cells infected with control (vector) or SPRY2-expressing retroviruses (SPRY2) were evaluated for apoptosis by annexin V–FITC and propidium iodide staining with flow cytometry. (C) Nalm-6 cells infected with a control (−) or SPRY2-expressing (+) retrovirus were evaluated for ERK1/2 phosphorylation after anti-IgM stimulation. Data are representative of 3 (B,C) or 4 (A) independent experiments.
SPRY2 overexpression decreases B-cell viability and reduces ERK1/2 signaling activity. (A) Nalm-6 cells infected with control (vector) or SPRY2-expressing retroviruses (SPRY2) were evaluated for viability by MTT assay over a 4-day time course. The data are from triplicate samples normalized to day 0 absorbance at 570 nm, which is set to 1.0. (B) Nalm-6 cells infected with control (vector) or SPRY2-expressing retroviruses (SPRY2) were evaluated for apoptosis by annexin V–FITC and propidium iodide staining with flow cytometry. (C) Nalm-6 cells infected with a control (−) or SPRY2-expressing (+) retrovirus were evaluated for ERK1/2 phosphorylation after anti-IgM stimulation. Data are representative of 3 (B,C) or 4 (A) independent experiments.
Inactivation of Spry2 increases ERK1/2-dependent B-cell proliferation
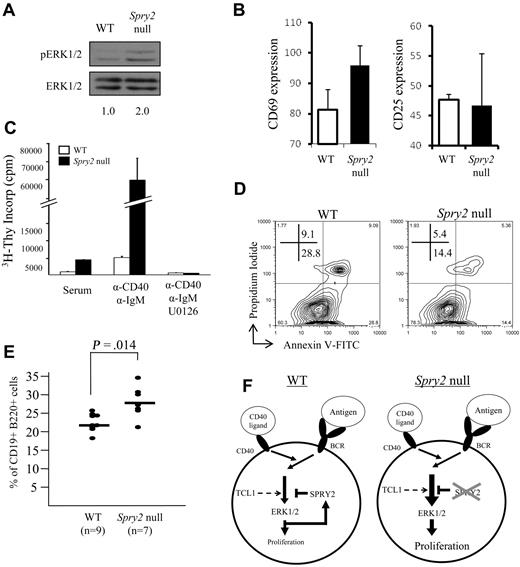
Enhanced SPRY2 expression caused decreased ERK1/2 phosphorylation and decreased B-cell viability, effects opposite those anticipated for Spry2 silencing. Consistent with this notion, human B-cell lines with dense SPRY2 promoter DNA methylation and repressed SPRY2 expression (Figure 4A) showed enhanced ERK1/2 phosphorylation with BCR stimulation compared with cell lines lacking DNA methylation that expressed abundant SPRY2 (Figure S4). Spry2 inactivation also resulted in 2-fold enhanced ERK1/2 phosphorylation in freshly isolated Spry2-null splenic B cells compared with WT control cells (Figure 7A). CD69, a glycoprotein whose surface expression increases with increased ERK1/2 activity resulting from BCR activation,48 was evaluated on splenic B cells from Spry2-null mice and WT littermates before and after 16 hours of CD40/BCR costimulation. As a control, CD25 surface expression was assessed because its expression increases with BCR activation independent of ERK1/2 pathway signaling. Spry2-null B cells showed a differential increase in CD69 but not CD25 expression compared with similarly costimulated WT B cells (Figure 7B), indicating that loss of Spry2 function results in an increase in ERK signaling activity.
Inactivation of Spry2 increases ERK1/2-dependent B-cell proliferation and survival. (A) Freshly isolated Spry2-null or WT splenic B cells from sex-matched littermates were assessed for ERK1/2 phosphorylation by Western blot. (B) Spry2-null or WT B cells were assayed for CD69 (ERK1/2-dependent) and CD25 (ERK1/2-independent) expression after 16 hours of costimulation with anti-CD40 and anti-IgM Abs. The mean fluorescence intensity for each B-cell activation marker from 3 separate experiments as measured by flow cytometry after costimulation is shown. (C) 3H-thymidine incorporation by Spry2-null or WT splenic B cells from sex-matched littermates, stimulated with serum alone or with serum plus anti-CD40 plus anti-IgM costimulation in the presence or absence of the MEK 1/2 inhibitor, U0126. 3H-thymidine was added to the medium at 44 hours of incubation for a final 16 hours of labeling (60-hour total culture time). (D) Spry2-null or WT splenic B cells from sex-matched littermates were anti-CD40 plus anti-IgM costimulated and evaluated for apoptosis by annexin V–FITC and propidium iodide staining with flow cytometry. (E) Percentage of CD19+B220+ B cells determined by flow cytometry from the bone marrows of 7 poly-dI:dC-injected Mx1-Cre × Spry2fl/fl (Spry2-null), 6 poly-dI:dC-injected Spry2fl/fl (WT), and 3 1× PBS, (pH 7.4)-injected Mx1-Cre × Spry2fl/fl mice (WT). (F) Model illustrating SPRY2-mediated regulation of the ERK signal transduction pathway in mature peripheral B cells. The diagram on the left (WT) shows that BCR/CD40 signaling results in activation of ERK1/2, which in turn stimulates B-cell proliferation. However, it also induces expression of SPRY2 protein, which subsequently represses ERK1/2 pathway signaling and prevents excessive cell proliferation. The diagram on the right (Spry2-null) illustrates the consequences of loss of SPRY2 function, by either dense promoter DNA hypermethylation or gene knockout: repression of ERK1/2 pathway signaling does not occur, resulting in increased B-cell proliferation. TCL1 also enhances the ERK signaling pathway in response to BCR stimulation in mature peripheral B cells, which results in an increase in Spry2 protein expression. Data are representative of 2 (A), 3 (C), or 4 (B,D) independent experiments.
Inactivation of Spry2 increases ERK1/2-dependent B-cell proliferation and survival. (A) Freshly isolated Spry2-null or WT splenic B cells from sex-matched littermates were assessed for ERK1/2 phosphorylation by Western blot. (B) Spry2-null or WT B cells were assayed for CD69 (ERK1/2-dependent) and CD25 (ERK1/2-independent) expression after 16 hours of costimulation with anti-CD40 and anti-IgM Abs. The mean fluorescence intensity for each B-cell activation marker from 3 separate experiments as measured by flow cytometry after costimulation is shown. (C) 3H-thymidine incorporation by Spry2-null or WT splenic B cells from sex-matched littermates, stimulated with serum alone or with serum plus anti-CD40 plus anti-IgM costimulation in the presence or absence of the MEK 1/2 inhibitor, U0126. 3H-thymidine was added to the medium at 44 hours of incubation for a final 16 hours of labeling (60-hour total culture time). (D) Spry2-null or WT splenic B cells from sex-matched littermates were anti-CD40 plus anti-IgM costimulated and evaluated for apoptosis by annexin V–FITC and propidium iodide staining with flow cytometry. (E) Percentage of CD19+B220+ B cells determined by flow cytometry from the bone marrows of 7 poly-dI:dC-injected Mx1-Cre × Spry2fl/fl (Spry2-null), 6 poly-dI:dC-injected Spry2fl/fl (WT), and 3 1× PBS, (pH 7.4)-injected Mx1-Cre × Spry2fl/fl mice (WT). (F) Model illustrating SPRY2-mediated regulation of the ERK signal transduction pathway in mature peripheral B cells. The diagram on the left (WT) shows that BCR/CD40 signaling results in activation of ERK1/2, which in turn stimulates B-cell proliferation. However, it also induces expression of SPRY2 protein, which subsequently represses ERK1/2 pathway signaling and prevents excessive cell proliferation. The diagram on the right (Spry2-null) illustrates the consequences of loss of SPRY2 function, by either dense promoter DNA hypermethylation or gene knockout: repression of ERK1/2 pathway signaling does not occur, resulting in increased B-cell proliferation. TCL1 also enhances the ERK signaling pathway in response to BCR stimulation in mature peripheral B cells, which results in an increase in Spry2 protein expression. Data are representative of 2 (A), 3 (C), or 4 (B,D) independent experiments.
Next, the effect of loss of Spry2 function on cell proliferation was assessed by measuring [3H]-thymidine incorporation (Figure 7C). An equal number of Spry2-null and WT B cells from sex-matched littermates were evaluated in triplicate in serum alone or with CD40/BCR costimulation. Spry2-null B cells showed a 4-fold increase in proliferation in serum alone and a 12-fold increase with CD40/BCR costimulation compared with WT B cells. Together, these data indicate that Spry2-null cells have increased ERK1/2 activity and cell proliferation. To determine whether the increased proliferation of Spry2-null B cells is dependent on the increased ERK1/2 activity, a nontoxic dose of U0126 was added to the costimulated cells to block ERK1/2 phosphorylation. Costimulated Spry2-null and WT B cells incubated with U0126 showed almost no proliferation, providing evidence that the increase in cell proliferation observed in Spry2-null versus WT B cells was the result of enhanced ERK1/2 activity. Finally, CD40/BCR costimulated Spry2-null B cells showed a 2-fold reduction in apoptosis compared with WT sex-matched littermates (Figure 7D), which was opposite the 2- to 3-fold enhanced apoptosis detected with SPRY2 overexpression (Figure 6B) and consistent with a cell-protective role for SPRY2 silencing in B cells. Finally, conditional Spry2 knockout resulted in an average increase of 20% up to more than 50% of CD19+B220+ B cells in vivo compared with WT sex-matched littermates (Figures 7E, S5), consistent with a combined increase in B-cell proliferation (Figure 7C) and reduced apoptosis (Figure 7D) from loss of Spry2 expression.
Discussion
The current study was aimed at determining whether epigenetic silencing of Spry2, a member of the sprouty family of RTK-negative regulators, plays a role in B-cell lymphomas. Our previous survey of genes in B-cell tumors from TCL1-tg mice suggested Spry2 as a candidate tumor suppressor whose promoter region contained at least some DNA methylation.12 Here we show that Spry2 is hypermethylated and that dense hypermethylation is associated with repression of Spry2 expression in many TCL1-tg and human B-cell lymphomas (Figures 1, 3) as well as in B-cell tumor cell lines from mice (Figure 2) and humans (Figure 4). Moreover, treatment of Spry2 hypermethylated tumor cell lines with a demethylating agent results in as much as a 100-fold increase in Spry2 expression (Figures 2, 4), whereas CpG methylation of SPRY2 promoter luciferase reporter constructs represses SPRY2 promoter activity 50- to 200-fold (Figure S2). These data indicate that extensive DNA methylation of the Spry2 promoter results in repression of Spry2 expression in B-cell tumors and raise the possibility that such repression contributes to B-cell lymphoma formation or progression, as has been suggested for other tumor types.27,,,–31
In approximately 20% to 30% of human cancers, an activating mutation in RAS or other ERK pathway MAPKs, such as BRAF, is thought to be a key event in the etiology of the tumor. However, mutations that activate the MAPK-ERK signaling pathway are rare in hematologic malignancies despite the apparent requirement for precise control over MAPK-ERK pathway activity to prevent immune cell transformation.49,–51 On the other hand, there is evidence that loss-of-function mutations in genes that encode negative regulators of MAPK-ERK pathway signaling promote lymphomas. For example, children with neurofibromatosis type 1, which is most often caused by inactivation of the NF1-negative regulator of RAS, have a 10-fold increased risk of developing a lymphoid malignancy compared with unaffected children.52 Our data indicate that epigenetic silencing of Spry2 in splenic B cells is far more common than loss of NF1 function or activating MAPK-ERK pathway mutations in lymphocyte malignancies, and that Spry2 functions as a negative feedback inhibitor of MAPK-ERK pathway signaling in B cells (Figures 6, 7, S4), as it does in many other cell types.3,53,54
There are various mechanisms by which loss of Spry2 expression in B cells could promote lymphomas via enhancement of MAPK-ERK pathway signaling. In one scenario, enhanced MAPK-ERK signaling interacts with elevated MYC expression during transformation, as is the case in many cell types, including rat embryonic fibroblasts. In those cells, neither constitutive activation of RAS nor overexpression of c-MYC alone causes transformation, but they are readily transformed by a combination of both genetic insults.55,56 In this context, it is interesting to note that we identified Spry2 expression to be decreased in more than 75% of human BL cell lines, a BLL from TCL1-tg mice, and a human BL patient sample, all of which display increased c-MYC expression.10
In the case of other B-cell lymphomas from TCL1-tg mice, in which c-MYC expression is not elevated, the increase in MAPK-ERK pathway signaling caused by repression of Spry2 may combine with positive effects of TCL1 expression on MAPK-ERK activity (Figure 5F) to provide sufficient MAPK-ERK pathway signaling for promoting lymphomas. This notion is consistent with recent studies showing that oncogene-induced stress, perhaps similar to that caused by aberrant TCL1 expression in GC B cells, activates a negative feedback signaling network, including robust induction of SPRY2, to prevent transformation of a precancerous state.32 Thus, loss of Spry2 function may participate in tumor progression in a subset of the GC B-cell lymphomas that develop in TCL1-tg mice at an average age of 12 to 15 months,7 and this loss may be part of a hypermethylation program that also epigenetically silences Epha7 expression.12
We have also studied the function of Spry2 in normal mouse splenic B cells in response to CD40/BCR costimulation. The results of this analysis indicate that Spry2 expression is induced within approximately 10 to 12 hours of BCR engagement, a response that is dependent on MAPK-ERK activation (Figure 5). MAPK-ERK pathway signaling initiates events that lead to B-cell proliferation57,58 and differentiation.59 Our data further suggest that SPRY2 protein expression subsequently represses MAPK-ERK signaling, thus establishing a SPRY2-mediated autoregulatory loop that dampens the antigen-induced expansion of mature B cells (see model, Figure 7F, WT). In support of this hypothesis, we show that complete loss of Spry2 function results in an increase phosphorylated ERK1/2, an increase in CD69 expression, a marker of MAPK-ERK pathway signaling, robust B-cell proliferation with reduced apoptosis, and an increase in the number of mature B cells in vivo (Figures 7, S5). Loss of this autoregulatory loop presumably reduces opposition to MAPK-ERK pathway activation and contributes to B-cell tumor progression47 and possibly transformation (see model, Figure 7F, Spry2-null). Beyond its role in controlling B-cell proliferation, SPRY2 probably has additional activities that regulate B-cell physiology, via its impact on the MAPK-ERK and possibly additional key signal transduction pathways, which remain to be investigated.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank members of the University of California–Los Angeles Flow Cytometry Core Facility and Joshua Troke for technical assistance and David Warburton (Childrens Hospital Los Angeles) for providing human SPRY2 promoter-reporter constructs.
This work was supported by National Institutes of Health (Bethesda, MD; grants T32-AI052031 and T32-GM08042, M.J.F.; grant CA78711, G.R.M.; and CA90571, CA107300, and PN2EY018228, M.A.T.). M.A.T. is a Scholar of the Leukemia & Lymphoma Society (White Plains, NY).
National Institutes of Health
Authorship
Contribution: M.J.F. and M.A.T. designed research; M.J.F., D.W.D., S.J.B., J.S.H., W.M.K., C.E.B., E.L.A., and R.R.S. performed research; L.X., W.M.K., D.B.-S., and G.R.M. provided essential reagents; M.J.F., S.J.B., D.B.-S., G.R.M., and M.A.T. interpreted results and discussed experimental strategies; and M.J.F., G.R.M., and M.A.T. wrote and edited the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michael A. Teitell, Department of Pathology and Laboratory Medicine, David Geffen School of Medicine at University of California–Los Angeles, 10833 Le Conte Avenue, Los Angeles, CA 90095-1732; e-mail: mteitell@ucla.edu.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal